Abstract
Sinorhizobium meliloti produces an exopolysaccharide called succinoglycan that plays a critical role in promoting symbiosis with its host legume, alfalfa (Medicago sativa). We performed a transposon mutagenesis and screened for mutants with altered succinoglycan production and a defect in symbiosis. In this way, we identified a putative two-component histidine kinase associated with a PAS sensory domain, now designated CbrA (calcofluor-bright regulator A). The cbrA::Tn5 mutation causes overproduction of succinoglycan and results in increased accumulation of low-molecular-weight forms of this exopolysaccharide. Our results suggest the cbrA::Tn5 allele leads to this succinoglycan phenotype through increased expression of exo genes required for succinoglycan biosynthesis and modification. Interestingly, CbrA-dependent regulation of exo and exs genes is observed almost exclusively during stationary-phase growth. The cbrA::Tn5 mutant also has an apparent cell envelope defect, based on increased sensitivity to a number of toxic compounds, including the bile salt deoxycholate and the hydrophobic dye crystal violet. Growth of the cbrA mutant is also slowed under oxidative-stress conditions. The CbrA-regulated genes exsA and exsE encode putative inner membrane ABC transporters with a high degree of similarity to lipid exporters. ExsA is homologous to the Escherichia coli MsbA protein, which is required for lipopolysacharide transport, while ExsE is a member of the eukaryotic family of ABCD/hALD peroxisomal membrane proteins involved in transport of very long-chain fatty acids, which are a unique component of the lipopolysaccharides of alphaproteobacteria. Thus, CbrA could play a role in regulating the lipopolysaccharide or lipoprotein components of the cell envelope.
The gram-negative soil bacterium Sinorhizobium meliloti establishes a symbiotic relationship with the alfalfa legume (Medicago sativa) by eliciting formation of a specialized plant organ, the nodule, within which bacteria establish a chronic intracellular infection that ultimately leads to nitrogen fixation (13). The successful symbiosis produces elongated nodules of the indeterminate type that reflect continuous bacterial invasion of new nodule cells within the plant. These nodules are also pink in coloration as a result of plant leghemoglobin production, which is responsive to the bacterial capacity to reduce dinitrogen. Nodule formation and bacterial nitrogen fixation is the result of a highly regulated and complex developmental process that requires continuous communication between the two symbiotic partners.
A number of bacterial requirements for the S. meliloti chronic host infection have been characterized (65). For instance, production of Nod factor in response to specific flavonoids exuded from host roots into the soil is essential for initial recognition and interaction between the bacterium and the root hairs of its host (36, 56). Ultimately, metabolic requirements for nitrogen fixation within the host depend in part on a group of fix and nif regulatory genes responsible for sensing the microaerobic environment within the nodule and activating genes that encode the nitrogenase enzyme and specialized respiratory complex (32). However, there are important developmental events between the initiation of nodule formation and the activation of nitrogenase gene expression that are less well understood.
Throughout symbiotic development, the S. meliloti cell surface is in intimate association with its host. Not surprisingly, then, several cell surface components play important roles in promoting nodulation, including the exopolysaccharides (EPSs), lipopolysaccharides (LPSs), K antigen, and cyclic β-(1, 2)-glucans (33). Succinoglycan, also called EPS I, is an exopolysaccharide that plays a critical role in S. meliloti nodulation; thus, the genes involved in the production, modification, and secretion of succinoglycan have been well characterized (40, 62). Succinoglycan is a polymer of repeating octasaccharide subunits (one galactose and seven glucose residues) that can be modified with succinyl, acetyl, and pyruvyl substituents (1, 69). The succinoglycan monomer is polymerized to various degrees, leading to production of low-molecular-weight (LMW) forms (monomers, dimers, and trimers) and high-molecular-weight (HMW) forms (several hundred monomers) of succinoglycan (47, 90). S. meliloti mutants unable to produce succinoglycan can elicit nodule organogenesis, but these aberrantly small and white nodules are devoid of bacteria (49). This is because succinoglycan is required for initiation and extension of the infection threads that allow bacterial invasion into the plant roots (19).
While the complete absence of succinoglycan production prevents nodulation, it has become clear that specific modifications to the succinoglycan are also important. For instance, an exoH mutant unable to succinylate the EPS is also incapable of promoting normal infection thread growth (19, 48). Similarly, an exoZ mutant defective in acetylation of succinoglycan (71) has decreased efficiency of infection thread initiation and elongation (19). It has also been suggested that LMW forms of succinoglycan are more effective at promoting root hair invasion than HMW forms (6, 82, 84). While succinoglycan production is critical during early root hair invasion, there are indications that succinoglycan biosynthetic genes are down-regulated during the later stages of bacterial development within plant cells (5, 70). Insight into how S. meliloti regulates the production and modification of succinoglycan is therefore fundamental to our understanding of the physiological requirements for symbiosis.
A component of the cell envelope that plays a later role in nodule development to promote intracellular bacterial persistence is the LPS of the outer membrane (50). S. meliloti mutants with LPS alterations in either the lipid A or the carbohydrate core can have a weakened or severely diminished capacity to establish a chronic infection (16, 17, 28, 29, 46, 63, 78). Modification of LPS by sulfation may also affect the symbiotic proficiency of S. meliloti (22, 43). Although the precise function of LPS in promoting symbiotic development remains unclear, defects in LPS sensitize bacteria to detergents and antimicrobial peptides, suggesting it could provide a barrier against environmental stress and host defense responses. In fact, there are indications that purified S. meliloti LPS may play a more active role by suppressing the oxidative burst of the plant innate immune response (76).
In the past, a number of genetic screens were performed with the goal of identifying S. meliloti mutants defective for symbiosis. For instance, Long et al. directly tested the symbiotic proficiencies of transposon mutants with alfalfa (57), and this intensive search yielded mutants involved in production of Nod factor. Others have taken the approach of isolating auxotrophic mutants and screening them for symbiotic phenotypes (30, 45, 58). Previously, we isolated mutants unable to produce succinoglycan (49), and this screen yielded mutants that have provided a wealth of information about succinoglycan production, as well as revealing the key role succinoglycan plays in establishing the symbiosis. These considerations led us to screen for mutants having increased or aberrant succinoglycan production with the goal of identifying unknown genes that play a role in symbiotic development.
Here, we describe our characterization of an important regulator of S. meliloti succinoglycan that also has a profound effect on alfalfa symbiosis. We have isolated a transposon mutation disrupted in a putative two-component sensor that we designated CbrA and for which we found homologs in the closely related plant pathogen Agrobacterium tumefaciens and animal pathogens, Brucella spp. The cbrA::Tn5 mutation results in overproduction of succinoglycan. By analyzing differential expression of genes in the exo/exs cluster required for succinoglycan biosynthesis, we observed that the cbrA::Tn5 mutation affects target gene expression predominantly during stationary phase, suggesting that a common CbrA regulatory signal exists under this particular ex planta growth condition and in the host environment. In addition, we found that the cbrA::Tn5 allele conferred growth defects in the presence of a variety of toxic compounds, such as deoxycholate (DOC) and indole. Our observations are consistent with CbrA functioning as a key regulator of S. meliloti physiology during free-living and in planta growth.
Although S. meliloti is a plant symbiont, it shares a close phylogenic relationship with animal and human pathogens among Brucella spp. (59). These bacteria have in common an intracellular chronic infection of their respective hosts within a host-derived membrane-bound compartment (81). It is becoming clear that some of the bacterial requirements for rhizobial persistence within the legume host are shared with the brucellae (27, 51, 52, 77). Thus, a broader implication of the identification of genes required for S. meliloti symbiotic development derives from the potential to also distinguish genes that aid in Brucella sp. pathogenesis based on common physiological requirements.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, and growth conditions.
Bacterial strains, generalized transducing phage, plasmids, and primer sequences are listed in Table 1. Bacteria were grown on either LB or LB/MC (LB supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2). Unless otherwise noted, S. meliloti strains were cultured at 30°C in LB/MC to logarithmic phase (approximate optical density at 600 nm [OD600] = 0.5) and stationary phase (approximate OD600 = 6.0) for physiological and gene expression assays. For calcofluor analyses of succinoglycan production, LB agar was buffered with 10 mM MES (morpholineethanesulfonic acid), pH 7.5, and calcofluor white MR2 (Cellufluor; Polysciences) was added at a final concentration of 0.02%. The following antibiotics were used: streptomycin (500 μg/ml), neomycin (200 μg/ml), gentamicin (12.5 μg/ml), spectinomycin (100 μg/ml), chloramphenicol (34 μg/ml), oxytetracycline (750 ng/ml), and tetracycline (10 μg/ml).
TABLE 1.
Bacterial strains, phage, plasmids, and PCR primers used in this study
| Strain/reagent | Relevant characteristics | Source or reference | ||
|---|---|---|---|---|
| Straina
|
||||
| MT616 | E. coli MM294; pRK600 Cmr | T. Finan | ||
| DH5α | E. coli endA1 hsdR17 supE44 thi-1 recA1 gyrArelA1 Δ(lacZYA-argG) U169 deoR | BRL Corp. | ||
| Rm1021 | SU47 Smr | F. Ausubel | ||
| SmGC24 | Original cbrA::Tn5 isolate | This study | ||
| KEG2016 | Transduced cbrA::Tn5 | This study | ||
| KEG2018 | exoY210::Tn5-233 | ΦM12 transduction from Rm7210 | ||
| KEG2019 | exoH::lacZ+-accC1 (Gmr) | ΦM12 transduction from RmI-H1 | ||
| KEG2020 | KEG1016 exoH::lacZ+-accC1 (Gmr) | This study | ||
| KEG2081 | KEG2016 exoY210::Tn5-233 | This study | ||
| KEG2098 | recA::Tn5-233 | ΦM12 transduction from Rm6062 | ||
| KEG2099 | KEG2016 recA::Tn5-233 | This study | ||
| KEG2101 | exoK445::Tn5-233 | ΦM12 transduction from Rm8826 | ||
| KEG2102 | exsH13::Tn5-132 | ΦM12 transduction from Rm8823 | ||
| KEG2103 | KEG2102 exoK445::Tn5-233 | This study | ||
| KEG2104 | KEG2016 exoK445::Tn5-233 | This study | ||
| KEG2105 | KEG2016 exsH13::Tn5-132 | This study | ||
| KEG2106 | KEG2098 pLAFR2070 | This study | ||
| KEG2107 | KEG2098 pLAFR2070ΔfbpB | This study | ||
| KEG2108 | KEG2098 pLAFR2070ΔcbrA | This study | ||
| KEG2115 | KEG2099 pLAFR2070 | This study | ||
| KEG2116 | KEG2099 pLAFR2070ΔfbpB | This study | ||
| KEG2117 | KEG2099 pLAFR2070ΔcbrA | This study | ||
| KEG2118 | KEG2105 exoK445::Tn5-233 | This study | ||
| KEG2124 | exsA::lacZ+-accC1 (Gmr) | ΦM12 transduction from RmH22a | ||
| KEG2125 | KEG2124 cbrA::Tn5 | This study | ||
| KEG2126 | KEG2124 exsE317::Tn5-233 | This study | ||
| KEG2127 | KEG2126 cbrA::Tn5 | This study | ||
| KEG2128 | KEG2102 exoK445::Tn5-233 | This study | ||
| KEG2129 | KEG2124 cbrA::Tn5 | This study | ||
| KEG2130 | exsE317::Tn5-233 | ΦM12 transduction from Rm8317 | ||
| KEG2131 | KEG2016 exsE317::Tn5-233 | This study | ||
| SmJL12 | pVO155 cbrA′-uidA+cbrA+ | This study | ||
| Phage
|
||||
| φM12 | Generalized transducing phage | T. Finan | ||
| Plasmids
|
||||
| pLAFR1 library | Genomic library in pLAFR1 cosmid | 34 | ||
| pLAFR2070 | cbrA::Tn5 complementation cosmid | This study | ||
| pLAFR2070ΔfbpB | pLAFR2070 except fbpB replaced with cat | This study | ||
| pLAFR2070ΔcbrA | pLAFR2070 except cbrA replaced with cat | This study | ||
| pJL1049 | pVO155 derivative with a cbrA′-uidA+ fusion | This study | ||
| PCR primers
|
||||
| cbrA5′ | 5′CGCAAACGATCCGTC | This study | ||
| cbrA3′ | 5′CAATTGGCCAAGAC | This study | ||
| fbpB5′ | 5′GTTAACTTGCACTTC | This study | ||
| fbpB3′ | 5′GTTAACCTATGAC | This study | ||
| cbrAup | 5′GTCGGGTCGAAAGAGAG | This study | ||
| cbrA353 | 5′GAACAGAATTGCCTCTTC | This study | ||
| cbrA-CATprime1 | 5′GTTGCTGGACCGGGCTGGGTATTAATAGAGCGTTAACGCGCAAACGATCCGTCGTGTAGGCTGGAGCTGCTTC | This study | ||
| cbrA-CATprime2 | 5′TCAATTGGCCAAGACGCGTTGTGAGGGAAAGGATATTTCCACCAGCGTGCCCTCATATGAATATCCTCCTTAG | This study | ||
| fbpB-CATprime1 | 5′CCTGCGGGAGCCAGTTTGTCGGTTCTTGAGGGTTGAGGAAACTGCGGGCTGTGTAGGCTGGAGCTGCTTCG | This study | ||
| fbpB-CATprime2 | 5′CTATGACCGGCGTTCGAGCGACCGGGACACCAGGATGACGGGGACCATCCCCATATGAATATCCTCCTTAG | This study | ||
| c1 | 5′TTATACGCAAGGCGACAAGG | 23 | ||
| c2 | 5′GATCTTCCGTCACAGGTAGG | 23 | ||
| cbrAgus5′ | 5′TCTTCGGTCGCGGTCTTCATCTTG | This study | ||
| cbrAgus3′ | 5′GGGGTACTGTCTTTCGGGCATAGG | This study | ||
All S. meliloti strains used in this study were derived from the wild-type Rm1021.
Genetic techniques.
Transductions with φM12 were performed as described previously (31). Random Tn5 mutagenesis and triparental matings were performed as described previously (49). Recombinant cosmids that complemented the cbrA::Tn5 mutation were isolated from a pLAFR1-derived S. meliloti genomic library generously provided by S. Long (34). The pLAFR1 cosmid library was mated into KEG2016, and cbrA::Tn5 complementing clones were selected for on LB containing antibiotics (streptomycin, neomycin, and tetracycline) and 10 mM deoxycholate.
DNA manipulations.
Genomic DNA was isolated as described previously (2). The cbrA::Tn5 transposon insertion in SmGC24 was cloned from an EcoRI/BamHI digest of genomic DNA into pUC19, and the resulting kanamycin-resistant plasmids were sequenced. pLAFR1 cosmid DNA was isolated as described previously (2). The presence of cbrA and fbpB in pLAFR1 complementing cosmids was determined by PCR using Thermalase polymerase (Invitrogen) and primer sets cbrA5′/cbrA3′ and fbpB5′/fbpB3′, respectively. The cbrA and fbpB open reading frames (ORFs) were disrupted within cosmid pLAFR2070 using the λRed method as described previously (23). We performed PCR using the pKD3 template with primer sets cbrA-CATprime1/cbrA-CATprime2 and fbpB-CATprime1/fbpB-CATprime2. The cbrA::cat and fbpB::cat deletions in pLAFR2070 were verified by sequencing PCR products obtained using primer c1 or c2 (23) in combination with either cbrA or fbpB gene-specific primers.
RNA isolation and RT-PCR analysis.
S. meliloti cultures were diluted in 15 ml of LB/MC to an OD600 of 0.1 and were immediately treated with 30 ml of RNAProtect Bacterial Reagent (QIAGEN) for 5 min at room temperature. The cells were centrifuged at 4°C for 10 min at 10,000 relative centrifugal force (RCF), and the supernatant was decanted. The cell pellet was immediately frozen in an ethanol/dry ice bath and either stored at −70°C or immediately processed using the MasterPure Complete RNA purification kit (Epicenter) following the manufacturer's instructions. One microgram of total RNA was analyzed by gel electrophoresis (0.37 M formaldehyde, 1.2% agarose) with MOPS (morpholinepropanesulfonic acid) running buffer (40 mM MOPS, pH 7.0, 10 mM NaAc, 1 mM Na2EDTA, pH 8.0, and 0.2 M formaldehyde) to judge sample integrity. To test for DNA contamination, we performed 25-μl PCRs with 0.5 μg of RNA and 0.1 μM of the primers cbrAup and cbrA353; purified genomic DNA was used as a positive control. For each RNA sample, a stock of 50 pg/μl of DNA-free total RNA was made in the SuperScript One Step RT-PCR (QIAGEN) reaction buffer, and aliquots of 20 μl were combined with a set of gene-specific primers at 0.1 μM. Reverse transcription (RT)-PCRs with primers specific for 23S RNA were used as a control to ensure equal amounts of RNA template between reactions, and the number of PCR cycles was varied in increments from 20 to 40 to determine the linear range for each target transcript. The PCR products were analyzed by 1% agarose Tris-acetate-EDTA electrophoresis in the presence of ethidium bromide and visualized with UV. Total RNA from each growth phase was independently isolated at least three times, and the RT-PCR data presented represent a typical result within the linear range for each transcript.
EPS isolation.
Stationary-phase cultures were centrifuged at 4°C for 30 min at 3,000 RCF, and the cleared supernatant was isolated. EPS present in the supernatant was isolated as described previously (25): EPS was precipitated away from the sugars present in LB medium using 0.3 volume of 1% centrimide, the pellet was resuspended in 10% NaCl, and the EPS was reprecipitated with 2 volumes of cold acetone. The concentration of total carbohydrate was determined using the anthrone-H2SO4 method (54); sterile LB/MC treated similarly gives no anthrone-positive signal. The amount of carbohydrate present in each supernatant was normalized to the protein concentration of its corresponding cell pellet, which was determined using the Bradford assay (Bio-Rad). The average and standard deviation for each strain was derived from the results of three independent cultures.
Protein isolation and Western blot analysis.
Bacterial cultures were centrifuged at 4°C for 10 min at 10,000 RCF, and the cleared supernatant was isolated. Proteins within the supernatant were precipitated by the addition of 0.2 volume of 100% trichloroacetic acid. The pellet was resuspended in 10 mM Tris, pH 8.0, and the protein concentration of each sample was determined using the Bradford assay (Bio-Rad). Ten micrograms of protein was subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore) for subsequent Western blot analysis. The polyvinylidene difluoride membrane was probed with both an anti-ExoK primary antibody (1:5,000 dilution) and an anti-ExsH primary antibody (1:10,000 dilution) (87) and subsequently probed with a horseradish peroxidase-conjugated anti-rabbit (Pierce) secondary goat antibody (1:10,000 dilution). The Western blot was developed using the SuperSignal West Dura kit (Pierce) and exposed to film.
Physiological-stress assays.
For the efficiency-of-plating (EOP) assays, cultures were grown to exponential phase (OD600, ∼0.5) in LB/MC medium and then diluted to an OD600 of 0.1 in LB. Each sample was serially diluted from 100 to 10−6 in LB, and 50 μl was spread onto LB agar containing either deoxycholate (Sigma) or indole (Sigma). After 4 to 5 days of growth at 30°C, the number of CFU was determined, with the exception of the cbrA::Tn5 mutant, which required an additional 48 h of growth at 30°C for colonies to appear on LB supplemented with 1 mM indole. The average and standard deviation for each strain were derived from three independent cultures, with each dilution spread onto two plates. The EOP assay to determine pLAFR2070 complementation was performed similarly with the exception that tetracycline was added to all of the growth media.
Nodulation assays.
Medicago sativa AS13R seeds were sterilized by treatment with 70% ethanol for 30 min, followed by treatment with 20% bleach for another 30 min. The seeds were washed in distilled water (dH2O) for 5 h and stored in dH2O at 4°C in the dark for approximately 2 days prior to germination. The seeds were germinated on 1% agar/dH2O upside down in the dark at room temperature overnight. Seedlings were transferred to buffered nodulation medium (BNM) agar plates (pH 6.0) (26) and allowed to grow for 4 to 5 days before bacterial inoculation to promote synchronous nodule development. The roots were directly inoculated with 100 μl of bacteria that had been cultured to logarithmic phase (OD600, ∼0.5) in LB/MC medium, spun down, washed in 0.5× BNM medium, and then diluted to an OD600 of 0.1 in 0.5× BNM medium. Plant photographs were taken and microscopic analysis of nodules (see Fig. 2) was performed at 28 days postinoculation. Nodule development (number of nodules/plant and percent pink nodules/total number of nodules per plant) and plant height were monitored for a period of 5 weeks (see Table 3). Plant height was assessed by measuring the length of the epicotyl stem. Nodules were surface sterilized with 70% ethanol for 30 s, followed by three water washes, and treatment with 10% bleach for 30 s, followed by three water washes. The nodules were then crushed with a sterile pestle in 100 μl of LB/MC medium containing 0.3 M glucose. The nodule suspension was serially diluted (100 to 10−6) and plated onto LB/MC medium containing 0.3 M glucose. When appropriate, the medium was supplemented with 0.02% calcofluor and 10 mM MES, pH 7.5, with or without neomycin.
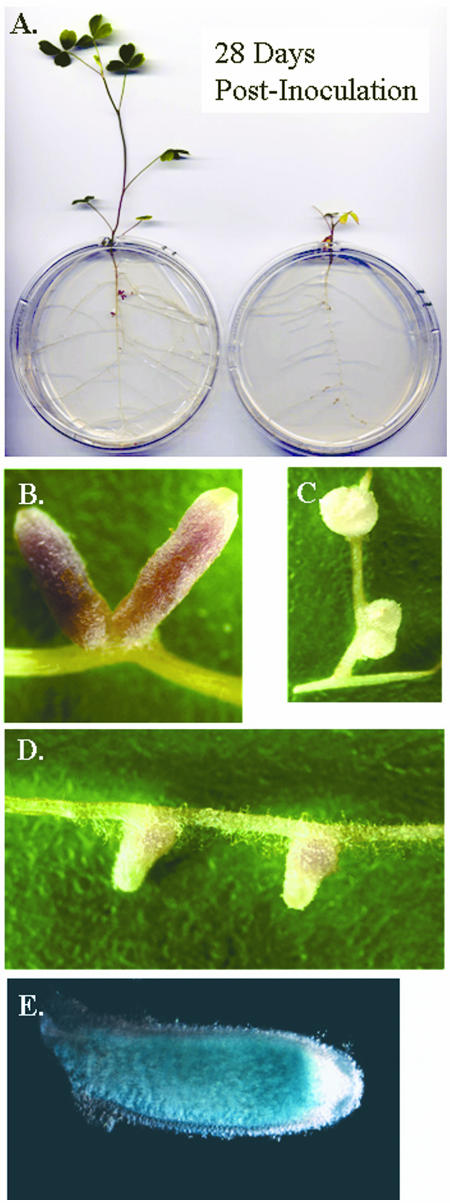
FIG. 2.
Plant growth and nodule morphology resulting from symbiosis of alfalfa. (A) Average plants inoculated with either wild-type or cbrA bacteria were photographed after 28 days of growth. (B) The type III nodule, elongated and pink in coloration (Fix+), represents 99% of nodules elicited by the wild type and 0 to 5% of nodules elicited by the cbrA mutant. (C) The type I nodule is small and white (Fix−) and represents on average 60% of the nodules elicited by the cbrA mutant. (D) The type II nodule is stunted in size with a slight pink coloration at the base of the nodule (Ndv−) and represents approximately 40% of the nodules elicited by the cbrA mutant. (E) A nodule from a plant inoculated with a strain containing a cbrA′-uidA+ fusion in a cbrA+ background was stained for β-glucuronidase activity.
TABLE 3.
pLAFR2070 complementation of the S. meliloti-alfalfa symbiosis
| Strain (genotype) | Plant hta | No. of nodules/planta | % Pink nodulesb |
|---|---|---|---|
| KEG2098 (recA) | 10 ± 1.7 | 8.6 ± 3 | 70 ± 10 |
| KEG2099 (recA cbrA) | 4.5 ± 0.88 | 12 ± 4 | 19 ± 11 |
| KEG2106 (recA pLAFR2070) | 11 ± 1.5 | 8.1 ± 5 | 81 ± 8 |
| KEG2107 (recA pLAFR2070ΔfbpB) | 12 ± 0.85 | 15 ± 3 | 70 ± 11 |
| KEG2108 (recA pLAFR2070ΔcbrA) | 11 ± 1.5 | 9.2 ± 3 | 76 ± 12 |
| KEG2115 (recA cbrA pLAFR2070) | 8.6 ± 3.1 | 11 ± 7 | 67 ± 8 |
| KEG2116 (recA cbrA pLAFR2070ΔfbpB) | 9 ± 3 | 11 ± 7 | 77 ± 12 |
| KEG2117 (recA cbrA pLAFR2070ΔcbrA) | 4 ± 1.1 | 14 ± 6 | 9.5 ± 8 |
Standard deviations are derived from a minimum of 10 plants.
Includes type II and type III nodules.
Expression analysis of cbrA in nodules.
A fragment containing the 5′ ends of both cbrA and the oppositely transcribed Smc00777 ORF was PCR amplified with primers cbrAgus5′/cbrAgus3′. The fragment was cloned into the uidA+ promoterless suicide plasmid pVO155 (3), resulting in plasmid pJL1049. pJL1049 was mated into Rm1021, and transconjugants were screened for the correct insertion. The resulting strain, SmJL12, contains a cbrA′-uidA+ transcription fusion, followed by the intact cbrA gene with its native promoter. Plants inoculated with SmJL12 showed a wild-type phenotype for nodulation. Four-week-old nodules were excised, sectioned, and stained with 1 mM 5-bromo-4-chloro-3-indoyl-β-d-glucuronide in 50 mM Na-phosphate buffer, pH 7, with 0.02% SDS. Stained sections were washed three times with Na-phosphate buffer, pH 7, and analyzed by microscopy.
RESULTS
Identification of the cbrA gene encoding a putative two-component histidine kinase affecting succinoglycan production, stress resistance, and symbiosis.
To identify genes involved in succinoglycan production, we performed a random Tn5 mutagenesis of the wild type, Rm1021. The screen was performed on LB containing calcofluor, a dye that fluoresces under UV light specifically when bound to certain β-linked polysaccharides, including succinoglycan (49). We chose to characterize mutants with a calcofluor-bright phenotype, which indicated succinoglycan overproduction or alteration in production. We then performed a secondary screen to identify calcofluor-bright mutants that were also defective for symbiosis with alfalfa, resulting in either a Nod− (no nodules), Fix− (white nodules), or Ndv− (defective nodule development) phenotype. Here, we describe our characterization of one Tn5 mutant with a calcofluor-bright phenotype (Fig. 1A) and a profound symbiotic deficiency (Nod+ with a mixture of Fix− and Ndv− nodules) (Fig. 2A to D). In contrast to other calcofluor-bright mutants (25, 61), this strain is not mucoid (Fig. 1A).
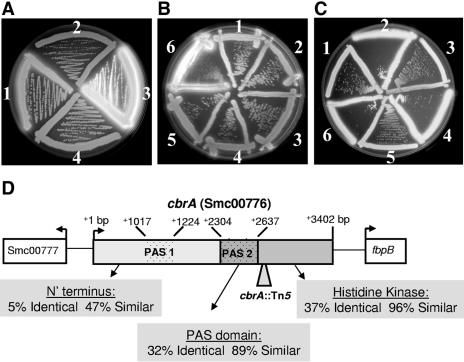
FIG. 1.
A calcofluor-bright transposon screen yields a succinoglycan overproducer disrupted for a putative histidine kinase sensor. Succinoglycan production was visualized on LB calcofluor medium. (A) 1, Rm1021 (wild type); 2, KEG2018 (exoY); 3, KEG2016 (cbrA); 4, KEG2081 (cbrA exoY). (B) 1, KEG2106 (recA pLAFR2070); 2, KEG2107 (recA pLAFR2070ΔfbpB); 3, KEG2108 (recA pLAFR2070ΔcbrA); 4, KEG2115 (cbrA recA pLAFR2070); 5, KEG2116 (cbrA recA pLAFR2070ΔfbpB); 6, KEG2117 (cbrA recA pLAFR2070ΔcbrA). (C) 1, Rm1021 (wild type); 2, KEG2016 (cbrA); 3, KEG2124 (exsA); 4, KEG2125 (cbrA exsA); 5, KEG2130 (exsE); 6, KEG2131 (cbrA exsE). (D) The calcofluor-bright Tn5 mutation is located within ORF Smc00776 (accession number CAC45293). The combined results of conserved domain database searches with NCBI rpsblast (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART database (http://smart.embl-heidelberg.de/) predict a two-component histidine kinase domain (E value = 1.2e−17), an associated histidine kinase ATPase domain (E value = 5.3e−24), and at least one sensory PAS domain: PAS domain 1 (E value = 2.2e+00) and PAS domain 2 (E value = 1.5e−10). Several CbrA orthologs were found in related alphaproteobacteria: Agrobacterium tumefaciens C58 (AAK86425), Brucella melitensis 16 M (AAL51598), B. suis 1330 (AAN30511), B. abortus biovar 1 strain 9-941 (YP_222279), Mesorhizobium loti (BAB52812), and Bradyrhizobium japonicum USDA 110 (BAC52661). The percent identity and percent similarity for each predicted domain (indicated by arrows below the ORF) were determined by aligning the CbrA orthologs using T-COFFEE (64); amino acid similarity was assigned based on ClustalW1.60.
We identified the genomic location of the Tn5 mutation by cloning the transposon with flanking S. meliloti genomic DNA into pUC19 and sequencing the genomic DNA adjacent to the transposon. The Tn5 insertion was found to disrupt a large ORF of 3.4 kb located on the S. meliloti chromosome annotated as Smc00776. Based on a conserved domain database search, this ORF is predicted to encode a soluble cytoplasmic two-component histidine kinase associated with at least one sensory PAS domain (66, 86) and a large N′-terminal sequence of unknown function that represents nearly two-thirds of the protein (Fig. 1D). We have designated this gene cbrA, for calcofluor-bright regulator A.
Putative CbrA orthologs were identified in several closely related alphaproteobacteria, including the plant pathogen Agrobacterium tumefaciens and the animal pathogens Brucella melitensis, Brucella abortus, and Brucella suis. The amino acid sequences of these proteins were aligned, revealing a 12.5% conservation of identity and a 53% conservation of similarity in amino acid sequence over the length of the protein (see Fig. S1 in the supplemental material). The most significant regions of similarity between CbrA orthologs are limited to the C′-terminal PAS and histidine kinase domains (Fig. 1D), providing little insight into the importance or function of the variable N′-terminal portion of CbrA. The putative kinase encoded by cbrA was not located beside or near a member of the response regulator family, leaving the identity of the presumed CbrA cognate response regulator unknown. Instead, cbrA is located 3′ of a divergently transcribed ORF encoding a conserved hypothetical protein annotated as Smc00777 and 5′ of the fbpB gene encoding a putative ABC iron transporter (Fig. 1D).
In contrast to the wild type, the cbrA::Tn5 mutant is unable to form single colonies on LB containing 10 mM DOC. Utilizing this DOC-sensitive phenotype, we isolated pLAFR1 cosmids from an S. meliloti genomic library that restored wild-type growth to the cbrA::Tn5 mutant. Fifteen independent pLAFR1 cosmids were isolated, and they each also restored a wild-type calcofluor phenotype to the cbrA::Tn5 mutant. The cosmids contained both cbrA and the downstream fbpB ORFs, as determined by PCR analysis (data not shown), so we used the λRed system in Escherichia coli to perform a directed deletion of either cbrA or fbpB in the complementing clone pLAFR2070 (23). The parent cosmid and each deletion derivative were introduced into the wild-type and the cbrA::Tn5 backgrounds, and effects on calcofluor fluorescence and DOC sensitivity were determined.
In a cbrA+ strain, pLAFR2070 (cbrA+ fbpB+) and the deletion derivatives pLAFR2070ΔfbpB and pLAFR2070ΔcbrA each present a wild-type calcofluor phenotype (Fig. 1B). Correspondingly, pLAFR2070 and its deletion derivatives support the same efficiency of plating (58 to 99% EOP) for the wild type on LB containing 10 mM DOC (Table 2) . In the cbrA::Tn5 mutant, only the pLAFR2070ΔcbrA cosmid resulted in a calcofluor-bright phenotype and a severely decreased EOP (0.0076%) on LB containing 10 mM DOC, indicating a loss of complementation relative to pLAFR2070. These complementation results suggest that the cbrA::Tn5 mutation is a recessive allele of cbrA, which is solely responsible for both the calcofluor-bright and DOC-sensitive phenotypes of the mutant. Therefore, we will refer to the cbrA::Tn5-containing strain as the cbrA mutant.
TABLE 2.
EOP assay for pLAFR2070 complementation of deoxycholate sensitivity
| Strain (genotype) | LB + Tet (CFU)a
|
EOP (%) | |
|---|---|---|---|
| 0 mM DOC | 10 mM DOC | ||
| KEG2106 (recA pLAFR2070) | 6.42 × 107 (±1.5 × 107) | 3.70 × 107 (±6.2 × 106) | 58 |
| KEG2107 (recA pLAFR2070ΔfbpB) | 7.44 × 107 (±2.1 × 107) | 7.33 × 107 (±7.3 × 106) | 99 |
| KEG2108 (recA pLAFR2070ΔcbrA) | 7.77 × 107 (±1.8 × 107) | 6.16 × 107 (±1.2 × 107) | 79 |
| KEG2115 (recA cbrA pLAFR2070) | 8.86 × 107 (±5.6 × 106) | 3.03 × 107 (±7.6 × 106) | 34 |
| KEG2116 (recA cbrA pLAFR2070ΔfbpB) | 5.23 × 107 (±1.0 × 107) | 2.52 × 107 (±9.3 × 106) | 48 |
| KEG2117 (recA cbrA pLAFR2070ΔcbrA) | 3.90 × 107 (±7.4 × 106) | 2.96 × 103 (±7.2 × 102) | 7.6 × 10−3 |
Numbers in parentheses are standard deviations derived from triplicate samples.
The cbrA mutant is ineffective at establishing alfalfa symbiosis.
To characterize the symbiotic deficiency associated with the cbrA mutant, alfalfa seedlings were either mock inoculated with 0.5× BMN medium or inoculated with several strains derived from the wild type, Rm1021. Alfalfa plants inoculated with the wild type grow to a height of 15 (±0.95) cm after 5 weeks of growth, and the leaves of the plants are a healthy green color (Fig. 2A). In contrast, plants inoculated with the cbrA mutant are significantly shorter, with an average height of 5.2 cm (±0.58), and the leaves are slightly yellowed compared to plants inoculated with the wild type (Fig. 2A). Plants that are mock inoculated grow only 2.4 cm (±0.87) and display sickly yellow leaves due to nitrogen starvation on the BMN medium used to test nodulation. The symbiotic deficiency of the cbrA mutant is fully complemented by the pLAFR2070 cosmid as long as the cbrA ORF is present, but not by the pLAFR2070ΔcbrA cosmid (Table 3).
We determined the efficiency of nodule formation throughout a 5-week period and observed that the numbers of nodules per plant elicited by the wild type (13 ± 5) and cbrA mutant (10 ± 4) were similar and that they appeared without an appreciable difference in timing (data not shown). As expected, nodules elicited by the wild type were elongated and pink in coloration (Fig. 2B). In contrast, nodules induced by the cbrA mutant were variable in both size and coloration. On plants inoculated with the cbrA mutant, nearly 60% of nodules were small and white (Fig. 2C, type I nodule), approximately 40% were small with a pale pink coloration at the base (Fig. 2D, type II nodule), and only a few (0 to 5%) nodules were indistinguishable in size and coloration from those elicited by the wild type (type III nodule). We assessed the level of cbrA expression in cbrA+ bacteria using a cbrA′-uidA+ fusion integrated at the gene locus and observed expression of cbrA throughout the nodule (Fig. 2E), consistent with the possibility that CbrA is present throughout symbiotic development and could potentially play a role in regulating bacterial physiology during multiple stages of nodulation.
We isolated bacteria from wild-type- and cbrA-induced nodules to assess whether the cbrA mutant was able to establish a chronic infection. The number of colonies recovered from nodules induced by the wild type was 6.4 × 105 (±8 × 104) per nodule. In contrast, we were unable to recover any bacteria from the predominant type I nodules elicited by the cbrA mutant. Recovery of bacteria from both of the cbrA-induced type II and type III nodules was highly variable (8.8 × 104 ± 3 × 104 per nodule), an indication that the mutant is compromised for colonization of its host.
We compared the calcofluor and DOC resistance phenotypes of colonies derived from cbrA-induced nodules to those of the wild type and cbrA mutant. Bacteria recovered from cbrA-induced type II and type III nodules were uniformly calcofluor bright and DOC sensitive in a manner indistinguishable from the original cbrA::Tn5 mutant (data not shown). However, it remained possible that the small number of type III nodules induced by the cbrA mutant represented extragenic suppressors and that neither the calcofluor-bright nor the DOC-sensitive phenotype was directly related to the symbiotic deficiency of the mutant. To test this, we reinoculated alfalfa with cbrA bacteria recovered from type III nodules. Plants inoculated with the nodule-recovered cbrA bacteria were indistinguishable from those inoculated with the original cbrA mutant, with an average plant height of 4.9 (±0.89) cm. Therefore, the cbrA mutant does not accumulate suppressors within the nodule but rather has a somewhat variable symbiosis phenotype on alfalfa.
Coinoculation experiments in which two strains are forced to compete for colonization of a single niche can often reveal defects not observed during single-strain inoculations. To determine the competitiveness of the cbrA mutant, we performed a coinoculation experiment with a 1:1 ratio of wild-type to cbrA bacteria. After 4 weeks of plant growth, we observed that the majority of nodules (87% on 20 plants) were indistinguishable from those elicited by the wild type alone. We isolated bacteria from coinoculated nodules and plated them onto LB calcofluor medium with or without neomycin. We observed a recovery rate of 3.9 × 105 (±2 × 104) per nodule for wild-type bacteria; however, we were unable to recover any calcofluor-bright or neomycin-resistant colonies indicative of the cbrA mutant. Thus, the cbrA mutant is unable to compete effectively with the wild type for colonization of the nodule, reflecting a crippled capacity for nodulation.
The cbrA mutant has a calcofluor-bright and extended-halo phenotype that reflects succinoglycan overproduction.
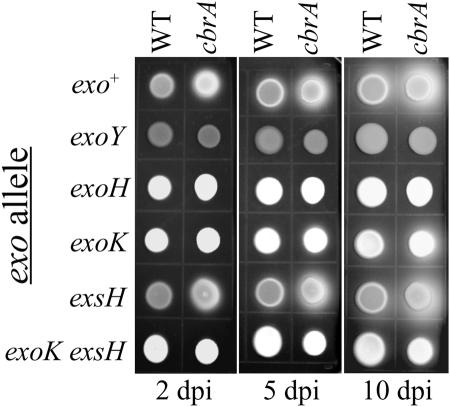
The S. meliloti exopolysaccharide succinoglycan is a symbiotically important macromolecule whose chemical modifications and polymerization state influence its symbiotic proficiency (19, 49). To distinguish whether the cbrA mutant calcofluor-bright phenotype reflected altered succinoglycan production or synthesis of a novel calcofluor-binding exopolysaccharide, we combined the cbrA allele with the well-characterized exoY (encoding a galactose-1-P-transferase) null mutation that completely blocks the first step of succinoglycan biosynthesis (15, 60). Like the exoY null mutant, the cbrA exoY double mutant is dark on LB calcofluor medium, demonstrating that the exoY mutation is epistatic to the cbrA mutation (Fig. 1A). Identical epistasis results were obtained with null mutations of exoB (encoding a UDP-glucose-4-epimerase) and the glycosyltransferase genes exoA and exoL, as well as exoF, exoP, exoQ, and exoT null mutations, i.e., the cbrA mutant is calcofluor dark when combined with any one of these exo mutations (data not shown). Thus, the calcofluor-bright phenotype of the cbrA mutant reflects altered succinoglycan production. In addition, the cbrA mutant rapidly develops a fluorescent halo on calcofluor medium after just 2 days of growth while the wild type requires approximately 5 days to produce a fluorescent halo (Fig. 3). The florescent halo makes visible the diffusion of LMW succinoglycan into the medium surrounding a colony (48, 88). Not surprisingly, halo formation by both the wild type and cbrA mutant is also dependent on exoY (Fig. 3).
FIG. 3.
The cbrA mutant has an extended-halo phenotype. The succinoglycan fluorescent-halo phenotype was assessed on LB calcofluor medium atr 2, 5, and 10 days postinoculation (dpi). The exo alleles transduced into the cbrA+ (wild-type [WT]) and cbrA− (cbrA::Tn5) backgrounds are indicated on the left.
To distinguish whether the cbrA extended-halo phenotype is dependent on the functions of known exo genes or is produced through a novel mechanism, we performed a series of genetic epistasis analyses with genes known to be involved in LMW succinoglycan production. For instance, halo formation is delayed in an exoK null mutant disrupted in one of the extracellular glycanases that degrade nascent HMW succinoglycan into LMW products (7) (Fig. 3). The cbrA exoK double mutant produces a halo that is also delayed relative to that of the cbrA single mutant. Interestingly, the exoK mutation is not completely epistatic to the cbrA allele, since the cbrA exoK double mutant produces a halo similar in timing and extent to that produced by the wild type rather than the exoK mutant. Null mutants disrupted for a second extracellular glycanase encoded by exsH were found to produce a halo indistinguishable from that of the wild type (88). Nevertheless, the exsH exoK double mutant completely lacks a halo, revealing that the delayed halo of the exoK mutant is exsH dependent (88). While the cbrA exsH double mutant produces an extended halo indistinguishable from that of the cbrA mutant, the moderate halo of the cbrA exoK double mutant is further delayed upon introduction of the exsH mutation. These results indicate that the activities of both extracellular glycanases contribute to production of the cbrA halo in a manner similar to that observed previously with the wild type.
The succinyltransferase ExoH is also required for production of LMW succinoglycan (48), since nonsuccinylated forms of this EPS are not substrates for either ExoK or ExsH degradation (89). Thus, an exoH null mutant was observed to completely lack a fluorescent halo (7, 47, 89) (Fig. 3). The cbrA exoH double mutant is defective for halo formation in a manner indistinguishable from the exoH single mutant, indicating that the cbrA mutant absolutely requires the succinylation of EPS monomers for halo production. Thus, the cbrA mutant extended-halo phenotype is dependent on the depolymerization of nascent secreted HMW succinoglycan based on the requirement for both ExoH-dependent succinylation and the extracellular glycanases ExoK and ExsH.
The extended-halo phenotype of the cbrA mutant suggests that succinoglycan production could be altered with a shift toward LMW exopolysaccharides at the expense of HMW forms. Alternatively, significant halo formation by the cbrA exoK mutant compared to the exoK mutant indicates that the cbrA allele could affect the absolute amount of succinoglycan produced and thereby lead to increased accumulation of LMW forms. Therefore, we quantified the production of total extracellular polysaccharides by analyzing the anthrone-positive carbohydrate content of conditioned LB/MC medium supernatant. When grown to stationary phase, the cbrA mutant produces approximately 23-fold more EPS than the wild type (Table 4). Under these growth conditions, the exoY mutant produces fourfold less EPS than the wild type. Importantly, the cbrA exoY double mutant produces the same low level of EPS observed with the exoY single mutant, demonstrating that the major extracellular polysaccharide being assayed under these growth conditions is succinoglycan. While the exoH and exoK single mutants produce wild-type levels of EPS, the levels of succinoglycan production in the exoH cbrA and exoK cbrA double mutants resemble that of the cbrA parent strain. Thus, exoH and exoK null mutations do not alter the amount of succinoglycan made but do alter the molecular-weight distribution of the molecule as reflected by fluorescent-halo formation. These results support the possibility that the cbrA mutant produces an extended-halo phenotype as a result of succinoglycan overproduction.
TABLE 4.
Quantitation of anthrone-positive EPSs from stationary-phase cultures
| Strain (genotype) | nM EPS/mg proteina | Ratio (strain/wild type)a |
|---|---|---|
| Rm1021 (wild type) | 15.3 ± 5.8 | 1 |
| KEG2018 (exoY) | 3.46 ± 0.9 | 0.23 |
| KEG2016 (cbrA) | 351 ± 11 | 23 |
| KEG2081 (cbrA exoY) | 5.34 ± 3.1 | 0.35 |
| KEG2019 (exoH) | 15.9 ± 2.2 | 1.03 |
| KEG2020 (cbrA exoH) | 395 ± 9.5 | 26 |
| KEG2101 (exoK) | 14.7 ± 4.8 | 0.96 |
| KEG2104 (cbrA exoK) | 381 ± 10 | 25 |
Standard deviations are derived from triplicate samples.
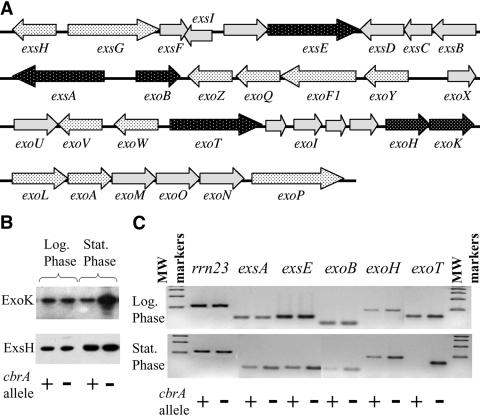
CbrA regulates the expression of genes within the succinoglycan biosynthetic cluster in a growth phase-dependent manner.
To understand the underlying mechanism for altered succinoglycan production in the cbrA mutant, we undertook an expression analysis of genes within the large exo/exs cluster required for production, modification, and secretion of succinoglycan (40) (Fig. 4A). We began by assaying secreted ExoK and ExsH protein levels, since these two extracellular glycanases contribute to the cbrA extended-halo phenotype (Fig. 3). The amounts of secreted ExoK and ExsH in LB/MC culture supernatants were determined by Western blot analysis of trichloroacetic acid-precipitated protein (Fig. 4B). Although we found no difference in secreted ExoK levels detected under exponential-growth conditions, we did observe an increase from the cbrA mutant relative to the wild type during stationary-phase growth. In contrast, the amount of secreted ExsH was unaltered by the presence of the cbrA allele during either exponential- or stationary-phase growth.
FIG. 4.
CbrA regulates the expression of a subset of exo/exs genes in a growth phase-dependent manner. (A) The exo/exs gene cluster on pSymB is required for succinoglycan biosynthesis. The stippled arrows represent genes that were assayed for CbrA-dependent regulation: those in black are derepressed in the cbrA mutant, while those in white are unaffected by cbrA. Genes represented by solid gray arrows were not assayed. (B) ExoK and ExsH levels were assayed by Western blot analysis of cbrA+ (wild type) and cbrA− (cbrA::Tn5) culture supernatants derived from either exponential- or stationary-phase-grown cells. (C) Total RNA isolated from cbrA+ (wild type) and cbrA− (cbrA::Tn5) strains grown to either exponential or stationary phase in LB/MC medium were subjected to RT-PCR analysis. The gene amplified in each RT-PCR is indicated above the gel; although the final products were analyzed in a single gel, a few intervening lanes were cropped out of the final image for clarity.
We wanted to further determine whether CbrA affects the expression of genes involved in succinoglycan production at the transcriptional level. ExoH and ExoT both contribute to the production of LMW succinoglycan (39, 47), while ExoY catalyzes the first step in monomer biosynthesis and is regulated by several transcription factors that modulate the level of succinoglycan production (11, 21, 67, 72, 85). We compared the expression levels of an exoH′-lacZ+, an exoT′-lacZ+, and an exoY′-lacZ+ fusion within cbrA+ and cbrA::Tn5 allelic backgrounds on LB medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The activity of β-galactosidase expressed from an exoY-′lacZ+ fusion is unchanged upon introduction of the cbrA mutation (data not shown). In contrast, the cbrA mutation results in increased β-galactosidase activity from the exoH′-lacZ+ and exoT′-lacZ+ fusions compared to the wild type (data not shown), suggesting that CbrA is able to alter gene expression at the level of either transcription or mRNA stability.
We therefore expanded our study of exo/exs gene expression by performing RT-PCR analyses to determine the extent to which CbrA regulates genes within the succinoglycan biosynthetic cluster. We primarily chose to analyze genes required for succinoglycan biosynthesis or modification, and particularly those that appear to represent the first gene within an operon (Fig. 4A). The wild type and cbrA mutant were grown in LB/MC medium under logarithmic- and stationary-phase growth conditions, and total RNA was isolated. The cbrA mutant displayed increased transcript levels of exsA, exsE, and exoB, as well as confirming the increased expression of exoH and exoT indicated by transcription fusions (Fig. 4C). Interestingly, the CbrA-dependent increase in expression of exsA, exsE, and exoB is strictly limited to stationary-phase growth conditions, as was observed with ExoK (Fig. 4B). Although we did observe a subtle, but consistent, cbrA-dependent increase in exoH and exoT transcripts during exponential growth, the more dramatic change in transcript levels is observed during stationary-phase growth. In contrast, the majority of gene transcripts assayed by RT-PCR (exsG, exsH, exoA, exoF, exoL, exoP, exoQ, exoV, exoW, exoY, exoZ, exoR, exoS, and chvI) are unaffected by the presence of the cbrA::Tn5 allele during stationary-phase growth (data not shown). In total, the results of our analyses indicate that CbrA regulates the expression of a very small subset of exo/exs genes, and primarily under stationary-phase growth conditions. Combined with our previous observations regarding halo formation, these data indicate that the cbrA allele may affect succinoglycan production in two distinct and genetically separable manners: increased biosynthesis of succinoglycan (through exoB regulation), as well as an altered molecular-weight distribution of the extracellular polysaccharide (through exoH, exoT, and ExoK regulation).
The CbrA-regulated exsA and exsE genes are predicted to function as inner membrane ABC transporters; however, their individual substrates have yet to be determined (9, 53). Null mutations in either exsA or exsE were shown to have no effect on calcofluor fluorescence (9, 55). We also found that these null mutations have little to no effect on the cbrA calcofluor-bright phenotype (Fig. 1C). Thus, in contrast to other cbrA-regulated genes, these transporters are not involved in the succinoglycan overproduction phenotype of the cbrA mutant.
The cbrA mutant is sensitive to deoxycholate, which is suggestive of a compromised cell envelope.
In addition to succinoglycan, another aspect of the cell surface that plays a role in symbiosis is the LPS of the outer membrane. Mutants with distinct LPS alterations, but a common sensitivity to detergents in the medium, are unable to establish a chronic intracellular host infection and rapidly degenerate (16, 29, 37). Given the cbrA mutant symbiosis deficiency, we were interested to determine whether the cbrA mutant had phenotypes characteristic of envelope defects. In contrast to the wild type, we observed that the cbrA mutant is unable to form single colonies when grown on LB plates supplemented with either the cationic detergent DOC (10 mM) or the hydrophobic dye crystal violet (1 μg/ml), suggestive of an alteration in the cell envelope of the cbrA mutant (data not shown). However, the cbrA mutant is not universally sensitive to the presence of membrane-disrupting agents in the medium. For instance, there was little to no sensitivity observed during growth in the presence of the cationic detergent sodium dodecyl sulfate or the cationic peptide melittin (data not shown). Therefore, the cbrA mutant appears to be sensitive to a certain spectrum of stress agents. We also observed that the cbrA exoY double mutant, which is unable to produce succinoglycan (Fig. 1A), is as sensitive to DOC as is the cbrA parent, while an exoY single mutant possesses a wild-type level of resistance to this agent (data not shown). Thus, the detergent sensitivity of the cbrA mutant appears to be independent of succinoglycan production.
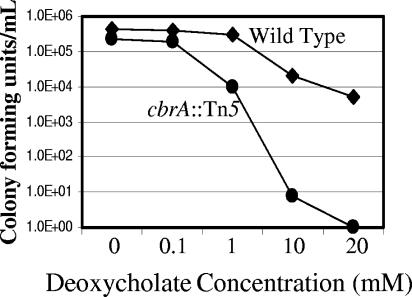
We quantified the DOC sensitivity of the wild type and the cbrA mutant by performing an EOP assay on LB medium containing various amounts of DOC (Fig. 5). With an initial inoculum at an OD600 of 0.1 (ca. 107 cells/ml), we observed the cbrA mutant to be approximately an order of magnitude more sensitive than the wild type to growth in the presence of as little as 1 mM DOC. A more pronounced cbrA mutant sensitivity was observed during growth in the presence of increasing amounts of DOC. We chose a concentration of 10 mM DOC for future studies in order to detect either an exacerbation or suppression of the sensitivity associated with the cbrA::Tn5 allele when combined with additional mutations.
FIG. 5.
The cbrA::Tn5 mutant is sensitive to deoxycholate treatment. Triplicate cultures of cbrA+ (wild-type) and cbrA− (cbrA::Tn5) strains were assayed for growth inhibition in the presence of various concentrations of deoxycholate. The number of CFU was determined after 4 to 5 days of growth at 30°C (further incubation of the plates did not result in the appearance of additional colonies).
Expression of the two ABC family transporters encoded by exsA and exsE is increased in the cbrA mutant. ExsA is related to the E. coli MsbA protein (64% similarity, 30% identity), which is required for transport of lipid A and phospholipids within the inner membrane (24, 92). It was reported that ExsE is a member of the BacA/hALD family of transporters involved in peroxisomal transport of very long-chain fatty acids (27, 53), and very long-chain fatty acids are a unique component of the lipid As in alphaproteobacteria (12, 14). Since the S. meliloti bacA gene is required for symbiosis (37), we looked for CbrA-dependent regulation of a bacA′-′phoA fusion but observed no difference in bacA expression when comparing the wild type to the cbrA mutant (data not shown). In addition, we found that the cbrA bacA double mutant was an order of magnitude more sensitive to DOC treatment than was either single mutant (data not shown), further suggesting that these two genes function independently to alter the cell envelope.
Since homology predicts that ExsA and ExsE could play roles in lipid trafficking and the exsA and exsE mutations had little effect on the cbrA succinoglycan phenotype, we assessed whether these genes play roles in the detergent sensitivity of the cbrA mutant. In comparison to the wild type, neither the exsE nor the exsA null mutant demonstrated altered growth or viability in the presence of 10 mM DOC (Table 5) . The exsE exsA double mutant also has a wild-type level of DOC resistance, indicating that these proteins do not perform a primary function in DOC import or export. In contrast, the cbrA mutant is several orders of magnitude more sensitive to 10 mM DOC than the wild type or exsA and exsE mutants. When either an exsA or exsE null allele is combined with the cbrA mutation, we found that each double mutant remained highly sensitive to DOC. In fact, the double mutants appear to have slightly increased DOC sensitivity relative to the cbrA mutant, indicating that neither exsA nor exsE is required to promote the DOC-sensitive physiology of the cbrA mutant. Thus, increased expression of exsA and exsE does not result in DOC sensitivity, indicating that there is an additional CbrA-regulated gene(s) to be discovered which plays a role in cell envelope integrity.
TABLE 5.
EOP assay for sensitivity to DOC
| Strain (genotype) | LBa
|
EOP (DOC/LB) (%) | |
|---|---|---|---|
| 0 mM DOC | 10 mM DOC | ||
| Rm1021 (wild type) | 1.02 × 108 (±2.2 × 106)* | 6.57 × 107 (±7.7 × 106) | 64 |
| KEG2016 (cbrA) | 5.48 × 107 (±1.2 × 106) | 2,847 (±711) | 5.2 × 10−3 |
| KEG2130 (exsE) | 1.03 × 108 (±6.8 × 106) | 5.90 × 107 (±5.0 × 106) | 57 |
| KEG2131 (cbrA exsE) | 3.51 × 107 (±2.7 × 106) | 223 (±39) | 6.4 × 10−4 |
| KEG2124 (exsA) | 9.29 × 107 (±1.7 × 107) | 5.43 × 107 (±1.7 × 106) | 58 |
| KEG2125 (cbrA exsA) | 5.55 × 107 (±5.8 × 106) | 233 (±113) | 4.2 × 10−4 |
| KEG2126 (exsA exsE) | 9.17 × 107 (±5.7 × 106) | 6.53 × 107 (±4.7 × 106) | 71 |
Numbers in parentheses are standard deviations derived from triplicate samples.
Growth of the cbrA mutant is reduced under oxidative-stress conditions.
During early nodule development, bacterial progression through the infection thread is concurrent with an oxidative burst derived from the plant (75), and mutants sensitive to oxidative stress were found to be symbiotically deficient (42, 74, 79). Since CbrA contains a PAS domain that may provide a redox sensory function (80), and the cbrA mutant is compromised for the symbiosis (Fig. 2A to D), we were interested in determining the role CbrA plays in bacterial resistance to oxidative stress. In contrast to the wild type, we observed that the cbrA mutant is unable to form single colonies when grown on LB containing the oxidative-stress agent hydrogen peroxide (1 mM), indole (1 mM), or cumene hydroperoxide (0.2 mM), as well as the redox-cycling agent plumbagin (100 μM) (data not shown).
To more quantitatively determine the role CbrA plays in protecting the cell from oxidative stress, we performed an EOP assay with the wild type and the cbrA mutant on LB medium containing 1 mM indole. Indole intercalates within the inner membrane and is thought to disrupt the respiratory apparatus, leading to the production of oxygen free radicals (35) and was chosen because of its relative chemical stability. With an initial inoculum at an OD600 of 0.1 (ca. 107 cells/ml), the wild type has an EOP of 102% and the cbrA mutant has a similar EOP of 83%. Unlike the wild type, however, the appearance of cbrA colonies on LB with 1 mM indole requires an additional 48 h of growth compared to LB alone. The delayed appearance of the cbrA mutant indicates that oxidative stress significantly impairs its growth. Thus, the cbrA mutant is compromised in its ability to combat oxidative stress, although not to the extent that viability is adversely affected.
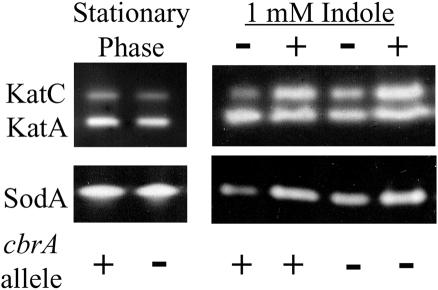
Given our observation that CbrA regulates the expression of several exo genes that alter succinoglycan production, we considered the possibility that CbrA plays a role in oxidative-stress resistance through regulation of one or more of the cellular proteins that combat this stress. To test this possibility, we assayed catalase and superoxide dismutase activities from whole-cell lysates that had been subjected to native PAGE (41, 74). We observed similar levels of SodB, KatA, and KatC activities in the cbrA mutant and the wild type during stationary-phase growth and during exponential growth in the presence of indole (Fig. 6). Thus, CbrA does not appear to play a critical role in regulating the activities of these proteins.
FIG. 6.
The oxidative-stress sensitivity of the cbrA mutant is not due to altered KatA/C or SodB expression. The cbrA+ (wild-type) and cbrA− (cbrA::Tn5) strains were grown to stationary phase in LB/MC medium or into logarithmic phase in LB/MC medium and subjected to 5 μl of 70% ethanol (no indole control) or 5 μl of 1 M indole (1 mM indole) as indicated above the gels. Whole-cell lysates were obtained using a French press and subjected to 10% acrylamide SDS-PAGE. Catalase activity was assayed as described previously (79) with 15 μg of lysate, and superoxide dismutase activity was assayed as described previously (73) with 12 μg of lysate.
DISCUSSION
Through a calcofluor-bright screen, we have isolated a previously unidentified regulator, CbrA, which is critically required for an effective symbiosis. CbrA is a putative two-component sensor kinase containing a PAS domain, and it controls aspects of ex planta cell physiology relevant to symbiotic development. The cbrA mutant is compromised for the symbiosis, eliciting nodules that are mostly Fix− or Ndv−, and it appears unable to effectively establish a chronic infection based on the decreased and highly variable numbers of bacteria recovered from cbrA-induced nodules. This defect is further exacerbated when the cbrA mutant is forced to compete with the wild type for nodule occupancy, resulting in the complete absence of cbrA bacteria within the nodules of coinoculated plants. This absence of cbrA bacteria within coinoculated nodules also indicates that wild-type bacteria are unable to rescue the symbiotic defect of the cbrA mutant, which implies that the cbrA defect cannot be explained simply as a lack of symbiotically active succinoglycan. Our identification of cbrA is particularly interesting, since there are sparingly few regulators, for instance, NodD (83) and Fix/Nif proteins (32), that have been described as being required for symbiosis, despite the fact that the developmental program dictating nodulation requires continual communication between the symbiotic partners. Characterization of NodD and Fix/Nif regulators over the past several decades has provided a wealth of information about the complex requirements for symbiosis, and we anticipate that additional insights into CbrA function will also prove important to understanding the rhizobium-legume symbiosis.
Consistent with the calcofluor-bright phenotype of the cbrA mutant, our quantitative measurements of secreted extracellular carbohydrate revealed a 23-fold overproduction of succinoglycan compared to the wild type. The enzymes responsible for succinoglycan production and modification have been identified and characterized in great detail (10, 40). The exoB gene is among the small subset of CbrA-regulated exo genes, and exoB encodes an epimerase required for formation of the UDP-galactose that serves as a substrate for the first step in monomer biosynthesis catalyzed by ExoY (15, 18, 72). In contrast, the galactose-1-P-transferase encoded by exoY and the glucosyl transferases encoded by exoA, -L, and -W, which function as core enzymes in the assembly of succinoglycan monomers (72), were not targets of CbrA regulation. Thus, the succinoglycan overproduction phenotype of the cbrA mutant appears to depend heavily on differential exoB expression.
The cbrA succinoglycan overproduction phenotype is accompanied by a calcofluor extended-halo phenotype. Each of these phenotypes requires exoY function, and in addition, the extended-halo phenotype requires ExoH, ExoK, and to a lesser extent ExsH. For reasons still not understood, exoT mutants are unable to secrete succinoglycan into the medium (72), so we were unable to test the genetic requirement for exoT in the cbrA mutant calcofluor-bright and extended-halo phenotypes.
While our genetic analyses show that the extended-halo phenotype of the cbrA mutant reflects an overproduction of LMW succinoglycan, it will be of great interest to biochemically determine the ratio of LMW to HMW succinoglycan and the relative degree of succinylation. However, we predict that the cbrA mutant specifically overproduces LMW forms of succinoglycan at the expense of HMW forms because of the limited number of exo genes whose expression is altered in the cbrA mutant. ExoH, ExoK, and ExoT each play a role in promoting LMW succinoglycan production (39, 47, 88), and the expression of these factors is increased in the cbrA mutant. We failed to observe CbrA-dependent regulation of the exoV and exoZ genes, which encode enzymes that add pyruvyl or acetyl substituents to the succinoglycan monomer (71, 72). Accordingly, it appears that CbrA plays a role in modulating two specific and related aspects of succinoglycan composition: monomer succinylation and polysaccharide molecular-weight distribution.
Much of our understanding of the role succinoglycan plays in promoting the symbiosis suggests that the symbiotically competent form of this exopolysaccharide must be succinylated (38, 47). In addition, it has been shown that LMW forms of succinoglycan are more effective than HMW forms at promoting infection thread growth and nodulation (6, 82, 84). Interestingly, the cbrA mutant overproduces LMW succinoglycan, and it seems likely that this exopolysaccharide is succinylated based on both the genetic requirement for exoH in extended-halo formation and the increased expression of exoH in the mutant. Thus, the cbrA mutant appears poised for efficient invasion of its host; however, cbrA bacteria were found to be defective for proper nodule development on alfalfa. Although this apparent contradiction is currently unresolved, it is possible that the cbrA regulatory mutant fails to produce the correct form of succinoglycan under the specific conditions present during nodule formation. Alternatively, an overproduction of LMW succinoglycan may itself provoke a nonproductive response from the plant. However, exoZ null mutants overproduce LMW succinoglycan and still elicit Fix+ nodules on alfalfa (71), despite a decreased efficiency of infection thread formation (19), making this possibility less likely. Perhaps the cbrA mutant makes symbiotically competent succinoglycan and the symbiotic defect of the mutant derives from an inability to modulate succinoglycan composition during later developmental stages.
Alternatively, the inability of the cbrA mutant to establish an effective symbiosis with alfalfa could be related to the physiological defects that express themselves through sensitivity to DOC and indole. In particular, the cbrA mutant may be less competent to withstand harsh aspects of the host environment, such as the oxidative burst, and these stress sensitivities could account for the inability of cbrA mutants to compete with the wild type for colonization of alfalfa nodules. It is tempting to speculate that the stress sensitivities of the cbrA mutant are related to changes in the composition of LPSs of the outer membrane. The exoB null mutant has an altered LPS profile suggestive of O-antigen modifications (47), so it is possible exoB overexpression in the cbrA mutant could affect the LPS carbohydrate content. However, it remains to be directly determined whether the cbrA mutant has alterations in its LPS composition or whether the DOC and indole sensitivities of the cbrA mutant derive from an alternative cell envelope component.
Prior to our identification of CbrA, a number of regulators were known to affect succinoglycan production in S. meliloti. However, to our knowledge, CbrA is the only regulator known to affect the expression of exoB rather than exoY. For instance, a null mutation in the LysR family transcription factor mucR results in succinoglycan overproduction with a shift toward LMW forms through regulation of exoF, exoH, exoK, exoX, and exoY expression (11, 44, 67). The two-component sensor ExoS also regulates succinoglycan levels (25), presumably via the cognate response regulator ChvI (20), but modulates expression of exoF, exoP, and exoY (21, 25, 85). Succinoglycan production is also regulated by ExoR and SyrA through unknown and indirect mechanisms (4, 21, 25, 67, 68). In addition to these regulatory factors, succinoglycan levels are responsive to the presence of noncarbon nutrients (25, 49), while the ratio of LWM and HMW succinoglycans reacts to osmotic conditions (91). Thus, regulation of succinoglycan production and composition is quite complex and is responsive to a number of environmental and intracellular sensory inputs.
Interestingly, the exo/exs genes subject to CbrA regulation appear to be primarily affected under stationary-phase growth conditions. The exact nature of the stationary-phase signal remains an outstanding question, but given that CbrA possesses a sensory PAS domain, it is possible that CbrA senses changes in intracellular redox as a result of altered metabolism. Given that CbrA plays a key role in establishing the symbiosis, this regulator is poised to provide bacteria important information about their location or development within the nodule. Ultimately, insights gained from characterizing how CbrA activity is modulated will be highly relevant to understanding bacterial perception of the host environment.
Supplementary Material
Acknowledgments
We are indebted to Anke Becker for providing the exo′-lacZ+ fusions used in this study (8, 9) and to A. Becker and Karsten Neihaus for helping to determine the Tn5 insertion site in the cbrA mutant. We thank Mary Lou Pardue for generously allowing us to use her microscope. We thank Veronica Godoy, Bryan Davies, Sanjay D'Souza, and Lyle Simmons for critical reading of the manuscript. Special thanks go to Hajime Kobayashi for careful reading and discussion of the manuscript on many occasions.
This work was supported by National Institutes of Health grant GM31010 (to G.C.W.), a National Science Foundation postdoctoral fellowship (to K.E.G.), and a Fullbright/Spanish Ministry of Education and Science postdoctoral fellowship (to J.L.). G.C.W. is an American Cancer Society Research Professor and a Howard Hughes Medical Institute Professor.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aman, P., M. McNeil, L. E. Franzen, A. G. Darvill, and P. Albersheim. 1981. Structural elucidation, using HPLC-MS and GLC-MS, of the acidic exopolysaccharide secreted by Rhizobium meliloti strain Rm1021. Carbohydr. Res. 95:263-282. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1991. Current protocols in molecular biology, vol. 1. John Wiley and Sons Inc., New York, N.Y.
- 3.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. BioTechniques 29:240-242, 244-245. [DOI] [PubMed] [Google Scholar]
- 4.Barnett, M. J., J. A. Swanson, and S. R. Long. 1998. Multiple genetic controls on Rhizobium meliloti syrA, a regulator of exopolysaccharide abundance. Genetics 148:19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battisti, L., J. C. Lara, and J. A. Leigh. 1992. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc. Natl. Acad. Sci. USA 89:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, A., A. Kleickmann, W. Arnold, and A. Puhler. 1993. Analysis of the Rhizobium meliloti exoH/exoK/exoL fragment: ExoK shows homology to excreted endo-beta-1,3-1,4-glucanases and ExoH resembles membrane proteins. Mol. Gen. Genet. 238:145-154. [DOI] [PubMed] [Google Scholar]
- 8.Becker, A., A. Kleickmann, M. Keller, W. Arnold, and A. Puhler. 1993. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol. Gen. Genet. 241:367-379. [DOI] [PubMed] [Google Scholar]
- 9.Becker, A., H. Kuster, K. Niehaus, and A. Puhler. 1995. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol. Gen. Genet. 249:487-497. [DOI] [PubMed] [Google Scholar]
- 10.Becker, A., S. Ruberg, B. Baumgarth, P. A. Bertram-Drogatz, L. Quester, and A. Puhler. 2002. Regulation of succinoglycan and galactoglucan biosynthesis in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 4:187-190. [PubMed] [Google Scholar]
- 11.Bertram-Drogatz, P. A., I. Quester, A. Becker, and A. Puhler. 1998. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 257:433-441. [DOI] [PubMed] [Google Scholar]
- 12.Bhat, U. R., H. Mayer, A. Yokota, R. I. Hollingsworth, and R. W. Carlson. 1991. Occurrence of lipid A variants with 27-hydroxyoctacosanoic acid in lipopolysaccharides from members of the family Rhizobiaceae. J. Bacteriol. 173:2155-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brewin, N. J. 1991. Development of the legume root nodule. Annu. Rev. Cell Biol. 7:191-226. [DOI] [PubMed] [Google Scholar]
- 14.Brozek, K. A., R. W. Carlson, and C. R. Raetz. 1996. A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium. J. Biol. Chem. 271:32126-32136. [DOI] [PubMed] [Google Scholar]
- 15.Buendia, A. M., B. Enenkel, R. Koplin, K. Niehaus, W. Arnold, and A. Puhler. 1991. The Rhizobium meliloti exoZ-exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol. Microbiol. 5:1519-1530. [DOI] [PubMed] [Google Scholar]
- 16.Campbell, G. R., B. L. Reuhs, and G. C. Walker. 2002. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. USA 99:3938-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell, G. R., L. A. Sharypova, H. Scheidle, K. M. Jones, K. Niehaus, A. Becker, and G. C. Walker. 2003. Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J. Bacteriol. 185:3853-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canter Cremers, H. C., M. Batley, J. W. Redmond, L. Eydems, M. W. Breedveld, L. P. Zevehuizen, E. Pees, C. A. Wijffelman, and B. J. Lugtenberg. 1990. Rhizobium leguminosarum exoB mutants are deficient in the synthesis of UDP-glucose 4′-epimerase. J. Biol. Chem. 265:21122-21127. [PubMed] [Google Scholar]
- 19.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng, H. P., and S. Y. Yao. 2004. The key Sinorhizobium meliloti succinoglycan biosynthesis gene exoY is expressed from two promoters. FEMS Microbiol. Lett. 231:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cronan, G. E., and D. H. Keating. 2004. Sinorhizobium meliloti sulfotransferase that modifies lipopolysaccharide. J. Bacteriol. 186:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doerrler, W. T., H. S. Gibbons, and C. R. Raetz. 2004. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279:45102-45109. [DOI] [PubMed] [Google Scholar]
- 25.Doherty, D., J. A. Leigh, J. Glazebrook, and G. C. Walker. 1988. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol. 170:4249-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrhardt, D. W., E. M. Atkinson, and S. R. Long. 1992. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256:998-1000. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson, G. P., A. Datta, J. Baumgartner, R. M. Roop II, R. W. Carlson, and G. C. Walker. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. USA 101:5012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferguson, G. P., A. Datta, R. W. Carlson, and G. C. Walker. 2005. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol. Microbiol. 56:68-80. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson, G. P., R. M. Roop II, and G. C. Walker. 2002. Deficiency of a Sinorhizobium meliloti BacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 184:5625-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferraioli, S., R. Tate, M. Cermola, R. Favre, M. Iaccarino, and E. J. Patriarca. 2002. Auxotrophic mutant strains of Rhizobium etli reveal new nodule development phenotypes. Mol. Plant-Microbe Interact. 15:501-510. [DOI] [PubMed] [Google Scholar]
- 31.Finan, T. M., E. Hartweig, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraysse, N., F. Couderc, and V. Poinsot. 2003. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur. J. Biochem. 270:1365-1380. [DOI] [PubMed] [Google Scholar]
- 34.Freidman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Use of cos derivatives for pRK290 in constructing a clone bank of Rhizobium meliloti DNA. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 35.Garbe, T. R., M. Kobayashi, and H. Yukawa. 2000. Indole-inducible proteins in bacteria suggest membrane and oxidant toxicity. Arch. Microbiol. 173:78-82. [DOI] [PubMed] [Google Scholar]
- 36.Geurts, R., E. Fedorova, and T. Bisseling. 2005. Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8:346-352. [DOI] [PubMed] [Google Scholar]
- 37.Glazebrook, J., A. Ichige, and G. C. Walker. 1993. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 7:1485-1497. [DOI] [PubMed] [Google Scholar]
- 38.Glazebrook, J., J. W. Reed, T. L. Reuber, and G. C. Walker. 1990. Genetic analyses of Rhizobium meliloti exopolysaccharides. Int. J. Biol. Macromol. 12:67-70. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez, J. E., C. E. Semino, L. X. Wang, L. E. Castellano-Torres, and G. C. Walker. 1998. Biosynthetic control of molecular weight in the polymerization of the octasaccharide subunits of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 95:13477-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez, J. E., G. M. York, and G. C. Walker. 1996. Rhizobium meliloti exopolysaccharides: synthesis and symbiotic function. Gene 179:141-146. [DOI] [PubMed] [Google Scholar]
- 41.Herouart, D., S. Sigaud, S. Moreau, P. Frendo, D. Touati, and A. Puppo. 1996. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J. Bacteriol. 178:6802-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamet, A., S. Sigaud, G. Van de Sype, A. Puppo, and D. Herouart. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant-Microbe Interact. 16:217-225. [DOI] [PubMed] [Google Scholar]
- 43.Keating, D. H., M. G. Willits, and S. R. Long. 2002. A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J. Bacteriol. 184:6681-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller, M., A. Roxlau, W. M. Weng, M. Schmidt, J. Quandt, K. Niehaus, D. Jording, W. Arnold, and A. Puhler. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant-Microbe Interact. 8:267-277. [DOI] [PubMed] [Google Scholar]
- 45.Kerppola, T. K., and M. L. Kahn. 1988. Symbiotic phenotypes of auxotrophic mutants of Rhizobium meliloti 104A14. J. Gen. Microbiol. 134:913-919. [DOI] [PubMed] [Google Scholar]
- 46.Lagares, A., G. Caetano-Anolles, K. Niehaus, J. Lorenzen, H. D. Ljunggren, A. Puhler, and G. Favelukes. 1992. A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J. Bacteriol. 174:5941-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leigh, J. A., and C. C. Lee. 1988. Characterization of polysaccharides of Rhizobium meliloti exo mutants that form ineffective nodules. J. Bacteriol. 170:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leigh, J. A., J. W. Reed, J. F. Hanks, A. M. Hirsch, and G. C. Walker. 1987. Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell 51:579-587. [DOI] [PubMed] [Google Scholar]
- 49.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lerouge, I., and J. Vanderleyden. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26:17-47. [DOI] [PubMed] [Google Scholar]
- 51.Lestrate, P., A. Dricot, R. M. Delrue, C. Lambert, V. Martinelli, X. De Bolle, J. J. Letesson, and A. Tibor. 2003. Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect. Immun. 71:7053-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LeVier, K., R. W. Phillips, V. K. Grippe, R. M. Roop II, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492-2493. [DOI] [PubMed] [Google Scholar]
- 53.LeVier, K., and G. C. Walker. 2001. Genetic analysis of the Sinorhizobium meliloti BacA protein: differential effects of mutations on phenotypes. J. Bacteriol. 183:6444-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loewus, F. A. 1952. Improvement in anthrone method for determination of carbohydrates. Anal. Chem. 24:219. [Google Scholar]
- 55.Long, S., J. W. Reed, J. Himawan, and G. C. Walker. 1988. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J. Bacteriol. 170:4239-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long, S. R. 1996. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 8:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long, S. R., G. B. Ruvkun, H. M. Meade, W. J. Buikema, S. E. Brown, A. M. Friedman, and F. M. Ausubel. 1983. Genetic analysis of symbiotic nitrogen fixation. Dev. Ind, Microbiol. 24:21-29. [Google Scholar]
- 58.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreno, E., E. Stackebrandt, M. Dorsch, J. Wolters, M. Busch, and H. Mayer. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172:3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller, P., M. Keller, W. M. Weng, J. Quandt, W. Arnold, and A. Puhler. 1993. Genetic analysis of the Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol. Plant-Microbe Interact. 6:55-65. [DOI] [PubMed] [Google Scholar]
- 61.Mulligan, J. T., and S. R. Long. 1989. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics 122:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niehaus, K., and A. Becker. 1998. The role of microbial surface polysaccharides in the Rhizobium-legume interaction. Subcell. Biochem. 29:73-116. [DOI] [PubMed] [Google Scholar]
- 63.Niehaus, K., A. Lagares, and A. Puhler. 1998. A Sinorhizobium meliloti lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol. Plant-Microbe Interact. 11:906-914. [Google Scholar]
- 64.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 65.Oke, V., and S. R. Long. 1999. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 2:641-646. [DOI] [PubMed] [Google Scholar]
- 66.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:R674-R677. [DOI] [PubMed] [Google Scholar]
- 67.Quester, I., and A. Becker. 2004. Four promoters subject to regulation by ExoR and PhoB direct transcription of the Sinorhizobium meliloti exoYFQ operon involved in the biosynthesis of succinoglycan. J. Mol. Microbiol. Biotechnol. 7:115-132. [DOI] [PubMed] [Google Scholar]
- 68.Reed, J. W., J. Glazebrook, and G. C. Walker. 1991. The exoR gene of Rhizobium meliloti affects RNA levels of other exo genes but lacks homology to known transcriptional regulators. J. Bacteriol. 173:3789-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reinhold, B. B., S. Y. Chan, T. L. Reuber, A. Marra, G. C. Walker, and V. N. Reinhold. 1994. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J. Bacteriol. 176:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reuber, T. L., J. Reed, J. Glazebrook, M. A. Glucksmann, D. Ahmann, A. Marra, and G. C. Walker. 1991. Rhizobium meliloti exopolysaccharides: genetic analyses and symbiotic importance. Biochem. Soc. Trans. 19:636-641. [DOI] [PubMed] [Google Scholar]
- 71.Reuber, T. L., and G. C. Walker. 1993. The acetyl substituent of succinoglycan is not necessary for alfalfa nodule invasion by Rhizobium meliloti Rm1021. J. Bacteriol. 175:3653-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269-280. [DOI] [PubMed] [Google Scholar]
- 73.Santos, R., S. Bocquet, A. Puppo, and D. Touati. 1999. Characterization of an atypical superoxide dismutase from Sinorhizobium meliloti. J. Bacteriol. 181:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santos, R., D. Herouart, A. Puppo, and D. Touati. 2000. Critical protective role of bacterial superoxide dismutase in rhizobium-legume symbiosis. Mol. Microbiol. 38:750-759. [DOI] [PubMed] [Google Scholar]
- 75.Santos, R., D. Herouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact. 14:86-89. [DOI] [PubMed] [Google Scholar]
- 76.Scheidle, H., A. Gross, and K. Niehaus. 2005. The lipid A substructure of the Sinorhizobium meliloti lipopolysaccharides is sufficient to suppress the oxidative burst in host plants. New Phytol. 165:559-565. [DOI] [PubMed] [Google Scholar]
- 77.Scupham, A. J., and E. W. Triplett. 1997. Isolation and characterization of the UDP-glucose 4′-epimerase-encoding gene, galE, from Brucella abortus 2308. Gene 202:53-59. [DOI] [PubMed] [Google Scholar]
- 78.Sharypova, L. A., K. Niehaus, H. Scheidle, O. Holst, and A. Becker. 2003. Sinorhizobium meliloti acpXL mutant lacks the C28 hydroxylated fatty acid moiety of lipid A and does not express a slow migrating form of lipopolysaccharide. J. Biol. Chem. 278:12946-12954. [DOI] [PubMed] [Google Scholar]
- 79.Sigaud, S., V. Becquet, P. Frendo, A. Puppo, and D. Herouart. 1999. Differential regulation of two divergent Sinorhizobium meliloti genes for HPII-like catalases during free-living growth and protective role of both catalases during symbiosis. J. Bacteriol. 181:2634-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor, B. L., and I. B. Zhulin. 1990. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ugalde, R. A. 1999. Intracellular lifestyle of Brucella spp. Common genes with other animal pathogens, plant pathogens, and endosymbionts.Microbes Infect. 1:1211-1219. [DOI] [PubMed] [Google Scholar]
- 82.Urzainqui, A., and G. C. Walker. 1992. Exogenous suppression of the symbiotic deficiencies of Rhizobium meliloti exo mutants. J. Bacteriol. 174:3403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Rhijn, P., and J. Vanderleyden. 1995. The Rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang, L. X., Y. Wang, B. Pellock, and G. C. Walker. 1999. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J. Bacteriol. 181:6788-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43:1115-1127. [DOI] [PubMed] [Google Scholar]
- 86.Wolanin, P. M., P. A. Thomason, and J. B. Stock. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3:3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.York, G. M., and G. C. Walker. 1998. The Rhizobium meliloti ExoK and ExsH glycanases specifically depolymerize nascent succinoglycan chains. Proc. Natl. Acad. Sci. USA 95:4912-4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.York, G. M., and G. C. Walker. 1997. The Rhizobium meliloti exoK gene and prsD/prsE/exsH genes are components of independent degradative pathways which contribute to production of low-molecular-weight succinoglycan. Mol. Microbiol. 25:117-134. [DOI] [PubMed] [Google Scholar]
- 89.York, G. M., and G. C. Walker. 1998. The succinyl and acetyl modifications of succinoglycan influence susceptibility of succinoglycan to cleavage by the Rhizobium meliloti glycanases ExoK and ExsH. J. Bacteriol. 180:4184-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zevehuizen, L. P. T. M., and A. R. W. van Neerven. 1983. (1-2)-β-d-Glucan and acidic oligosaccharides produced by Rhizobium meliloti. Carbohydr. Res. 118:127-134. [Google Scholar]
- 91.Zevenhuizen, L. P., and P. Faleschini. 1991. Effect of the concentration of sodium chloride in the medium on the relative proportions of poly- and oligo-saccharides excreted by Rhizobium meliloti strain YE-2SL. Carbohydr Res. 209:203-209. [DOI] [PubMed] [Google Scholar]
- 92.Zhou, Z., K. A. White, A. Polissi, C. Georgopoulos, and C. R. Raetz. 1998. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273:12466-12475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.