Abstract
High segregational stability of the streptococcal plasmid pSM19035 is achieved by the concerted action of systems involved in plasmid copy number control, multimer resolution, and postsegregational killing. In this study, we demonstrate the role of two genes, δ and ω, in plasmid stabilization by a partition mechanism. We show that these two genes can stabilize the native pSM19035 replicon as well as other θ- and σ-type plasmids in Bacillus subtilis. In contrast to other known partition systems, in this case the two genes are transcribed separately; however, they are coregulated by the product of the parB-like gene ω. Analysis of mutants of the parA-like gene δ showed that the Walker A ATPase motif is necessary for plasmid stabilization. The ParB-like product of the ω gene binds to three regions containing repeated WATCACW heptamers, localized in the copS (regulation of plasmid copy number), δ, and ω promoter regions. We demonstrate that all three of these regions can cause partition-mediated incompatibility. Moreover, our data suggest that each of these could play the role of a centromere-like sequence. We conclude that δ and ω constitute a novel type of plasmid stabilization system.
Bacterial plasmids are usually stably maintained in their hosts (23, 29, 41). High-copy-number plasmids can rely on random distribution among daughter cells. However, when the plasmid is present in only a few copies, a mechanism that ensures better-than-random distribution is required. The stability can be increased by the activity of site-specific recombinases (resolvases) that resolve plasmid oligomers formed due to plasmid replication or recombination (41). Other types of mechanisms allowing high plasmid stability in the bacterial population are postsegregational killing (PSK) systems (21, 31) and partition systems (3, 19, 23, 29, 38). Plasmid partitioning, which ensures a better-than-random plasmid distribution, guarantees the presence of at least one plasmid molecule in the future daughter cell by a mitosis-like event, whereas postsegregational killing systems prevent the appearance of plasmid-free cells in the bacterial population. A PSK system comprises a labile antidote and a more stable toxin. When the cell carries the plasmid, both proteins are produced and the antidote prevents killing of the cell by the toxin. When a daughter cell does not inherit the plasmid, it is killed by the toxin, while the antidote is degraded or no longer produced (27, 55).
Plasmid partition systems are generally composed of two genes which are organized in one operon encoding a ParA protein (ATPase) and a DNA-binding ParB protein. The third element of the system is a centromere-like region to which the ParB protein binds (3). The partition systems are classified into two distinct groups based on gene organization, the type of ATPase encoded by the parA gene, and the location of the centromere-like sequence (19).
Type I ParA proteins include ATPases containing Walker motifs, whereas type II proteins include actin-like ATPases (19, 20). Further classification of partitioning systems, based on the organization of the transcriptional unit and its regulation, distinguishes two subgroups of group I. Subgroup Ia is represented by plasmids such as F and P1, where the parAB operon is regulated by the ParA protein and the parS sequence is localized downstream of the operon. The characteristic feature of the ParA proteins of type Ia partition systems is the presence of an N-terminal DNA-binding domain (3). Hence, these proteins can bind to their promoters and regulate the transcription of the par operon (28, 43). In subgroup Ib systems, the ParB protein binds upstream of the operon and regulates its transcription (32). Type II partition systems are regulated in a similar way to Ib systems, where a ParB homolog binds to a centromere-like sequence located in the par operon promoter. The other factor which distinguishes subgroups Ia and Ib is the size of the encoded proteins. Subgroup Ia ParA proteins vary in size from 251 to 420 amino acids (aa), while the ParB proteins vary from 182 to 336 aa; both types are larger than proteins from subgroup Ib (ParA, 208 to 227 aa; ParB, 46 to 113 aa) (3).
In contrast to ParA proteins, ParB proteins cannot be classified according to their sequence similarities (19). However, they share some characteristic features, including DNA binding, dimerization, and interaction with ParA (13, 35, 54). ParB can also polymerize on DNA near the centromeric sequence and therefore silence genes by reducing their accessibility to cellular components (13, 36, 45).
Generally, ParB binding sequences are present only once within a plasmid. However, there are exceptions, such as the KorB protein (a ParB homolog) encoded by the broad-host-range plasmid RK2, which has dual functions as a “global” transcriptional repressor and a partitioning protein which binds to 12 sites within the plasmid (39, 42). Only one of these binding sites, OB3, is presumably required for partitioning, whereas other KorB-binding sites cause destabilization of the plasmid when OB3 is not present (53). An unusual organization of the centromere-like sites was also found for the linear prophage N15. Four SopB (a ParB homolog) binding sites are dispersed in the genome (44), and each of them can act as the centromere-like sequence (24).
The majority of the partition systems studied so far are encoded by plasmids from gram-negative bacteria, and only a few from gram-positive bacteria have been characterized.
The first well-characterized partition system from gram-positive bacteria is encoded by the pCI2000 plasmid from Lactococcus lactis (33). Its ParA protein is similar to partitioning Walker-type ATPases without the N-terminal regulatory part. A region upstream of parA contains repeated sequences constituting the putative centromere-like element. However, parA and the centromere-like sequence are not sufficient for efficient plasmid stabilization, which suggests that other elements (possibly a small orf1 located downstream) are needed (33).
Recently, a partition system of a novel type was found, encoded by the pSK1 plasmid from Staphylococcus aureus (49). The pSK1 partition seems to be very unusual; in contrast to other systems, only one gene—parA—is needed for plasmid stabilization. The ParA protein from pSK1 has no sequence similarity to type I or II ATPases, but the gene shares similarity with plasmid genes of unknown function from Staphylococcus, Streptococcus, Lactococcus, Lactobacillus, Clostridium, and Tetragenococcus (16, 49). Its homolog encoded by the Tetragenococcus halophilus plasmid pUCL287 was shown to influence the stability and plasmid copy number (2). A putative partition system from gram-positive bacteria is also encoded by the stable pAW63 plasmid from Bacillus thuringiensis (52).
One of the best-studied examples of stably maintained plasmids from gram-positive bacteria is the low-copy-number streptococcal plasmid pSM19035 (37). The high segregational stability of the pSM19035 replicon in Bacillus subtilis cells (10, 11) was shown to be ensured by the multimer resolution system encoded by the β gene (SegA region in Fig. 1) (46) and by strict regulation of the plasmid copy number (5-7, 14, 50). Stabilization is also ensured by the activity of the postsegregational killing system encoded by the ɛ and ζ genes (SegB region in Fig. 1) (9, 56). There are two other genes within the SegB region of pSM19035, localized upstream of the ɛ and ζ genes. The first one, δ, encodes a putative ATPase containing characteristic Walker motifs (11), and the second encodes the protein Omega, which was shown to be a DNA-binding protein and to act as the global regulator of pSM19035 gene expression (14, 50). Omega represses transcription by binding to repeated heptamers (WATCACW) located upstream of its own gene as well as upstream of the copS (copy number control) and δ (putative ATPase) genes. Omega links random (SegA region) and better-than-random (SegB region) plasmid distributions (14, 15, 50).
FIG. 1.
Localization of genes carried by one of the repeated arms of pSM19035. Regions SegA and SegB are involved in plasmid maintenance. SegA controls random plasmid distribution (resolvase β and topoisomerase γ). The SegB region contains genes encoding the plasmid addiction system (ɛ and ζ), a global regulator of plasmid gene expression (ω), and a putative ATPase (δ). Repeated sequences (WATCACW) in the promoter regions of copS, δ, and ω are denoted by arrows.
Because of the homology of Delta to ParA ATPases, its regulation by the closely located gene ω, and the presence of Omega binding sites, which could play the role of centromeric sequences, we hypothesized that the δ and ω genes could act together as a plasmid partitioning system of a novel type. This study presents evidence to support this hypothesis.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this work are listed in Table 1. Plasmids pBT205O+E and pOSE10 were integrated into the B. subtilis amyE locus via double crossover to generate strains YB1015/O+E and YB1015/OSE, respectively.
TABLE 1.
Bacterial strains used in this work
| Strain | Genotype and description | Reference |
|---|---|---|
| Escherichia coli strain | ||
| DH5α | F−gyrA96 recA1 relA1 endA1 thi-1 hsdR17 supE44 deoR Δ(lacZYA-argF)U169 φ80dlacZΔM15 | 26 |
| Bacillus subtilis strains | ||
| YB1015 | amyE metB5 trpC2 xin-1 attSPβ recA4 | 17 |
| YB1015/δ::lacZ | YB1015 derivative carrying integrated δ-lacZ fusion (pDB101 coordinates 4693 to 5798)a | 14 |
| YB1015/O+E | YB1015 derivative carrying integrated ω-ɛ-lacZ fusion (pDB101 coordinates 6305 to 6915)a | This work |
| YB1015/OSE | Same as above, but with inactivated ω gene (stop codon within sixth triplet) (pDB101 coordinates 6305 to 6894)a | This work |
pDB101 is a pSM19035 derivative which contains the two long inverted repeats and possesses all functions associated with replication, copy control, stable maintenance, and antibiotic resistance (accession no. X66468).
Plasmid construction.
Plasmids used for integration into the B. subtilis chromosome were constructed as follows. To generate pBT205O+E, the ω gene, its promoter sequence, and part of the ɛ gene were amplified by PCR with primers CG1 and CG2 and cloned into the pBT205 plasmid. As the first step in the construction of pOSE10, we generated plasmid pOSZ5. A HindIII/SspI fragment of pBT346-5 and an SspI/EcoRI fragment of pBT346 were simultaneously ligated into HindIII/EcoRI-digested pHP13 vector. pOSZ5 contains a fragment encompassing the region from the ω promoter to the end of the ζ gene, with a stop codon within the ω gene. A HindIII/HindIII fragment containing the ω promoter, inactivated ω, and part of the ɛ gene was subcloned into pBT205 to generate plasmid pOSE10.
We created a series of plasmids for stability and partition-mediated incompatibility tests that carried the following replicons: (i) the native θ pSM19035 replicon (pUSE2 and its derivatives), (ii) a nonnative θ replicon (pUHC13 and its derivatives), and (iii) a σ replicon (pHP13 and its derivatives). The pDO plasmid series served as a source of fragments from the δ-ω region for subsequent cloning. The plasmids and primers used in this work are listed in Table 2 and Table 3, respectively.
TABLE 2.
Plasmids used in this worka
| Plasmid | Relevant features | Reference |
|---|---|---|
| pBT205 | Vector for integration into B. subtilis chromosome, promoterless lacZ gene, ColE1 ori Apr Cmr | 14 |
| pBT205O+E | pBT205 with ω gene with its promoter, Pω, and part of the ɛ gene | This work |
| pBT233 | pSM19035 derivative, accession no. X64695 | 11 |
| pBT346 | pHP13 with δ, ω, ɛ, and ζ genes (pDB101 coordinates 4696 to 8271), HincII-HincII pBT233 fragment cloned into SmaI siteb | 11 |
| pBT346-5 | pHP13 with ω gene (pDB101 coordinates 6312 to 6894) (sixth triplet mutated into a stop codon)b | 14 |
| pDO1000 | pUC18 with δ and ω genes (pDB101 coordinates 5382 to 6740)b | This work |
| pDO1030 | pUC18 with δ and ω genes, with δ truncated after the 23rd codon | This work |
| pDO1040 | pUC18 with δ and ω genes, with δ truncated after the 40th codon | This work |
| pDO1212 | pUC18 with δ gene promoter (Pδ) | This work |
| pDO1340 | pUC18 with δ and ω genes, with δ's Walker motif A deleted | This work |
| pDO3000 | pUC18 with δ and ω genes, with δ promoter (Pδ) replaced with copS promoter (PcopS) | This work |
| pDO3212 | pUC18 with copS gene promoter (PcopS) | This work |
| pDO7000 | pUC18 with δ and ω genes, with δ promoter (Pδ) replaced with ω promoter (Pω) | This work |
| pDO7212 | pUC18 with ω gene promoter (Pω) | This work |
| pHP13 | Shuttle vector for E. coli/B. subtilis, with high copy number in E. coli and low copy number in B. subtilis, ColE1 ori/pTA1060 ori Err Cmr | 25 |
| pHP1000 | pHP13 with δ and ω genes | This work |
| pHP1340 | pHP13 with δ and ω genes, with δ's Walker motif A deleted | This work |
| pHV1436 | Shuttle vector for E. coli/B. subtilis, with high copy number in E. coli and low copy number in B. subtilis, ColE1 ori/pTB19 ori Apr Cmr | 30 |
| pOSE10 | pBT205 with inactivated ω gene with its promoter, Pω, and part of the ɛ gene | This work |
| pOSZ5 | pHP13 with inactivated ω gene with its promoter, Pω, and the ɛ and ζ genes | This work |
| pRC1 | pDB101 derivative carrying copS with its promoter and the repS and erm genes | 14 |
| pRP346-5 | pHP13 with δ, ω, ɛ, and part of the ζ gene (pDB101 coordinates 4693 to 7251)b | Laboratory collection |
| pUC18 | E. coli general cloning vector, ColE1 ori Apr | 51 |
| pUCRC | pUC18 with pRC1 fragment encompassing repS, α, and β genes (pDB101 coordinates 20 to 4103)b | This work |
| pUCRS | pUCRC derivative with HindIII site in repS gene converted into a StuI site | This work |
| pUHC13 | Shuttle vector for E. coli/B. subtilis, with high copy number in E. coli and low copy number in B. subtilis, ColE1 ori/pTB19 ori Apr Cmr | This work |
| pUHC1000 | pUHC13 with δ and ω genes | This work |
| pUHC1340 | pUHC13 with δ and ω genes, with δ's Walker motif A deleted | This work |
| pUHC1212 | pUHC13 with δ gene promoter (Pδ) | This work |
| pUHC3212 | pUHC13 with copS gene promoter (PcopS) | This work |
| pUHC7212 | pUHC13 with ω gene promoter (Pω) | This work |
| pUSE2 | Shuttle vector for E. coli/B. subtilis, with high copy number in E. coli and low copy number in B. subtilis, ColE1 ori/pSM19035 ori Apr Emr | This work |
| pUSE1000 | pUSE2 with δ and ω genes | This work |
| pUSE1030 | pUSE2 with δ and ω genes, with δ truncated after the 23rd codon | This work |
| pUSE1040 | pUSE2 with δ and ω genes, with δ truncated after the 40th codon | This work |
| pUSE1340 | pUSE2 with δ and ω genes, with δ's Walker motif A deleted | This work |
| pUSE3000 | pUSE2 with δ and ω genes, with δ promoter (Pδ) replaced with copS promoter (PcopS) | This work |
| pUSE7000 | pUSE2 with δ and ω genes, with δ promoter (Pδ) replaced with ω promoter (Pω) | This work |
Plasmid construction details are given in the text.
pDB101 is a pSM19035 derivative which contains the two long inverted repeats and possesses all functions associated with replication, copy control, stable maintenance, and antibiotic resistance (accession no. X66468).
TABLE 3.
Primers used in this work
| Primer | Sequence (5′-3′)a | pDB101 coordinates |
|---|---|---|
| RepSF | CAGAATTTAAATCTGTCAAAGC | 1113-1134 |
| RepH-SR | CCCAgGCcTTGCAAAGAATGG | 1649-1629 |
| RepH-SF | CCATTCTTTGCAAgGCcTGGG | 1629-1649 |
| RepNR | GGGCGTTCTGCTAGCTTG | 2082-2065 |
| FHindIII | gggaagcttGGAAAAGGGATATATTG | 5382-5398 |
| RPstI | gggctgcagTTTTTTGTATAGTAATATTGTATC | 5549-5526 |
| FPstI | gggctgcAGAATGGGGCGTAGTTATGG | 5550-5569 |
| RSalI | GCAAACATAGTcGAcAATTTTGACTTTCC | 5696-5668 |
| FSalI | GGAAAGTCAAAATTgTCgACTATGTTTGC | 5668-5696 |
| RXbaI | CCGATCATATCTAGaCCAGGGTTAAATTG | 6170-6142 |
| FXbaI | CAATTTAACCCTGGtCTAGATATGATCGG | 6142-6170 |
| CG2 | cCggAAttCGGCATTACAGAGCATAAAGG | 6305-6332 |
| RBamHI | cccggatccCTATTCTTTTTCGTTTTC | 6420-6403 |
| FBamHI | gggggatccAATCACAAATCACAAGTG | 6421-6438 |
| RKpnI | gggggtacCTTCTTACTCTTATTTTATCATC | 6495-6473 |
| FKpnI | gggggtAccAAATAGAAAGAAGTGAGTG | 6493-6514 |
| OmegaER | ccggaATTCTCACTCCTTTTAAAGTTTGTC | 6740-6716 |
| CG1 | GATTTACACGTTTTGCAAGCT | 6915-6895 |
| #3 | CCAGTCCAATAATCGTCG | 18977-18994 |
| #4 | GCTAGTTCCAAGTTTCTCCCCC | 19194-19173 |
Introduced mutations are shown as lowercase letters, and restriction sites are underlined.
(i) Construction of pDO plasmids.
The pDO1000 plasmid (see Fig. 3), which served as a source of δ-ω genes for several plasmids constructed in this work, is the result of sequential cloning of PCR products encompassing the region. As the first step, a fragment encompassing Pω (the ω promoter) was amplified using primers FBamHI and RKpnI and cloned into appropriate sites of pUC18. The resulting plasmid served as a backbone for cloning the ω sequence (primers FKpnI and OmegaER were used for amplification) between the KpnI and EcoRI sites. The next steps of construction involved cloning of four other fragments encompassing the distal, middle, and front parts of the δ sequence and its promoter. Fragments were amplified with the respective primer pairs FXbaI-RBamHI, FSalI-RXbaI, FPstI-RSalI, and FHindIII-RPstI and cloned into the appropriate sites of the plasmid. The plasmid carrying the δ and ω genes devoid of the Pδ (the δ promoter) fragment was named pDO1120. The primer pairs used for the construction of pDO1000 are listed in Table 3 and marked in Fig. 3. The pDO1000 plasmid derivatives pDO1030 and pDO1040 (Fig. 3) were the results of single digestion of pDO1000 with MunI and SalI, respectively, and religation after T4 DNA polymerase treatment of the 5′ protruding ends. The pDO1340 plasmid (Fig. 3) is a pDO1000 derivative that lacks the entire MunI-SalI fragment. Plasmids pDO3000 and pDO7000 (Fig. 3) carry the δ and ω genes, with Pδ (δ promoter) replaced with PcopS (copS promoter) and Pω (ω promoter), respectively. Fragments containing PcopS or Pω were cut out of plasmids pDO3212 and pDO7212 with HindIII and PstI and cloned into HindIII/PstI-digested pDO1120.
FIG. 3.
Schematic representation of fragments from δ-ω region carried by plasmids used in stabilization studies. Pδ and Pω indicate the δ and ω promoters, respectively. Primers used for amplification of DNA fragments are indicated by numbered arrows, as follows: 1, FHindIII; 2, RPstI; 3, FPstI; 4, RSalI; 5, FSalI; 6, RXbaI; 7, FXbaI; 8, RBamHI; 9, FBamHI; 10, RKpnI; 11, FKpnI; 12, OmegaER. The ATPase Walker A, A′, B, and C motifs are indicated as white boxes. Gray crosses indicate stop codons.
(ii) Construction of a plasmid with a native replicon and its derivatives.
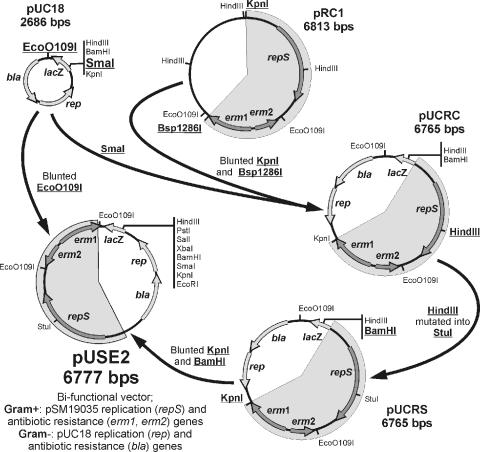
To construct the pUSE2 vector (Fig. 2), the KpnI-Bsp1286I DNA fragment from pRC1 (14, 50) was blunted with T4 DNA polymerase and cloned into the pUC18 vector, yielding plasmid pUCRC. This plasmid was subjected to site-specific mutagenesis by overlap extension (48), using primers RepSF, RepH-SR, RepH-SF, and RepNR, resulting in a pUCRS plasmid with the HindIII sequence within repS converted into a StuI restriction site. The KpnI-BamHI DNA fragment encompassing the engineered repS and erm genes was blunted and cloned into the filled-in EcoO109I site of pUC18.
FIG. 2.
Schematics of pUSE2 vector construction. Restriction sites used for construction and further cloning are indicated.
The shuttle vector pUSE2 was used as a backbone for all constructs containing the pSM19035 replicon, and all plasmids derived from it contain pUSE in their designations. DNA fragments from plasmids pDO1000, pDO1030, pDO1040, pDO1340, pDO3000, and pDO7000 were recloned into the HindIII and EcoRI sites of vector pUSE2 to produce plasmids pUSE1000, pUSE1030, pUSE1040, pUSE1340, pUSE3000, and pUSE7000, respectively. pUSE plasmids carry various fragments of the δ-ω region, as shown in Fig. 3.
(iii) Construction of a plasmid with a nonnative θ replicon and its derivatives.
The pUHC13 shuttle vector was used as a backbone for all constructs containing the nonnative θ replicon, and all plasmids derived from it contain pUHC in their designations. The vector was constructed by insertion of the EcoO109I DNA fragment (∼4.7 kb) of plasmid pHV1436 (carrying a gram-positive origin of replication from pTB19 [30]) into the EcoO109I site of pUC18. Plasmids pUHC1000 and pUHC1340 were constructed by insertion of the EcoO109I fragment carrying the gram-positive origin of replication of pHV1436 into the pDO1000 and pDO1340 plasmids, respectively (Fig. 3).
(iv) Construction of plasmids carrying a σ replicon.
HindIII-EcoRI fragments from plasmids pDO1000 and pDO1340 were cloned into the pHP13 vector to yield plasmids pHP1000 and pHP1340, respectively (Fig. 3).
(v) Construction of plasmids used for incompatibility tests.
Plasmids carrying Pδ (δ promoter), PcopS (copS promoter), and Pω (ω promoter) sequences were constructed as follows. Pδ was amplified with primers FHindIII-RPstI and cloned into the pUC18 HindIII and PstI restriction sites, resulting in plasmid pDO1212 (Fig. 3). The PcopS and Pω promoters were amplified with primer pairs #3-#4 and FBamHI-RKpnI, respectively, and cloned into the blunted SphI site of pUC18, resulting in plasmids pDO3212 and pDO7212, respectively (Fig. 3). Plasmids pUHC1212, pUHC3212, and pUHC7212 were derived from plasmids pDO1212, pDO3212, and pDO7212, respectively, and carry a gram-positive origin of replication cut out by EcoO109I from plasmid pHV1436 cloned into their EcoO109I sites.
DNA manipulations.
Plasmid DNA was isolated as described previously (48). For plasmid DNA isolation from B. subtilis, solution I was supplemented with fresh lysozyme (5 mg/ml), cells were incubated at 37°C until the suspension became viscous, and isolation was continued according to the protocol. Restriction endonucleases, polymerases, and modifying enzymes were used as recommended by their suppliers (MBI Fermentas, New England Biolabs, and Roche). Linear DNA fragments were gel purified after enzymatic reactions or PCR amplification (A&A Biotechnology gel extraction kit).
Bacterial growth and transformation.
Escherichia coli and B. subtilis were routinely grown in a rich medium (liquid L broth or L broth solidified with 1.5% agar [48]) at 37°C. Minimal media used for B. subtilis growth were GMI and GMII (47). When necessary, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 50 to 100 μg/ml (E. coli) or 5 μg/ml (B. subtilis); and chloramphenicol, 10 μg/ml (E. coli) or 5 μg/ml (B. subtilis).
E. coli and B. subtilis cells were transformed as described previously (47, 48) and plated onto L-agar plates with an appropriate antibiotic.
Isolation of total RNA from B. subtilis cells.
RNAs for transcriptional analysis were isolated from 25 ml of B. subtilis culture harvested in mid-exponential phase by a modified method described by Chomczynski (12).
RT-PCR.
For reverse transcription-PCR (RT-PCR), the first-strand reaction was performed as recommended by the Superscript II RT manufacturer (Invitrogen) with primers RBamHI and OmegaER, homologous to the δ and ω genes, respectively. RNAs were isolated from B. subtilis cells carrying plasmid pUSE1000 or pBT233 grown under conditions promoting partition (GMI medium at 30°C). Products of the first-strand reactions were used as templates for PCRs with pairs of primers allowing separate amplification of the δ (FHindIII/RBamHI) and ω (FKpnI/OmegaER) genes or amplification of both genes (FHindIII/OmegaER). In negative-control PCRs, we used products of first-strand reactions generated without the reverse transcriptase or primer as templates. In positive-control PCRs, the cDNAs were mixed with plasmid DNA to confirm that a lack of PCR product was not due to reaction inhibition by the first-strand reaction.
Northern hybridization and ECL system.
RNAs separated in an agarose-formamide gel without ethidium bromide were transferred to a nylon membrane as described previously (48). To visualize nucleic acids, the membrane was stained with methylene blue and destained with diethyl pyrocarbonate-treated water. A labeled fragment from pRP346-5 (250 to 500 ng) served as a probe. Hybridization, washes, and detection were carried out as described by the manufacturer of the ECL system (Amersham).
Plasmid stability and incompatibility tests.
B. subtilis cells carrying plasmids to be tested for stability were grown overnight in GMI medium supplemented with an appropriate antibiotic (erythromycin or chloramphenicol) at 30°C. An overnight culture was considered generation 0 and diluted 1:1,000 in fresh GMI medium without antibiotic for further growth. To test the number of plasmid-containing cells at the start point, serial dilutions were plated on L-agar without antibiotics. One hundred colonies were replica plated on L-agar with an appropriate antibiotic. Such dilutions and plating were repeated every 24 h, which represented an interval of about 10 generations. Plasmid stability was measured as the percentage of antibiotic-resistant clones. For the partition-mediated incompatibility test, cells containing the pUSE1000 plasmid were transformed with a plasmid containing the tested centromeric sequences. B. subtilis cells containing the two plasmids were grown overnight in GMI supplemented with erythromycin and chloramphenicol (generation 0), diluted 1:1,000 in fresh GMI medium with chloramphenicol, and plated on L-agar with the same antibiotic. One hundred colonies were replica plated on L-agar with erythromycin and chloramphenicol. Such dilutions and plating were repeated every 24 h. The plasmid destabilization effect was expressed as the percentage of erythromycin-resistant bacterial clones. The plasmid loss rate was calculated according to the equation (1 − a1/n) × 100, where a is the percentage of plasmid-carrying cells after n generations.
Stability and incompatibility experiments were performed independently at least three times for each plasmid tested.
RESULTS
δ and ω genes constitute a partitioning system with unusual organization.
Bioinformatic analysis of genes within the SegB region revealed that the δ gene encodes a putative ATPase which shares homology with the ParA family of partitioning proteins. It was shown previously that δ and the downstream ω gene are linked by a common regulator, Omega (14, 50). Based on the presence of the clearly defined, regulated ω promoter, de la Hoz et al. (14) postulated that ω is a separate transcription unit independent of the δ gene and suggested that it may form a transcription unit with downstream ɛ and ζ genes. Further functional analyses of ω involvement in plasmid stability supported this thesis and have shown that transcription from Pω is necessary for full activity of the PSK system (50) encoded by ɛ and ζ (9, 56). The ɛ and ζ genes had earlier been shown to belong to the same transcriptional unit (10).
To confirm the presence of a transcript encompassing the ω and ɛ genes, we performed Northern analysis of transcription within the studied region. As a source of transcripts from the SegB region, we used RNAs isolated from B. subtilis strains carrying promoters from the SegB region integrated into the chromosome and fused with the lacZ reporter gene to detect promoter activity (data not shown). Strain YB1015δ::lacZ carries the promoter of δ (Pδ), whereas strains YB1015/O+E and YB1015/OSE both contain the ω promoter (Pω), the ω coding sequence, and part of the ɛ gene, but the latter contains a single stop mutation within the sixth ω triplet. Because of the lack of ω gene product, its promoter is not repressed, which leads to a higher level of ω transcript and consequently facilitates its detection by hybridization. A DNA fragment carrying intact δ, ω, and ɛ genes served as a detection probe.
After hybridization, only transcripts of >3 kb were detected in lanes with RNAs isolated from strains YB1015/OSE and YB1015/δ::lacZ, which correlates with the summed lengths of ω, part of ɛ, and the lacZ gene or δ with lacZ, respectively (Fig. 4). A comparison of the strengths of the ω and δ promoters correlated with the results obtained in experiments with reporter genes, where the δ promoter was the strongest (14, 50).
FIG. 4.
Northern analysis of transcription from the δ and ω promoters. (A) Methylene blue-stained RNAs on nitrocellulose membrane. (B) Chemiluminescent detection of transcripts.
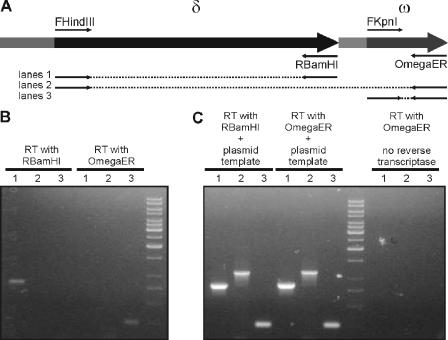
The presence of two independent transcription units, i.e., δ and ω-ɛ-ζ, does not exclude the theoretical possibility that transcription from the very strong Pδ generates a transcript also containing ω. Despite the presence of a putative rho-independent terminator (FindTerm; Softberry) (positions 6281 to 6337 in pBT233), we wanted to confirm that δ and ω transcripts are generated exclusively from their own promoters. Using RNAs isolated from cells carrying the pBT233 or pUSE1000 plasmid (native θ replicon), we generated cDNAs in reverse transcription reactions with the primers OmegaER (complementary to the end of the ω gene) and RBamHI (complementary to the end of the δ gene). If the transcript from the δ promoter also encompasses ω, it should be possible to generate not only ω but also δ and δ-ω amplicons in downstream PCRs using cDNA generated with the Omega ER primer. Therefore, for downstream PCR amplifications, we selected the pairs of primers in a way to prime amplification of DNA fragments encompassing δ and ω separately or both these genes together (Fig. 5A). In the case where the OmegaER-derived cDNA was used as a template, we observed exclusive amplification of a single DNA fragment of about 250 bp, as expected for the ω amplicon (Fig. 5B). PCRs with primer pairs allowing amplification of a δ or δ-ω fragment did not yield any product (Fig. 5B). The presence of the δ transcript in the analyzed RNA was confirmed by a PCR with the use of RBamHI-derived cDNA as the template (Fig. 5B). PCR controls with plasmid DNA added (to confirm that the lack of PCR amplicon was not due to inhibition by the first-strand reaction products) were positive (Fig. 5C). The absence of DNA contaminants in RNA samples was confirmed by the lack of PCR products when no enzyme or no primer was added to the reverse transcription reaction (Fig. 5C). RT-PCRs were performed with identical results for RNAs isolated from cells carrying each of the plasmids tested (data for pBT233 are not shown). These results confirm that δ and ω are transcribed independently.
FIG. 5.
RT-PCR analysis of the region encompassing the δ and ω genes. (A) Schematic representation of δ-ω region, with positions of primer sites used for amplification. (B) PCR amplification of δ (lanes 1), δ-ω (lanes 2), or ω (lanes 3) from cDNA templates generated with primers RBamHI and OmegaER. (C) Positive controls for PCR (plasmid DNA added to the template generated by RT) and negative controls for RT reactions (lack of reverse transcriptase in the RT reaction mixture). The products in lanes 1 to 3 in panels B and C were generated with the same primers.
Stabilization of θ and σ replicons by the δ and ω genes.
Previous analysis of the stabilization of pBT233 (pSM19035 replicon) and its deletion derivatives suggested that the δ gene was not involved in plasmid maintenance, and mutation of this gene did not result in any specific phenotype (10). However, all stabilization experiments were performed in rapidly dividing cells grown in rich medium. Recent observations concerning partitioning of IncP plasmids (RK2 and R751) and the Pseudomonas putida chromosome (4, 34) revealed that partition systems could be active only at certain physiological stages of cell growth and could be observed in slowly growing cells. Therefore, we tested the stability of the pSM19035 replicon carrying the δ and ω genes in slowly growing B. subtilis cells in minimal GMI medium at 30°C. Plasmid pUSE1000 carries the δ and ω genes cloned into the unstable pUSE2 vector (pSM19035 replicon). The stabilization test showed that after 40 generations, the pUSE1000 plasmid was maintained in almost 100% of the bacterial population, whereas pUSE2 without the insert was present in <60% of cells (Fig. 6), with a plasmid loss rate of 0.02 ± 0.01 per generation for the plasmid carrying both genes, in contrast to 1.56 ± 0.5 per generation for the vector. These results indicate that the δ and ω genes can stabilize their cognate replicon in slowly growing bacteria.
FIG. 6.
Influence of δ and ω genes on stabilities of different θ (pUSE and pUHC series) and σ (pHP series) replicons in slowly growing B. subtilis cells. •, pUSE2 vector; ▪, pUSE1000 carrying the δ and ω genes; gray circles, pUHC13 vector; gray squares, pUHC1000 carrying the δ and ω genes; ○, pHP13 vector; □, pHP1000 carrying the δ and ω genes. Plasmid stability is shown as the percentage of erythromycin (pUSE and pHP series)- or chloramphenicol (pUHC series)-resistant cells after growth under nonselective conditions. The plasmid loss rate per generation is indicated next to the appropriate curve on the graph.
In order to check if the stabilization effect of the δ and ω genes is specific only to the native replicon, we tested the ability of the studied genes to stabilize another θ-type replicon. The same DNA fragment carrying the δ and ω genes was cloned into the unstable vector pUHC13 (pTB19 origin of replication) to yield plasmid pUHC1000. The stabilities of pUHC1000 and the pUHC13 vector were tested in slowly growing B. subtilis cells (Fig. 6). pUHC13 was almost completely lost after 40 generations, whereas pUHC1000 was present in 90% of the bacterial population. Values for plasmid loss rates obtained for the two plasmids were 0.25 ± 0.04 (δ-ω) and 7.43 ± 0.39 (vector) per generation.
Since the activity of the δ and ω genes was not correlated with a specific type of θ replicon, we also tested a σ replicon. The pHP13 vector was unstable in B. subtilis cells and was maintained in <20% of the bacterial population after 40 generations of slow growth. However, insertion of the δ and ω genes (plasmid pHP1000) resulted in stabilization of this σ replicon, and pHP1000 was present in almost 90% of cells after 40 generations, with the plasmid loss rate changing from 3.72 ± 0.38 per generation (vector) to 0.2 ± 0.07 per generation for the plasmid carrying the δ-ω cassette (Fig. 6).
These results demonstrate that the δ and ω genes can stabilize segregationally unstable θ and σ replicons in slowly growing B. subtilis cells.
Stabilization of unstable replicons depends on the δ gene.
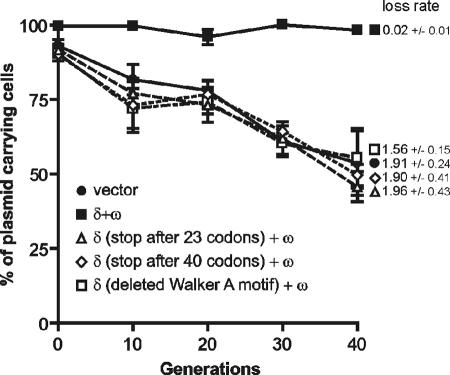
Further analysis of the role of the δ and ω genes in stable plasmid maintenance required an analysis of the consequences of their inactivation. Inactivation of ω in the presence of a fully functional δ gene was not possible due to the toxic effect of Delta overproduction on bacterial cells (unpublished results); therefore, only mutations within the δ sequence were analyzed. Since the three tested replicons (pUSE2, pUHC13, and pHP13) behaved in similar ways while carrying the stabilizing δ and ω genes, only the pUSE2 vector (with the native pSM19035 replicon) was chosen for mutational analysis. Plasmids pUSE1030 and pUSE1040, encoding Delta variants of 23 and 40 amino acids, respectively, were constructed, and their stabilities were compared with the stability of pUSE1000. pUSE1000 carrying intact δ and ω genes was present in almost 100% of cells after 40 generations, whereas plasmids pUSE1030 and pUSE1040 were present in <60% of B. subtilis cells after the same number of divisions (Fig. 7), with plasmid loss rates of 1.91 ± 0.24 and 1.9 ± 0.51 per generation, respectively. In the next step, we analyzed the stability of pUSE1340 lacking the sequence for the ATPase Walker A motif (deletion of amino acids VILNNYFKGGVGKSKL), with the rest of the protein sequence unchanged. Western blot analysis confirmed the production of a shorter Delta protein in B. subtilis cells carrying plasmid pUSE1340, as expected (data not shown). Plasmid pUSE1340 was present in <60% of bacterial cells grown for 40 generations (Fig. 7), with a plasmid loss rate of 1.96 ± 0.53 per generation. Additionally, two other replicons (pUHC13 and pHP13) carrying δ deprived of the Walker A motif (plasmids pUHC1340 and pHP1340) were tested for stability in B. subtilis cells. Plasmids pUHC1340 and pHP1340 were lost from cells at rates comparable to those of their respective vectors (data not shown). These observations strongly suggest that the δ gene is one of the components of the stabilization system.
FIG. 7.
Effects of mutations within the δ gene on stabilities of plasmids. The experiment was carried out in slowly growing B. subtilis cells. Plasmid stability is shown as the percentage of erythromycin-resistant cells after growth under nonselective conditions. •, pUSE2 vector; ▪, pUSE1000 carrying the δ and ω genes; ▵, pUSE1030 encoding a 23-amino-acid Delta protein; ⋄, pUSE1040 encoding a 40-amino-acid Delta protein; □, pUSE1340 with deletion of the Walker A ATPase motif. The plasmid loss rate per generation is indicated next to the appropriate curve on the graph.
All three Omega-binding promoters may possibly act as centromeric sequences.
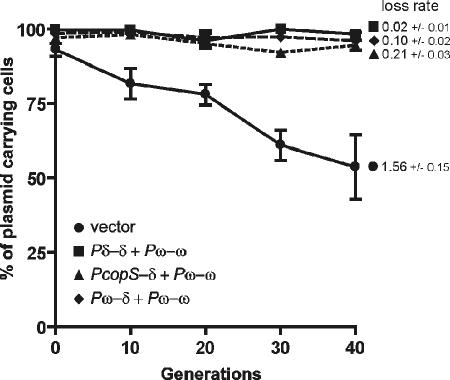
In classic plasmid partition systems, the centromere-like sequence parS, which is the ParB binding site, is located either upstream or downstream of the par operon. The Omega protein binds to sequences located in the promoter regions of both the δ and ω genes as well as in the copS gene promoter. Because δ and ω do not form a single operon and in this regard cannot be considered a classic partition system, it was necessary to test which of the Omega-binding sites can act as the centromere-like sequence. The promoter preceding the δ gene was a likely candidate for the primary centromere. To check this possibility, the δ promoter (Pδ) was replaced by the two other promoters containing Omega-binding sequences. The resulting plasmids, pUSE3000 and pUSE7000 (containing the copS [PcopS] and ω [Pω] promoters upstream of δ, respectively), were tested for stability in B. subtilis cells. Surprisingly, replacement of the Omega-binding site upstream of δ with the ω or copS gene promoter did not result in a significant loss of plasmid stability (Fig. 8). The plasmid loss rate compared with the value for the construct carrying the native sequence was higher for both tested plasmids, with values of 0.21 ± 0.03 and 0.10 ± 0.02 per generation, respectively, but was lower than that for the vector without an insert (1.59 ± 0.15 per generation). This observation suggests that all three of these regions can possibly play the role of a centromere-like sequence. To further investigate this possibility, Pδ was replaced by the other pSM19035 promoter, Pα. This promoter was chosen because it does not contain the repeated sequences present in putative centromeric sequences and therefore is not a target of Omega. Moreover, the strength of this promoter, as assessed in vivo (Pα-lacZ fusion) by a β-galactosidase assay, is similar to that of Pδ in repressed form or of Pω (unpublished results). Our attempts to measure the stability of this plasmid were mostly unsuccessful and hard to explain. Bacteria carrying this construct plated on solid medium yielded small colonies that were unable to grow in liquid medium in the presence of antibiotic. These observations suggest a loss of the plasmid from cells (loss of antibiotic resistance) and that the proper level of Delta in the cell may be necessary for correct functioning of the system. The other possibility is that partitioning in this system requires more than one centromeric sequence.
FIG. 8.
Effect on plasmid stability of replacement of the δ promoter (Pδ) with the copS promoter (copSp) or ω promoter (Pω). •, pUSE2 vector; ▪, pUSE1000 carrying the δ and ω genes; ▴, pUSE3000 carrying the copS promoter (PcopS) upstream of the δ gene; ⧫, pUSE7000 carrying the ω promoter (Pω) upstream of the δ gene. The plasmid loss rate per generation is indicated next to the appropriate curve on the graph.
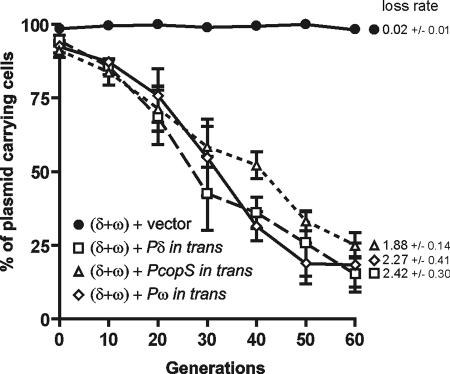
To further investigate the function of the Omega-binding sequences as centromeres, we tested their ability to cause a phenomenon known as partition-mediated incompatibility. This incompatibility occurs when one bacterial cell carries two compatible plasmids, with the first being stabilized by a partition system and the second, unstable plasmid containing only the centromere of the same partition system (1). The mechanism of this incompatibility is not fully understood: probably the additional copy of the centromere-like sequence titrates the ParB-type protein or/and paring of the two plasmids results in their random distribution into daughter cells. To test the hypothesis that all three regions, i.e., Pδ, PcopS, and Pω, can play the role of the centromere-like sequence and will mediate incompatibility, each of these sequences was cloned into the unstable pUHC13 vector. The resulting plasmids, pUHC1212, pUHC3212, and pUHC7212, respectively, as well as the vector pUHC13 were used for transformation of B. subtilis cells carrying the stable plasmid pUSE1000, which contains both the δ and ω genes and their promoters. Bacterial cells carrying a pair of plasmids, namely, the stable plasmid pUSE1000 and the pUHC13 derivative, which provided a region with repeated sequences (Pδ, PcopS, or Pω), were grown with selection for the unstable plasmid. The incompatibility effect was measured as the percentage of cells which had lost the plasmid carrying the functional partition system (Fig. 9). The plasmid loss rate during the experiment varied from 0.02 ± 0.01 per generation for the pUSE1000-pUHC13 pair to 2.42 ± 0.30, 1.88 ± 0.14, and 2.27 ± 0.51 per generation for plasmids delivering the δ, copS, and ω promoters, respectively.
FIG. 9.
Effect of repeated heptamers provided in trans on stability of plasmid pUSE1000 carrying the δ and ω genes. The heptamers provided in trans came from the δ promoter (plasmid pUHC1212) (□), the copS promoter (plasmid pUHC3212) (▵), and the ω promoter (plasmid pUHC7212) (⋄). As a control, the vector pUHC13 (•) was used. Plasmid stability is shown as the percentage of erythromycin- and chloramphenicol-resistant cells (carrying both plasmids) after growth with selection for chloramphenicol resistance (pUHC series plasmids). The plasmid loss rate per generation is indicated next to the appropriate curve on the graph.
The presence of any of the sequences containing the repeated heptamers abolished the stabilization effect of the partition system. This experiment shows that each of the studied putative centromeric sequences confers partition-mediated incompatibility and suggests that any of the three sequences can play the role of the centromere.
DISCUSSION
Partitioning of plasmid molecules into daughter cells after division is one of the mechanisms that ensure that plasmids are not lost from a bacterial population. In contrast to broadly studied systems from gram-negative bacteria, plasmid partitioning in gram-positive bacteria has not been investigated in detail. Only a few examples of gram-positive systems have been described to date (33, 49, 52).
The stability of plasmid pSM19035 and its derivatives has been studied extensively. Its high segregational stability is ensured by the toxin-antidote system (56), the plasmid resolution system (46), and coordinated copy number control (14, 50). In this work, we describe a third mechanism used by this plasmid to improve stability, namely, partitioning.
The results presented in this study demonstrate that the δ and ω genes constitute a novel plasmid partition system. We show that the studied pair of genes can stabilize their native replicon as well as unrelated θ and σ replicons in B. subtilis cells. A stabilization phenotype was observed only in slowly growing cells, a phenomenon described previously for Pseudomonas (4, 34) and for the pAW63 plasmid (52).
As described herein, the Delta-Omega system is a novel type of stabilizing system with properties not described so far. With only one example of an unusual partition system, encoded by plasmid pSK1 from S. aureus, the organization of the pSM19035 system differs dramatically from that of other plasmid partition systems studied so far. In all partition systems, the genes parA and parB are organized in one operon, which is regulated by the ParA and/or ParB protein. Proper balance between the amounts of ParA and ParB, which is crucial for proper functioning of the partitioning system (18), is achieved by the production of one transcript encoding both proteins. The unusual feature of the δ-ω partition system is the separation of transcription of both genes. The δ gene constitutes an independent monocistronic transcription unit, whereas the ω gene, which encodes the ParB-like component, is transcribed together with the ɛ and ζ genes, encoding the antidote and the toxin of the postsegregational killing system, respectively. However, the expression of the δ and ω genes is coordinated because they both are under control (repression) of the Omega protein (14, 50). This probably mimics the effect of a single transcription unit and ensures a proper balance between the products of both genes. The lack of stability of a plasmid with the δ promoter replaced by an unregulated one (α promoter) seems to support the hypothesis that δ and ω transcription must be precisely regulated for the activity of the system.
Production of the Delta protein is a factor necessary for plasmid stability, and hence plasmids carrying shortened versions of δ are lost from the bacterial population. The deletion of the fragment encoding the Walker motif of Delta suggests that the ATPase function of the protein may play a role in stabilization; however, we have no data to confirm that this deletion does not promote changes in protein folding and structure and therefore in its function.
The Omega protein, the postulated ParB protein in the described system, belongs to the MetJ/Arc superfamily of transcriptional regulators. The characteristic feature of this family is a ribbon-helix-helix structure in the C-terminal region, with characteristic DNA binding via a β-strand (40). The only other ParB-like protein gene belonging to the same family was described for plasmid TP228 by Golovanov and coworkers (22).
Omega not only regulates the transcription of genes encoding the partition system but is also the global regulator of other important plasmid functions: it is involved in the coordination of plasmid replication and copy number control by regulation of transcription of the copS gene. This global regulator also represses transcription of the ɛ and ζ genes, which encode the postsegregational killing system (9, 14, 50, 56). A similar situation was found for the plasmid RK2, where the ParB-like protein KorB is the global regulator which coordinates the expression of genes involved in plasmid replication, stable maintenance, and conjugative transfer (39).
Many plasmids have been shown to carry both a partition system and a postsegregational killing system, but the two were always separated in their organization and regulation. Therefore, the presence of ω in one operon with the ɛ and ζ genes and, in parallel, its involvement in partitioning are rather unusual. The advantage of the combination of a partition system and a postsegregational killing system was shown by Brendler and coworkers (8). It was proposed that the action of the PSK system in addition to the partition system would eliminate the plasmid-free cells which might have appeared despite the presence of the partition system. On the other hand, the partition system could minimize the effect of reduction of the bacterial population growth rate resulting from the death of plasmid-free cells caused by the PSK system.
The ParB-like protein Omega binds to three regions which contain repeated heptamers (WATCACW) and are located in the promoter regions upstream of the genes copS, δ, and ω (14, 50). In most partition systems, the ParB protein binds to the centromere-like sequence located either upstream or downstream of the par operon. According to this model, the repeated heptamers upstream of the δ gene would constitute the centromere region. Pδ is the strongest of the three Omega-binding promoters in vivo, and the repeats preceding the δ gene have a different organization from that of the repeats preceding the ω and copS genes (14, 50), which would suggest its different function. However, we showed in our study that replacement of the heptamers from the δ gene promoter with those from the copS and ω promoters does not result in a loss of plasmid stability, which suggests that all of these sequences can act as the centromere, despite their varied strengths or organization. Partition-mediated incompatibility tests have shown that all three studied regions containing the repeated heptamers (from the promoters of copS, δ, and ω), when provided in trans, can destabilize a plasmid carrying a functional δ-ω-encoded plasmid partition system. In vitro analysis has shown the affinities of the Omega protein to be similar for the three promoter regions (14, 50). Moreover, providing more than four heptads does not increase the Omega affinity for the promoter (15). A plasmid partition system with more than one centromere region was observed on the linear prophage N15. Any of the four parS sequences cloned separately can enhance the plasmid stability but is not fully efficient. Insertion of two of these parS sequences increases the stabilization effect, and each parS sequence can destabilize a stable mini-N15 derivative but not the whole N15 prophage. A proposed role of multiple centromere-like sequences is the prevention of simultaneous loss of all centromeres during recombination and ensuring the redundancy of partition function (24).
Summary.
The results presented in this study demonstrate that the δ and ω genes encode a novel partition system in gram-positive bacteria. This system differs from other known systems in its organization: the two genes are transcribed separately, and transcription of the parB-like gene is linked with transcription of the genes encoding the PSK system. The preliminary data suggest that three Omega (ParB-like) binding sequences can act as centromeric regions and are incompatibility determinants.
Acknowledgments
We are grateful to Izabela Kern-Zdanowicz and Urszula Zielenkiewicz for many helpful discussions and comments on the manuscript. We also thank Grażyna Jagura-Burdzy and Jacek Bardowski for critical readings of the manuscript.
This work was supported by the State Committee for Scientific Research (KBN, Poland) by grant no. 6 P04B 008 20 to P.C.
REFERENCES
- 1.Austin, S., and K. Nordstrom. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351-354. [DOI] [PubMed] [Google Scholar]
- 2.Benachour, A., J. Frere, S. Flahaut, G. Novel, and Y. Auffray. 1997. Molecular analysis of the replication region of the theta-replicating plasmid pUCL287 from Tetragenococcus (Pediococcus) halophilus ATCC33315. Mol. Gen. Genet. 255:504-513. [DOI] [PubMed] [Google Scholar]
- 3.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 4.Bignell, C. R., A. S. Haines, D. Khare, and C. M. Thomas. 1999. Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol. Microbiol. 34:205-216. [DOI] [PubMed] [Google Scholar]
- 5.Brantl, S. 1994. The copR gene product of plasmid pIP501 acts as a transcriptional repressor at the essential repR promoter. Mol. Microbiol. 14:473-483. [DOI] [PubMed] [Google Scholar]
- 6.Brantl, S., and D. Behnke. 1992. Copy number control of the streptococcal plasmid pIP501 occurs at three levels. Nucleic Acids Res. 20:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brantl, S., and D. Behnke. 1992. The amount of RepR protein determines the copy number of plasmid pIP501 in Bacillus subtilis. J. Bacteriol. 174:5475-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brendler, T., L. Reaves, and S. Austin. 2004. Interplay between plasmid partition and postsegregational killing systems. J. Bacteriol. 186:2504-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camacho, A. G., R. Misselwitz, J. Behlke, S. Ayora, K. Welfle, A. Meinhart, B. Lara, W. Saenger, H. Welfle, and J. C. Alonso. 2002. In vitro and in vivo stability of the epsilon2zeta2 protein complex of the broad host-range Streptococcus pyogenes pSM19035 addiction system. Biol. Chem. 383:1701-1713. [DOI] [PubMed] [Google Scholar]
- 10.Ceglowski, P., A. Boitsov, S. Chai, and J. C. Alonso. 1993. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene 136:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Ceglowski, P., A. Boitsov, N. Karamyan, S. Chai, and J. C. Alonso. 1993. Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035-derived plasmids in Bacillus subtilis. Mol. Gen. Genet. 241:579-585. [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski, P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 15:532-537. [PubMed] [Google Scholar]
- 13.Davey, M. J., and B. E. Funnell. 1997. Modulation of the P1 plasmid partition protein ParA by ATP, ADP, and P1 ParB. J. Biol. Chem. 272:15286-15292. [DOI] [PubMed] [Google Scholar]
- 14.de la Hoz, A. B., S. Ayora, I. Sitkiewicz, S. Fernandez, R. Pankiewicz, J. C. Alonso, and P. Ceglowski. 2000. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl. Acad. Sci. USA 97:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Hoz, A. B., F. Pratto, R. Misselwitz, C. Speck, W. Weihofen, K. Welfle, W. Saenger, H. Welfle, and J. C. Alonso. 2004. Recognition of DNA by Omega protein from the broad-host range Streptococcus pyogenes plasmid pSM19035: analysis of binding to operator DNA with one to four heptad repeats. Nucleic Acids Res. 32:3136-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth, N., S. Apisiridej, T. Berg, B. A. O'Rourke, S. Curnock, K. G. Dyke, and R. A. Skurray. 2000. Replication of staphylococcal multiresistance plasmids. J. Bacteriol. 182:2170-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, B. M., and R. E. Yasbin. 1983. The genetics and specificity of the constitutive excision repair system of Bacillus subtilis. Mol. Gen. Genet. 190:481-486. [DOI] [PubMed] [Google Scholar]
- 18.Funnell, B. E. 1988. Mini-P1 plasmid partitioning: excess ParB protein destabilizes plasmids containing the centromere parS. J. Bacteriol. 170:954-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes, K., J. Moller-Jensen, and J. R. Bugge. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 20.Gerdes, K., J. Moller-Jensen, G. Ebersbach, T. Kruse, and K. Nordstrom. 2004. Bacterial mitotic machineries. Cell 116:359-366. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golovanov, A. P., D. Barilla, M. Golovanova, F. Hayes, and L. Y. Lian. 2003. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 50:1141-1153. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 24.Grigoriev, P. S., and M. B. Lobocka. 2001. Determinants of segregational stability of the linear plasmid-prophage N15 of Escherichia coli. Mol. Microbiol. 42:355-368. [DOI] [PubMed] [Google Scholar]
- 25.Haima, P., D. van Sinderen, H. Schotting, S. Bron, and G. Venema. 1990. Development of a beta-galactosidase alpha-complementation system for molecular cloning in Bacillus subtilis. Gene 86:63-69. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 27.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 28.Hayes, F., and S. J. Austin. 1993. Specificity determinants of the P1 and P7 plasmid centromere analogs. Proc. Natl. Acad. Sci. USA 90:9228-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiraga, S. 1992. Chromosome and plasmid partition in Escherichia coli. Annu. Rev. Biochem. 61:283-306. [DOI] [PubMed] [Google Scholar]
- 30.Janniere, L., C. Bruand, and S. D. Ehrlich. 1990. Structurally stable Bacillus subtilis cloning vectors. Gene 87:53-61. [DOI] [PubMed] [Google Scholar]
- 31.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 32.Kalnin, K., S. Stegalkina, and M. Yarmolinsky. 2000. pTAR-encoded proteins in plasmid partitioning. J. Bacteriol. 182:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, R. A., C. R. Bignell, W. Zeng, A. C. Jones, and C. M. Thomas. 2002. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology 148:537-548. [DOI] [PubMed] [Google Scholar]
- 35.Lukaszewicz, M., K. Kostelidou, A. A. Bartosik, G. D. Cooke, C. M. Thomas, and G. Jagura-Burdzy. 2002. Functional dissection of the ParB homologue (KorB) from IncP-1 plasmid RK2. Nucleic Acids Res. 30:1046-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch, A. S., and J. C. Wang. 1995. SopB protein-mediated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl. Acad. Sci. USA 92:1896-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malke, H. 1974. Genetics of resistance to macrolide antibiotics and lincomycin in natural isolates of Streptococcus pyogenes. Mol. Gen. Genet. 135:349-367. [DOI] [PubMed] [Google Scholar]
- 38.Moller-Jensen, J., R. B. Jensen, and K. Gerdes. 2000. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 8:313-320. [DOI] [PubMed] [Google Scholar]
- 39.Motallebi-Veshareh, M., D. Balzer, E. Lanka, G. Jagura-Burdzy, and C. M. Thomas. 1992. Conjugative transfer functions of broad-host-range plasmid RK2 are coregulated with vegetative replication. Mol. Microbiol. 6:907-920. [DOI] [PubMed] [Google Scholar]
- 40.Murayama, K., P. Orth, A. B. de la Hoz, J. C. Alonso, and W. Saenger. 2001. Crystal structure of Omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 A resolution. J. Mol. Biol. 314:789-796. [DOI] [PubMed] [Google Scholar]
- 41.Nordstrom, K., and S. J. Austin. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 42.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 43.Radnedge, L., B. Youngren, M. Davis, and S. Austin. 1998. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 17:6076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravin, N., and D. Lane. 1999. Partition of the linear plasmid N15: interactions of N15 partition functions with the sop locus of the F plasmid. J. Bacteriol. 181:6898-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodionov, O., M. Lobocka, and M. Yarmolinsky. 1999. Silencing of genes flanking the P1 plasmid centromere. Science 283:546-549. [DOI] [PubMed] [Google Scholar]
- 46.Rojo, F., and J. C. Alonso. 1994. A novel site-specific recombinase encoded by the Streptococcus pyogenes plasmid pSM19035. J. Mol. Biol. 238:159-172. [DOI] [PubMed] [Google Scholar]
- 47.Rottlander, E., and T. A. Trautner. 1970. Genetic and transfection studies with B. subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol. Gen. Genet. 108:47-60. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Simpson, A. E., R. A. Skurray, and N. Firth. 2003. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J. Bacteriol. 185:2143-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sitkiewicz, I. 2002. The regulatory gene omega of plasmid pSM19035. Ph.D. thesis. Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland.
- 51.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 52.Wilcks, A., L. Smidt, O. A. Okstad, A. B. Kolsto, J. Mahillon, and L. Andrup. 1999. Replication mechanism and sequence analysis of the replicon of pAW63, a conjugative plasmid from Bacillus thuringiensis. J. Bacteriol. 181:3193-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, D. R., D. P. Macartney, and C. M. Thomas. 1998. The partitioning activity of the RK2 central control region requires only incC, korB and KorB-binding site O(B)3 but other KorB-binding sites form destabilizing complexes in the absence of O(B)3. Microbiology 144:3369-3378. [DOI] [PubMed] [Google Scholar]
- 54.Yarmolinsky, M. 2000. Transcriptional silencing in bacteria. Curr. Opin. Microbiol. 3:138-143. [DOI] [PubMed] [Google Scholar]
- 55.Zielenkiewicz, U., and P. Ceglowski. 2001. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48:1003-1023. [PubMed] [Google Scholar]
- 56.Zielenkiewicz, U., and P. Ceglowski. 2005. The toxin-antitoxin system of the streptococcal plasmid pSM19035. J. Bacteriol. 187:6094-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]