Abstract
Butane monooxygenase (BMO) from Pseudomonas butanovora has high homology to soluble methane monooxygenase (sMMO), and both oxidize a wide range of hydrocarbons; yet previous studies have not demonstrated methane oxidation by BMO. Studies to understand the basis for this difference were initiated by making single-amino-acid substitutions in the hydroxylase α subunit of butane monooxygenase (BMOH-α) in P. butanovora. Residues likely to be within hydrophobic cavities, adjacent to the diiron center, and on the surface of BMOH-α were altered to the corresponding residues from the α subunit of sMMO. In vivo studies of five site-directed mutants were carried out to initiate mechanistic investigations of BMO. Growth rates of mutant strains G113N and L279F on butane were dramatically slower than the rate seen with the control P. butanovora wild-type strain (Rev WT). The specific activities of BMO in these strains were sevenfold lower than those of Rev WT. Strains G113N and L279F also showed 277- and 5.5-fold increases in the ratio of the rates of 2-butanol production to 1-butanol production compared to Rev WT. Propane oxidation by strain G113N was exclusively subterminal and led to accumulation of acetone, which P. butanovora could not further metabolize. Methane oxidation was measurable for all strains, although accumulation of 23 μM methanol led to complete inhibition of methane oxidation in strain Rev WT. In contrast, methane oxidation by strain G113N was not completely inhibited until the methanol concentration reached 83 μM. The structural significance of the results obtained in this study is discussed using a three-dimensional model of BMOH-α.
Pseudomonas butanovora utilizes an alkane monooxygenase, commonly referred to as butane monooxygenase (BMO), to initiate growth on alkanes C2 to C9. BMO, like soluble methane monooxygenase (sMMO), consists of three protein components: a hydroxylase (BMOH) consisting of three unique subunits (α2, β2, and γ2), a reductase (BMOR), and an effector protein (BMOB). The genes carrying bmoX, bmoY, and bmoZ, the α, β, and γ subunits of BMOH, have 65, 42, and 38% amino acid identity with the corresponding subunits of MMOH from M. capsulatus (Bath) (26). There is 86.3% sequence identity among the MMOH-α subunits of six strains of sequenced methanotrophs (reviewed in reference 15). Although BMO and sMMO share extended substrate ranges, including alkanes, alkenes, aromatics, and chlorinated xenobiotics, BMO is the only member of the sMMO subfamily of soluble diiron monooxygenases in which methane oxidation has not been observed. Because BMO and sMMO comprise a group of powerful oxidative systems capable of activating highly stable hydrocarbons, they garner serious attention for their potential in bioremediation and bioindustrial catalysis (17, 27).
Investigation of the crystal structure of MMOH has led to identification of five hydrophobic cavities that extend from the surface of the α subunit to the diiron active site (21). Cavity 1 is the hydrophobic pocket containing the active site, and cavity 2 comprises a substantial pocket extending from the active site. Evidence that these cavities bind substrates and provide passage to the active site was provided by crystallization of MMOH in the presence of the methane surrogate xenon or dibromomethane (40). Since longer chained halogenated alcohols were also found to bind in cavities 1 and 2 of MMOH-α, it is presumed that the hydrophobic cavities provide passage to and from the active site (25). MMOH-α residues L110, T213, and F188 are conserved residues that contribute to the formation of the “leucine gate,” which apparently allows substrate access to the active site from hydrophobic cavity 2 (21). Crystallization of oxidized and reduced MMOH and alcohol-soaked structures revealed changes in the rotameric conformations of L110 and T213 (21, 25). Alteration of Thr213 to Ser in MMOH-α resulted in lower specific activity toward different substrates, including methane (28). DNA shuffling and saturation mutagenesis of the corresponding gating residues in the more distantly related toluene ortho-monooxygenase (TOM), toluene para-monooxygenase (TpMO), and toluene-o-xylene monooxygenase (ToMO) resulted in variants that have new regiospecificities (4, 9, 36).
Sequence alignment of BMOH-α and MMOH-α allowed a comparison to be made of residues lining hydrophobic cavities 1 and 2. All but 5 of the 19 residues lining cavity 2 are identical in BMOH-α and MMOH-α (26). Although the residues coordinating the diiron center and comprising the leucine gate in BMOH-α are identical to those in MMOH-α, several residues adjacent to the active site are different. The amino acid differences in the hydrophobic cavities and adjacent to the active site provided clear targets for exploration of the fundamental differences in substrate specificity between BMO and sMMO. In analogy to sMMO, the effector protein of BMO (BMOB) is likely to affect the rate and regioselectivity of hydroxylation as well as the redox potential of the active site (19). Site-directed mutagenesis of the gene encoding MMOB and in vitro analysis has continued to clarify the influence of the effector protein (3, 5, 37). Although crystal structures of bacterial multicomponent monooxygenases in complex have not been solved, specific charged residues were determined to be involved in the interaction of MMOH-α and MMOB (3). We hypothesized that altering BMOH-α surface residues involved in BMOB-BMOH-α interaction could change substrate access or alter regions of structural flexibility such that methane oxidation or another alteration to substrate catalysis would be observed. Sequence alignment of BMOH-α and MMOH-α in conjunction with a spatial model describing the hydroxylase-effector protein complex for MMO (3) revealed BMOH-α surface residues that were of particular interest for study.
The development of a bacterial system to investigate the roles of individual residues within MMOH-α has been challenging due to enzyme instability or low enzyme activity in Escherichia coli expression (13, 38). One promising system that has circumvented this problem utilizes a plasmid-based expression system for sMMO in Methylosinus trichosporium, thus avoiding the use of a heterologous host (28). In this article, we describe another approach using a homologous expression background by which single-amino-acid substitutions were created in BMOH-α of P. butanovora. Five of these P. butanovora mutants permitted investigation of several structural regions of the BMO hydroxylase. The results of these whole-cell experiments were applied to a model of BMO derived from MMO crystal structures, and the two enzymes were structurally compared. The mutant phenotypes should be valuable in furthering our understanding of the recently discovered complexities in BMO regulation and downstream alkane metabolism (7, 23).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The plasmids and bacterial strains used in this study are described in Table 1. P. butanovora wild-type and mutant strains were cultured at 30°C in sealed 160 ml vials containing 33 ml of liquid medium with 10 ml n-butane, 15 ml propane, or 25 ml ethane gas (Airgas, Inc., Randor, Pa.) (99.0%) added as overpressure. Liquid alkanes (pentane, hexane, or octane) (15 mM) or 1- or 2-butanol (5 mM) were added directly to a mineral salts medium which was previously described (23) except that 30 mM KNO3 was substituted for NH4Cl. Alternatively, when BMO expression was not required, strains were grown in 30 to 100 ml of the sterile medium described above with 10 mM sodium lactate as the carbon source.
TABLE 1.
Plasmids and bacterial strains used in this study
| Plasmid or strain | Description | Source or reference |
|---|---|---|
| Plasmids | ||
| pGEM-T Easy | PCR product TA cloning vector; Ampr | Promega |
| pGBKB | pGEM-T Easy carrying the 1,973-bp PCR product BmoUP-Kan-BmoDN (bmoX partially deleted and Kanr inserted into the deleted region) | This study |
| pBluescript II SK | Cloning vector, 3.0 kb; AprlacZ′ | Stratagene |
| pBSbmoxyb | pBluescript carrying a 3.68-kb bmoXYZ PCR product cloned with KpnI | This study |
| Strains | ||
| P. butanovora | ATCC 43655; n-butane-assimilating bacteria | 30 |
| PBKB | Mutant strain with bmoX partially deleted and Kanr inserted into the deleted region; created by homologous recombination of BmoUP-Kan-BmoDN from pGBKB with P. butanovora | This study |
| Rev WT | Butane-oxidizing control strain used for comparison of P. butanovora mutants containing single amino-acid substitutions in BMOH-α; created by homologous recombination of bmoXYB from pBSbmoxyb with P. butanovora PBKB replacing Kanr with wild-type bmoX sequence | This study |
| E. coli | ||
| JM109 | Cloning host strain; Ampr | 41 |
| DH5α | Cloning host strain; Ampr | Invitrogen |
For growth experiments, ethane-grown cells were obtained from early stationary phase, diluted, and grown again to stationary phase for 48 h. A 3% inoculum was transferred to fresh medium containing the specific alkane of interest. Samples were taken periodically using sterile technique for determination of optical density at 600 nm (OD600).
Engineering P. butanovora bmoX mutants.
A P. butanovora bmoX mutant host strain (P. butanovora PBKB) was constructed for use in mutagenesis experiments. Primers used in construction of P. butanovora PBKB are listed in Table 2. By the use of PCR ligation mutagenesis (14), a 273 bp fragment of bmoX containing 161 bp flanking DNA upstream of ATG was amplified by PCR from P. butanovora genomic DNA (“BmoUP”). A second fragment 1,212 bp in length and containing a kanamycin resistance cassette was amplified by PCR from the pUC4K plasmid (31). Both fragments were XbaI digested, ligated, and PCR amplified to yield “BmoUP-Kan” (1,485 bp). A third fragment that was 488 bp in length containing 165 bp of flanking DNA downstream of TGA was PCR amplified from bmoX (“BmoDN”). “BmoUP-Kan” and “BmoDN” were digested with SacII, ligated, and PCR amplified to yield the 1,973 bp full-length fragment (“BKB”). “BKB” was cloned into pGEM-EZ to form pGBKB (Table 1). Following electroporation (1 mm electrode gap cuvette; ISC BioExpress cat no. E-5010-1) (1,600 V, 150 Ω, 50 μF) of pGBKB into wild-type P. butanovora, the 1,185 bp fragment of bmoX was replaced with the Kanr cassette from pGBKB. P. butanovora PBKB was selected by growth in 30 ml of the lactate medium described above containing 25 μg/ml kanamycin.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| Used in generating pGBKB | |
| LBMOUPSac130 | GGAGCGGCCAAGAGCtCCGCACCTTGCGGC |
| RBMOUPXba129 | GATACTTCGCGGtCtAGACCTTGAAATCC |
| LKanXba130 | CAGTTGGTGATTCTAGACTTTTGCTTTGCC |
| RKanSac229 | TCATTAGGCACCCGCGGCTTTACACTTTA |
| LBMODNSac220 | TATCAAGCCGCGGTTCACTG |
| RBMODNKpn129 | GCTTGGTTTCCAGCGGtaCCTTGGGTACG |
| Used in generating pBSbmoxyz | |
| P1UPX | CACACGCCTGGAGCGGtCtAGAGCCCCGCACCTTG |
| P4DNB | TCCAATTGTATTCCTTCTaGAGATCGTACGTCGTG |
| Used in mutagenesis of pBSbmoxyz | |
| T148C top | AAATCCGCCATGTGAACCAGTGCGCGTACGTGAAT |
| T148C bot | CTGGTTCACATGGCGGATTTCGTCGATCACCTG |
| Q320K top | TGGCTGGGCCGACTGCAGAAGTTCGGCGTCAAAACG |
| Q320K bot | CTGCAGTCGGCCCAGCCAGATGCCAGCCCA |
| F321Y top | CTGGGCCGACTGCAGCAGTACGGCGTCAAAACGCCA |
| F321Y bot | CTGCTGCAGTCGGCCCAGCCAGATGCCAGC |
| G113N top | GAGACCGGCGAATACAACGCAATCGCCGGTTCTGCT |
| G113N bot | CTCTGGCCGCTTATG |
| L279F top | GACGCAGCACAAGTTCTTCACCCCCTTCGTCGGGG |
| L279F bot | CTGCGTCGTGTTCAAG |
Restriction sites are underlined. Changes to the P. butanovora sequence are in lowercase characters. Codons encoding substituted amino acids are bold.
Site-directed mutagenesis was performed using a GeneTailor site-directed mutagenesis system (Invitrogen, Carlsbad, CA). Single specific mutations were introduced into bmoX by use of pBSbmoxyz (Table 1) as template DNA with pairs of mutagenic primers (Table 2). The reaction mixtures were transformed into competent E. coli DH5α cells. Mutations in pBSbmoxyz were verified by DNA sequencing (Center for Genome Research and Biocomputing Core Laboratory, Oregon State University).
Plasmids containing site-directed mutations of bmoX were electroporated into the mutant host strain P. butanovora PBKB under the conditions described above. pBSbmoxyb with no mutation was also electroporated into the mutant host strain to recover the wild-type genotype (P. butanovora Rev WT) (Table 1). Following electroporation, mutant strains were immediately transferred to 30 ml P. butanovora medium containing 1 mM citrate for recovery. Butane (10 ml) was added as overpressure for selection of mutant strains with and without altered amino acids. Growth was observed after 10 to 30 days. Liquid cultures were plated on LB agar and single colonies picked for clonal cell lines. The entire bmoX gene was amplified using high fidelity PCR (Platinum Pfx; Invitrogen), and sequence changes for each P. butanovora mutant cell line were verified by DNA sequencing (Center for Genome Research and Biocomputing Core Laboratory, Oregon State University).
BMO induction.
Lactate-grown cells were grown overnight and harvested at late-exponential phase (OD600, 0.8 to 1.0). Cells were washed three times in phosphate buffer (25 mM KH2PO4·25 mM Na2HPO4, pH 7.2) and resuspended to an OD600 of 0.65 to 0.75 in growth medium with 1 mM 1-butanol added to induce expression of BMO (23). Following incubation at 30°C with shaking for 3 h, cells were harvested for analysis of BMO activity by use of an ethylene oxidation assay (12). Briefly, following harvest, cells were exposed to ethylene (an alternative substrate for BMO) and ethylene oxide accumulation was measured by gas chromatography (GC).
Butane oxidation.
Butane consumption was measured using a 1 ml syringe assay (1). Briefly, a 0.025 ml cell suspension was added to 0.75 ml O2-saturated phosphate buffer and 0.1 ml butane-saturated phosphate buffer. A glass bead within the chamber mixed the contents of the syringe. Samples (5 μl) were taken periodically, and butane concentrations were measured by GC.
Regiospecificity of BMO mutants toward butane and propane.
Butane-grown cells were harvested by centrifugation for analysis at late-exponential phase (OD600, 0.60 to 0.75). Cells were washed three times in phosphate buffer and resuspended as a concentrated cell suspension (5 mg total protein/ml). Accumulation of 1- and 2-butanol was measured using 1-propanol or 2-pentanol as an inhibitor of butanol consumption (1). Vials (7 ml) containing 0.8 ml phosphate buffer were capped and sealed, and 1.5 ml butane was added as overpressure to the headspace. Vials were placed in a 30°C water bath with constant shaking. Assays were initiated by the addition of 0.2 ml concentrated cell suspension, and liquid samples (1 μl) were removed periodically for measurement of butanol accumulation by gas chromatography. Rates of 1- and 2-butanol accumulation were linear during the first 8 and 40 min. We verified that the differences in the rates of 1- and 2-butanol production in the P. butanovora bmoX mutants compared to strain Rev WT results were attributable to the single-amino-acid alterations and not to differences in expression levels of BMOH-α. Butane-grown mutant strains were harvested as described above, and the relative amounts of the different BMO subunits were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). No apparent differences in expression levels were observed.
To determine regiospecificity of propane oxidation, accumulation of propionate and acetone as downstream metabolites of 1- and 2-propanol, respectively, was monitored. In this case, cells were grown in the presence of ethane because the enzymatic pathways required for propionate consumption are not expressed (7). Propionate accumulates as the product of 1-propanol transformation by alcohol and aldehyde dehydrogenases (7, 32), and no propionaldehyde accumulation was measurable during incubations. Similarly, if propane is oxidized subterminally to 2-propanol, acetone should accumulate. Vials were prepared as described above but with 2.0 ml propane added instead of butane, and assays proceeded as described above.
Methane oxidation.
Vials (7.7 ml) containing 0.8 ml phosphate buffer, 5 mM lactate, and 3 ml methane gas were incubated at 30°C with shaking. Washed cell suspensions (5 mg total protein/ml) were added to the vials to initiate the assays, and liquid samples (1 μl) were removed for measurement of methanol by GC.
Analytical techniques.
Ethylene oxide (100 μl headspace samples), methanol, butane, acetone, and propionate (1 μl liquid-phase samples) were analyzed by GC with a Shimadzu GC-8A chromatograph (Kyoto, Japan) equipped with a flame ionization detector and a stainless steel column packed with Porapak Q (Alltech, Deerfield, Ill) (80/100 mesh). Butanol accumulation (1 μl liquid-phase samples) was monitored using GC, a flame ionization detector, and a CarboWax 1500 column (Alltech) (6 ft by 2 mm). Compounds were identified by correspondence to retention times of standards. Compounds were quantified by comparison of peak areas or peak heights obtained from known quantities of standard solutions.
Structural modeling.
The BMOH-α subunit was modeled using the Mod Web Server (8, 18, 22) based on MMOH-α templates (PDBs 1FZ1B, 1FZHA, 1FZ8A, 1FZ2A, and 1FZ0B) from studies of Methylococcus capsulatus (Bath) (39, 40) and viewed using a Deep View Swiss-Pdb viewer and Pymol (6). BMOH-α with the G113N substitution was modeled using 1MTY (21). Torsion angles for F185 in the BMOH-α and G113N models were calculated using the Deep View program, and those for MMOH-α F188 were calculated using the Deep View program from the coordinates of PDB 1FZ8A. The methanol-bound MMOH crystal structure (PDB 1FZ6) was used for cavity 1 comparisons.
RESULTS
Mutagenesis of P. butanovora bmoX.
Investigations of regions of the hydroxylase component of BMO were initiated by engineering nine single-amino-acid substitutions in BMOH-α of P. butanovora. By comparison to crystal structures of MMOH-α and sequence alignment with BMOH-α, residues targeted for substitution reside in three key regions: the area adjacent to or contributing to formation of the active site (G113, T148, and P179); hydrophobic cavity 2, which leads to the active site (V181, L287, and L279); and the surface of BMOH-α that interacts with the effector protein, BMOB (Q319, Q320, and F321). BMOH-α residues were changed to residues occupying the equivalent positions in MMOH-α; for example, for P. butanovora mutant strain T148C, the Thr148 in wild-type BMOH-α was changed to the corresponding Cys in MMOH-α.
First, mutant strain P. butanovora PBKB was constructed containing a selectable kanamycin resistance cassette within partially deleted bmoX (Table 1). Homologous recombination of bmoX sequences flanking the kanamycin cassette facilitated replacement of the kanamycin marker in P. butanovora PBKB with plasmid-borne bmoX sequences containing site-directed mutations. Because we were interested in studying the roles of individual amino acids in BMO catalysis, mutants were selected that maintained butane oxidation. To verify that the double-crossover event was successful, the entire bmoX gene from each recovered clonal line was sequenced. P. butanovora mutants were kanamycin sensitive, and PCR analysis using external bmoX primers and internal kan primers verified removal of kan and recovery of full-length bmoX (data not shown). Of the nine mutant strains engineered, five were recovered by growth on butane. Mutant strains Q319G, P179K, V181M, and L287F were not recovered and therefore may have yielded altered BMOH-α subunits that could not support sufficient butane oxidation activity for growth on butane or produced products that could not be metabolized further. It is also possible that each of these residues is essential for butane oxidation or that recombination was incomplete. Finally, to ensure that any observed phenotypic differences were not artifacts of the mutagenesis procedures but were instead attributable to the single-amino-acid changes, one strain with no amino acid changes was recovered for use as the control “wild-type” BMOH-α phenotype (strain Rev WT; Table 1). The five mutant strains were studied for the mechanistic and physiological implications of single-amino-acid substitutions within BMOH-α.
Growth of P. butanovora mutants on alkanes.
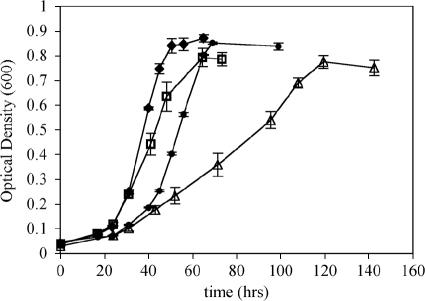
The growth characteristics of the P. butanovora mutants on butane were examined (Fig. 1). Mutant strains L279F and G113N showed both distinctly longer lag phases and slower growth rates, reflecting a diminished ability to grow on butane. All mutant strains reached ODs similar to that seen with the Rev WT control strain. Generation times of butane-grown mutant strains Q320K and F321Y were not significantly different from that of strain Rev WT; however, the remaining three mutant strains showed generation times at least 0.75 h longer than that of strain Rev WT (Table 3). Because BMOH-α was altered to reflect the corresponding residues in MMOH-α, we were interested in determining whether growth of the mutant strains on either shorter- or longer-chain alkanes was different from strain Rev WT growth rates. While the generation time of Rev WT was longer on ethane than on butane (4.5 h versus 3.6 h), the slower growth of the mutants on ethane was exacerbated by the single-amino-acid substitutions present in the mutant strains. The lag phase of mutant strain G113N when grown on ethane was about twice as long as that seen with butane (data not shown). The generation time of mutant strain F321Y was not significantly different from that of Rev WT when grown on butane, but this strain was able to maintain a shorter generation time than Rev WT when grown on propane and ethane. The growth rate of mutant strain T148C was as slow as that of strain L279F on ethane but was nearly the same as that of Rev WT on propane. While all other mutant strains grew on propane to a final OD with growth rates similar to that seen with Rev WT, mutant strain G113N showed only a slight increase in OD (0.10 to 0.15) after nearly 8 days. All mutants grew to similar ODs on alkanes C5 to C8. Generation times for wild-type and mutant P. butanovora strains increased during growth on C4+ alkanes. Furthermore, differences in lag phases and growth rates for the mutant strains relative to strain Rev WT on alkanes C5 to C8 did not reveal other notable phenotypic differences (data not shown).
FIG. 1.
Growth of P. butanovora mutant strains on butane. Data are presented as the mean OD600 at each time point for three replicate growth curves ± standard deviations. Symbols: ⧫, strain Rev WT; □, T148C; •, L279F; ▵, G113N.
TABLE 3.
Generation times for P. butanovora mutants grown on butane, propane, and ethanea
| P. butanovora strain | Generation time (h ± SD) in growth substrate:
|
||
|---|---|---|---|
| Butane | Propane | Ethane | |
| Rev WT | 3.6 ± 0.2 | 4.0 ± 0.4 | 4.5 ± 0.3 |
| T148C | 4.4 ± 0.3 | 4.2 ± 0.4 | 7.0 ± 0.1 |
| Q320K | 4.1 ± 0.3 | 3.9 ± 0.1 | 5.9 ± 0.1 |
| F321Y | 3.4 ± 0.3 | 3.4 ± 0.2 | 3.8 ± 0.1 |
| L279F | 5.7 ± 0.1 | 4.5 ± 0.1 | 7.9 ± 0.6 |
| G113N | 9.1 ± 0.4 | NAb | 13.1 ± 0.5 |
Generation times were determined based on exponential increases in optical density of three replicate growth curves.
NA, not applicable.
BMO-specific activity of mutant BMOH-α strains.
We chose to alter amino acids in BMOH-α in key areas affecting substrate binding, active site geometry or chemistry, and interaction with the effector protein with the expectation that they would influence butane oxidation activity. Although growth on butane was diminished in most of the mutant strains (Fig. 1 and Table 3), changes in the specific activity of BMO could have been masked by changes in the amount of total BMO produced. To generate cells with identically induced BMO, cultures were grown on lactate and then washed and exposed to 1-butanol to induce BMO expression (23). Specific activities of the altered BMOs were lower than that of strain Rev WT. Mutant strains L279F and G113N had specific activities at least sevenfold lower than Rev WT, corroborating the more dramatically diminished growth rates observed for these mutant strains. We also used SDS-PAGE to evaluate the relative amounts of the BMO subunits produced by each of the mutant strains following growth to an OD600 of 0.75. No differences in protein levels were observed in BMOH subunits, BMOR, or BMOB (data not shown), providing further evidence that the differences in growth rates were due to changes in the specific activity of BMO.
Regiospecificity of mutant BMOH-α strains.
Previous research has shown that alterations to residues within multicomponent monooxygenases can result in specific catalytic effects, including changes in the rate of substrate turnover, substrate specificity, and position of oxygen insertion into the substrate (references 9, 28, and 33 to 35 and others). Wild-type P. butanovora predominantly oxidizes butane at the terminal carbon (1); likewise, strain Rev WT oxidized butane to 1-butanol and 2-butanol in a 24:1 ratio. The rates of 1- and 2-butanol accumulation were determined for each of the P. butanovora mutant strains (Table 4). Substantial differences from strain Rev WT were observed. Most notably, 92 percent of the product of butane oxidation by mutant strain G113N was 2-butanol. The ratios of the rates of 2- to 1-butanol accumulation for mutant strains L279F and G113N were 5.5-fold and 278-fold higher than those seen with strain Rev WT. Wild-type P. butanovora can grow on both 1- and 2-butanol, although the generation time for growth on 2-butanol (5 mM) is 3.5 times longer than that seen with growth on 1-butanol (data not shown). Mutant strain F321Y was the only mutant in which 2-butanol production was not measurable (detection limit, 0.0015 nmol min−1 [mg protein−1]).
TABLE 4.
Regiospecificity of butane oxidation by P. butanovora mutant strains
| P. butanovora strain | Rate of product accumulation (nmol min−1) (mg protein−1) ± SD
|
Ratio of 2-butanol to 1-butanol accumulation | Fold increase over Rev WT accumulation | |
|---|---|---|---|---|
| 1-Butanol | 2-Butanol | |||
| Rev WT | 74.0 ± 3.9 | 2.70 ± 0.91 | 0.04 | |
| T148C | 41.3 ± 4.0 | 6.14 ± 0.89 | 0.15 | 3.7 |
| Q320K | 50.4 ± 7.8 | 10.3 ± 0.53 | 0.20 | 5.6 |
| F321Y | 102.4 ± 5.7 | NDa | NAb | NA |
| L279F | 47.0 ± 7.1 | 10.45 ± 4.2 | 0.22 | 5.5 |
| G113N | 1.77 ± 0.06 | 19.67 ± 2.4 | 11.1 | 277.5 |
ND, none detected.
NA, not applicable.
To further investigate the extent of subterminal oxidation of alkanes by the P. butanovora mutant strains, the regiospecificity of propane oxidation was also determined. When butane is the growth substrate, the subsequent oxidation of 1-butanol yields butyraldehyde that is transformed to butyrate, while oxidation of 2-butanol yields butanone (1). When propane is the growth substrate, its terminal oxidation yields propionate, and the subterminal oxidation of propane yields propanone (commonly known as acetone). In P. butanovora the further metabolism of the products of propane oxidation, namely, propionate and acetone, requires induction of metabolic pathways that are not expressed in lactate-, ethane-, or butane-grown cells (7). Therefore, cells with BMO induced by ethane and subsequently exposed to propane should initially accumulate propionate or acetone, since the downstream enzymes required for their metabolism are not expressed. Washed ethane-grown cells were exposed to propane, and rates of propionate and acetone accumulation were measured. The rates of propionate and acetone accumulation were linear over the 60 min time course. The ratio of propionate to acetone accumulation by strain Rev WT was 17:1 (Table 5). In sharp contrast, mutant strain G113N oxidized propane exclusively to acetone at a rate of 11.3 nmol min−1 (mg protein−1), and no propionate was detected during the 60 min incubation (detection limit, 0.0033 nmol min−1 [mg protein−1]). Because mutant strain G113N oxidized propane to the subterminal oxidation product and yet did not grow on propane, we considered the possibility that although wild-type P. butanovora grows on propane and 1-propanol, it may not grow on acetone. Indeed, no increase in OD600 was measured when wild-type P. butanovora was inoculated in liquid medium with 5 mM, 10 mM, or 25 mM acetone. To verify that acetone is not toxic to P. butanovora at these concentrations, we inoculated cells in normal growth medium with acetone (0, 5, 10, and 25 mM) and butane. All cultures grew to similar final ODs. Interestingly, 2-butanone (5 mM) and 2-pentanone (5 mM) were growth substrates for P. butanovora (data not shown).
TABLE 5.
Accumulation of propionate and acetone in butane-grown P. butanovora mutant strains following removal of butane and exposure to propane
| P. butanovora strain | Rate of product accumulation (nmol min−1) (mg total protein−1) ± SD
|
|
|---|---|---|
| Propionate | Acetone | |
| Rev WT | 70.0 ± 3.0 | 4.2 ± 1.2 |
| L279F | 53.5 ± 2.3 | 4.4 ± 1.1 |
| F321Y | 74.2 ± 3.5 | 5.4 ± 0.09 |
| G113N | NDa | 11.3 ± 2.1 |
ND, none detected.
Propane oxidation by mutant strains L279F and F321Y did not result in enrichment of the subterminal oxidation product compared to strain Rev WT results. Although acetone was shown to be consumed by P. butanovora at approximately 9 nmol min−1 (mg protein−1) (reference 1 and this study), its consumption was acetylene sensitive and can be attributed to turnover by BMO (data not shown). Nevertheless, the concentration of propane in the propane oxidation assay was high enough so that it outcompeted acetone for the active site of BMO.
Methane oxidation.
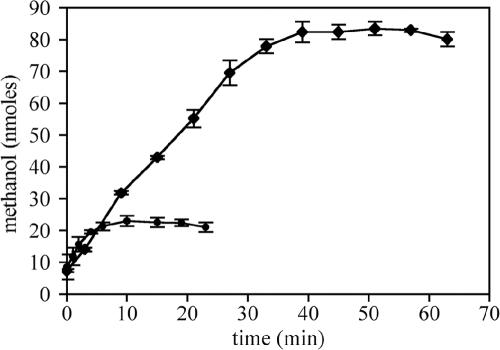
Given that five specific amino acid substitutions in BMOH-α were changed to reflect the corresponding residues of MMOH-α, it was of interest to determine whether rates of methane oxidation by the BMOH-α mutant strains were changed relative to strain Rev WT rates. Methanol consumption by wild-type P. butanovora and Rev WT was not detected at low concentrations in the presence of methane, obviating a need for an inhibitor of methanol consumption. In this study we measured an initial rate of methanol accumulation of 3.6 ± 0.6 nmol min−1 (mg protein−1) for Rev WT (first 2 min; Fig. 2), about 5% of the rate of 1-butanol production. However, unlike butane oxidation to 1-butanol, methanol accumulation stopped after 10 min at a final concentration of 23 μM. With the exception of mutant strain G113N, all other BMOH-α mutant strains exhibited kinetic properties of methane oxidation similar to Rev WT. In contrast, mutant strain G113N oxidized methane to methanol at a slower initial rate (2.1 ± 0.1 nmol min−1 [mg protein−1]), and yet methanol accumulated to a final concentration 3.5-fold higher than the Rev WT level (83 ± 2.2 μM versus 23 ± 1.7 μM).
FIG. 2.
Time course of methane oxidation to methanol. Washed butane-grown cells (1 mg total protein) were exposed to 3 ml methane in 7.7 ml sealed vials. Data are presented as the means of the results obtained with at least three replicate experiments ± standard deviation. Symbols: •, strain Rev WT; ⧫, strain G113N.
To determine whether methanol accumulation ceased due to enzyme inactivation or methanol inhibition of methane oxidation, strain Rev WT and mutant strain G113N cells that were exposed to methane were washed three times and exposed again to methane as described above. The rate and extent of methanol accumulation in Rev WT cells were equivalent to those shown in Fig. 2, indicating that methanol reversibly inhibits methane oxidation by BMO. Although only 75% of the methanol oxidation activity was recovered in mutant strain G113N, mechanical disturbance due to prolonged incubation and wash procedures was previously shown to reduce BMO activity (11). We also determined whether methanol inhibition is turnover dependent. The methanol accumulation assay was repeated with Rev WT and mutant G113N strain cells, and methanol (20 μM for Rev WT and 80 μM for G113N) was added to the reaction mixtures at time 0. No additional methanol accumulated over a 30 min incubation period for either P. butanovora strain. Furthermore, methanol (20 μM) did not inhibit butane or ethylene oxidation by Rev WT cells (data not shown).
DISCUSSION
We have demonstrated that BMO from P. butanovora can be altered directly within the native host organism, allowing valuable mechanistic and physiologic studies. Five mutant strains of P. butanovora were engineered to initiate investigations of the fundamental differences in substrate specificity between the two closely related enzymes BMO and MMO. Effects of the single-amino-acid substitutions in BMOH-α on growth rates and downstream metabolism in addition to hydrocarbon oxidation were evaluated.
Altered BMOH-α residue T148C resulted in slightly slower growth on alkanes and reduced specific activities. T148 corresponds to C151 in MMOH-α and Q141 in toluene 4-monooxygenase (T4MO) from Pseudomonas mendocina KR1. Although T148C had little effect on the regiospecificity of propane and butane oxidation, Q141C in T4MO resulted in changes to regiospecificity of m- and p-xylene oxidation (20). Site-directed mutagenesis of Cys 151 in MMOH-α to Glu or Tyr yielded mutant strains of M. trichosporium OB3b that were unable to support growth on methane under conditions that favored expression of the mutant enzyme (28). However, there is no evidence about the activity of these altered enzymes toward methane since the α, β, and γ subunits of MMOH could not be visualized by SDS-PAGE analysis, suggesting that the altered enzymes were unstable (28). Because neither mutant strain T148C nor Q141C in T4MO (29) promoted methane oxidation, we can conclude that simple acquisition of the Cys residue is not sufficient for active site chemistry facilitating methane oxidation.
Mutant strain F321Y oxidized butane even more selectively to 1-butanol than the Rev WT strain, as no 2-butanol was detected during the butane regiospecificity assay. It is possible that the specificity of strain F321Y allowed it to keep pace with the growth of Rev WT on alkanes, even though its specific activity was diminished as measured by the ethylene oxide assay. However, propane oxidation by strain F321Y yielded both terminal and subterminal oxidation products. It remains to be seen whether the terminal oxidation regiospecificity exhibited by strain F321Y with butane extends to other longer-chained alkanes. In methane oxidizers, MMOB is known to affect the regiospecificity of the enzyme complex (reviewed in reference 19). In vitro experiments revealed that the product distribution of sMMO-catalyzed butane oxidation is 44% 2-butanol and 56% 1-butanol. The product distribution of butane oxidation shifts to 95% 2-butanol in the absence of MMOB (10). The strict terminal oxidation of butane by mutant strain F321Y is consistent with the idea that the interaction between BMOH-α and BMOB was affected. Complementary mutational analysis of BMOB would lead to more-detailed understanding of the role of BMOB in affecting the regiospecificity of substrate oxidation.
Two residues, G113 and L279, within hydrophobic cavities 1 and 2 in BMOH-α were found to have distinct roles in hydrocarbon oxidation. Striking differences in specific activities and regiospecificity were observed for mutant strains G113N and L279F. The predominantly subterminal oxidation of propane and butane by strain G113N suggests that the single-amino-acid substitution caused a significant alteration of BMOH-α active site geometry. Likewise, the ratio of the rates of 2-butanol to 1-butanol production by strain L279F was increased 5.5-fold relative to the wild-type phenotype. It is also possible that the altered regiospecificities of mutant strains G113N and L279F were conferred by modified interactions with BMOB.
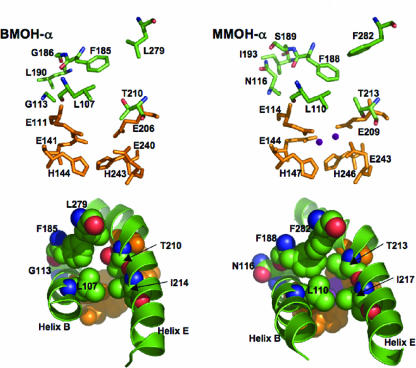
With the availability of site-directed bmoX mutants that demonstrate interesting changes in enzyme properties, we were in a position to spatially compare the geometry of BMOH-α with MMOH-α. We have used MMOH-α crystal structures as templates to model BMOH-α and focused attention on the area immediately surrounding the active site and including the leucine gate (Fig. 3). The crystal structure of MMOH-α reveals that residue F282 contributes to both the strained conformation of gating residue F188 (χ1 and χ2 torsion angles are −98° and −168°, respectively) and the reduction in the size of cavity 1. In the same plane in MMOH-α, N116 borders cavity 1 on the opposite side of helix B from E114 (Fig. 3, top right). The two residues packing to either side of N116 are S189 and I193. These three residues interact with P58 and W83 of MMOH-β. In BMOH-α, the two residues flanking G113 are G186 and L190 (Fig. 3, top left), which probably interact with N57 and Y82 from the BMOH-β subunit. Neither of these residues has space-filling potential substantially different from that of P58 and W83 from MMOH-β. In BMOH-α, the missing or reduced side chains of G113, G186, and L190 on one side of F185, and of L279 on the other, may allow F185 to adopt the more relaxed rotameric conformation present in the BMOH-α model (predicted χ1 and χ2 torsion angles are −53° and −156°, respectively) (Fig. 3). The position of F185 shifts the geometry of the active site and the leucine gate to a more open position relative to that of MMOH-α. This opening may favor entry of butane, BMO's natural substrate, to the active site. Likewise, the more restricted access to the active site in MMOH-α appears to favor its natural substrate, methane. We also modeled BMOH-α with the G113N substitution by use of the MMOH-α crystal structure (not shown). Indeed, the predicted χ1 and χ2 torsion angles for F185 in the G113N model (−85° and −124°, respectively) are intermediary between those of the wild-type BMOH-α model and MMOH-α crystal structure and result in partial closing of the leucine gate. The model of BMOH-α may suggest a relationship between substrate size and specificity. Recognizing that in the MMO system the effector protein influences substrate access (37) as well as active site geometry (10) and chemistry (42), the results obtained with the single-amino-acid substitutions in BMOH-α studied here reiterate the importance of the finely tuned structures at the active site of the hydroxylase.
FIG. 3.
Model of BMOH-α (left) compared to the MMOH-α crystal structure (right). (Top panels) BMOH-α residues G113, L279, G186, and L190 affect the conformation of residue F185, opening the leucine gate and shifting the geometry of the active site relative to MMOH-α. The atoms of residues are colored by type, where green is carbon, red is oxygen, blue is nitrogen, and purple is iron (MMOH-α only), except for residues coordinating the active site, which are colored orange. (Bottom panels) Space-filling representations. The view is through the leucine gate toward the diiron center (depicted in MMOH-α only). Atom colors are the same as described for the top panel. The figure was created using Pymol software.
We believe that the sensitivity of methane oxidation by BMO to methanol may have significant implications associated with product release from the active site. Product release is considered to be rate limiting in the sMMO catalytic cycle (16, 42). Altered MMOB proteins revealed that MMOB affects the rates of specific steps in the catalytic cycle, including product release (37). Product release may comprise more than one definable step, or these multicomponent monooxygenases may have regions of flexibility that accommodate unique postures during different steps of the cycle (2). Product-bound MMOH crystal structures revealed positional changes of I217 in addition to the leucine gate residues (25, 40). In the methanol-bound MMOH crystal structure, we note that L110 moves 0.8 Å away from F188 and 2.7 Å closer to Fe1 compared to native MMOH positions. It was previously suggested that these changes are necessary to allow residence of the oxidation product (25). The 3.5-fold increase in methanol accumulation by P. butanovora mutant strain G113N compared to that of strain Rev WT suggests that geometric alterations to cavity 1 and the leucine gate may make the active site less accommodating to the product, thus encouraging its release. In MMOH-α, the strained coordination of F188 held in place by the counterforces of F282 and N116 and its interacting residues could provide the necessary constraints within cavity 1 such that methanol is released.
The results obtained from investigations of the site-directed bmoX mutants of P. butanovora support the idea that the geometric intricacies of the leucine gate influence catalysis at the active site. Furthermore, BMO's striking sensitivity of methane oxidation to methanol inhibition provides a new avenue for exploration of the mechanistic differences between BMO and sMMO. The inability of P. butanovora to metabolize acetone exposed vulnerability in its metabolic flexibility. The mutant strains should prove valuable in unraveling the complexities associated with downstream metabolism of products generated from a broad substrate range monooxygenase.
Acknowledgments
This research was funded through a grant from the National Institutes of Health (5R01GM56128-06).
We gratefully acknowledge P. Andrew Karplus at Oregon State University for helpful discussions regarding protein chemistry and structure.
REFERENCES
- 1.Arp, D. J. 1999. Butane metabolism by butane-grown Pseudomonas butanovora. Microbiology 145:1173-1180. [DOI] [PubMed] [Google Scholar]
- 2.Brazeau, B. J., and J. D. Lipscomb. 2003. Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B. Biochemistry 42:5618-5631. [DOI] [PubMed] [Google Scholar]
- 3.Brazeau, B. J., B. J. Wallar, and J. D. Lipscomb. 2003. Effector proteins from P450(cam) and methane monooxygenase: lessons in tuning nature's powerful reagents. Biochem. Biophys. Res. Commun. 312:143-148. [DOI] [PubMed] [Google Scholar]
- 4.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, S. L., B. J. Wallar, J. D. Lipscomb, and K. H. Mayo. 2001. Residues in Methylosinus trichosporium OB3b methane monooxygenase component B involved in molecular interactions with reduced- and oxidized-hydroxylase component: a role for the N-terminus. Biochemistry 40:9539-9551. [DOI] [PubMed] [Google Scholar]
- 6.DeLano, W. L. 2002. The PyMol Molecular Graphics System. DeLano Scientific, San Carlos, CA.
- 7.Doughty, D. M., L. A. Sayavedra-Soto, D. J. Arp, and P. J. Bottomley. 2006. Product repression of alkane monooxygenase expression in Pseudomonas butanovora. J. Bacteriol. 188:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiser, A., R. K. Do, and A. Sali. 2000. Modeling of loops in protein structures. Protein Science 9:1753-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fishman, A., Y. Tao, L. Rui, and T. K. Wood. 2005. Controlling the regiospecific oxidation of aromatics via active site engineering of toluene para-monooxygenase of Ralstonia pickettii PKO1. J. Biol. Chem. 280:506-514. [DOI] [PubMed] [Google Scholar]
- 10.Froland, W. A., K. K. Andersson, S. K. Lee, Y. Liu, and J. D. Lipscomb. 1992. Methane monooxygenase component-B and reductase alter the regioselectivity of the hydroxylase component-catalyzed reactions—a novel role for protein-protein interactions in an oxygenase mechanism. J. Biol. Chem. 267:17588-17597. [PubMed] [Google Scholar]
- 11.Halsey, K. H., L. A. Sayavedra-Soto, P. J. Bottomley, and D. J. Arp. 2005. Trichloroethylene degradation by butane-oxidizing bacteria causes a spectrum of toxic effects. Appl. Microbiol. Biotechnol. 68:794-801. [DOI] [PubMed] [Google Scholar]
- 12.Hamamura, N., R. T. Storfa, L. Semprini, and D. J. Arp. 1999. Diversity in butane monooxygenases among butane-grown bacteria. Appl. Environ. Microbiol. 65:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahng, D., and T. K. Wood. 1994. Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 60:2473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 15.Leahy, J. G., P. J. Batchelor, and S. M. Morcomb. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27:449-479. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. K., J. C. Nesheim, and J. D. Lipscomb. 1993. Transient intermediates of the methane monooxygenase catalytic cycle. J. Biol. Chem. 268:21569-21577. [PubMed] [Google Scholar]
- 17.Lipscomb, J. D. 1994. Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol. 48:371-399. [DOI] [PubMed] [Google Scholar]
- 18.Marti-Renom, M. A., A. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291-325. [DOI] [PubMed] [Google Scholar]
- 19.Merkx, M., D. A. Kopp, M. H. Sazinsky, J. L. Blazyk, J. Muller, and S. J. Lippard. 2001. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew. Chem. Int. Ed. Engl. 40:2782-2807. [DOI] [PubMed] [Google Scholar]
- 20.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry 36:9283-9289. [DOI] [PubMed] [Google Scholar]
- 21.Rosenzweig, A. C., H. Brandstetter, D. A. Whittington, P. Nordlund, S. J. Lippard, and C. A. Frederick. 1997. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins 29:141-152. [PubMed] [Google Scholar]
- 22.Sali, A., and T. L. Blundell. 1993. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 23.Sayavedra-Soto, L. A., D. M. Doughty, E. G. Kurth, P. J. Bottomley, and D. J. Arp. 2005. Product and product-independent induction of butane oxidation in Pseudomonas butanovora. FEMS Microbiol. Lett. 250:111-116. [DOI] [PubMed] [Google Scholar]
- 24.Sazinsky, M. H., J. Bard, A. Di Donato, and S. J. Lippard. 2004. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J. Biol. Chem. 279:30600-30610. [DOI] [PubMed] [Google Scholar]
- 25.Sazinsky, M. H., and S. J. Lippard. 2005. Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the alpha-subunit. J. Am. Chem. Soc. 127:5814-5825. [DOI] [PubMed] [Google Scholar]
- 26.Sluis, M. K., L. A. Sayavedra-Soto, and D. J. Arp. 2002. Molecular analysis of the soluble butane monooxygenase from “Pseudomonas butanovora.” Microbiology 148:3617-3629. [DOI] [PubMed] [Google Scholar]
- 27.Smith, T. J., and H. Dalton. 2004. Biocatalysis by methane monooxygenase and its implications for the petroleum industry, p. 177-192. In R. Vazquez-Duhalt and R. Qintero-Ramirez (ed.), Studies in surface science and catalysis, vol. 151. Elsevier B.V., Amsterdam, The Netherlands. [Google Scholar]
- 28.Smith, T. J., S. E. Slade, N. P. Burton, J. C. Murrell, and H. Dalton. 2002. Improved system for protein engineering of the hydroxylase component of soluble methane monooxygenase. Appl. Environ. Microbiol. 68:5265-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steffan, R. J., and K. R. McClay. 2000. Preparation of enantio-specific epoxides. Patent WO/2000/073425.
- 30.Takahashi, J., Y. Ichikawa, H. Sagae, I. Komura, H. Kanou, and K. Yamada. 1980. Isolation and identification of n-butane-assimilating bacterium. Agric. Biol. Chem. 44:1835-1840. [Google Scholar]
- 31.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vangnai, A. S., D. J. Arp, and L. A. Sayavedra-Soto. 2002. Two distinct alcohol dehydrogenases participate in butane metabolism by Pseudomonas butanovora. J. Bacteriol. 184:1916-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vardar, G., Y. Tao, J. Lee, and T. K. Wood. 2005. Alanine 101 and alanine 110 of the alpha subunit of Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase influence the regiospecific oxidation of aromatics. Biotechnol. Bioeng. 92:652-658. [DOI] [PubMed] [Google Scholar]
- 34.Vardar, G., and T. K. Wood. 2005. Alpha-subunit positions methionine 180 and glutamate 214 of Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase influence catalysis. J. Bacteriol. 187:1511-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vardar, G., and T. K. Wood. 2005. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for enhanced chlorinated ethene degradation and o-xylene oxidation. Appl. Microbiol. Biotechnol. 68:510-517. [DOI] [PubMed] [Google Scholar]
- 36.Vardar, G., and T. K. Wood. 2004. Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for synthesizing 4-methylresorcinol, methylhydroquinone, and pyrogallol. Appl. Environ. Microbiol. 70:3253-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallar, B. J., and J. D. Lipscomb. 2001. Methane monooxygenase component B mutants alter the kinetics of steps throughout the catalytic cycle. Biochemistry 40:2220-2233. [DOI] [PubMed] [Google Scholar]
- 38.West, C. A., G. P. Salmond, H. Dalton, and J. C. Murrell. 1992. Functional expression in Escherichia coli of proteins B and C from soluble methane monooxygenase of Methylococcus capsulatus (Bath). J. Gen. Microbiol. 138:1301-1307. [DOI] [PubMed] [Google Scholar]
- 39.Whittington, D. A., and S. J. Lippard. 2001. Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometrical variability at the dinuclear iron active site. J. Am. Chem. Soc. 123:827-838. [DOI] [PubMed] [Google Scholar]
- 40.Whittington, D. A., A. C. Rosenzweig, C. A. Frederick, and S. J. Lippard. 2001. Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase. Biochemistry 40:3476-3482. [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, H., and J. D. Lipscomb. 2006. Regulation of methane monooxygenase catalysis based on size exclusion and quantum tunneling. Biochemistry 45:1685-1692. [DOI] [PubMed] [Google Scholar]