Abstract
The opportunistic pathogen Bacteroides fragilis is a commensal organism in the large intestine, where it utilizes both dietary and host-derived polysaccharides as a source of carbon and energy. In this study, a four-gene operon required for starch utilization was identified. The operon also was found to be oxygen responsive and thus was designated osu for oxygen-induced starch utilization. The first three genes in the operon were predicted to encode outer membrane proteins involved in starch binding, and a fourth gene, osuD, encoded an amylase involved in starch hydrolysis. Insertional mutation of the osuA gene (ΩosuA) resulted in the inability to utilize starch or glycogen and an insertional mutation into the osuD gene (ΩosuD) was severely impaired for growth on starch media. Transcriptional studies indicated that maltose, maltooligosaccharides, and starch were inducers of osu expression and that maltose was the strongest inducer. A transcriptional activator of osuABCD, OsuR, was identified and found to mediate maltose induction. The ΩosuA and ΩosuD mutants were able to grow on maltose but not starch, whereas a mutation in osuR abolished growth on both substrates, indicating that additional genes under the control of OsuR are needed for maltose utilization. The osuABCD operon also was induced by exposure to oxygen and was shown to be part of the oxidative stress response important for aerotolerance of B. fragilis. Transcriptional analyses showed that osuA was induced 20-fold by oxygen, but OsuR was not required for this activation. Analysis of osu mutants suggested that expression of the operon was important for survival during oxygen exposure but not to hydrogen peroxide stress.
The Bacteroides species are gram-negative, obligate anaerobes that form an integral component of the indigenous microflora in the human gastrointestinal tract. The Bacteroides organisms comprise approximately 30% of the total bacterial population of the lower intestine, and as commensal organisms, they benefit their hosts by aiding in digestion of complex carbohydrates, in the biotransformation of bile acids, vitamin synthesis, and development of the immune system (21, 37, 44). The Bacteroides spp. also can be opportunistic pathogens, and Bacteroides fragilis is the most commonly isolated organism from anaerobic infections such as abdominal abscesses and postoperative wound infections (11). This organism is also commonly found to be associated with bacteremia, abscesses of the female genital tract and pelvis, brain abscesses, diabetic foot ulcers, appendicitis, and diverticulitis abscesses in elderly patients (11, 13, 18-20, 27). The success of B. fragilis as an opportunistic pathogen has been attributed in part to the production of several virulence factors including the complex polysaccharide capsule that has been shown to induce abscess formation and have antiphagocytic properties (23, 44). The prolonged aerotolerance of B. fragilis and its ability to survive oxidative stress also may play an important role in pathogenesis or in other extraintestinal situations.
B. fragilis has evolved a complex oxidative stress response to allow it to combat the toxic effects of oxygen exposure. Although not capable of replication in an aerobic environment, it has been well established that this organism can synthesize new RNA and upregulate a distinct set of more than 28 proteins in response to oxygen exposure (32). These include catalase (encoded by katB), superoxide dismutase (sod), alkyl hydroperoxide reductase (ahpCF), thioredoxin peroxidase (tpx), and the nonspecific DNA binding protein (dps), all of which play a role in detoxifying reactive oxygen intermediates and protecting cellular components (14, 15, 32, 35). Most of these detoxification enzymes (AhpC, Dps, Tpx, and KatB) are controlled at the transcriptional level by the redox-sensitive regulator OxyR, which modulates their expression in response to peroxide (31). Another level of oxidative stress control has been demonstrated for Dps, which can be induced during oxygen exposure in an OxyR mutant, although the levels of induction are lower than in the wild-type strain (30, 42).
Compared to the detoxification arm of the oxidative stress response, other aspects are not as well understood. In a study on the related organism, B. thetaiotaomicron, it was found that two key enzymes of central metabolism were rapidly inactivated by oxygen exposure and that this was in part responsible for aerobic growth inhibition (24); however, at the same time, glucose uptake is stimulated by oxygen (17). In another study with B. fragilis, it was reported that the genes for several metabolic enzymes were induced by aerobic exposure (42). The induced genes encoded an aerobic ribonucleotide reductase, a cation efflux pump, an aspartate decarboxylase and a starch binding outer membrane protein similar to SusC. Thus, there is likely a shift in metabolism that occurs during aerobiosis which remodels cellular physiology to help deal with the increased oxygen stress. Consistent with this is the ability of B. fragilis to consume oxygen when present in the nanomolar range by using a cytochrome bd oxidase (2). In order to better understand the role of this aerobically induced metabolism, we have begun to characterize the novel multigene operon that encodes the SusC-like outer membrane protein. In B. thetaiotaomicron, the SusC gene is part of a seven-gene starch utilization locus and is required for starch binding on the cell surface. Moreover, SusC also was the first described member of a larger orthologous family of outer membrane proteins found extensively in the Bacteroides and related genera. In fact, there are nearly 80 paralogous genes within B. thetaiotaomicron (The Institute for Genomic Research [http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=ntbt01]) and 54 related genes in B. fragilis (The Wellcome Trust Sanger Institute [http://www.sanger.ac.uk/Projects/B_fragilis/]). Except for the case with the SusC gene, the functions of these genes have not been determined, but it is generally expected that most will be found to be involved in some aspect of nutrient binding and uptake.
In the present study, the oxygen-induced B. fragilis SusC gene homolog was found to be in a four-gene operon. This operon was shown to be necessary for starch utilization during anaerobic growth, as well as being oxygen responsive. Two of the genes have significant homology to genes in the sus operon of B. thetaiotaomicron, but the types of genetic organization of these two operons are quite different. Given the propensity of Bacteroides to switch off central metabolism upon oxygen exposure, it was curious that the expression of a metabolic operon was strongly up-regulated under such conditions. In this study, we begin to elucidate the regulation and control of this operon and determine its role and contribution to starch utilization and to the oxidative stress response in this organism.
MATERIALS AND METHODS
Strains and growth conditions.
The B. fragilis strains used in this study (Table 1) were routinely grown anaerobically in brain heart infusion broth supplemented with hemin, cysteine, and NaHCO3 (BHIS) (43). B. fragilis ADB77 was grown in BHIS supplemented with 50 μg thymine ml−1. For carbohydrate utilization studies, cells were grown in a defined medium as described previously (SDM [36]), supplemented with 0.05% tryptone and 0.3% glucose, 0.3% maltose, or 0.5% starch as the sole carbon source. Growth was measured as an increase in A550 versus time. In experiments in which the levels of induction of gene expression by different oligosaccharides were measured, cells were transferred from an overnight SDM culture supplemented with 0.3% glucose to SDM supplemented with one of the following as the sole carbohydrate: glucose, 0.3%; maltose, 0.3%; maltotriose, 0.3%; maltoheptaose, 0.3%; maltopentaose, 0.3%; and starch, 0.5%. All carbohydrates were obtained from Sigma Chemical Co. (St. Louis, Mo.). Cells used for transcriptional studies of oxidative stress induction were grown anaerobically to mid-exponential phase (A550 of 0.35) and then divided. Half was shaken aerobically at 250 rpm at 37°C for 1 h in an Erlenmeyer flask with at least a 1:20 liquid-to-headspace ratio. The remaining anaerobic half was immediately harvested by centrifugation. For viability assays, cells were grown to mid-exponential phase in BHIS and then exposed to oxygen by shaking in air as described above. Viable cell counts were determined at indicated time points by plating appropriate dilutions on Wilkins-Chalgren agar (Difco Laboratories, Detroit, MI) in triplicate and allowing anaerobic recovery for 24 to 72 h.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference(s) and/or source |
|---|---|---|
| Strains | ||
| B. fragilis | ||
| 638R | Clinical isolate; wild-type strain; Rfr | 26 |
| IB298 | 638R ΔoxyR::tetQ; Rfr Tcr | 31 |
| IB367 | 638R ΩosuA, contains insertion of pFD1038 in osuA; Rfr Emr | This study |
| IB371 | 638R ΩosuD, contains insertion of pFD1039 in osuD; Rfr Emr | This study |
| ADB77 | TM4000 (638R) ΔthyA1; Rfr Tpr | 1 |
| IB393 | ADB77 ΔosuR, thyA+; Rfr | This study |
| IB442 | ADB77 minus 19 bp of osuC-osuD intergenic region; Rfr Tpr | This study |
| E. coli DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araΔ139 Δ(ara leu)7697 galU galK λ−rpsL (Str) nupG | Invitrogen |
| Plasmids | ||
| pFD842 | Suicide vector derived from pFD516, 8.8 kb; xylB, tetX, aad9, ermF; (Tcr) (Spr) Emr | 31, 46; this study |
| pFD1038 | A 650-bp osuA internal gene fragment cloned into PstI/SmaI sites of pFD842; (Tcr) (Spr) Emr | This study |
| pFD1039 | An 800-bp osuD internal gene fragment cloned into SalI/SphI sites of pFD842; (Tcr) (Spr) Emr | This study |
| pYT102 | Suicide vector, 8.2 kb; p15A ori, RP4 oriT; thyA, tetQ, cat; (Cmr) Tcr | This study |
| pYTosuR | osuR gene fragment with in-frame deletion cloned into BamHI/HindIII sites of pYT102; (Cmr) Tcr | This study |
| pYTstmlp | A 1.6-kb gene fragment from osuC-osuD intergenic region containing the 19-bp stem-loop deletion cloned into the BamHI/HindIII sites of pYT102; (Cmr) Tcr | This study |
| pFD972 | Expression vector, 9.1 kb; derived from pFD340 by EcoRI deletion of ermF and replacement with a 2.6-kb SstI fragment containing tetQ; (Apr) Tcr | 45; this study |
| pFD1040 | A 1.2-kb osuR gene inserted into BamHI/SmaI site of pFD972; (Apr) Tcr | This study |
| pFD1077 | A 1.8-kb osuD gene cloned into BamHI/SmaI site of pFD972; (Apr) Tcr | This study |
Emr, erythromycin resistance; Rfr, rifampin resistance; Tcr, tetracycline resistance; Spr, spectinomycin resistance; Cmr, chloramphenicol resistance; Apr, ampicillin resistance; Tpr, trimethoprim resistance. Parentheses around an antibiotic resistance phenotype indicate that it is expressed only in E. coli. If there are no parentheses, then the resistance phenotype is expressed only in B. fragilis.
Amylase activity on starch azure plates was determined by spotting 10 μl of a BHIS overnight culture onto an SDM agar plate containing 0.5% xylose, 0.3% starch, and 0.4% starch azure (potato starch linked to remazole brilliant blue [Sigma Chemical Co., St. Louis, Mo.]). Plates were incubated anaerobically at 37°C for 24 to 48 h and then examined for a zone of clearing. Amylase activity also was measured in cell extracts by measuring the increase in reducing sugar formation from starch by using the dinitrosalicylic acid assay with glucose as the standard (8). Cell extracts were prepared in TSD buffer (50 mM Tris, 150 mM NaCl, 100 μM dithiothreitol [pH 8]) as described previously (33), and protein was measured with the Bradford reagent, using lysozyme as the standard (5). One unit of amylase activity liberates 1 μmol equivalent of glucose from starch per min.
Mutant construction.
By use of routine procedures (39) and the primers listed in Table 2, a 650-bp internal fragment of osuA and an 800-bp fragment of osuD was amplified from the 638R chromosome by PCR and cloned into the suicide vector pFD842. These constructs, pFD1038 and pFD1039, were then mobilized from Escherichia coli DH10B into B. fragilis 638R by an aerobic triparental mating procedure (41). Transconjugants were selected on BHIS agar containing rifampin (20 μg/ml), gentamicin (100 μg/ml), and erythromycin (10 μg/ml). Colony PCR was used to confirm single-crossover disruption of the target genes. Briefly, a single colony was picked using a sterile pipette tip and boiled for 5 min in a 1.5-ml tube containing 38 μl H2O. Twelve microliters of a PCR supermix containing 2.5 U Platinum Taq DNA polymerase, 1 × PCR buffer, 50 mM MgCl2, 10 mM dNTPs (Invitrogen, Carlsbad, CA), and gene-specific primers was then added to a total volume of 50 μl, and PCR was performed as follows: 95°C for 3 min (1 time), then 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 1 min (35 times), and then 72°C for 10 min (1 time).
TABLE 2.
Oligonucleotide primers
| Designation | 5′ to 3′ sequencea | Useb |
|---|---|---|
| osuAPstI | GTGCCTGCAGATAACACCGAGATC | ΩosuA mutant |
| osuASmaI | GTCTCCCGGGTGCTTCATATAC | ΩosuA mutant |
| amy1 | GGAATCTATCGTTCTCGTCAG | ΩosuD mutant |
| amy2 | CCATTGACGGATCGAAGGTG | ΩosuD mutant |
| osuRBam | CCATGGATCCCTTCAAACTCTTTGG | osuR mutant |
| osuRPst1 | CTACTCCTGCAGACTGACCGGACGGAA | osuR mutant |
| osuRPst2 | CTAACTGCAGGATGATGACCCCGAT | osuR mutant |
| osuRHind | GTCGGAAGCTTAGGTAGCCATACC | osuR mutant |
| osuRBamC | CACTGAACTTTTCAGGATCCTTTAC | osuR in pFD1040 |
| osuRSmaC | GCTTTGAACCCGGGAGGCTATCT | osuR in pFD1040 |
| osuDBam | GGAAGATACTTCGGATCCCAACCTGC | ousD in pFD1077 |
| osuDSma | GCTACACCTTCCCGGGTAAAGG | osuD in pFD1077 |
| susCupst1 | CCAGCAATTCCTGGTAATGG | RT-PCR |
| pex1 | GCATAAGCGCTCAATGAC | RT-PCR |
| fwd1 | CCGTGTATTCATGATCGGCC | RT-PCR |
| rev4B | CCGTAGGTCAAATAGAAGCC | RT-PCR |
| rtsusD/Ffwd | CCGATTTGACATTGAACTTC | RT-PCR |
| rtsusD/Frev | CGGTTTCACCCAATAAGACAG | RT-PCR |
| rtsusE/afwd | GGTGCCGATATGAAGAAGC | RT-PCR |
| rtsusE/arev | CGCTACGGCTGTCATAATC | RT-PCR |
| amyendfwd | GGAGTCTGGACAAACTATCTGG | RT-PCR |
| amyendrev | CCATGTATCAGGGAGACAAACG | RT-PCR |
| raceosu1 | GACATGGATGAACCTCCA | 5′ RACE |
| raceosu2 | GTAATCACACTGACACC | 5′ RACE |
| suscdwnst1 | GGAGCGGCTTTCATTTCAAC | 5′ RACE |
| osuA1 | GCAGCTATCGGTCTGGCTAC | Real-time PCR |
| osuA2 | GCCGATATCAATCGGAGAAG | Real-time PCR |
| osuD1 | GTCGTCATCCTGGAGCATTT | Real-time PCR |
| osuD2 | GTTTGACGGATACCCCATTG | Real-time PCR |
| 16S fwd | GCGTTCCATTAGGTTGTTG | Real-time PCR |
| 16S rev | CAGTGCTGCCTCCCGTAG | Real-time PCR |
| stmlp1bam | CTGAGGATCCATAATTACGGAGCGGTATCC | Construction of IB442 |
| stmlp1pst | CATGCTGCAGAAGTATCTTCCTGGCATTT | Construction of IB442 |
| stmlp2pst | CATGCTGCAGGTTGGCATCCTTTTTTATA | Construction of IB442 |
| stmlp2hind | CATGAAGCTTATAGCGTCAATTGCCTGTCA | Construction of IB442 |
Bold letters indicate restriction enzyme recognition sites.
The intended use of each primer as described in the text.
An osuR mutant (IB393) was made by a two-step allelic exchange procedure using the suicide vector pYTosuR (Table 1) in strain ADB77 (1). By use of the primers described in Table 2, an osuR gene fragment with a 597-bp internal, in-frame deletion was constructed in a stepwise fashion and cloned into pYT102 and then mobilized into ADB77. Following resolution, the mutant was restored to thymine prototrophy by a single-step marker exchange with pYT102. Colony PCR was used to confirm the deletion of the internal fragment. The same approach was used to construct the mutant IB442 with a 19-bp deletion of the stem-loop structure in the intergenic region between osuC and osuD. Primers described in Table 2 were used in the construction of IB442.
For complementation of IB393, the osuR gene was PCR amplified using the primers described in Table 2 and was cloned into an expression vector (pFD972) with BamHI and SmaI, resulting in plasmid pFD1040. Similarly, pFD1077 was constructed to be used to complement ΩosuD mutants.
Northern hybridizations and 5′ RACE.
Total RNA isolation using the hot phenol method and Northern blotting were performed as described previously (34). osuA and osuD gene-specific probes were generated by PCR amplification of 638R genomic DNA and labeled with [α-32P]dCTP by using the Prime-a-Gene labeling system (Promega, Madison, WI). RNA used for 5′ rapid amplification of cDNA ends (RACE) was further purified on an RNeasy column (QIAGEN Inc., Valencia, CA), followed by DNase I treatment according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). The 5′ RACE was performed as described by the manufacturer (Invitrogen, Carlsbad, CA), using raceosu1 and raceosu2 primers (Table 2) to generate 5′-specific cDNA products. Nested PCR amplification was performed using an abridged universal amplification primer (Invitrogen, Carlsbad, CA) and suscdwnst1 (Table 2). The final amplified 5′ RACE products were sequenced at the Molecular Biology Resources Facility at the University of Tennessee, Knoxville. Twenty clones with 5′ RACE fragments generated from anaerobic conditions and 20 clones with 5′ RACE fragments generated from oxidative stress conditions were sequenced.
RT-PCR and real-time RT-PCR.
Total RNA was purified and DNase I treated as described above. One hundred nanograms of RNA template and appropriate primer sets (Table 2) were used in a one-tube reverse transcription-PCR (RT-PCR) with the Accessquick RT-PCR system (Promega, Madison, WI). Reaction conditions were as follows: 45°C for 45 min (1 time), 95°C for 3 min (1 time), then 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 1 min (35 times), and then 72°C for 10 min (1 time). cDNA was synthesized by reverse transcriptase with random hexamers. Real-time RT-PCR amplification was performed using gene-specific primers (Table 2) with product sizes of approximately 150 bp. The PCR efficiency of each primer set was determined with a standard curve using serially diluted BamHI-digested 638R genomic DNA. The cDNA templates were used in a reaction mixture containing iQ SYBR green supermix (Bio-Rad, Hercules, CA) and gene-specific forward and reverse primers. Reaction conditions were 95°C for 3 min (1 time), then 95°C for 10 seconds, 55°C for 15 seconds, and 72°C for 15 seconds (40 times). Gene-specific products were quantified as a measure of incorporated SYBR green fluorescent dye. 16S rRNA was amplified to serve as an internal reference point against which expression of the genes of interest were normalized. Relative expression levels were determined in both anaerobic growth and oxygen exposure using the equation described previously (25). Results were expressed as n-fold induction of expression upon exposure to oxygen, with 1 representing anaerobic expression. For expression analysis with SDM supplemented with different carbohydrates, results were expressed as n-fold induction of expression relative to expression during growth in glucose medium.
Disk inhibition assays.
One hundred microliters of an overnight BHIS culture was spread onto Wilkens-Chalgren agar plates. A 6-mm sterile disk was placed in the center of the plate, and 10 μl of either 3% menadione in dimethyl sulfoxide, 3% hydrogen peroxide aqueous solution, or 0.5% t-butyl hydroperoxide aqueous solution was applied to the disk. Plates were incubated anaerobically at 37°C for 24 h and the growth inhibition zones around the paper disks were measured and reported in millimeters. For menadione sensitivity, plates were first incubated aerobically for 6 h before being placed in the anaerobe chamber for recovery. Diameters of the zones of growth inhibition were measured in millimeters. All assays were performed in triplicate.
RESULTS
Characterization of the osu locus.
In a previous study to screen for mRNAs induced by oxidative stress, we identified an mRNA encoding a protein that had significant homology to the large orthologous SusC family of outer membrane proteins involved in nutrient binding in Bacteroides (42). A physical map of this region was determined from the B. fragilis 638R genome sequence, and we identified a putative four-gene operon that has been designated osuABCD, for oxygen-induced starch utilization (Fig. 1A). This operon was highly conserved in the genome sequences of other B. fragilis strains (ATCC 25285 and YCH46) with greater than 99% nucleotide identity for each of the four Osu open reading frames (ORFs). The first gene, osuA, is a 3,006-bp ORF with significant nucleotide homology to the B. thetaiotaomicron SusC gene (50% identity) (28). Analysis of osuB (1,617 bp) and osuC (1,596 bp) suggested that they encode starch binding proteins similar to B. thetaiotaomicron SusD (34% identity) and SusF (24% identity over C-terminal half), respectively (29). Further analysis of OsuC by using position-specific iterated BLAST detected some similarity to α-glucosidase, amylase, and pullulanase enzymes from a variety of organisms, but this similarity was not detected in SusF. The osuD gene (2,859 bp) encodes a putative amylase, and analysis of the protein sequence for conserved domains detected strong matches to α-amylase (Pfam database accession number pfam00128.11), pullulanase (COG database accession number COG1523.1), and a signal peptide sequence of 23 amino acid residues (4). Database searches only showed sequence homology to amylase, pullulanase, and other enzymes in the glycosidase 13 family.
FIG. 1.
Functional map and operon structure of the osu locus. A. Functional map derived from base pairs 3709740 to 3720756 of the B. fragilis 638R genome sequence. The approximate sizes of osu mRNA transcripts are shown by arrows below the map and RT-PCR primer sets 1 through 5 are shown above the map. B. RT-PCR analysis of the osuABCD operon. Agarose gel showing RT-PCR products resulting from primer pairs shown in panel A. Lane designations are as follows: Mw, 1-kb molecular mass standard; lane 1, primer set 1; lane 2, primer set 2; lane 3, primer set 3; lane 4, primer set 4; lane 5, primer set 5. Lanes 6 and 7 were positive controls using primer sets 1 and 5, respectively, in PCRs with wild-type strain 638R DNA.
Directly upstream, in the opposite orientation from osuA, was a fifth gene, osuR (1,011 bp), which encodes a protein with a helix-turn-helix motif at its carboxy-terminal end and overall sequence similarity to transcriptional regulators in the LacI-type regulator family, including the Bacillus subtilis ccpA gene for the catabolite control protein A (30% identity). The deduced amino acid sequence of the OsuR protein showed no homology to SusR and MalR, the two known regulators of starch utilization in B. thetaiotaomicron.
To determine if the osu operon was required for starch utilization two mutations, ΩosuA (IB367) and ΩosuD (IB371), were constructed by insertional inactivation. These single-crossover disruptions of the target genes by the suicide plasmid should, in the case of ΩosuA, have a polar effect on the osuABCD operon. When compared to wild-type 638R in SDM-glucose or -maltose medium over 24 h, neither mutant displayed a significant defect in growth (Fig. 2A and B). However, both mutants were severely impaired in medium with starch as the sole carbon/energy source. The ΩosuA mutant was completely unable to utilize starch, but the ΩosuD mutant, although severely hampered, maintained a slow growth rate over 24 h (Fig. 2C). ΩosuD eventually reached wild-type growth density after approximately 70 h in starch medium, but ΩosuA never recovered (data not shown). The ΩosuD strain was complemented by a plasmid containing osuD under the control of a weak constitutive promoter. As shown in Fig. 2C, this complemented strain grew at a greater rate than the mutant in medium with starch, but it did not reach wild-type growth rates. In contrast, the ΩosuA mutant strain still failed to grow when complemented with osuD. These findings indicate that the osuABCD operon is the primary starch utilization operon in B. fragilis. The ΩosuA mutant also was unable to utilize glycogen as a carbon source (data not shown), suggesting that this operon also is required for utilization of this more highly branched glucose polymer.
FIG. 2.
Growth of osu mutant strains (osuA and osuD) in SDM supplemented with glucose, maltose, or starch. Wt, wild type. A. SDM supplemented with 0.3% glucose. B. SDM supplemented with 0.3% maltose. C. SDM supplemented with 0.5% starch. Data are presented as the means of three independent experiments performed in triplicate.
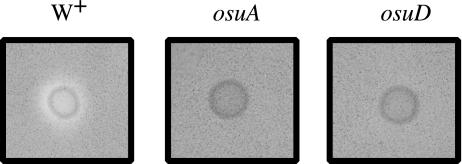
Analysis of the OsuD gene nucleotide sequence suggested that it encoded an α-amylase. To determine if OsuD had amylase activity, overnight BHIS cultures of 638R, ΩosuA and ΩosuD were spotted onto starch azure amylase indicator plates containing the nonrepressing carbon source xylose and starch. Following incubation, a zone of clearing was observed around strain 638R but not surrounding the two mutant strains (Fig. 3). Amylase activity in cell extracts showed that there was a decrease in total amylase activity in ΩosuD from 1.45 U to 0.55 U, and this was restored in part in the osuD-complemented strain (1.12 U). It is not clear why the starch azure plates did not show a clear zone for ΩosuD but it could indicate a difference in substrate specificity for the different amylolytic enzymes. As implied by the starch azure plates, some OsuD seemed to release from the cells and analysis of culture supernatants were consistent with this observation (data not shown). However, this may just represent loss from lysed cells as suggested for B. thetaiotaomicron (9). Overall, these results are consistent with osuD encoding an enzyme with amylase activity, but the gene clearly is not responsible for all B. fragilis amylase activity.
FIG. 3.
Amylase activity of the B. fragilis osu mutant strains on starch azure plates. Ten microliters of an overnight BHIS culture of wild-type strain 638R (W+), IB367 (ΩosuA), and IB371 (ΩosuD) were spotted onto starch azure plates and incubated at 37°C. Starch azure plates contained 0.5% xylose, 0.3% starch, and 0.4% starch azure to allow for growth of the mutant strains.
Transcription of the osuABCD operon.
RT-PCR was used to show that the osuABCD operon was transcribed as a polycistronic message (Fig. 1B). Total RNA was isolated from strain 638R after anaerobic growth in SDM with 0.5% starch, then cDNA was synthesized, and gene-specific primers were used to amplify junctions between genes within the operon. Lanes 2, 3, and 4 of Fig. 1B show the presence of an amplified PCR product which is consistent with an RNA message encoding all four genes in the operon. The absence of an amplified product for the region between osuA and osuR and for the region downstream of osuD suggests that osu is a four-gene operon beginning with osuA and ending with osuD.
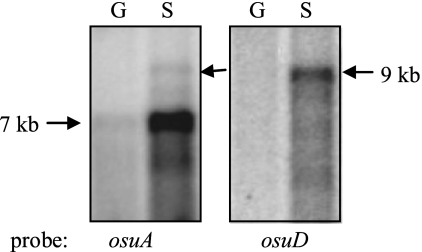
Northern hybridization analysis indicated that osu transcription was not as straightforward as first implied by the RT-PCR analysis. When cells were grown anaerobically with starch as the sole carbon/energy source, two distinct mRNA species were observed (Fig. 4). The predominant mRNA was approximately 7 kb in size, and it hybridized to an osuA probe but not to an osuD probe. This mRNA was predicted to encode the first three genes of the operon. The second mRNA species was approximately 9 kb and was observed in cells grown with starch when it was probed with either an osuA or osuD probe. Based on the size and its homology to both probes, this mRNA was predicted to include the entire osuABCD operon. Interestingly, this larger mRNA species was only about 1/18 as abundant as the 7-kb mRNA (based on densitometry analysis); this difference in abundance may have been overlooked in the RT-PCR results due to the large number of cycles used in the PCR.
FIG. 4.
Autoradiographs of Northern blots probed with internal fragments of osuA or osuD. Total RNA was isolated from mid-log-phase cultures of strain 638R grown anaerobically in SDM supplemented with either 0.3% glucose (G) or 0.5% starch (S) as the sole carbon/energy source. All lanes contain 30 μg of total RNA. Approximate sizes of the hybridizing transcripts were extrapolated from a molecular mass ladder in an adjacent lanes.
Inspection of the DNA sequence between osuC and osuD revealed a 104-bp intergenic region with a strong (19.3 kcal/mol) hairpin loop structure (Fig. 5A). It seemed plausible that the low abundance of the 9-kb mRNA might be due to a termination mechanism mediated by this secondary structure. To test this idea, the secondary structure was disrupted by a 19-bp deletion encompassing half of the palindrome. Northern analysis of the mutant showed an increase in the level of the 9-kb mRNA and real-time RT-PCR data also indicated that the ratio of the larger mRNA to the smaller one was greater in the stem loop mutant than in the parent strain (Fig. 5B and data not shown). Starch azure plates indicated that there was a concomitant increase in amylase activity (Fig. 5C). Finally, when amylase activity of the two strains was examined by an amylase assay measuring the production of reducing sugars from starch, there was nearly a threefold increase in activity of the mutant strain IB442 with 19 bp deleted (3.9 U/mg) compared to the wild type (1.4 U/mg).
FIG. 5.
Role of secondary structure in the expression of osuD. A. Sequence of the intergenic region between osuC and osuD. The inverted repeat is shown by the arrows over the sequence and the 19-bp deletion disrupting the structure is shown by the dashed line under the sequence. B. Phosphorimager image of Northern blot analysis of wild-type 638R (W+) and the mutant IB442 (with the 19-bp deletion) (Δ19 bp) grown on SDM with starch as the sole carbon/energy source. The probe was an internal osuA gene fragment. The arrows to the right of the autoradiograph show the location of the 7- and 9-kb mRNA species. C. Amylase activity on starch azure plates. Ten microliters of overnight cultures grown in BHIS were spotted on starch azure plates as described in the legend to Fig. 3. Incubation was for 24 h at 37°C.
Induction of osu by carbon source.
Real-time RT-PCR was used to measure relative expression levels of osuA and osuD in the wild-type strain. Cells were grown anaerobically in SDM supplemented with glucose, starch, and a range of maltooligosaccharides as potential inducers of the system (Fig. 6). In these experiments, the n-fold induction relative to growth in glucose was expressed, and both osu genes were both strongly induced by growth by maltose. A 12-fold increase in osuA expression and a 28-fold increase in osuD expression relative to expression in glucose were seen. Maltotriose and starch also were strong inducers of expression. There was a somewhat greater increase in osuD induction compared to osuA, but this might be misleading when relative levels of osuA and osuD expression in glucose medium are taken into account (Fig. 4).
FIG. 6.
Induction of osuA and osuD expression by maltooligosaccharides in the wild-type strain 638R. Relative expression was determined by real-time RT-PCR during anaerobic growth in SDM supplemented with glucose (G1), maltose (G2), maltotriose (G3), maltopentaose (G5), maltoheptaose (G7) or starch (STA). Real-time RT-PCR results were normalized to the amount of 16S rRNA in each sample and then expressed as n-fold induction relative to the expression during growth in glucose medium. Data are presented as the means from three independent experiments.
Regulation of the osu operon.
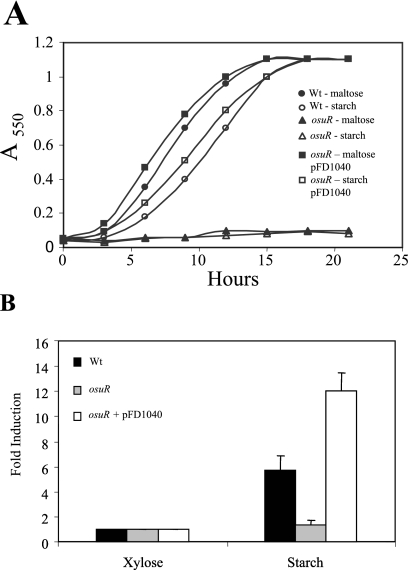
The proximity of osuR to the osuABCD operon suggested a possible link to regulation. To determine the role of OsuR, a mutant with an internal in-frame deletion of 200 amino acids was constructed and tested for growth in SDM supplemented with glucose, maltose, or starch. There was no significant difference between the osuR mutant and the wild-type strain in glucose medium (data not shown); however, the osuR mutant was unable to grow with either maltose or starch as the sole carbon source (Fig. 7A). To confirm these results, the osuR gene was cloned into an expression vector under the control of a constitutive promoter. Complementation of osuR resulted in wild-type growth rates in both maltose and starch media, suggesting that OsuR has a role in regulation of maltose and possibly starch utilization.
FIG. 7.
Role of OsuR in growth and gene expression. Wt, wild type (638R); osuR, osuR mutant. A. Effect of osuR mutation on growth in SDM supplemented with starch or maltose. B. Effect of starch on the expression of osuA in wild-type and osuR mutant strains. Relative expression levels were determined during anaerobic growth in a xylose-containing basal medium supplemented with or without 0.5% starch. Results are expressed as n-fold induction relative to expression levels in basal xylose medium without starch. Strains were grown anaerobically to mid-log phase and then 0.5% starch was added to induce osuABCD expression. Total RNA was isolated 1 h after starch addition and expression of osuA was analyzed by real-time RT-PCR. Data are presented as the means from three independent experiments.
To determine if OsuR controlled osu expression, real-time RT-PCR was used to examine the induction of osuA by starch in the osuR mutant strain background. Since an osuR mutant will not grow on starch and glucose is a repressing carbon source, this analysis was performed using medium containing the nonrepressing carbon source xylose (34). These studies revealed that osuA was not induced in the osuR mutant, but induction with starch was restored in the complemented strain (Fig. 7B). In fact, there was a greater increase in expression of osuA in the complemented strain than in wild-type cells.
Induction of osu by oxidative stress.
The osuA gene was initially identified as an oxidative stress gene and Northern hybridization analysis revealed that the 7-kb osuA-containing transcript was highly induced upon exposure to atmospheric oxygen (Fig. 8A) (42). Similar levels of osu induction were observed for an oxyR deletion mutant, suggesting that this is an OxyR-independent response. The dominant mRNA observed was the 7-kb species, just as seen with starch induction (Fig. 4), and the larger species was not readily detected by Northern hybridization. Real-time RT-PCR analysis showed that there was nearly a 20-fold increase in osuA expression upon oxygen exposure, whereas osuD expression was only increased fivefold (Fig. 8B). The induction of osuD was somewhat less compared to that observed for starch induction (Fig. 6).
FIG. 8.
Oxygen-induced expression of the osu operon. A. Autoradiograph of Northern blot probed with internal osuA gene fragment. Total RNA was isolated from mid-log-phase cultures of strains 638R (wild type [wt]) and IB298 (oxyR) grown anaerobically in BHIS (−) or stressed by aerobic incubation for 1 h (+). All lanes contain 30 μg of total RNA. Approximate size of the hybridizing transcript, shown in kb, was extrapolated from a molecular mass ladder in an adjacent lane. B. Effect of oxygen stress on induction of osuA and osuD expression in the wild-type strain. Relative expression levels were determined by real-time RT-PCR for both anaerobic growth and oxygen exposure. Wild-type strain 638R was grown anaerobically to mid-log phase in BHIS and then exposed to oxygen for 1 h. Total RNA was isolated, and the expression of osuA and osuD was analyzed, with the results expressed as n-fold induction upon exposure to oxygen relative to anaerobic expression. Data are presented as the means from three independent experiments.
Another important observation was that the oxygen induction shown in Fig. 8 occurred in BHIS medium which contains 0.2% glucose which is normally a repressing condition for osu. Northern hybridization analysis showed that oxygen induction of osuA also occurred in SDM-glucose medium, which is a highly repressing growth medium under anaerobic conditions (data not shown).
Contribution of the osu operon to the oxidative stress response.
To test the hypothesis that expression of osu is important for survival during oxidative stress, sensitivity to oxygen was tested in the ΩosuA and osuR mutants (Fig. 9A). When compared to the parent strain, there was a significant effect of oxygen exposure on the viability of the ΩosuA mutant. Viability decreased during the first 24 h of aeration dropping more than two logs in 24 h. Interestingly, an osuR mutant showed no significant difference in viability over 72 h compared to the wild type. There was no difference in viable counts of anaerobic cultures for these strains (data not shown).
FIG. 9.
Effect of osu on survival during oxidative stress. A. Survival of wild-type (wt)(•), ΩosuA (▪) and osuR (▴) strains following exposure to atmospheric oxygen. Mid-log-phase cells (A550 of 0.35) grown in BHIS were exposed to oxygen by shaking at 250 rpm in air at 37°C. Viable cell counts were determined over 72 h. Inset, real-time RT-PCR analysis of osuA expression during oxygen exposure (ox) or under anaerobic conditions (an). The relative n-fold increase in expression is compared to anaerobic conditions, which were normalized to 1. B. Sensitivity of ΩosuA and osuR mutant strains to oxidizing agents. Disk diffusion assays were used to compare wild-type (638R), osuR (IB393), and ΩosuA (IB367) strains. Results are reported as the diameter of growth inhibition around filters treated with the following oxidizing agents: 3% menadione (MD), 3% hydrogen peroxide (H2O2), 0.5% t-butyl hydroperoxide (t-butyl). Values are the means from three independent experiments performed in triplicate.
The ΩosuA mutant also showed increased sensitivity to menadione exposure in growth inhibition assays but it was not markedly affected by exposure to the oxidizing agents H2O2 or t-butyl hydroperoxide (Fig. 9B). An osuR mutant showed no significant difference from the wild type in sensitivity to any of the oxidizing agents. The different survival phenotypes of ΩosuA compared to the osuR mutant suggested that oxygen stress controls osu expression by an OsuR independent mechanism. Consistent with this idea, Real-time RT-PCR analysis of oxygen induced osuA expression was similar for both the osuR mutant and wild-type cells (Fig. 9A, inset).
To determine if different promoters were being used for transcription during oxidative stress compared to anaerobic growth on starch, 5′ RACE was used to map the transcription start site of the osu operon. Under anaerobic conditions with starch induction, the osu 5′ end mapped 42 bp upstream from the predicted translational start site for OsuA in 70% of the sequenced clones (14/20) (Fig. 10). The remaining clones obtained from anaerobic conditions had 5′ ends at apparently random positions. Under conditions of oxygen stress, transcription was initiated 44 bp from the translational start site in 100% of sequenced clones (20/20). Inspection of the DNA sequence adjacent to the transcription start sites revealed two potential B. fragilis promoters similar to the consensus sequence (TAnnTTTG) in the −7 region (3). Also, about 69 bp upstream from the −33 region of the promoter, there was a conserved inverted repeat usually associated with binding of the LacI type regulator family proteins (Fig. 10). Further, within this inverted repeat was a 14/15 match to the consensus binding site for B. subtilis CcpA which is a LacI family regulatory protein capable of both repression and activation of transcription (22, 48).
FIG. 10.
Identification of the osuABCD operon transcription initiation site. By use of 5′ RACE analysis, the transcription start site was located 42 bp upstream from the translation start site of OsuA during anaerobic growth conditions with starch as the sole carbon source. During oxygen stress transcription was initiated 44 bp from the translational start site. Upstream from the start sites, a conserved inverted repeat putative binding site for the LacI type regulator family is shown in bold with arrows over the sequence. The consensus cre binding sequence for CcpA is shown below the conserved LacI binding site; W, A or T; N, any nucleotide; ST, starch. Lines below and above the sequence indicate the putative −7 and −33 promoter consensus sequences.
DISCUSSION
Evidence presented in this paper indicates that the four-gene osu operon encodes the primary starch utilization system in B. fragilis. Support for this is that the polar ΩosuA mutant completely inhibited the ability to grow on starch medium (Fig. 2). The first two gene products in the operon, OsuA and OsuB, were similar to SusC and SusD, respectively, which are required for starch utilization in B. thetaiotaomicron, where it has been shown that they interact to form a starch binding complex on the cell surface (7, 28, 29, 40). The C-terminal half of the OsuC gene product had some similarity to SusF. SusF together with SusE are two additional members of the Sus outer membrane complex, but they are not required for binding and SusF may even inhibit binding (7).
The last gene in the operon, osuD, encodes an enzyme with conserved domains for α-amylase and pullulanase, and it likely has an important although not exclusive role in starch hydrolysis. Consistent with this prediction, the ΩosuD mutant was impaired for growth on starch, and this growth inhibition was largely overcome by plasmid complementation (Fig. 2). The lack of full complementation may have resulted from inappropriate regulation and the uncoupling of osuD from its posttranscriptional control. In B. thetaiotaomicron, SusA is a neopullulanase responsible for the majority of starch hydrolysis and like ΩosuD, disruption of susA did not completely abolish growth on starch (9). A second B. thetaiotaomicron amylase, SusG, is located in the outer membrane and has been reported to be required for growth on starch. Interestingly, there is no homology between OsuD and either SusA or SusG, but based on the growth characteristics and enzyme assays with ΩosuD, there is at least one more amylolytic enzyme in B. fragilis that can partially compensate the loss of osuD. In fact, an examination of the B. fragilis genome reveals five additional ORFs with amylase motifs.
Starch and glycogen are cleaved by α-amylase to yield maltose which was shown to be a strong inducer of osu expression (Fig. 6). Starch itself and the maltooligosaccharides also induced the system, but it is likely that it was their digestion to maltose that resulted in induction. Free maltose would not likely be present in most environments where B. fragilis is found, so this would be an ideal inducer, as it would be one of the first signals indicating the presence of starch. Similarly, the B. thetaiotaomicron starch utilization system also is induced by maltose, but the regulatory proteins are different, as discussed below (6, 10). Induction of the osu operon resulted in the appearance of two mRNA species as shown by Northern analysis (Fig. 4). The small species was predominant and corresponds to the osuABC genes, whereas the larger species encompassed all four osu genes but was much less abundant. An explanation for this observation is that the intergenic region between osuC and osuD contains considerable secondary structure elements that could act as a rho-independent transcriptional terminator or other signal that might affect mRNA stability. A deletion construct that removed the stem-loop structure led to an increase in the level of the larger transcript and an increase in amylase activity (Fig. 5). This posttranscriptional regulation would allow higher levels of osuABC expression than amylase. This strategy would maximize the ability to bind and transport starch which in the gastrointestinal-tract environment would be the rate-limiting steps. Presumably, there would be sufficient amylase present in the periplasm to degrade all substrate that became available.
Induction of the osu operon was mediated at the level of transcription by OsuR, which appears to function as a transcriptional activator responsive to maltose and related maltooligosaccharides. An osuR mutant was unable to grow in medium with starch or maltose as the sole carbon source (Fig. 7A), although conversely, both ΩosuA and ΩosuD demonstrated wild-type growth in maltose medium (Fig. 2). This suggests that OsuR is involved in regulation of a maltose utilization operon as well as the osu operon and supports the hypothesis that maltose is the inducer of the osu operon. The inability of an osuR mutant to grow on starch was not entirely due to the inability to utilize maltose but also resulted from a lack of transcriptional activation of the osuABCD operon (Fig. 7B). In B. thetaiotaomicron, there are two transcriptional regulators, MalR and SusR, which control expression of the sus genes in response to the presence of starch or maltose in the medium (6, 10). Unlike the B. fragilis OsuR regulator, SusR is responsible for the majority of sus transcriptional activation but it has no effect on maltose utilization and susR mutants grow well on maltose. Mutations in the second regulator, MalR, also affect sus transcription and show some defects in maltose and maltotriose utilization but can grow well on starch medium. A double malR and susR mutant does not grow on starch, maltotriose, or maltose; hence, there are at least two regulators responsible for starch utilization in B. thetaiotaomicron. It is interesting that, like with SusR and MalR, overexpression of OsuR on a plasmid resulted in enhanced growth on starch and higher expression of the starch utilization genes (Fig. 7). This suggests that the concentration of these regulators is critical for normal control of their regulons.
A unique observation investigated in this report was that the osuABCD operon was oxygen responsive (Fig. 8) and that expression was induced approximately 20-fold upon exposure to air. The Bacteroides spp. are some of the more aerotolerant anaerobic bacteria, and this is due in part to oxygen-induced synthesis of new proteins that protect the cell against toxic oxygen radicals, but there is little information on how oxygen exposure alters their metabolism to deal with the consequences of oxygen exposure (30, 32). The oxygen-induced expression of osuABCD appeared to be independent of the presence of peroxide, and OxyR did not control the response. Further experiments showed that oxygen induction also was independent of the starch/maltose regulation mediated by OsuR. As shown in Fig. 9, there were nearly wild-type levels of oxygen-induced osuA transcription in the OsuR mutant. There must be another level of control during the oxidative stress response that controls this operon. Concurrent with these results, there was a 2-bp shift in the transcriptional start site when cells were moved from anaerobic growth on starch to oxygen stress. This shift may not be significant, but it could indicate a shift in the binding of RNA polymerase that resulted from a change in the regulator.
It was not immediately clear why starch utilization would be induced by oxygen, but additional studies suggested that it was important for survival. The ΩosuA mutant was impaired in its ability to survive oxygen exposure over 72 h (Fig. 9A), and it was more sensitive than the parent strain to the superoxide-generating agent menadione (Fig. 9B). This mutant did not show sensitivity to the oxidizing agents H2O2 or t-butyl hydroperoxide, which is consistent with the osuABCD operon being independent of OxyR control. Conversely, an osuR mutant had wild-type sensitivity in all of the oxygen stress assays we performed. This was expected, since OsuR was not involved in oxygen regulation of the operon. The mechanism that mediates this protection is not understood and will be the focus of future work.
Polysaccharide utilization is an important activity in the lower intestine and the ability of resident bacteria to utilize different polysaccharides provides a distinct competitive advantage and may explain why Bacteroides spp. are the numerically predominant genus in the intestine. Recently, Sonnenberg et al. (47), described the capacity of B. thetaiotaomicron to redirect its carbohydrate utilizing abilities from dietary to host polysaccharides according to nutrient availability. The Bacteroides species have also been shown to cleave l-fucose moieties from host cell surfaces and internalize them for use as an energy source (16), as well as scavenging a wide variety of other host cell glycans both in vitro and in vivo (38). In the pathogenic environment, outside of the anaerobic colon, B. fragilis is exposed to a vast array of host cell polysaccharides, as well as to oxidative stress. It has been proposed that the B. fragilis neuraminidase is required during the course of infection to provide carbon sources from host tissues (12). It is tempting to speculate that the Osu starch utilization system is important in a similar way during infection and that the ability to induce these genes in the presence of oxygen may provide an advantage that will help the organisms to survive.
Acknowledgments
This research was supported in part by Public Health Service grant AI40588 to C.J.S. from the National Institutes of Health.
We thank C. Sund for useful discussions and input on the real-time RT-PCR.
REFERENCES
- 1.Baughn, A. D., and M. H. Malamy. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 99:4662-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 3.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cho, K. H., D. Cho, G. R. Wang, and A. A. Salyers. 2001. New regulatory gene that contributes to control of Bacteroides thetaiotaomicron starch utilization genes. J. Bacteriol. 183:7198-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, K. H., and A. A. Salyers. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 183:7224-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotta, M. A., and T. R. Whitehead. 1993. Regulation and cloning of the gene encoding amylase activity of the ruminal bacterium Streptococcus bovis. Appl. Environ. Microbiol. 59:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Elia, J. N., and A. A. Salyers. 1996. Contribution of a neopullulanase, a pullulanase, and an α-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J. Bacteriol. 178:7173-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Elia, J. N., and A. A. Salyers. 1996. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 178:7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finegold, S. M., and W. L. George. 1998. Anaerobic infections in humans. Academic Press, New York, N.Y.
- 12.Godoy, V. G., M. M. Dallas, T. A. Russo, and M. H. Malamy. 1993. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect. Immun. 61:4415-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein, E. J. 1996. Anaerobic bacteremia. Clin. Infect. Dis. 23(Suppl. 1):S97-S101. [DOI] [PubMed] [Google Scholar]
- 14.Gregory, E. M. 1985. Characterization of the O2-induced manganese-containing superoxide dismutase from Bacteroides fragilis. Arch. Biochem. Biophys. 238:83-89. [DOI] [PubMed] [Google Scholar]
- 15.Herren, C. D., E. R. Rocha, and C. J. Smith. 2003. Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene 316:167-175. [DOI] [PubMed] [Google Scholar]
- 16.Hooper, L. V., J. Xu, P. G. Falk, T. Midtvedt, and J. I. Gordon. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 96:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hylemon, P. B., J. L. Young, R. F. Roadcap, and P. V. Phibbs. 1977. Uptake and incorporation of glucose and mannose by whole cells of Bacteroides thetaiotaomicron. Appl. Environ. Microbiol. 34:488-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingham, H. R., J. B. Selkon, and C. M. Roxby. 1978. The role of Bacteroides fragilis in abscesses of the central nervous system: implications for therapy. J. Antimicrob. Chemother. 4:283-284. [DOI] [PubMed] [Google Scholar]
- 19.Ingham, H. R., J. B. Slekon, and C. M. Roxby. 1977. Bacteriological study of otogenic cerebral abscesses: chemotherapeutic role of metronidazole. Br. Med. J. 2:991-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louie, T. J., J. G. Bartlett, F. P. Tally, and S. L. Gorbach. 1976. Aerobic and anaerobic bacteria in diabetic foot ulcers. Ann. Intern. Med. 85:461-463. [DOI] [PubMed] [Google Scholar]
- 21.Mazmanian, S. K., C. H. Liu, A. O. Tzianabos, and D. L. Kasper. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107-118. [DOI] [PubMed] [Google Scholar]
- 22.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onderdonk, A. B., D. L. Kasper, R. L. Cisneros, and J. G. Bartlett. 1977. The capsular polysaccharide of Bacteroides fragilis as a virulence factor: comparison of the pathogenic potential of encapsulated and unencapsulated strains. J. Infect. Dis. 136:82-89. [DOI] [PubMed] [Google Scholar]
- 24.Pan, N., and J. A. Imlay. 2001. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol. Microbiol. 39:1562-1571. [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Privitera, G., A. Dublanchet, and M. Sebald. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 139:97-101. [DOI] [PubMed] [Google Scholar]
- 27.Redondo, M. C., M. D. Arbo, J. Grindlinger, and D. R. Snydman. 1995. Attributable mortality of bacteremia associated with the Bacteroides fragilis group. Clin. Infect. Dis. 20:1492-1496. [DOI] [PubMed] [Google Scholar]
- 28.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves, A. R., G. R. Wang, and A. A. Salyers. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocha, E. R., C. D. Herren, D. J. Smalley, and C. J. Smith. 2003. The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe 9:165-173. [DOI] [PubMed] [Google Scholar]
- 31.Rocha, E. R., G. Owens, Jr., and C. J. Smith. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha, E. R., T. Selby, J. P. Coleman, and C. J. Smith. 1996. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J. Bacteriol. 178:6895-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha, E. R., and C. J. Smith. 1995. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J. Bacteriol. 177:3111-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocha, E. R., and C. J. Smith. 1997. Regulation of Bacteroides fragilis katB mRNA by oxidative stress and carbon limitation. J. Bacteriol. 179:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocha, E. R., and C. J. Smith. 1999. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 181:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocha, E. R., and C. J. Smith. 2004. Transcriptional regulation of the Bacteroides fragilis ferritin gene (ftnA) by redox stress. Microbiology 150:2125-2134. [DOI] [PubMed] [Google Scholar]
- 37.Salyers, A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293-313. [DOI] [PubMed] [Google Scholar]
- 38.Salyers, A. A., and M. Pajeau. 1989. Competitiveness of different polysaccharide utilization mutants of Bacteroides thetaiotaomicron in the intestinal tracts of germfree mice. Appl. Environ. Microbiol. 55:2572-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Shipman, J. A., J. E. Bargeman, and A. A. Salyers. 2000. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 182:5365-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoemaker, N. B., C. Getty, J. F. Gardner, and A. A. Salyers. 1986. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J. Bacteriol. 165:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smalley, D., E. R. Rocha, and C. J. Smith. 2002. Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J. Bacteriol. 184:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, C. J. 1985. Characterization of Bacteroides ovatus plasmid pBI136 and structure of its clindamycin resistance region. J. Bacteriol. 161:1069-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, C. J., E. R. Rocha, and B. J. Palter. 2004. The medically important Bacteroides spp. in health and disease. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. Springer Verlag, New York, N.Y.
- 45.Smith, C. J., M. B. Rogers, and M. L. McKee.1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141-154. [DOI] [PubMed] [Google Scholar]
- 46.Smith, C. J., L. A. Rollins, and A. C. Parker. 1995. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid 34:211-222. [DOI] [PubMed] [Google Scholar]
- 47.Sonnenburg, J. L., J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler, and J. I. Gordon. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955-1959. [DOI] [PubMed] [Google Scholar]
- 48.Stulke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]