Abstract
Escherichia coli DNA polymerase IV incorporated 2-hydroxy-dATP opposite template guanine or thymine and 8-hydroxy-dGTP exclusively opposite adenine in vitro. Mutator phenotypes in sod/fur strains were substantially diminished by deletion of dinB and/or umuDC. DNA polymerases IV and V may be involved in mutagenesis caused by incorporation of the oxidized deoxynucleoside triphosphates.
Excess oxidation is a major threat to the genomic integrity of most living organisms. Reactive oxygen species oxidize deoxynucleoside triphosphates (dNTPs), as well as DNA, and some of the oxidized dNTPs have been shown to be mutagenic when they are incorporated in DNA. 8-Oxo-7,8-dihydro-2′-deoxyguaniosine 5′-triphosphate (8-OH-dGTP) leads to A · T-to-C · G transversions when it is incorporated opposite adenine (A) in the template (5, 14). To counteract the mutagenic 8-OH-dGTP, Escherichia coli has a sanitizing enzyme, MutT, that hydrolyzes 8-OH-dGTP (20). When the mutT gene is inactivated, the frequency of mutation of A · T to C · G increases more than a thousandfold compared with the wild-type frequency (35). In the case of 2-oxo-1,2-dihydro-2′-deoxyadenosine 5′-triphosphate (2-OH-dATP), G · C-to-T · A transversions occur when it is incorporated opposite guanine (G) in the template (14, 16). Another sanitizing enzyme, Orf135, degrades 2-OH-dATP in E. coli, and G · C-to-T · A mutations occur in an orf135-deficient strain more frequently than in the wild-type strain (15, 17).
The members of the Y family of DNA polymerases (DNA Pols) are involved in error-free and error-prone translesion synthesis (TLS) of damaged template DNA in various species, including humans (13, 26). Recently, involvement of Y-family DNA polymerases in the incorporation of damaged dNTPs was suggested by in vitro experiments performed with purified DNA Pols (28). The archaeal Y-family DNA Pols from Sulfolobus sp. and the human DNA Pols exclusively incorporate 8-OH-dGTP opposite A in the template DNA and incorporate 2-OH-dATP opposite G and thymine (T). Thus, it would be interesting to examine the in vivo roles of Y-family DNA Pols in the incorporation of mutagenic dNTPs into DNA. Escherichia coli strain QC1736 seems to be an appropriate background to investigate the roles of Y-family DNA Pols (DNA Pol IV and Pol V encoded by dinB and umuDC, respectively) in the mutagenesis caused by oxidized nucleotides. Iron metabolism is deregulated in this strain due to the lack of the Fur protein, a negative regulator of iron uptake (29). This strain also lacks both superoxide dismutases (SodA and SodB), which catalyze the breakdown of the superoxide anion. Thus, both iron overload and superoxide stress occur in strain QC1736, which leads to high rates of spontaneous mutation from A · T to C · G and from G · C to T · A (24). The hot spots and sequence contexts of A · T-to-C · G mutations are almost identical to those in a mutT strain (25). In contrast, the hotspots of G · C-to-T · A mutations are very different from those in mutM mutY strains, in which 8-OH-G in DNA acts as a major mutagenic lesion. Expression of the cDNA of the human counterpart of E. coli MutT, MTH1, which hydrolyzes both 8-OH-dGTP and 2-OH-dATP (12), suppresses the mutator phenotype of the strain. Thus, it has been concluded that the targets contributing to the oxidative mutagenesis in the sod/fur mutant are oxidized dNTPs, such as 8-OH-dGTP and 2-OH-dATP, rather than DNA (25).
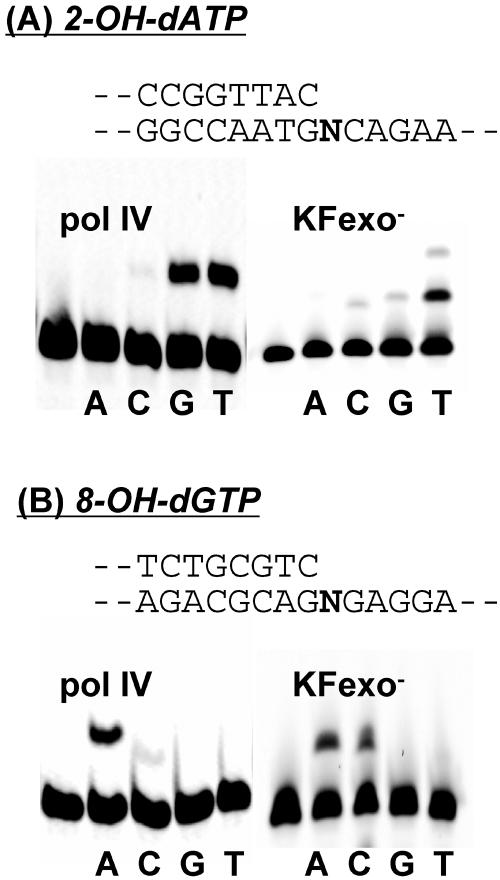
We first examined the specificity with which the purified native form of DNA Pol IV (31) incorporates 2-OH-dATP and 8-OH-dGTP in vitro. DNA Pol IV predominantly incorporated 2-OH-dATP opposite template G and T, and the frequency of incorporation opposite G was almost equal the frequency of incorporation opposite T (Fig. 1 A). In contrast, Klenow fragment exo− (KF exo−) (New England Biolabs, Massachusetts) predominantly incorporated 2-OH-dATP opposite T. DNA Pol IV almost exclusively incorporated 8-OH-dGTP opposite A, and KF exo− incorporated 8-OH-dGTP opposite A and cytosine (C) (Fig. 1 B). These in vitro results suggest possible involvement of Y-family DNA polymerases in oxidative mutagenesis through misincorporation of the oxidized dNTPs during DNA synthesis in E. coli.
FIG. 1.
Incorporation of oxidized nucleotides by DNA polymerases. The incorporation of 2-OH-dATP (A) and 8-OH-dGTP (B) into DNA by DNA Pol IV and KF exo− of E. coli was assayed as described previously (28). Cy3-conjugated primer, annealed to the template at a 1:1 ratio (0.1 μM), was incubated with DNA Pol IV (0.1 μM) or KF exo− (0.02 U), and then 50 μM 2-OH-dATP (A) or 50 μM 8-OH-dGTP (B) was added. No other dNTPs were added to the reaction mixtures. All the reactions were carried out at room temperature for 30 min. The reaction products were analyzed on 15% denatured polyacrylamide gels, and the bands were visualized using a Molecular Imager FX Pro system (Bio-Rad, Richmond, CA). The oligonucleotide sequences of the primer and template were 5′-Cy3-CGCGCGAAGACCGGTTAC-3′ and 5′-GAAGGGATCCTTAAGACNGTAACCGGTCTTCGCGCG-3′, respectively, for 2-OH-dATP and 5′-Cy3-CGGAGCTCGGTCGGCGTCTGCGTC and 5′-AGCCGCAGGAGNGACGCAGACGCCGACCGAGCTCCG-3′, respectively, for 8-OH-dGTP (N = A, C, G, or T). Parts of the sequences of the primer and template are shown. The unlabeled lanes on the left indicate the positions of Cy3-labeled primers without extension.
To examine the in vivo roles of Y-family DNA Pols, mutation frequencies were compared for sod/fur strains with and without Pol IV and Pol V (Table 1). Both A · T-to-C · G and G · C-to-T · A transversion frequencies were reduced by 80 to 90% by deletion of either dinB or umuDC or both in the sod/fur strains (Table 2). Interestingly, the double mutants (ΔdinB ΔumuDC) exhibited levels of mutation frequency similar to those of single mutants (ΔdinB or ΔumuDC). These results suggest that the base substitutions by erroneous incorporation of 2-OH-dATP or 8-OH-dGTP require both DNA Pol IV and DNA Pol V functions.
TABLE 1.
Strains used in this studya
| Strain | Characteristics | P1 transduction or conjugation | Reference or source |
|---|---|---|---|
| CC101 | Derivative of strain P90C [araA(lac proB)xIII] carrying F′ lacIZ- proB+; lacZ has a mutation (GAG to TAG) at codon 461 | 7 | |
| CC104 | Derivative of strain P90C [araA(lac proB)xIII] carrying F′ lacIZ- proB+; lacZ has a mutation (GAG to GCG) at codon 461 | 7 | |
| AR30 | ΔdinB61::ble sulA211 | 4 | |
| DE2302 | thr-1 ara-14 leuB6 Δ(gpt proA)62 lacY1 tsx-33 supE44 galK2 hisG4 rpsL31 xyl-5 mtl-1 arg3 thi-1 uvrA6 Δ(umuDC)595::cat fadR615::Tn10 purB58 | 34 | |
| EC8 | thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4 rpsL31 xyl-5 mtl-1 argE3 thi-1 uvrA6 Δ(umuDC)596::ermGT fadR+ purB+ | 11 | |
| KY1056sFtet101 | AB1157 derivative; harboring F′ derived from CC101, which has Tn10 in it for selection of F′ | K. Yamamoto | |
| KY1056sFtet104 | AB1157 derivative; harboring F′ derived from CC104, which has Tn10 in it for selection of F′ | K. Yamamoto | |
| YG6125A | AB1157 derivative; harboring F′ derived from CC101, which has Tn10 in it for selection of F′ and ΔdinB::kan | This study | |
| YG6125B | AB1157 derivative; harboring F′ derived from CC104, which has Tn10 in it for the selection of F′ and ΔdinB::kan | This study | |
| QC1736 | Δ(argF-lac)U169 rpsL ΔsodA3 sodB::MudPR fur::kan; Cmr Kmr | 29 | |
| YG6177 | Like QC1736 but ΔdinB61::ble; Cmr Kmr Zcr | AR30 (P1) → QC1736 | This study |
| YG6180 | Like QC1736 but ΔumuDC(596)::ermGT; Cmr Kmr | DE2302/EC8 (P1) → QC1736 | This study |
| YG6124 | Like QC1736 ΔdinB61::ble; ΔumuDC(596)::ermGT; Cmr Kmr Zcr | DE2302/EC8 (P1) → YG6177 | This study |
| YG6175b | Like QC1736 but harboring F′ from CC101; Cmr Kmr Tcr | KY1056sFtet101 → QC1736 | This study |
| YG6176b | Like QC1736 but harboring F′ from CC104; Cmr Kmr Tcr | KY1056sFtet104 → QC1736 | This study |
| YG6178b | Like QC1736 but ΔdinB61::ble and harboring F′ from CC101; Cmr Kmr Tcr Zcr | YG6125A → YG6177 | This study |
| YG6179b | Like QC1736 but ΔdinB61::ble and harboring F′ from CC104; Cmr Kmr Tcr Zcr | YG6125B → YG6177 | This study |
| YG6181b | Like QC1736 but ΔumuDC(596)::ermGT and harboring F′ derived from CC101; Cmr Kmr Tcr | KY1056sFtet101 → YG6180 | This study |
| YG6182b | Like QC1736 but ΔumuDC(596)::ermGT and harboring F′ from CC104; Cmr Kmr Tcr | KY1056sFtet104 → YG6180 | This study |
| YG6126b | Like QC1736 but ΔdinB61::ble and ΔumuDC(596)::ermGT and harboring F′ from CC101; Cmr Kmr Tcr Zcr | YG6125A → YG6124 | This study |
| YG6127b | Like QC1736 but ΔdinB61::ble and ΔumuDC(596)::ermGT and harboring F′ from CC104; Cmr Kmr Tcr Zcr | YG6125B → YG6124 | This study |
The deletion strains for dinB encoding DNA Pol IV were constructed by P1 transduction as indicated. The umuDC deletion encoding DNA Pol V was introduced into QC1736 and YG6177 by two-step P1 transduction (11). (P1) indicates that P1vir phage lysate was prepared in the strain. F′ with a mutation for specific detection of changes from G · C to T · A or from A · T to C · G was separately introduced by conjugation as indicated. The arrows indicate the directions of transfer for P1 transduction and conjugation. Chloramphenicol, kanamycin, tetracycline, and zeocin were used at concentrations of 10 μg/ml, 25 μg/ml, 10 μg/ml, and 50 μg/ml, respectively. Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Zcr, zeocin resistance.
Strain used for the LacZ reversion assay.
TABLE 2.
Mutation frequencies for the sodAB fur strains with and without DNA Pol IV and DNA Pol Va
| Expt | Mutation frequency (10−6)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Pol IV+/Pol V+ (YG6176) | Pol IV−/Pol V+ (YG6179) | Pol IV+/Pol V− (YG6182) | Pol IV−/Pol V− (YG6127) | Pol IV+/Pol V+ (YG6175) | Pol IV−/Pol V+ (YG6178) | Pol IV+/Pol V− (YG6181) | Pol IV−/Pol V− (YG6126) | |
| G · C → T · A | ||||||||
| Expt I | 28.2 ± 3.5 | 3.66 ± 0.32 | 2.72 ± 0.20 | 3.57 ± 0.42 | ||||
| Expt II | 35.6 ± 2.5 | 2.61 ± 0.12 | 2.82 ± 0.15 | 3.23 ± 0.30 | ||||
| A · T → C · G | ||||||||
| Expt I | 12.8 ± 0.61 | 2.49 ± 0.18 | 3.24 ± 0.25 | 2.43 ± 0.15 | ||||
| Expt II | 10.0 ± 0.60 | 2.08 ± 0.16 | 3.7 ± 0.21 | 1.86 ± 0.19 | ||||
| Expt III | 9.1 ± 0.80 | 2.38 ± 0.47 | ||||||
The mutagenicity assay was carried out as described previously (32). Briefly, a single colony was inoculated into 2 ml of M9-glucose minimal medium, and then the overnight culture was diluted 1,000-fold. Eight to twelve diluted cultures were prepared, and they were cultivated overnight. One milliliter of each culture was harvested and washed twice with phosphate buffer (pH 7.4), and then the cell pellet was suspended in phosphate buffer. All of the suspension was spread on one plate for mutation, and a portion of the diluted suspension was used for determining survival. Twenty amino acids were added in the assays (both liquid medium and plates) for growth of sodAB fur strains because the production of amino acids is inhibited by oxygen radicals (3). The values are means ± standard errors.
The dNTP pool and DNA are continuously exposed to a variety of exogenous and endogenous damaging agents, including reactive oxygen species, and the incorporation of oxidized dNTPs into DNA is a major source of spontaneous mutagenesis and carcinogenesis (1). Here we obtained biochemical and genetic evidence that DNA Pol IV and Pol V may be involved in oxidative mutagenesis through misincorporation of altered nucleotides (i.e., 2-OH-dATP and 8-OH-dGTP) during DNA synthesis. This is consistent with the report by Satou et al. (27) that DNA Pol IV promotes mutation of G · C to T · A in E. coli when 2-OH-dATP is directly introduced into cells by CaCl2 treatment. It has also been suggested that SOS-inducible polymerases, including Pol IV and Pol V, are involved in mutagenesis caused by increases in the normal levels of dNTPs (33). It has been reported that more than one DNA polymerase is involved in mutagenesis when the Y-family DNA polymerases are involved in TLS. For benzo[a]pyrene-induced mutagenesis, both Pol IV and Pol V are required for a −1 frameshift TLS (23). DNA lesions induced by other chemicals, including 3-methylcholanthrene or dimethylbenzo[a]anthracene, also require both DNA Pol IV and Pol V for a −2 frameshift in a CG repetitive sequence in Salmonella enterica serovar Typhimurium (18, 21). Thus, we speculate that DNA Pol IV and Pol V are involved in sequential biochemical steps, such as incorporation and extension of oxidized dNTPs during chromosome replication. One of these polymerases might incorporate oxidized dNTPs into DNA in an erroneous manner, and the other might extend the mutagenic primer termini containing the oxidized deoxynucleoside monophosphate, thereby inducing base substitutions. It is obvious, however, that more experiments are needed to elucidate the precise mechanisms.
DNA Pol IV is controlled by σs, and the level of expression of Pol IV in the stationary phase decreases significantly when the rpoS gene encoding σs is defective (10, 19). Thus, Pol IV appears to be regulated not only by the SOS response but also by the σs-dependent stress response. In stationary-phase cells, the amount of cellular mismatch repair proteins decreases at least 10-fold (8). Hence, the error-prone nature of Pol IV is expected to be more significant. In fact, DNA Pol IV is responsible for some of the adaptive mutations in stationary-phase cells (9, 22), Interestingly, adaptive mutagenesis is approximately fourfold more frequent in a sodA sodB strain than in the parental strain, and this mutagenesis is suppressed under anaerobic conditions (2). Therefore, DNA Pol IV might be involved in stationary-phase mutagenesis by either incorporation of oxidized dNTPs or extension of primers having oxidized deoxynucleoside monophosphates or both, although it is possible that DNA Pol IV induces mutations by error-prone bypass across oxidized bases in template DNA.
The oxidized nucleotide pools also cause a problem in mammalian cells. Spontaneous tumorigenesis in lungs, livers, and stomachs is enhanced in mice that are deficient in Mth1 (30). In addition, a recent study suggested that the majority of mutations in human cells that are deficient in mismatch repair do not arise from spontaneous replication errors but from the incorporation of oxidized dNTPs (6). Thus, it might be interesting to examine the roles of mammalian Y-family DNA Pols in genome instability caused by oxidation of the nucleotide pool.
Acknowledgments
We thank Roger Woodgate and Kazuo Yamamoto for providing E. coli strains.
Part of this study was financially supported by the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology Japan. This work was also supported by grant-in-aid for international collaborative research SH34407from the Japan Health Science Foundation.
REFERENCES
- 1.Ames, B. N., and L. S. Gold. 1991. Endogenous mutagens and the causes of aging and cancer. Mutat. Res. 250:3-16. [DOI] [PubMed] [Google Scholar]
- 2.Benov, L., and I. Fridovich. 1996. The rate of adaptive mutagenesis in Escherichia coli is enhanced by oxygen (superoxide). Mutat. Res. 357:231-236. [DOI] [PubMed] [Google Scholar]
- 3.Benov, L., and I. Fridovich. 1999. Why superoxide imposes an aromatic amino acid auxotrophy on Escherichia coli. The transketolase connection. J. Biol. Chem. 274:4202-4206. [DOI] [PubMed] [Google Scholar]
- 4.Borden, A., P. I. O'Grady, D. Vandewiele, A. R. Fernandez de Henestrosa, C. W. Lawrence, and R. Woodgate. 2002. Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn cyclobutane dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J. Bacteriol. 184:2674-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, K. C., D. S. Cahill, H. Kasai, S. Nishimura, and L. A. Loeb. 1992. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J. Biol. Chem. 267:166-172. [PubMed] [Google Scholar]
- 6.Colussi, C., E. Parlanti, P. Degan, G. Aquilina, D. Barnes, P. Macpherson, P. Karran, M. Crescenzi, E. Dogliotti, and M. Bignami. 2002. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol. 12:912-918. [DOI] [PubMed] [Google Scholar]
- 7.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, G., H. C. Tsui, and M. E. Winkler. 1996. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J. Bacteriol. 178:2388-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, P. L. 2000. Adaptive mutation in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 65:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, P. L. 2005. Stress responses and genetic variation in bacteria. Mutat. Res. 569:3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, E. G., M. Gonzalez, D. G. Ennis, A. S. Levine, and R. Woodgate. 1996. In vivo stability of the Umu mutagenesis proteins: a major role for RecA. J. Bacteriol. 178:3550-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujikawa, K., H. Kamiya, H. Yakushiji, Y. Fujii, Y. Nakabeppu, and H. Kasai. 1999. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 274:18201-18205. [DOI] [PubMed] [Google Scholar]
- 13.Goodman, M. F. 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71:17-50. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, M., H. Kamiya, K. Fujikawa, Y. Ootsuyama, N. Murata-Kamiya, T. Osaki, K. Yasumoto, and H. Kasai. 1998. Induction of chromosomal gene mutations in Escherichia coli by direct incorporation of oxidatively damaged nucleotides. New evaluation method for mutagenesis by damaged DNA precursors in vivo. J. Biol. Chem. 273:11069-11074. [DOI] [PubMed] [Google Scholar]
- 15.Kamiya, H., E. Iida, N. Murata-Kamiya, Y. Yamamoto, T. Miki, and H. Harashima. 2003. Suppression of spontaneous and hydrogen peroxide-induced mutations by a MutT-type nucleotide pool sanitization enzyme, the Escherichia coli Orf135 protein. Genes Cells 8:941-950. [DOI] [PubMed] [Google Scholar]
- 16.Kamiya, H., and H. Kasai. 2000. 2-Hydroxy-dATP is incorporated opposite G by Escherichia coli DNA polymerase III resulting in high mutagenicity. Nucleic Acids Res. 28:1640-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamiya, H., N. Murata-Kamiya, E. Iida, and H. Harashima. 2001. Hydrolysis of oxidized nucleotides by the Escherichia coli Orf135 protein. Biochem. Biophys. Res. Commun. 288:499-502. [DOI] [PubMed] [Google Scholar]
- 18.Kokubo, K., M. Yamada, Y. Kanke, and T. Nohmi. 2005. Roles of replicative and specialized DNA polymerases in frameshift mutagenesis: mutability of Salmonella typhimurium strains lacking one or all of SOS-inducible DNA polymerases to 26 chemicals. DNA Repair 4:1160-1171. [DOI] [PubMed] [Google Scholar]
- 19.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355:273-275. [DOI] [PubMed] [Google Scholar]
- 21.Matsui, K., M. Yamada, M. Imai, K. Yamamoto, and T. Nohmi. 2006. Specificity of replicative and SOS-inducible DNA polymerases in frameshift mutagenesis: mutability of Salmonella typhimurium strains overexpressing SOS-inducible DNA polymerases to 30 chemical mutagens. DNA Repair 5:465-478. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie, G. J., P. L. Lee, M. J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell. 7:571-579. [DOI] [PubMed] [Google Scholar]
- 23.Napolitano, R., R. Janel-Bintz, J. Wagner, and R. P. Fuchs. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19:6259-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunoshiba, T., F. Obata, A. C. Boss, S. Oikawa, T. Mori, S. Kawanishi, and K. Yamamoto. 1999. Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions, and mutagenesis in Escherichia coli. J. Biol. Chem. 274:34832-34837. [DOI] [PubMed] [Google Scholar]
- 25.Nunoshiba, T., T. Watanabe, Y. Nakabeppu, and K. Yamamoto. 2002. Mutagenic target for hydroxyl radicals generated in Escherichia coli mutant deficient in Mn- and Fe-superoxide dismutases and Fur, a repressor for iron-uptake systems. DNA Repair 1:411-418. [DOI] [PubMed] [Google Scholar]
- 26.Ohmori, H., E. C. Friedberg, R. P. Fuchs, M. F. Goodman, F. Hanaoka, D. Hinkle, T. A. Kunkel, C. W. Lawrence, Z. Livneh, T. Nohmi, L. Prakash, S. Prakash, T. Todo, G. C. Walker, Z. Wang, and R. Woodgate. 2001. The Y-family of DNA polymerases. Mol. Cell 8:7-8. [DOI] [PubMed] [Google Scholar]
- 27.Satou, K., M. Yamada, T. Nohmi, H. Harashima, and H. Kamiya. 2005. Mutagenesis induced by oxidized DNA precursors: roles of Y family DNA polymerases in Escherichia coli. Chem. Res. Toxicol. 18:1271-1278. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu, M., P. Gruz, H. Kamiya, S. R. Kim, F. M. Pisani, C. Masutani, Y. Kanke, H. Harashima, F. Hanaoka, and T. Nohmi. 2003. Erroneous incorporation of oxidized DNA precursors by Y-family DNA polymerases. EMBO Rep. 4:269-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuzuki, T., A. Egashira, H. Igarashi, T. Iwakuma, Y. Nakatsuru, Y. Tominaga, H. Kawate, K. Nakao, K. Nakamura, F. Ide, S. Kura, Y. Nakabeppu, M. Katsuki, T. Ishikawa, and M. Sekiguchi. 2001. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc. Natl. Acad. Sci. USA 98:11456-11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, J., S. Fujii, P. Gruz, T. Nohmi, and R. P. Fuchs. 2000. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe, M., T. Nohmi, and T. Ohta. 1994. Effects of the umuDC, mucAB, and samAB operons on the mutational specificity of chemical mutagenesis in Escherichia coli. I. Frameshift mutagenesis. Mutat. Res. 314:27-37. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler, L. J., I. Rajagopal, and C. K. Mathews. 2005. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair 4:1450-1456. [DOI] [PubMed] [Google Scholar]
- 34.Woodgate, R. 1992. Construction of a umuDC operon substitution mutation in Escherichia coli. Mutat. Res. 281:221-225. [DOI] [PubMed] [Google Scholar]
- 35.Yanofsky, C., E. C. Cox, and V. Horn. 1966. The unusual mutagenic specificity of an E. coli mutator gene. Proc. Natl. Acad. Sci. USA 55:274-281. [DOI] [PMC free article] [PubMed] [Google Scholar]