Abstract
The vitamin B6 biosynthetic pathway in Sinorhizobium meliloti is similar to that in Escherichia coli K-12; in both organisms this pathway includes condensation of two intermediates, 1-deoxy-d-xylulose 5-phosphate and 4-phosphohydroxy-l-threonine (4PHT). Here, we report cloning of a gene designated pdxR that functionally corresponds to the pdxB gene of E. coli and encodes a dye-linked flavin adenine dinucleotide-dependent 4-phospho-d-erythronate (4PE) dehydrogenase. This enzyme catalyzes the oxidation of 4PE to 3-hydroxy-4-phosphohydroxy-α-ketobutyrate and is clearly different in terms of cofactor requirements from the pdxB gene product of E. coli, which is known to be an NAD-dependent enzyme. Previously, we revealed that in S. meliloti IFO 14782, 4PHT is synthesized from 4-hydroxy-l-threonine and that this synthesis starts with glycolaldehyde and glycine. However, in this study, we identified a second 4PHT pathway in S. meliloti that originates exclusively from glycolaldehyde (the major pathway). Based on the involvement of 4PE in the 4PHT pathway, the incorporation of different samples of 13C-labeled glycolaldehyde into pyridoxine molecules was examined using 13C nuclear magnetic resonance spectroscopy. On the basis of the spectral analyses, the synthesis of 4PHT from glycolaldehyde was hypothesized to involve the following steps: glycolaldehyde is sequentially metabolized to d-erythrulose, d-erythrulose 4-phosphate, and d-erythrose 4-phosphate by transketolase, kinase, and isomerase, respectively; and d-erythrose 4-phosphate is then converted to 4PHT by the conventional three-step pathway elucidated in E. coli, although the mechanism of action of the enzymes catalyzing the first two steps is different.
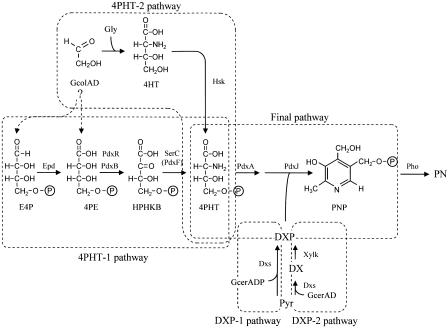
Two divergent pathways for vitamin B6 biosynthesis are known to exist in nature (11). One pathway exists in prokaryotes such as Escherichia coli, Salmonella, and many other gram-negative bacteria that have pdxA/pdxJ gene homologs; this pathway has been well studied in E. coli (19) (Fig. 1). The other pathway exists in eukaryotes, some prokaryotes, and plants, although it has not yet been elucidated (2, 12, 30). In the former group, the pdxA and pdxJ genes are involved in the final pathway that starts with two building blocks, 4-phosphohydroxy-l-threonine (4PHT) and 1-deoxy-d-xylulose 5-phosphate (DXP), and results in the formation of pyridoxine 5′-phosphate (PNP) (4, 21, 23). 4PHT is synthesized by the 4PHT-1 pathway, which starts with d-erythrose 4-phosphate (E4P) and forms 4-phospho-d-erythronate (4PE), 3-hydroxy 4-phosphohydroxy-α-ketobutyrate (HPHKB), and 4PHT in three reactions that are catalyzed by E4P dehydrogenase (Epd), 4PE dehydrogenase (PdxB), and 3-phosphoserine (or 4PHT) transaminase (SerC or PdxF), respectively (7, 9, 25, 33, 47, 49). DXP is synthesized from pyruvate and d-glyceraldehyde 3-phosphate by DXP synthase (Dxs; DXP-1 pathway) (32, 43).
FIG. 1.
Proposed vitamin B6 pathway in E. coli and S. meliloti. Abbreviations: GcolAD, glycolaldehyde; Gly, glycine; Pyr, pyruvate; GcerADP, d-glyceraldehyde 3-phosphate; GcerAD, d-glyceraldehyde. The 4PHT-1, 4PHT-2, DXP-1, DXP-2, and final pathways are pathways that result in the synthesis of 4PHT from E4P (E. coli), the synthesis of 4PHT from glycolaldehyde and glycine via 4HT (S. meliloti), the synthesis of DXP from pyruvate and d-glyceraldehyde 3-phosphate (E. coli), the synthesis of DXP from pyruvate and d-glyceraldehyde via DX (S. meliloti), and the synthesis of PNP from 4PHT and DXP, respectively; these pathways are indicated by the areas enclosed by the dashed lines.
Sinorhizobium meliloti, formerly known as Rhizobium meliloti (6), belongs to the group of organisms with pdxA/pdxJ gene homologs (15), and the vitamin B6 biosynthetic pathway has been elucidated in this organism by incorporation experiments using 13C- or 15N-labeled precursors of pyridoxine (PN) or its intermediates (38). In this study, we hypothesized that PN is synthesized from two precursors, 4-hydroxy-l-threonine (4HT), which is synthesized from glycolaldehyde/glycine, and 1-deoxy-d-xylulose (DX), which is synthesized from pyruvate/d-glyceraldehyde. We purified the enzymes involved in the synthesis of PN from DX and 4HT (39, 40). As a result, five enzymes were purified. Four of these enzymes were predicted to be xylulose kinase, homoserine kinase, 4PHT dehydrogenase (PdxA), and PNP synthase (PdxJ) based on a protein database search with the peptide sequences of the purified enzymes (5). The fifth enzyme was identified as a novel PNP phosphatase (41). It was observed that these enzymes participated in the following successive reactions (40). DX and 4HT were converted to DXP and 4PHT by xylulose kinase (46) and homoserine kinase (14, 35), respectively. 4PHT was further oxidized by PdxA. Subsequently, the product of this oxidation was combined with DXP by PdxJ to form PNP in a reaction sequence similar to that observed in E. coli. The pathway that starts with pyruvate/d-glyceraldehyde and results in the formation of DXP via DX is designated the DXP-2 pathway, while the pathway that starts with glycolaldehyde/glycine and results in the formation of 4PHT via 4HT is designated the 4PHT-2 pathway. The pathway in which DXP and 4PHT are condensed to form PNP is called the final pathway (Fig. 1).
In this paper, we describe genetic and biochemical analyses of the pdxR gene that complements the vitamin B6 auxotrophy of a vitamin B6-deficient mutant that has a mutation in the 4PHT pathway. Elucidation of the function of the pdxR gene product revealed new aspects of the 4PHT pathway in S. meliloti, including the synthesis of PN from two different substrate systems, DX/glycolaldehyde (the major 4PHT pathway) and DX/glycolaldehyde/glycine (the minor 4PHT pathway), during intact cell reactions with S. meliloti IFO 14782. In addition, on the basis of 13C nuclear magnetic resonance (NMR) spectral analyses of the PN isolated from DX and different 13C-labeled samples of glycolaldehyde, we identified a pathway for the synthesis of 4PHT originating exclusively from glycolaldehyde (the major 4PHT pathway).
MATERIALS AND METHODS
13C, 1H, and 31P NMR and fast atom bombardment mass spectrometry experiments.
13C, 1H, and 31P NMR experiments were performed with a Jeol JNM GSX-400 spectrometer (Jeol Ltd., Tokyo, Japan), and all compounds were dissolved in heavy water (D2O). Fast atom bombardment (FAB)-mass spectrometry (MS) experiments were performed with a Jeol SX-102/102 mass spectrometer equipped with a Hewlett-Packard Apollo series 400 data system (Jeol Ltd., Tokyo, Japan). m-Nitrobenzyl alcohol was used as the liquid matrix for the FAB-MS experiments.
Materials.
Cytochrome c from equine heart, glutamate dehydrogenase (type III) from bovine liver, E4P, and phosphohydroxypyruvate dimethyl ketal, which was hydrolyzed according to the manufacturer's recommendations, were purchased from Sigma-Aldrich, St. Louis, MO. 2,6-Dichlorophenolindophenol (DCIP), ferricyanide, and 2-hydroxy acid compounds were purchased from Wako Pure Chemical, Osaka, Japan. [1,2-13C]glycolaldehyde (99 atom% 13C) and [1-13C]glycolaldehyde (99 atom% 13C) were purchased from Cambridge Isotope Laboratories, Andover, MA. 4PE was synthesized from E4P using a procedure described by Woodruff and Wolfenden (45), and it was purified using a Sephadex LH 20 column (ratio of methanol to H2O, 1:1; Amersham Biosciences, Uppsala, Sweden). The product had an Rf of 0.26 on DEAE-cellulose thin-layer chromatography plates (n-butanol/acetic acid/H2O ratio, 50:15:35; Tokyo Kasei Kogyo, Japan), which were sprayed with Hanes-Isherwood reagent (17) and then heated for 1 min at 85°C and exposed to UV radiation for 10 min (1) (cf. E4P, whose Rf was 0.05). The product was also analyzed by MS. The FAB-MS negative ion mode of the product was 215 (M − H)−. d-Erythronate was prepared from d-erythrono-γ-lactone (Fluka, Buchs, Switzerland) by the method described by Hill et al. (18). 13C NMR (100 MHz, D2O) δ values: 61.9 (C-4), 73.1, 74.0 (C-2 and C-3), and 178.5 (C-1). 4HT and DX were prepared by the methods described previously (28, 48). Chromatographic media and prepacked columns for purification of the PdxR and SerC proteins were purchased from Amersham Biosciences, Uppsala, Sweden. QIAEXII that was used to recover DNA from agarose gels was purchased from QIAGEN, GmbH, Germany.
Microorganisms and plasmids.
S. meliloti IFO 14782 was purchased from the Institute for Fermentation, Osaka, Japan. Saccharomyces carlsbergensis ATCC 9080 was used to estimate the amounts of vitamin B6 in the agar and intact cell reaction mixtures (42). E. coli WG1012 [lacY or lacZ62 tsx-65 glnV44(AS) gal-6 λ− hisG1(Fs) pdxB5 rpsL104 or rps157(Strr)] and E. coli ED8767 (29) were purchased from the E. coli Genetic Stock Center, Yale University. E. coli HB101 and E. coli JM109 were obtained from Takara Bio Inc., Shiga, Japan. Plasmids pVK100 (22) and pRK2013 were purchased from ATCC, while plasmid pKK223-3 was obtained from Amersham Biosciences Corp., New Jersey. Plasmid pSUP2021 (36) was a gift from A. Pühler. Plasmid pCR-TOPO was obtained from Invitrogen Japan K.K.
Media and growth conditions.
Luria-Bertani (LB) broth and LB medium supplemented with 0.061% MgSO4 · 7H2O and 0.036% CaCl2 · 2H2O (LBMC) were used as the growth media for E. coli and S. meliloti, respectively. Vitamin B6-free Edinburgh minimal medium (EMM) contained 1% glucose, 0.8% vitamin-free Casamino Acids, inorganic salts, thiamine, and biotin. Vitamin B6 assay agar comprised 5% glucose, 14 amino acids instead of Casamino Acids, inorganic salts, thiamine, biotin, and 1.5% agar. A productivity test medium, glucose peptone yeast extract medium, containing 4% glucose, 4% peptone, 0.8% yeast extract, 0.05% MgSO4 · 7H2O, 0.05% MnSO4 · 5H2O, and 0.001% FeSO4 · 7H2O, was used for cell harvesting. The cultivation temperature for E. coli was 37°C, while that for S. meliloti and S. carlsbergensis ATCC 9080 was 30°C. Tetracycline (10 μg/ml), kanamycin (50 μg/ml), nalidixic acid (20 μg/ml), neomycin (50 μg/ml), and ampicillin (100 μg/ml) were added to the media as indicated below.
Construction of a genomic library of S. meliloti IFO 14782 in E. coli.
Chromosomal DNA from S. meliloti IFO 14782 was prepared using QIAGEN Genomic-tips (QIAGEN GmbH, Germany). The partially digested DNA was subjected to agarose gel electrophoresis on 0.6% (wt/vol) gels, and 15- to 35-kb DNA fragments were recovered from the gel by electroelution.
Simultaneously, plasmid pVK100 was completely digested using EcoRI and dephosphorylated with alkaline phosphatase. The treated pVK100 was ligated with the chromosomal DNA fragments of S. meliloti IFO 14782 mentioned above using a ligation kit (Takara Bio Inc., Shiga, Japan). The DNA was packaged into a phage. Subsequently, E. coli ED8767 was harvested during the exponential growth phase and was infected with the phage containing various DNA fragments of S. meliloti IFO 14782. The infected E. coli was spread on LB medium containing tetracycline. After incubation for 17 h, all the colonies that appeared on the agar were scraped off and incubated in LB medium for 1 h with shaking. They were then dispensed into small vials and stored at −120°C.
Triparental conjugal mating.
Triparental conjugal mating was performed as previously described (16). For Tn5 mutagenesis, S. meliloti IFO 14782 (a recipient strain) was inoculated into liquid LBMC, and E. coli HB101 harboring pRK2013 (a helper strain) and E. coli HB101 harboring pSUP2021 (a Tn5 donor strain) were inoculated into LB medium containing kanamycin. These three strains were incubated with shaking at 140 rpm. After incubation for 16 h, the three cultures were transferred to identical fresh media and were again incubated for 6 h. Each strain was harvested, and the cells were mixed at a 1:1:4 (vol/vol/vol) ratio. The mixture was placed on a nitrocellulose filter that was placed on LBMC agar. After this plate was incubated for 20 h at 30°C, the cells on the filter were scraped off and suspended in sterile 0.85% saline.
During the transfer of the genomic library of S. meliloti IFO 14782 from E. coli ED8767 to S. meliloti 16C18, S. meliloti 16C18 (a recipient strain) and E. coli HB101 harboring pRK2013 were cultured using the procedure described above. Frozen stored E. coli ED8767 harboring the genomic library of S. meliloti IFO 14782 (a donor strain) was cultured with shaking for 2 h in LB medium containing tetracycline. After each strain was harvested, the strains were mixed at a 1:1:1 (vol/vol/vol) ratio, and mating was performed on a nitrocellulose filter placed on LBMC agar. After mating, the cells on the filter were scraped off and suspended in sterile 0.85% saline. During transfer of the plasmid carrying a DNA fragment containing the pdxR gene from E. coli HB101 to S. meliloti 16C18, triparental conjugal mating was carried out using the procedure described above.
PCR amplification of pdxR.
The pdxR gene was amplified from 100 ng of S. meliloti IFO 14782 chromosomal DNA by using 10 pmol of two oligonucleotide primers (5′-GAATTCATGGCCATCGGCA-3′ and 5′-CCACTTCCCTTGTAGTACGAGCT-3′). The reaction conditions were as follows: 94°C for 15 s, followed by 25 cycles of 94°C for 15 s, 50°C for 15 s, and 68°C for 3 min.
Determination of DNA sequences.
DNA sequences were determined with an ALFred DNA sequencer supplied by Amersham Biosciences, Corp., New Jersey.
Preparation of cell extract.
S. meliloti strains were cultivated in glucose peptone yeast extract medium for 72 h. After centrifugation of the culture broth, the cells were washed twice with a 0.85% NaCl solution and suspended in 10 mM Tris-HCl buffer (buffer A) (pH 7.5). The suspension was passed through a French pressure cell. After centrifugation at 37,000 × g for 90 min, the supernatant was dialyzed against buffer A and stored at −30°C until use. The protein concentration was determined by the method of Lowry et al. (26).
Assay of 4PE dehydrogenase activity in S. meliloti.
The reaction mixtures (150 μl) used for assaying 4PE dehydrogenase activity contained 33 mM potassium phosphate buffer (pH 7.0), 2.3 mM 4PE, an electron acceptor, and cell extract (0.16 mg protein) or 2.4 μg purified proteins. Either 0.091 mM DCIP, 1.3 mM potassium ferricyanide, or 80 μM cytochrome c was used as the electron acceptor. The enzyme activity was estimated spectroscopically by measuring the change in absorbance at 600 or 420 nm due to the reduction of DCIP or potassium ferricyanide. In the case of cytochrome c, the spectrum in the range from 400 to 650 nm was monitored using a recording spectrophotometer.
Purification of pdxR-encoded 4PE dehydrogenase.
A cell extract (3,055 mg protein) of S. meliloti 16C18/pVK-pdxR was loaded onto a Q Sepharose FF column. The column was eluted with a linear gradient of KCl (0.1 to 0.4 M) in buffer A. The desalted active fractions (58 mg protein) were applied to a prepacked Resource Q column. The column was eluted with a linear gradient of KCl (0.1 to 0.4 M) in buffer A. Active fractions (21.6 mg protein) were dialyzed against 50 mM Tris-HCl buffer (pH 7.5) containing 150 mM KCl (buffer B), and these fractions were concentrated to small volumes by ultrafiltration. A sample was applied to a prepacked HiPrep Sephacryl S-200 HR column that had been previously equilibrated using buffer B. After elution with the same buffer, the activity of 4PE dehydrogenase in the desalted samples of each fraction was determined using DCIP. Subsequently, 4PE dehydrogenase was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% (wt/vol) polyacrylamide gel using the method of Laemmli (24) and was stained with Coomassie brilliant blue R-250. The molecular mass of the protein was estimated by comparison with a commercial mixture of molecular mass standards. The molecular mass of the native 4PE dehydrogenase was calculated using a HiPrep Sephacryl S-200 HR column calibrated with molecular mass standards (vitamin B12 [1.35 kDa], equine myoglobin [17 kDa], chicken ovalbumin [44 kDa], bovine γ-globulin [158 kDa], and thyroglobulin [670 kDa]; Bio-Rad Laboratories, Richmond, CA).
Identifying the product of 4PE dehydrogenase in sequential reactions with 4PE dehydrogenase and 3-phosphoserine transaminase (SerC).
The serC gene was generated from the chromosomal DNA of E. coli K-12 with two primers, 5′-TCCCGGGAGGGGAAATGGCT-3′ and 5′-ACCCGGGCAAAATTTCGGCA-3′. The amplified PCR product (ca. 1.1 kb) was directly cloned by using a TOPO TA cloning kit. The sequence of the amplified region was confirmed to be identical to the sequence of the coding sequence region of the serC gene (accession number NC_000913). The product was digested with SmaI and inserted downstream of the tac promoter of the pKK223-3 expression vector. After transformation of E. coli HB101 with the pKK-serC plasmid, the recombinant was cultured for 16 h in LB medium. Bacterial cells harboring SerC were suspended in buffer A, passed through a French pressure cell, and centrifuged. 3-Phosphoserine transaminase was purified from the supernatant by three chromatography steps using Q Sepharose FF, Resource Q, and Sephacryl S-200 HR columns. The reaction mixture (12.5 ml) that was used for isolation of the oxidation product of 4PE and subsequent transamination contained 33 mM potassium phosphate buffer (pH 8.2), 2.3 mM 4PE, 2.5 mM DCIP, 4 mM potassium fluoride, 0.25 mM ammonium acetate, 0.125 mM potassium glutamate, 2.5 mM NADH, and 2 mg each of PdxR, SerC, and glutamate dehydrogenase. After a 1-h incubation at 28°C, the reaction mixture was passed through an ultrafiltration membrane, and the filtrate was applied to a Dowex 1 (acetate form) column. Elution was carried out with a linear gradient of NH4HCO3 (10 to 750 mM). A ninhydrin-positive product (5.4 mg) was eluted in tubes 10 to 13 and was found to have an Rf of 0.14 on a DEAE-cellulose thin-layer chromatography plate (n-butanol/acetic acid/H2O ratio, 4:2:2). This product was then subjected to MS with an FAB ion mode, 1H NMR, and 31P NMR.
FAB-MS (negative): 214 (M − H)−, 1H NMR (400 MHz, D2O) δ values: 4.09 (1H, mult.), 4.15 (1H, mult.), 4.34 (1H, d, J = 2.4 Hz), 4.54 (1H, d, J = 2.9 Hz) and 31P NMR (200 MHz, D2O) δ value: 1.85 (t, JH-P = 6.05 Hz).
Formation of vitamin B6 from DX and glycolaldehyde and/or glycine and isolation of vitamin B6 from reaction mixtures using a 13C-labeled precursor.
Cells of S. meliloti IFO 14782/pVK-pdxJ (20) and strain 16C18/pVK-pdxJ were harvested from a 3-day culture broth by centrifugation, washed twice with a sterile 0.85% NaCl solution, and suspended in a small volume of sterile water. Ten milliliters of the substrate mixture (containing 1.5 mM DX, 40 mM glycolaldehyde, and 32 mM glycine or 1.5 mM DX and 40 mM glycolaldehyde), 200 mM Tris-HCl buffer (pH 7.6), and cells of each strain (final A600, 20) were shaken in a tube at 28°C. After 22 h, the reaction mixture was centrifuged, and the vitamin B6 present in the supernatant was subsequently assayed using S. carlsbergensis ATCC 9080.
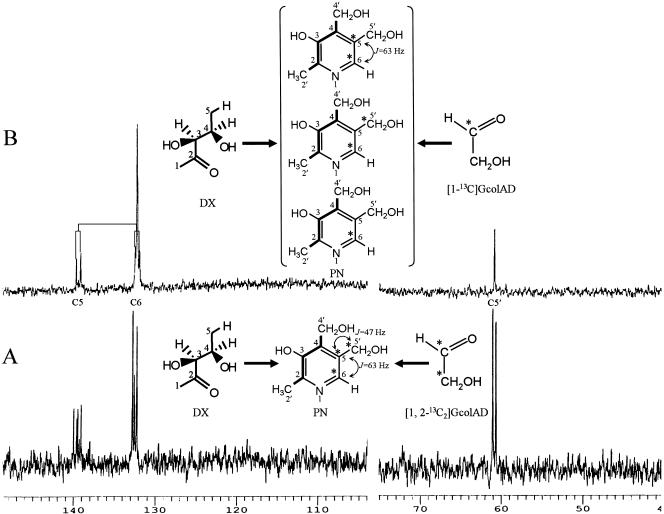
In order to isolate the vitamin B6 synthesized during the intact cell reaction with a labeled substrate, 20 tubes (total volume, 20 ml) containing 1 ml of the mixture along with 1.5 mM DX and 40 mM [1,2-13C]glycolaldehyde or 40 mM [1-13C]glycolaldehyde, 200 mM Tris-HCl buffer (pH 7.6), and washed cells of strain IFO 14782/pVK-pdxJ were shaken for 22 h with a shaker. The reaction mixture was centrifuged and chromatographed on an Amberlite CG 120 (H+) column using a 5% ammonium hydroxide solution for developing the chromatogram. The fractions containing a peak that corresponded to vitamin B6 were concentrated under reduced pressure. The concentrate was dissolved in a small volume of water, applied to a Dowex 1 (OH−) column, and developed by using a 0.2 N HCl solution. The vitamin B6 fractions were concentrated under reduced pressure and subjected to 13C NMR structural analysis. 13C NMR (100 MHz, D2O) of PN isolated from DX and [1,2-13C]glycolaldehyde: 60.9 (d, J2 = 47 Hz), 132.5 (d, J1 = 63 Hz), 139.5 (dd, J1 = 63 Hz, J2 = 47 Hz). 13C NMR (100 MHz, D2O) of PN isolated from DX and [1-13C]glycolaldehyde: 60.9 (s), 132.5 (s) (d, J = 63 Hz), 139.5 (d, J = 63 Hz).
RESULTS AND DISCUSSION
Isolation of a vitamin B6-deficient S. meliloti mutant and cloning of the pdxR gene complementing the mutation.
Vitamin B6 auxotrophs were screened from the Tn5 insertion mutants of S. meliloti IFO 14782. A cell suspension of Tn5-mutagenized S. meliloti IFO 14782 that was obtained under the triparental conjugal mating conditions described in Materials and Methods was diluted appropriately and spread on LBMC agar with nalidixic acid (to select for Sinorhizobium) and neomycin (to select for Tn5). After incubation for 4 days, only nalidixic acid- and neomycin-resistant colonies appeared on the agar. Tn5 was introduced into S. meliloti IFO 14782 at a frequency of 4.2 × 10−8/recipient. These colonies were streaked onto LBMC agar containing the same antibiotics and were incubated for 2 days. The cells grown on LBMC agar containing nalidixic acid and neomycin were used to screen the vitamin B6 auxotrophs. The production of vitamin B6 was assayed in 10,000 transconjugants using the vitamin B6 assay agar and a vitamin B6 indicator strain, S. carlsbergensis ATCC 9080. One colony that produced no halo was obtained in this way. This strain, which was designated S. meliloti 16C18, did not grow on vitamin B6-free EMM agar and DX-containing agar; however, it grew on 4HT- or PN-containing agar (Fig. 2). Therefore, we concluded that strain 16C18 had a mutation in the 4PHT pathway that is involved in vitamin B6 biosynthesis in S. meliloti.
FIG. 2.
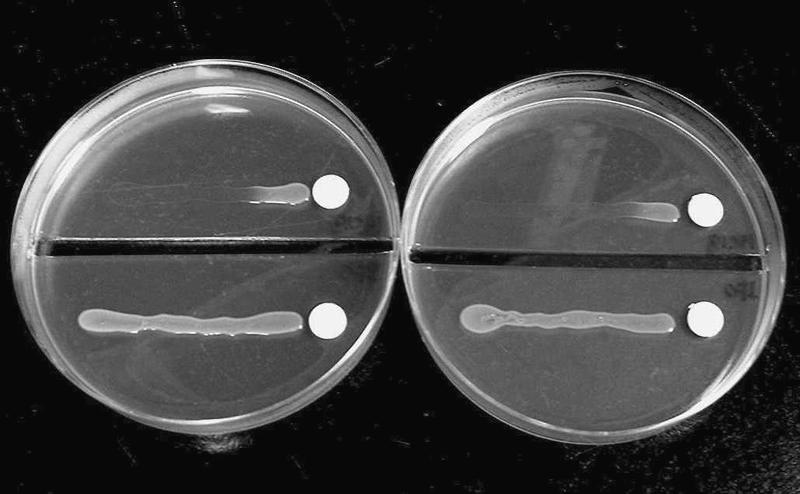
Growth of S. meliloti IFO 14782 and strain 16C18 on vitamin B6-free EMM agar after 16 h of incubation. (Left plate) Paper disks containing 150 ng pyridoxine; (right plate) paper disks containing 1.5 μg 4-hydroxy-l-threonine. The upper parts of the plates contained cultures of the 16C18 strain, while the lower parts of the plates contained cultures of the IFO 14782 strain.
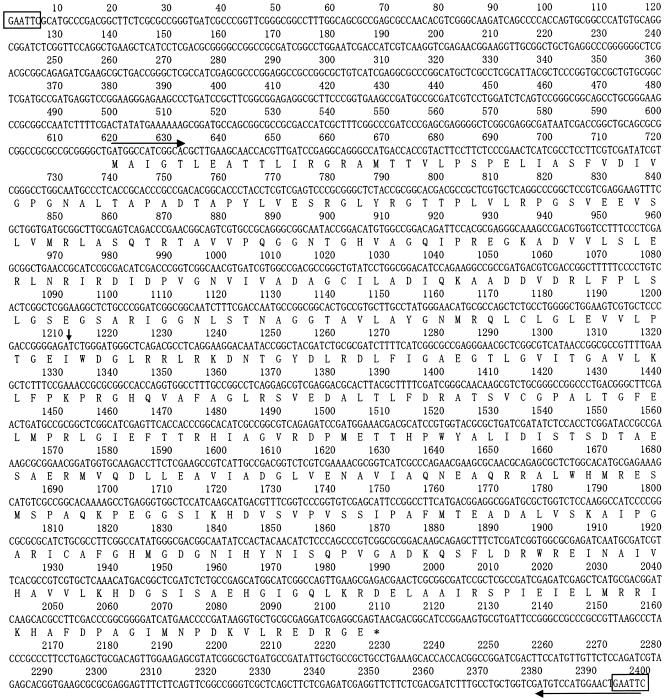
In order to isolate the DNA fragment that complements the mutation in strain 16C18, the genomic library of S. meliloti IFO 14782 (see Materials and Methods) was transferred into strain 16C18 by triparental conjugal mating. Subsequently, the transconjugants containing the target gene were selected on EMM agar containing tetracycline; the vitamin B6 auxotrophs of S. meliloti could not grow on this agar. After incubation for 8 days, four transconjugants appeared on the plates. The plasmid patterns of these transconjugants on agarose gels were analyzed. A common 2.5-kb DNA fragment was observed for all plasmids when they were digested with EcoRI. We examined whether this fragment complemented the mutation in strain 16C18; it was inserted into pVK100, and the resulting plasmid was transferred into S. meliloti 16C18. All the transconjugants obtained in this way grew on EMM agar, indicating that the 2.5-kb fragment contained a gene that complemented the mutation in S. meliloti 16C18. We then sequenced the DNA of the 2.5-kb fragment. In the sequence, there was a 1,491-bp open reading frame, which was designated pdxR (Fig. 3). The sequence of the open reading frame was identical to that of the coding sequence region of SMc00985 (positions 951316 to 952806, complement; accession number NC_003047).
FIG. 3.
Sequence of a 2.5-kb DNA fragment that complements a mutation in S. meliloti 16C18 and the deduced amino acid sequence of a 1,491-bp open reading frame in PdxR. The boxes indicate the EcoRI site. The horizontal and vertical arrows indicate the region used for PCR primers and the Tn5 insertion point, respectively.
The DNA fragment containing pdxR and a flanking region was amplified, as described in Materials and Methods. A 2.2-kb PCR product was inserted into a pCRII-TOPO vector. The amplified DNA sequence was confirmed to be identical to the original sequence. A 1.7-kb fragment of DNA corresponding to the amplified region was recovered from the agarose gels using QIAEXII, and it was ligated to the pKK223-3 expression vector. After transformation of E. coli JM109 with the ligation mixture, recombinant plasmid pKK-pdxR was obtained. In this recombinant plasmid, the pdxR gene was inserted into the EcoRI site of pKK223-3 in the same direction as the tac promoter on the vector.
After plasmid pKK-pdxR was digested using BamHI, the resulting 2-kb fragment containing the tac promoter and pdxR was purified from the agarose gel and then ligated to pVK100 using BglII. Recombinant plasmid pVK-pdxR, in which the tac promoter and the pdxR gene were inserted at the BglII site of pVK100 in the direction opposite the direction of the kanamycin resistance gene, was obtained. The resulting plasmid, pVK-pdxR, was introduced into S. meliloti 16C18 by triparental conjugal mating, and clones with pVK-pdxR were obtained on EMM agar containing tetracycline, on which S. meliloti 16C18/pVK100 did not grow.
In order to determine the insertion point of Tn5, the region around Tn5 in the genome of strain 16C18 was subcloned and then sequenced. The results indicated that in S. meliloti 16C18 Tn5 was inserted between nucleotides 1211 and 1212 (Fig. 3).
Complementation of an E. coli pdxB mutant with pdxR.
An S. meliloti DNA sequence alignment search performed using the BLAST and Swiss-Prot databases predicted that the pdxR gene encodes an oxidoreductase that catalyzes the oxidation of 2-hydroxy acids, such as d-lactate and glycolate (5). In the vitamin B6 biosynthetic pathways in E. coli, particularly the pathway resulting in the formation of 4PHT, only one reaction step is catalyzed by a 2-hydroxy acid dehydrogenase (49). This step involves the oxidation of 4PE that is catalyzed by NAD-dependent 4PE dehydrogenase (PdxB) (Fig. 1). Plasmid pKK-pdxR was therefore transferred to a pdxB mutant, E. coli WG1012 (8), to examine whether pdxR complements the pdxB mutation. The resultant transformants and E. coli WG1012/pKK223-3 (vector control) were streaked onto EMM agar containing ampicillin with and without PN. After incubation for 16 h, all the transformants tested could grow on the EMM agar containing ampicillin without PN, on which E. coli WG1012/pKK223-3 could not grow (data not shown). This indicated that the pdxR gene product in S. meliloti performs the same function that PdxB performs in E. coli.
Assay of 4PE oxidoreductase activity in cell extracts.
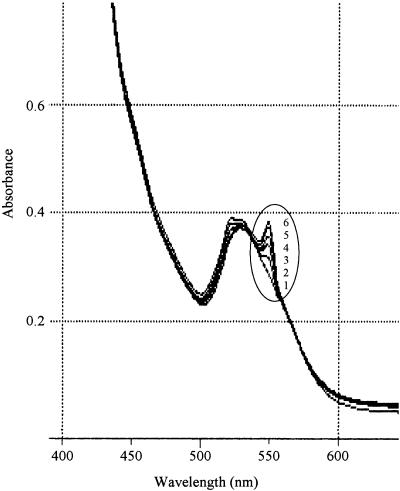
Cell extracts of S. meliloti 16C18 and 16C18/pVK-pdxR were used to examine the catalytic activities of oxidase and dehydrogenase during the synthesis of 4PE in the presence of oxygen, NAD+, and NADP+. However, no activity was found in either of the cell extracts. When DCIP or ferricyanide was added to the reaction mixture containing 4PE and the cell extract of the pdxR recombinant, rapid reduction of the electron acceptors was observed. Furthermore, when the natural electron acceptor cytochrome c, instead of DCIP or ferricyanide, was added to the mixture with the pdxR recombinant, a new absorption band at 550 nm appeared in the spectrum; the height of this band increased with time, suggesting that the reduced form of cytochrome c was being produced (Fig. 4). These results indicated that the pdxR gene probably encodes a dehydrogenase that uses electron acceptors rather than an oxidase. Therefore, we concluded that the pdxR gene product is a 2-hydroxy acid dehydrogenase, although the pdxR gene showed no homology to the E. coli pdxB gene.
FIG. 4.
Time course of the absorption spectra of cytochrome c in the reaction mixture containing 4-phospho-d-erythronate and a cell extract of S. meliloti 16C18/pVK-pdxR in buffer. Curve 1, zero time; curve 2, after 2 min; curve 3, after 5 min; curve 4, after 10 min; curve 5, after 15 min; curve 6, after 20 min.
Purification and partial biochemical characterization of pdxR-encoded 4PE dehydrogenase and identification of the reaction product of this enzyme.
The pdxR-encoded 4PE dehydrogenase was purified to electrophoretic homogeneity from the cell extract of S. meliloti 16C18/pVK-pdxR in three chromatography steps (see Materials and Methods). The SDS-PAGE patterns of the purified enzyme are shown in Fig. 5. The molecular mass (53 kDa) of the purified PdxR as determined by SDS-PAGE corresponded to a mass of 53,189, which had been predicted from the pdxR DNA sequence. The protein was estimated by gel filtration chromatography to be a homodimer with a native molecular mass of 106 kDa.
FIG. 5.
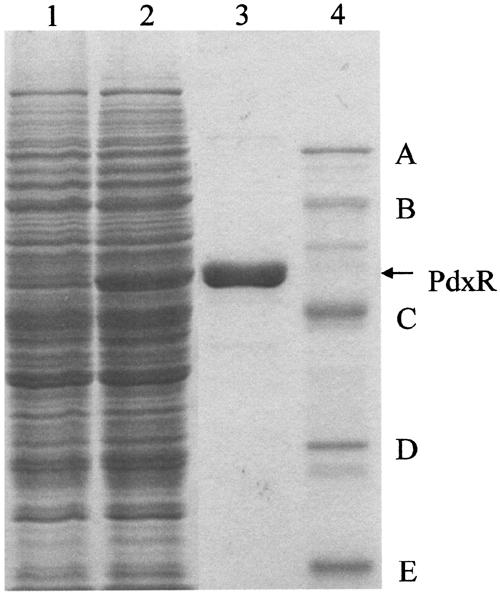
SDS-PAGE of cell extracts of S. meliloti and purified PdxR. Lane 1, 11.2 μg of a cell extract of S. meliloti 16C18; lane 2, 11.4 μg of a cell extract of S. meliloti 16C18/pVK-pdxR; lane 3, 2.3 μg of purified PdxR; lane 4, molecular mass markers (A, phosphorylase B [92.5 kDa]; B, bovine serum albumin [66.2 kDa]; C, ovalbumin [45 kDa]; D, carbonic anhydrase [31 kDa]; and E, soybean trypsin inhibitor [21.5 kDa].
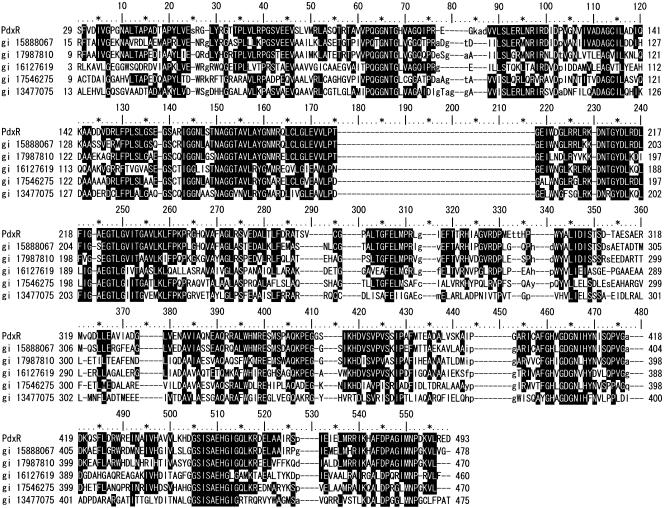
Since the purified protein was reddish yellow and an S. meliloti DNA sequence BLAST search predicted that PdxR exhibited homology with flavin adenine dinucleotide (FAD)-dependent, FAD-binding, or FAD/flavin mononucleotide (FMN)-containing oxidoreductases in many bacteria (27), the conserved domains of the PdxR protein were searched in the Conserved Domain Database. This search resulted in the most significant alignment with FAD/FMN-containing dehydrogenases involved in energy production and conversion (Fig. 6). In order to determine the binding in the PdxR protein, it was treated with (NH4)2SO4 using the method described by Cammack (3). A purified PdxR sample (1 ml, containing 4.74 mg protein) was dialyzed overnight against 1 liter of an 80% (wt/vol) (NH4)2SO4 solution. The resulting precipitate was collected by centrifugation, dissolved in a small quantity of buffer A, and desalted. The reduction of DCIP was tested in a reaction mixture (150 μl) containing 2.3 mM 4PE, 0.091 mM DCIP, 2.4 μg of desalted proteins, and various concentrations of FMN or FAD in 33 mM potassium phosphate buffer (pH 7.0) by monitoring the change in the absorbance at 600 nm. Reduction of DCIP was observed only when 0.014 μM FAD, and not when FMN, was added to the reaction mixture and reached a maximum speed at 3.8 μM or more. We also examined the substrate specificity of the enzyme for nonphosphorylated and phosphorylated 2-hydroxy acid compounds, such as glycolic acid, d-glyceric acid, l-lactic acid, dl-lactic acid, dl-isocitric acid, l-malic acid, d-erythronate, phosphoglycolate, 3-phospho-d-glycerate, and 4PE. After the reaction mixture (150 μl) containing 0.091 mM DCIP and purified PdxR (2.4 μg proteins) in 33 mM potassium phosphate buffer (pH 7.0) was preincubated for 1 min at room temperature, the reaction was started by addition of each substrate at a concentration of 2.3 mM. The decrease in the absorbance at 600 nm was monitored for 1 min by using a spectrophotometer. Of all the compounds tested, the protein showed activity only against 4PE (946 U/min/mg protein). One unit of enzyme activity was defined as the amount of enzyme that reduced 1 nmol of DCIP in 1 min. Based on these results, we concluded that the pdxR-encoded protein was an FAD-dependent 4PE dehydrogenase. We consider this protein a unique 2-hydroxy acid dehydrogenase that oxidizes only 4PE in the presence of the natural electron acceptor, cytochrome c.
FIG. 6.
Amino acid alignment of Sinorhizobium PdxR with 4PE dehydrogenase and FMN/FAD-containing oxidoreductase homologs from other organisms. Identical and similar sequences present in three or more sequences are indicated by a black background. Abbreviations: PdxR, 4PE dehydrogenase (S. meliloti IFO 14782); gi 15888067, hypothetical protein AGR_C_1314 (Agrobacterium tumefaciens str. C58); gi 17987810, glycolate oxidase subunit GlcD (Brucella melitensis 16 M); gi 16127619, FAD-binding oxidoreductase (Caulobacter crescentus CB15); gi 17546275, probable oxidoreductase protein (Ralstonia solanacearum GMI1000); gi 13477075, similar to actin-interacting protein (Mesorhizobium loti MAFF303099).
The reaction rate of this dehydrogenase appeared to be quite high in the presence of an electron acceptor. However, the oxidation product was isolated neither in S. meliloti nor in E. coli. The product was therefore identified as HPHKB from the reactions with 4PE dehydrogenase and, subsequently, 3-phosphoserine (or 4PHT) transaminase (SerC or PdxF). Modifying a procedure described by Drewke et al. (9) (see Materials and Methods), we coupled the 4PHT transaminase reaction with glutamate dehydrogenase, which catalyzed the conversion of glutamate to α-oxoglutarate. After the reactions, the product was purified and identified as 4PHT by MS and 1H and 31P NMR analyses (35). The results indicated that 4PE was probably oxidized to HPHKB by PdxR and was subsequently aminated in a reaction catalyzed by 4PHT transaminase to form 4PHT; a similar reaction is presumed to occur in E. coli. It is possible that the 4PE oxidation product might be labile and similar to the 3-phosphoglycerate oxidation product in the serine phosphorylation pathway (31).
Synthesis of PN from DX and glycolaldehyde and incorporation of 13C-labeled glycolaldehyde into PN molecules.
In our previous studies involving the biosynthesis of vitamin B6 in S. meliloti (38, 39), we proposed that the 4PHT-2 pathway starts with glycolaldehyde and glycine and that the resulting 4HT is converted to 4PHT by homoserine kinase, as shown in Fig. 1. However, as mentioned above, pdxR-encoded 4PE dehydrogenase, which converts 4PE to HPHKB, is involved in de novo 4PHT synthesis. These results raise the following questions. What is the origin of 4PE, and how are glycolaldehyde and glycine, which were previously proposed to be the precursors of 4PHT, involved in vitamin B6 biosynthesis in S. meliloti? We therefore examined vitamin B6 synthesis from DX/glycolaldehyde/glycine and from DX/glycolaldehyde in the vitamin B6 biosynthetic pathway by using an intact cell system with S. meliloti IFO 14782 and 16C18 recombinants containing the amplified pdxJ gene, which is located downstream of the pdxR gene. As shown in Table 1, the IFO 14782 recombinant produced large amounts of PN not only from DX/glycolaldehyde/glycine but also from the mixture containing DX and glycolaldehyde without glycine (86% of the total PN was produced via the major pathway). However, the 16C18 recombinant produced no PN from DX/glycolaldehyde and produced small amounts of PN from DX/glycolaldehyde/glycine, and the small amounts of PN corresponded to the amounts produced via the minor pathway. Furthermore, the IFO 14782 recombinant produced no PN from DX and glycine, glycolate, glyoxylate, d-erythrose, or d-erythronate instead of glycolaldehyde (data not shown). These results suggest that S. meliloti has two pathways for 4PHT synthesis; one pathway starts exclusively with glycolaldehyde and leads to the synthesis of 4PHT via 4PE (major pathway), and the other pathway involves the synthesis of 4PHT starting with the conversion of glycolaldehyde/glycine to 4HT and is catalyzed by homoserine kinase (minor pathway) (Fig. 1). A pdxR mutant produces vitamin B6 if it is incubated in a medium supplemented with DX/glycolaldehyde/glycine (Table 1). However, vitamin B6 auxotrophy in the pdxR mutant could not be rescued when the mutant was inoculated onto EMM agar containing sufficient levels of glycolaldehyde/glycine. This implies that the minor pathway may be functional in Sinorhizobium.
TABLE 1.
Pyridoxine synthesis from precursors in intact cell reactions for pdxJ recombinants of S. meliloti IFO 14782 and 16C18
| Strain | Precursors | Amt of pyridoxine formed (μg/ml) |
|---|---|---|
| S. meliloti IFO14782/pVK-pdxJ | DX + GcolADa | 32.3 |
| DX + GcolAD + glycine | 37.6 | |
| S. meliloti 16C18/pVK-pdxJ | DX + GcolAD | 0 |
| DX + GcolAD + glycine | 6.3 |
GcolAD, glycolaldehyde.
The C5 (C-2′, -2, -3, -4, and -4′) and NC3 (N-1 and C-6, -5, and -5′) units of PN molecules are derived from the C5 unit of DX and the NC3 unit of 4HT (and 4PHT), respectively, as demonstrated previously (38). These results imply that if our hypotheses are correct, all the carbons (C-6, -5, and -5′) in the latter unit of the PN skeleton must have originated only from glycolaldehyde. Therefore, 13C-labeled glycolaldehyde was incorporated into PN molecules using an intact cell system with the IFO 14782/pdxJ recombinant cells, DX, and [1,2-13C]glycolaldehyde. The PN formed in this way was purified using cation- and anion-exchange columns and was then analyzed using a 13C NMR spectrometer. The spectrum of the purified PN contained three signals which could be assigned to the C-5′, C-6, and C-5 carbons in the PN skeleton (Fig. 7A). A signal at 139.5 ppm appeared as a double doublet due to 13C enrichment in the three contiguous carbon atoms (C-6, C-5, and C-5′) of the PN skeleton, while the other two signals appeared as two doublets due to 13C enrichment in the two adjacent carbon atoms (C-5 and C-5′ and C-5 and C-6) of the skeleton. This result implies that the labeled carbon of glycolaldehyde was incorporated into C-5′, C-5, and C-6 of the PN molecules, in which three carbons (C-2, C-3, and C-4) of 4PHT are incorporated.
FIG. 7.
13C NMR spectra of pyridoxine hydrochloride isolated from intact cell reaction of S. meliloti IFO 14782/pVK-pdxJ with 1-deoxy-d-xylulose and [1,2-13C2]glycolaldehyde (A) or 1-deoxy-d-xylulose and [1-13C]glycolaldehyde (B). Abbreviation: GcolAD, glycolaldehyde. An asterisk indicates a 13C-labeled carbon.
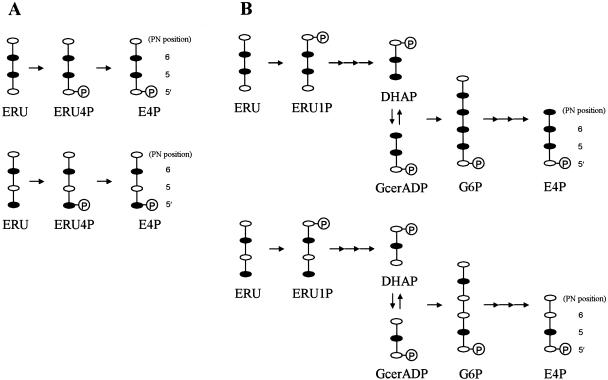
It was also of interest to determine whether E4P is involved in major 4PHT synthesis from glycolaldehyde (Fig. 1). We examined the incorporation of a specific 13C-labeled sample of glycolaldehyde into PN molecules further. The 13C NMR spectrum of PN formed from DX and [1-13C]glycolaldehyde contained complicated but reproducible signals comprising two doublet peaks corresponding to the 13C enrichment of two adjacent carbon atoms (C-5 and C-6) of the PN skeleton, a large singlet peak assigned to C-6, and a smaller singlet peak assigned to C-5′ (Fig. 7B). Based on the spectral analysis, 4PHT synthesis from glycolaldehyde is most likely to start with a transketolase reaction, which catalyzes the formation of d-erythrulose (ERU) only from glycolaldehyde, which acts as both an acceptor and a donor substrate, as reported by Sevostyanova et al. (34); the C-1 label of the added glycolaldehyde is mainly incorporated into the C-6 of PN molecules. In addition, the aldehyde group of C-1 in glycolaldehyde molecules, which functions as an acceptor, undergoes intramolecular rearrangement to form C-2; this reaction is catalyzed by an isomerase-like enzyme that results in the formation of [1-13C]glycolaldehyde and [2-13C]glycolaldehyde. ERU is further metabolized to E4P. There are two possible pathways for the synthesis of E4P from ERU; one is a pathway leading to ERU 4-phosphate and E4P catalyzed by a kinase and an isomerase (10), and the other leads to E4P via ERU 1-phosphate, dihydroxyacetone 3-phosphate, d-glyceraldehyde 3-phosphate, and glucose 6-phosphate by the erythritol catabolic pathway found in Propionibacterium pentosaceum and Brucella abortus (37, 44) and the Embden-Meyerhof-Parnas and pentose-phosphate pathways (Fig. 8). E4P is probably synthesized by the former pathway because the 13C NMR spectrum has no singlet peak corresponding to the C-5 of PN molecules (Fig. 7B). The singlet peak assigned to C-6 had an inexplicable signal corresponding to C-6 (s) minus C-5′ (s). Intracellular endogenous glycolaldehyde might be an acceptor in the transketolase reaction. The E4P is then converted to 4PHT in three reactions, probably catalyzed by aldehyde dehydrogenase, FAD-dependent 4PE dehydrogenase (PdxR), and serine aminotransferase (SerC). In E. coli, 4PE is formed mainly from E4P by NAD-dependent E4P dehydrogenase that is encoded by epd (49). However, NAD- or NADP-dependent E4P dehydrogenase was absent in S. meliloti (38). An unidentified NAD-independent aldehyde dehydrogenase might be involved in the conversion of E4P to 4PE.
FIG. 8.
Fate of the 13C label of glycolaldehyde in the synthesis of d-erythrose 4-phosphate from d-erythrulose in two possible pathways. Abbreviations: ERU4P, d-erythrulose 4-phosphate; ERU1P, d-erythrulose 1-phosphate; DHAP, dihydroxyacetone 3-phosphate; GcerADP, d-glyceraldehyde 3-phosphate; G6P, glucose-6-phosphate. (A) Pathway leading to E4P via d-erythrulose 4-phosphate. (B) Pathway leading to E4P via d-erythrulose 4-phosphate, dihydroxyacetone 3-phosphate, d-glyceraldehyde 3-phosphate, and glucose-6-phosphate by the erythritol catabolic pathway and Embden-Meyerhof-Parnas and pentose phosphate pathways. Solid symbols indicate 13C-labeled carbon atoms.
The major 4PHT pathway in S. meliloti was studied by incorporating different samples of 13C-labeled glycolaldehyde into PN molecules. The results demonstrate that glycolaldehyde is metabolized to E4P through a pathway different from the pentose phosphate pathway and that E4P is then converted to 4PHT by the same three reactions that occur in E. coli; however, the mechanism of action of the enzymes catalyzing the first two steps is different from the mechanism of action in E. coli. Rhizobia, including S. meliloti, utilize carbohydrates, such as pentoses and hexoses, in catabolic pathways, including the pentose phosphate and Entner-Doudoroff pathways (13). It is a reasonable assumption that E4P, as a metabolic intermediate in the pentose phosphate pathway, is a substrate of 4PHT in common carbohydrate metabolism.
Acknowledgments
We are grateful to A. Pühler of Universität Bielefeld Biologie VI (Genetik) for providing the pSUP2021 plasmid. We thank H. P. Hohmann of DSM Nutritional Products Ltd. Biotechnology Research and Development for his helpful advice and critical comments.
REFERENCES
- 1.Bandurski, R. S., and B. Axelrod. 1951. The chromatographic identification of some biologically important phosphate esters. J. Biol. Chem. 193:405-410. [PubMed] [Google Scholar]
- 2.Burns, K. E., Y. Xiang, C. L. Kinsland, F. W. McLafferty, and T. P. Begley. 2005. Reconstitution and biochemical characterization of a new pyridoxal-5′-phosphate biosynthetic pathway. J. Am. Chem. Soc. 127:3682-3683. [DOI] [PubMed] [Google Scholar]
- 3.Cammack, R. 1968. Flavoprotein properties of d-2-hydroxyacid dehydrogenase purified from rabbit kidney. Biochem. J. 109:45-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cane, D. E., S. Du, J. K. Robinson, Y. Hsiung, and I. D. Spenser. 1999. Biosynthesis of vitamin B6: enzymatic conversion of 1-deoxy-d-xylulose-5-phosphate to pyridoxol phosphate. J. Am. Chem. Soc. 121:7722-7723. [Google Scholar]
- 5.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dréano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thébault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Lajudie, P., A. Willems, B. Pot, D. Dewettinck, G. Maestrojuan, M. Neyra, M. D. Collins, B. Dreyfus, K. Kersters, and M. Gillis. 1994. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int. J. Syst. Bacteriol. 44:715-733. [Google Scholar]
- 7.Dempsey, W. B. 1987. Synthesis of pyridoxal phosphate, p. 539-543. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 8.Dempsey, W. B. 1969. Characterization of pyridoxine auxotrophs of Escherichia coli: chromosomal position of linkage group I. J. Bacteriol. 100:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drewke, C., M. Klein, D. Clade, A. Arenz, R. Müller, and E. Leistner. 1996. 4-O-Phosphoryl-l-threonine, a substrate of the pdxC (serC) gene product involved in vitamin B6 biosynthesis. FEBS Lett. 390:179-182. [DOI] [PubMed] [Google Scholar]
- 10.Ebner, M., and A. E. Stütz. 1997. Glucose isomerase catalysed isomerisation reactions of (2R,3R)-configured aldofuranoses into the corresponding open-chain 2-ketoses. Carbohydr. Res. 305:331-336. [Google Scholar]
- 11.Ehrenshaft, M., P. Bilski, M. Y. Li, C. F. Chignell, and M. E. Daub. 1999. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc. Natl. Acad. Sci. USA 96:9374-9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrenshaft, M., and M. E. Daub. 2001. Isolation of PDX2, a second novel gene in the pyridoxine biosynthesis pathway of eukaryotes, archaebacteria, and a subset of eubacteria. J. Bacteriol. 183:3383-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkan, G. H., and C. R. Bunn. 1994. The rhizobia, p. 2197-2213. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 3. Springer-Verlag, New York, N.Y. [Google Scholar]
- 14.Farrington, G. K., A. Kumar, S. L. Shames, J. I. Ewaskiewicz, D. E. Ash, and F. C. Wedler. 1993. Threonine synthase of Escherichia coli: inhibition by classical and slow-binding analogues of homoserine phosphate. Arch. Biochem. Biophys. 307:165-174. [DOI] [PubMed] [Google Scholar]
- 15.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dréano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F.-J. Vörholter, S. Weidner, D. H. Wells, K. Wong, K.-C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 16.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 17.Hanes, C. S., and F. A. Isherwood. 1949. Separation of the phosphoric esters on the filter paper chromatogram. Nature 164:1107-1112. [DOI] [PubMed] [Google Scholar]
- 18.Hill, R. E., K. Himmeldirk, I. A. Kennedy, R. M. Pauloski, B. G. Sayer, E. Wolf, and I. D. Spenser. 1996. The biogenetic anatomy of vitamin B6. J. Biol. Chem. 271:30426-30435. [DOI] [PubMed] [Google Scholar]
- 19.Hill, R. E., and I. D. Spenser. 1996. Biosynthesis of vitamin B6, p. 695-703. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 20.Hoshino, T., Y. Nagatani, K. Ichikawa, and M. Tazoe. April 2004. W.O. patent 029270 A2.
- 21.Kennedy, I. A., R. E. Hill, R. M. Pauloski, B. G. Sayer, and I. D. Spenser. 1995. Biosynthesis of vitamin B6: origin of pyridoxine by the union of two acyclic precursors, 1-deoxy-d-xylulose and 4-hydroxy-l-threonine. J. Am. Chem. Soc. 117:1661-1662. [Google Scholar]
- 22.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 23.Laber, B., W. Manurer, S. Scharf, K. Stepusin, and F. S. Schmidt. 1999. Vitamin B6 biosynthesis: formation of pyridoxine 5′-phosphate from 4-(phosphohydroxy)-l-threonine and 1-deoxy-d-xylulose-5-phosphate by PdxA and PdxJ protein. FEBS Lett. 449:45-48. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lam, H.-M., and M. E. Winkler. 1990. Metabolic relationships between pyridoxine (vitamin B6) and serine biosynthesis in Escherichia coli K-12. J. Bacteriol. 172:6518-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 27.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell, R. E., E. J. Frey, and M. H. Benn. 1986. Rhizobitoxine and l-threo-hydroxythreonine production by the plant pathogen Pseudomonas andropogonis. Phytochemistry 25:2711-2715. [Google Scholar]
- 29.Murray, N. E., W. J. Brammar, and K. Murray. 1977. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol. Gen. Genet. 150:53-61. [DOI] [PubMed] [Google Scholar]
- 30.Osmani, A. H., G. S. May, and S. A. Osmani. 1999. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J. Biol. Chem. 274:23565-23569. [DOI] [PubMed] [Google Scholar]
- 31.Pizer, L. I. 1963. The pathway and control of serine biosynthesis in Escherichia coli. J. Biol. Chem. 238:3934-3944. [PubMed] [Google Scholar]
- 32.Rohmer, M., M. Knani, P. Simonin, B. Sutter, and H. Sahm. 1993. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoenlein, P. V., B. B. Roa, and M. E. Winkler. 1989. Divergent transcription of pdxB and homology between the pdxB and serA gene products in Escherichia coli K-12. J. Bacteriol. 171:6084-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevostyanova, I. A., O. N. Solovjeva, and G. A. Kochetov. 2004. A hitherto unknown transketolase-catalyzed reaction. Biochem. Biophys. Res. Commun. 313:771-774. [DOI] [PubMed] [Google Scholar]
- 35.Shames, S. L., D. E. Ash, F. C. Wedler, and J. J. Villafranca. 1984. Interaction aspartate and aspartate-derived antimetabolites with the enzymes of the threonine biosynthetic pathway of Escherichia coli. J. Biol. Chem. 259:15331-15339. [PubMed] [Google Scholar]
- 36.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 37.Sperry, J. F., and D. C. Robertson. 1975. Erythritol catabolism by Brucella abortus. J. Bacteriol. 121:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tazoe, M., K. Ichikawa, and T. Hoshino. 2000. Biosynthesis of vitamin B6 in Rhizobium. J. Biol. Chem. 275:11300-11305. [DOI] [PubMed] [Google Scholar]
- 39.Tazoe, M., K. Ichikawa, and T. Hoshino. 2002. Biosynthesis of vitamin B6 in Rhizobium: in vitro synthesis of pyridoxine from 1-deoxy-d-xylulose and 4-hydroxy-l-threonine. Biosci. Biotechnol. Biochem. 66:934-936. [DOI] [PubMed] [Google Scholar]
- 40.Tazoe, M., K. Ichikawa, and T. Hoshino. Unpublished results.
- 41.Tazoe, M., K. Ichikawa, and T. Hoshino. 2005. Purification and characterization of pyridoxine 5′-phosphate phosphatase from Sinorhizobium meliloti. Biosci. Biotechnol. Biochem. 69:2277-2284. [DOI] [PubMed] [Google Scholar]
- 42.Tazoe, M., K. Ichikawa, and T. Hoshino. 1999. Production of vitamin B6 in Rhizobium. Biosci. Biotechnol. Biochem. 63:1378-1382. [DOI] [PubMed] [Google Scholar]
- 43.Thérisod, M., J. C. Fischer, and B. Estramareix. 1981. The origin of the carbon chain in the thiazole moiety of thiamine in Escherichia coli: incorporation of deuterated 1-deoxy-d-threo-2-pentulose. Biochem. Biophys. Res. Commun. 98:374-379. [DOI] [PubMed] [Google Scholar]
- 44.Wawszkiewicz, E. J., and H. A. Barker. 1968. Erythritol metabolism by Propionibacterium pentosaceum. J. Biol. Chem. 243:1948-1956. [PubMed] [Google Scholar]
- 45.Woodruff, W. W., III, and R. Wolfenden. 1979. Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate. J. Biol. Chem. 254:5866-5867. [PubMed] [Google Scholar]
- 46.Wungsintaweekul, J., S. Herz, S. Hecht, W. Wisenreich, R. Feicht, F. Rohdich, A. Bacher, and M. H. Zenk. 2001. Phosphorylation of 1-deoxy-d-xylulose by d-xylulokinase of Escherichia coli. Eur. J. Biochem. 268:310-316. [DOI] [PubMed] [Google Scholar]
- 47.Yang, Y., G. Zhao, T.-K. Man, and M. E. Winkler. 1998. Involvement of the gapA- and epd (gapB)-encoded dehydrogenases in pyridoxal 5′-phosphate coenzyme biosynthesis in Escherichia coli K-12. J. Bacteriol. 180:4294-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yokota, A., and K. Sasajima. 1984. Formation of 1-deoxy-d-threo-pentulose and 1-deoxy-l-threo-pentulose by cell-free extracts of microorganisms. Agric. Biol. Chem. 48:149-158. [Google Scholar]
- 49.Zhao, G., A. J. Pease, N. Bharani, and M. E. Winkler. 1995. Biochemical characterization of gapB-encoded erythrose 4-phosphate dehydrogenase of Escherichia coli K-12 and its possible role in pyridoxal 5′-phosphate biosynthesis. J. Bacteriol. 177:2804-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]