Abstract
The StcE zinc metalloprotease is secreted by enterohemorrhagic Escherichia coli (EHEC) O157:H7 and contributes to intimate adherence of this bacterium to host cells, a process essential for mammalian colonization. StcE has also been shown to localize the inflammatory regulator C1 esterase inhibitor (C1-INH) to cell membranes. We tried to more fully characterize StcE activity to better understand its role in EHEC pathogenesis. StcE was active at pH 6.1 to 9.0, in the presence of NaCl concentrations ranging from 0 to 600 mM, and at 4°C to 55°C. Interestingly, antisera against StcE or C1-INH did not eliminate StcE cleavage of C1-INH. Treatment of StcE with the proteases trypsin, chymotrypsin, human neutrophil elastase, and Pseudomonas aeruginosa elastase did not eliminate StcE activity against C1-INH. After StcE was kept at 23°C for 65 days, it exhibited full proteolytic activity, and it retained 30% of its original activity after incubation for 8 days at 37°C. Together, these results show the StcE protease is a stable enzyme that is probably active in the environment of the colon. Additionally, kcat/Km data showed that StcE proteolytic activity was 2.5-fold more efficient with the secreted mucin MUC7 than with the complement regulator C1-INH. This evidence supports a model which includes two roles for StcE during infection, in which StcE acts first as a mucinase and then as an anti-inflammatory agent by localizing C1-INH to cell membranes.
Enterohemorrhagic E. coli (EHEC) O157:H7 strains cause infections that result in diarrhea and hemorrhagic colitis, which can progress to hemolytic-uremic syndrome (17). These bacteria belong to a group of pathogens that form attaching and effacing (A/E) lesions, which are characterized by intimate adherence to the host cells and effacement of the microvilli (9, 17). The StcE protease produced by EHEC is secreted by the type II secretion system (15) that is encoded 3′ of stcE on the pO157 virulence plasmid (2). Our laboratory has recently demonstrated that StcE contributes to intimate adherence of EHEC to host cells (12), a process essential for colonization of the terminal rectum of the bovine host (18). We have also shown that StcE cleaves C1 esterase inhibitor (C1-INH), a regulator of multiple inflammatory pathways (15), and localizes it to host cell surfaces (14).
The current model for StcE activity (12) proposes that initially, StcE allows passage of EHEC through the oral cavity by cleaving the salivary glycoproteins that are responsible for bacterial aggregation. Similarly, in the colon, StcE cleaves the glycoproteins that protect the intestinal epithelial surface, allowing EHEC to come into close contact with host cell membranes, where components of the locus of enterocyte effacement mediate formation of the characteristic A/E lesions. Later during infection, Shiga toxins and other virulence factors compromise the intestinal epithelial and endothelial barriers, allowing blood with its complement effectors into the intestinal lumen (9, 17). At this point, StcE localizes C1-INH to membranes, thereby reducing complement-mediated lysis of the bacteria and host cells (14).
Based on this model for StcE activity, several predictions can be made. As a secreted protein, StcE is likely to be resistant to most proteases encountered during passage through the body and at the site of EHEC infection in the colon. The model suggests that mucins comprising the glycocalyx and mucus layer should be cleaved efficiently because intimate adherence should be established quickly for EHEC so that it is not flushed out of the colon. StcE cleavage of C1-INH is likely to be less efficient than cleavage of mucus-associated glycoproteins. The result could be increased localization of C1-INH around the bacteria and host cells to reduce complement deposition on the cell membrane, thereby protecting cells from complement-mediated lysis.
The data presented here describe StcE protease activity in a variety of temperature, pH, and salt conditions. After treatment with other proteases or after treatment with polyclonal antisera against it, StcE remained active. StcE exhibited activity after weeks of incubation at 23°C and after days of incubation at 37°C. These findings show that StcE is probably active during infection in the conditions presented by environments such as the colon. Kinetic studies demonstrated that StcE activity for MUC7 was 2.5-fold more efficient than StcE activity for C1-INH. These results both broaden our understanding of StcE proteolytic activity and support the model for StcE activity in EHEC infection.
MATERIALS AND METHODS
Production of StcE protease.
The recombinant secreted form of StcE (rStcE′) was produced as described previously (12). Briefly, the protein was expressed as a fusion protein using the IMPACT system (New England Biolabs, Beverly, Mass.). The fusion was then purified with a chitin column, and rStcE′ was eluted by inducing proteolytic cleavage of the fusion protein. A proteolytically inactive mutant, E435D rStcE′, was produced using the same method.
Metal ion analysis.
rStcE′ and E435D rStcE′ were each dialyzed into phosphate-buffered saline (PBS) or 20 mM Tris (pH 7.5). The metal contents of the dialyzed samples and the corresponding buffer controls were determined at the Soil Science Laboratory (University of Wisconsin-Madison) by inductively coupled plasma mass spectrometry using VG PlasmaQuad PQ2 Turbo Plus. The metals examined included calcium, copper, nickel, magnesium, and iron.
Purification of MUC7.
Whole human saliva was collected and boiled in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample loading buffer (15 mM Tris-HCl, 2% SDS, 10% glycerol, 70 mM 2-mercaptoethanol, 100 mM bromophenol blue). The solution was loaded onto a 6% acrylamide SDS-PAGE gel using a one-well setup. The edges of the gel were stained with Coomassie brilliant blue to visualize the banding pattern, which allowed the MUC7 band to be excised from the unstained gel. The gel fragments containing MUC7 were placed in an electroelution chamber, and the protein was electroeluted overnight at 100 V in buffer (192 mM glycine, 25 mM Tris, 0.1% Tween 20; pH 7.5). The detergent was removed from the eluted protein by extraction using Extract-Gel D detergent removal gel (Pierce, Rockford, IL).
Antibodies and antisera.
Polyclonal antiserum (rabbit) against C1-INH was purchased from Serotec (Raleigh, NC). Monoclonal antibodies (mouse) 3C7 and 4C3 against C1-INH were kind gifts from M. Schapira and P. Patston (6).
Monospecific polyclonal antisera were raised in rabbits against synthetic peptides representing StcE residues 80 to 98 (N-terminal peptide) and 872 to 886 (C-terminal peptide) by Open Biosystems (Huntsville, AL). Polyclonal antiserum against rStcE′ was raised in rabbits (Chemicon, Temecula, CA).
StcE ELISA activity.
C1-INH (100 ng; Complement Technologies, Tyler, TX) and rStcE′ (50 ng) were combined in a polypropylene 96-well plate (Nalge Nunc International, Naperville, Ill.) in various buffers (final volume, 100 μl). Reactions were stopped at 0, 20, 40, and 60 min by adding EDTA (50 μl of a 50 mM solution), and the mixtures were transferred to a 96-well enzyme-linked immunosorbent assay (ELISA) plate (Medisorp; Nalge Nunc International) overnight at 4°C. Wells were emptied and blocked with 1% bovine serum albumin (BSA) (Sigma, St. Louis, MO) in PBS for 1 h at room temperature (23°C). Wells were emptied and washed with PBS with 0.1% Tween 20 three times for 1 min each time. Wells were probed with the 3C7 monoclonal antibody against C1-INH that did not recognize StcE-cleaved C1-INH. Wells were washed again and probed with goat anti-mouse antibody conjugated with horseradish peroxidase (HRP) (1:3,000; Bio-Rad) for 30 min. The ELISA plates were developed with the TMB substrate (Bio-Rad), and the results were visualized with a 96-well plate reader (Molecular Dynamics, Sunnyvale, CA). This method was used for the salt and pH activity experiments.
Temperature experiments.
C1-INH (12 μg) and rStcE′ (3 μg) were equilibrated at various temperatures for 1 h in separate tubes containing 50 mM Tris (pH 7.5). C1-INH and StcE were combined and aliquoted into separate samples for each time (0, 20, 40, and 60 min). At each time, EDTA (final concentration, 25 mM) was added to samples to stop the reaction. Samples were transferred to nitrocellulose using a slot blot apparatus. Full-length C1-INH was detected with the 3C7 monoclonal antibody (1:250, 1 h), followed by HRP-conjugated goat anti-mouse antibody (1:1,000, 30 min). Blots were visualized with a phosphorimager (Typhoon 8600; Amersham Biosciences). The intensities of bands were quantified with the ImageQuant software (GE Healthcare, Little Chalfont, United Kingdom) to determine the loss of full-length C1-INH.
Stability experiments.
The rStcE′ protease was repeatedly frozen in an acetone-dry ice bath and thawed in 37°C water, and samples were taken after various cycles. Aliquots of freeze-thawed rStcE′ or control rStcE′ (1 μg) were added to PBS containing C1-INH (1 μg) and incubated for 30 and 60 min. The reactions were stopped by adding SDS-PAGE sample loading buffer containing EDTA and boiling the preparations for 5 min. Proteins were separated by SDS-PAGE and blotted onto nitrocellulose. The blots were probed with rabbit antiserum against C1-INH (1:2,000; Serotec) and HRP-conjugated goat anti-rabbit antibodies (1:2,000; Bio-Rad). The blots were analyzed with the ImageQuant software to determine the loss of full-length C1-INH.
The rStcE′ protease was also incubated at room temperature for 8, 18, or 65 days or at 37°C for 8 or 18 days. Aliquots were assayed for activity by the method used for the stability experiments described above.
Protease treatment experiments.
BSA was used as a control for the activity of the proteases tested. The proteases tested were trypsin (Sigma), chymotrypsin (Sigma), Pseudomonas aeruginosa elastase (Calbiochem), human neutrophil elastase (Crescent Chemicals), and proteinase K (Sigma). The treatments were sufficient to degrade BSA (2 μg) completely, as visualized on a Coomassie blue-stained SDS-PAGE gel after various times (Table 1 shows specific incubation times for various proteases). All reactions took place at 37°C in PBS. C1-INH and rStcE′ were each treated with the proteases for 2 h and also for 4 h as a control. rStcE′ was treated with a protease for 2 h, and then C1-INH was added and the preparation was incubated for an additional two hours. Conversely, C1-INH was treated with a protease for 2 h, and then rStcE′ was added and the preparation was incubated for an additional two hours. A control reaction mixture containing rStcE′ and C1-INH was also incubated for 2 h. All reactions were stopped by adding SDS-PAGE sample loading buffer containing EDTA and boiling the mixtures for 5 min. Proteins were separated on an 8% acrylamide gel, blotted onto nitrocellulose, and probed with antiserum against C1-INH (polyclonal, 1:2,000) and secondary antibodies conjugated with HRP. The C1-INH immunoblots were analyzed to determine whether addition of C1-INH to protease-treated rStcE′ resulted in C1-INH fragments that matched the fragments obtained in the StcE-cleaved C1-INH control reaction, which were indicative of StcE activity.
TABLE 1.
StcE activity after treatment with proteases
| Protease | Amt (μg) | Incubation time (h) | StcE activity | BSA degraded |
|---|---|---|---|---|
| Trypsin | 1 | 1 | + | + |
| Chymotrypsin | 2 | 1 | + | + |
| Neutrophil elastase | 1 | 2 | + | + |
| P. aeruginosa elastase | 0.1 | 1 | + | + |
| Proteinase K | 0.1 | 0.5 | − | + |
Anti-StcE antiserum experiments.
Purified rStcE′ (0.5 μg) was incubated with antiserum against the N-terminal peptide (1:60), antiserum against the C-terminal peptide (1:300), and polyclonal antisera against rStcE′ (1:1,500) for 2 h at 23°C in PBS. The corresponding control sera were also tested. Following this incubation, human C1-INH (0.5 μg) was added to each sample and incubated for 2.5 h at 23°C in a 10-μl (total volume) mixture. Samples were boiled in SDS-PAGE sample loading buffer for 5 min. Antibody inhibition of rStcE′ proteolytic activity against C1-INH was examined by SDS-PAGE and immunoblot analysis using polyclonal goat antiserum against human C1-INH (1:2,000; Cedarlane, Hornby, Canada) and an HRP-conjugated secondary antibody (1:2,000; Bio-Rad).
Anti-C1-INH antiserum experiments.
The effect of antibodies on StcE-mediated cleavage of C1-INH was determined by incubating C1-INH (0.5 μg) with 3C7 (1:250), 4C3 (1:50), or goat anti-human C1-INH polyclonal antibodies (1:100) at 23°C in PBS. After this incubation, rStcE′ (0.5 μg) was added to each mixture, and the samples were incubated overnight at 23°C in 10-μl (final volume) mixtures. Proteins were boiled for 5 min in SDS-PAGE sample loading buffer and analyzed by SDS-PAGE and immunoblot analysis using rabbit anti-human C1-INH polyclonal antibody (1:2,000) and an HRP-conjugated secondary antibody (1:2,000; Bio-Rad).
Mucin immunoblots.
Cell fractions of Hep-2, Caco-2, and LS174T cells were prepared as described previously (12). Briefly, S1 fractions contained proteins secreted by cells into PBS, S2 fractions were cell lysates extracted with saponin, and S3 fractions were lysates extracted with saponin and digitonin. Cell fractions (30 μl for S2 and S3 and 300 μl for S1) were treated with rStcE′ (3 μg) or 20 mM Tris (pH 7.5) at 37°C for 4 h (3 h for S1 fractions, which were subsequently precipitated with trichloroacetic acid). Control reactions demonstrated that rStcE′ was proteolytically active against C1-INH in the lysis buffers (data not shown). Reactions were stopped by adding SDS-PAGE sample loading buffer containing EDTA and boiling the mixtures for 5 min. Proteins were separated on an agarose gel (1%) and blotted onto nitrocellulose as described previously (12). Each blot was probed and stripped several times. The antibodies and antisera used as probes recognized MUC5AC (1:500; US Biologicals,) and MUC2 (polyclonal [1:500; Biomeda] and monoclonal [1:500; US Biologicals]). The results for the MUC5AC immunoblots were corroborated by the results of similar experiments performed with sputum from persons with cystic fibrosis (11).
Kinetic activity assays.
C1-INH and MUC7 (700 ng) were treated with rStcE′ (molar ratios, 1:20, 1:40, and 1:80) for 5 to 7 h in 20 mM Tris (pH 7.5) at 37°C. Reactions were stopped at various times with SDS-PAGE sample loading buffer containing EDTA. Proteins obtained at the different times were separated by SDS-PAGE and stained with Pro-Q Emerald 300 glycoprotein stain (Molecular Probes, Eugene, OR). Gel images were captured with a Gel-Doc transilluminator equipped with a charge-coupled device camera (Bio-Rad, Hercules, CA). Gel images were processed with the ImageQuant software to quantify protein bands. For these experiments, 1 U of product was defined as loss of 1 U of full-length substrate. Kinetic curve data were processed using the KaleidaGraph software (Synergy Software, Reading, PA) and were fitted to the integrated Michaelis-Menten equation (28): t = p/kcatE0 + (Km/kcatE0) × ln[p∞/(p∞ − p)].
The catalytic efficiencies were compared by averaging the kcat/Km ratios of three independent experiments. The Vmax was calculated using the first several times to determine the amount of turnover of substrate per minute and then dividing by the amount of protease (in micrograms). The apparent Km for each reaction was calculated by dividing the Vmax by kcat/Km.
RESULTS
StcE contains zinc but not calcium.
The StcE primary sequence contains the zinc metalloprotease consensus sequence, HEXXH (15). Other zinc proteases contain additional metals that have structural roles, most commonly calcium (10). To determine which metals were present in StcE, the metal contents of samples of rStcE′ and E435D rStcE′ diluted in either PBS or 20 mM Tris (pH 7.5) were analyzed using inductively coupled plasma mass spectrometry. The molar ratio of zinc to protein was 0.7 for rStcE′ and 0.9 for E435D rStcE′. These values were less than the expected value (1.0) probably because some protein in the preparations was misfolded or degraded. For other metals, including calcium, we did not obtain consistent molar ratios that correlated with the concentrations of rStcE′ or E435D rStcE′ compared to the buffer controls. This indicates that only zinc is associated with the StcE protease (16).
StcE is active under a variety of conditions.
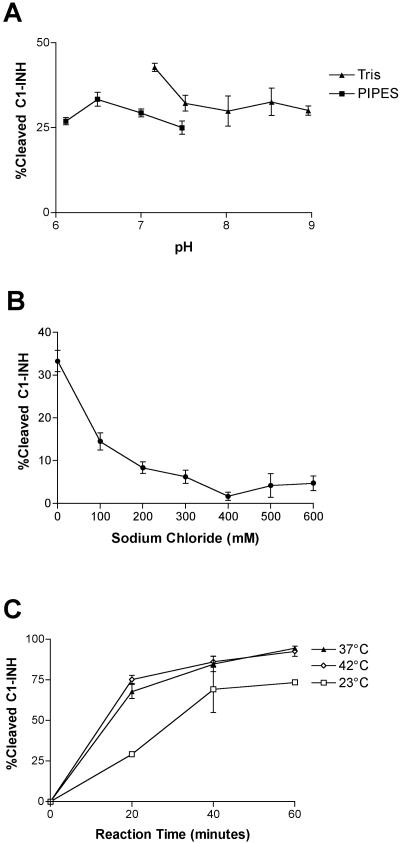
EHEC grows or persists in many different environments, including the bovine rumen and intestines, ground beef, apple cider, and the human oral cavity, stomach, and colon. We wanted to determine the range of conditions under which the StcE protease was active, as measured by C1-INH cleavage. Proteolytic activity was observed at pH 6.1 to 9.0. The optimum pH, pH 6.5 to 7.0, resulted in only 5 to 10% additional cleavage of C1-INH compared with the cleavage at other pHs (Fig. 1A). rStcE′ was most active in the absence of sodium chloride, although activity was observed with 600 mM NaCl in Tris buffer (Fig. 1B). Several repetitions of the experiment showed that the rStcE′ activity in the presence of 400 mM NaCl was lower than the rStcE′ activity in the presence of either 300 mM or 500 mM NaCl. These anomalous results may reflect some structural effect at a salt concentration of 400 mM. rStcE′ activity with C1-INH was determined at a range of temperatures. The maximum activity was observed at 37°C and 42°C, and more moderate activity was observed at 23°C (Fig. 1C). Low levels of activity were observed at 4°C and 55°C, as determined by the appearance of StcE-cleaved bands that were the size of C1-INH bands on an immunoblot (data not shown). rStcE′ was also active in water, in two tissue culture media, RPMI and Eagle's minimal essential medium supplemented with 10% fetal bovine serum, and in buffers containing detergents, such as saponin (2%), digitonin (0.5%), and SDS (0.02%) (data not shown).
FIG. 1.
Characterization of rStcE′ activity with C1-INH in various reaction conditions. The activity of rStcE′ was examined in Tris or piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (50 mM) at pH 6.1 to 9.0 (A), in the presence of various NaCl concentrations (0 to 600 mM) (B), and at various temperatures (C).
StcE activity is stable after prolonged incubation and freeze-thawing.
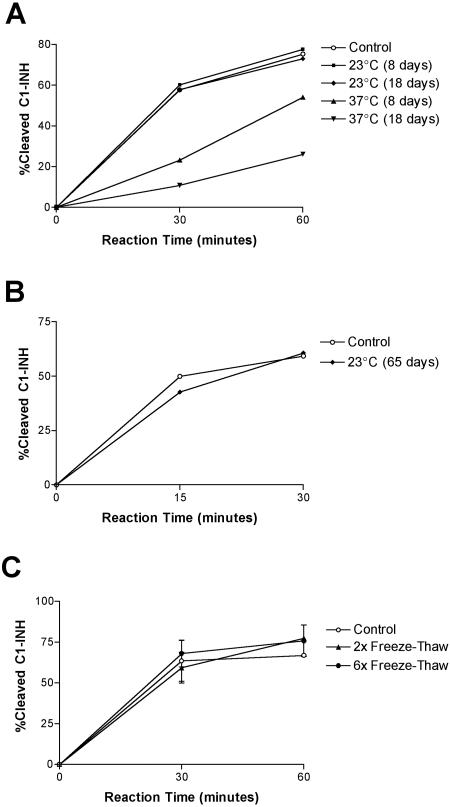
Using experiments similar to the experiments described above, we examined the practical aspects of handling the StcE protease in the laboratory setting. A test of the stability of rStcE′ protease activity produced some surprising results. Incubation for 8 or 18 days at 23°C did not result in a loss of StcE activity against C1-INH (Fig. 2A). After incubation at 37°C rStcE′ retained its 30% activity after 8 days, and after 18 days about 15% of the activity remained (Fig. 2A). Immunoblots of rStcE′ samples stored at 37°C revealed that there was significant loss of full-length StcE (data not shown). After 65 days of incubation at room temperature (23°C), rStcE′ was as active as the control protein stored at −20°C (Fig. 2B). Freezing and thawing seemed to have little effect on rStcE′ activity (Fig. 2C). Immunoblots revealed that full-length rStcE′ protein was still present after rStcE′ was stored at 23°C for 53 days and after rStcE′ was subjected to 11 cycles of freezing and thawing (data not shown).
FIG. 2.
Determination of rStcE′ activity after prolonged incubation or freeze-thaw cycles. A sample of rStcE′ in Tris buffer was incubated at room temperature (23°C) or at 37°C, and activity assays were performed with C1-INH (A and B). Additional enzyme samples were subjected to no, two, or six freeze-thaw cycles before activity assays (C). The activity of rStcE′ after various treatments was compared to the activity of control rStcE′, which was obtained from a thawed stock solution and was used immediately in the assays.
StcE retains activity after treatment with proteases.
As an enteric pathogen, EHEC faces proteolytic activity throughout its passage from the oral cavity through the colon, both from host-produced proteases and from microbial proteases. Since StcE is a secreted protein, we examined whether human or microbial proteases eliminated StcE activity.
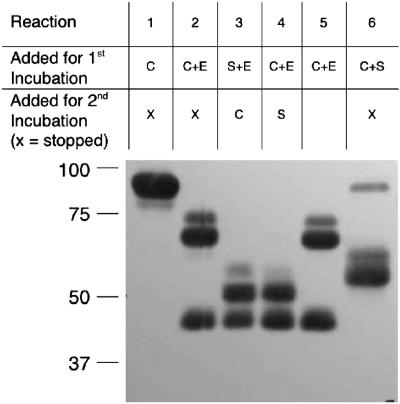
To determine whether rStcE′ activity was protease resistant, rStcE′ was treated with a protease, and then C1-INH was added to observe whether StcE-cleaved fragments that were the size of C1-INH fragments were produced. Figure 3 shows the results of these experiments for neutrophil elastase. Table 1 shows a summary of rStcE′ activity after treatment with other proteases. The incubation times and concentrations of the proteases were sufficient to degrade BSA. The reaction mixtures were separated by SDS-PAGE, blotted, and probed with antiserum against C1-INH. Treatment of C1-INH with the proteases resulted in a unique pattern of breakdown products. Conversely, rStcE′ was relatively resistant to proteolytic digestion, except for digestion with proteinase K. Addition of C1-INH to protease-treated rStcE′ resulted in C1-INH fragments different than the fragments obtained for protease-treated C1-INH, as well as some fragments whose sizes were similar to the sizes of StcE-cleaved C1-INH fragments (Fig. 3). Control reaction mixtures contained C1-INH that was treated with proteases for an extended incubation time, and the results demonstrated that the C1-INH breakdown products created by addition of rStcE′ were unique and not due to continued breakdown of C1-INH with the proteases. C1-INH cleavage activity was observed after rStcE′ was treated with any of four proteases, trypsin, chymotrypsin, neutrophil elastase, or P. aeruginosa elastase. Proteinase K, however, degraded StcE enough that proteolytic activity with C1-INH was eliminated.
FIG. 3.
Determination of rStcE′ activity with C1-INH after treatment with proteases. rStcE′ was treated with various proteases at levels that completely degraded BSA (not shown). The results of a representative experiment performed with neutrophil elastase are shown. All reaction mixtures were incubated for 2 h at 37°C; then either the reaction was stopped or there was a second 2-h incubation. Reaction 1, untreated C1-INH; reaction 2, C1-INH treated with neutrophil elastase; reaction 3, rStcE′ treated with neutrophil elastase prior to addition of C1-INH; reaction 4, C1-INH treated with elastase prior to treatment with rStcE′; reaction 5, C1-INH treated with neutrophil elastase for 4 h; reaction 6, C1-INH treated with rStcE′. All reaction mixtures were separated by SDS-PAGE, blotted, and probed with polyclonal antiserum against C1-INH. The blots were exposed for 5 to 10 times the normal time to detect all breakdown products. The molecular masses of markers (in kilodaltons) are indicated on the left. C, C1-INH; E, neutrophil elastase; S, rStcE′.
Antibodies against StcE or C1-INH do not eliminate StcE activity with C1-INH.
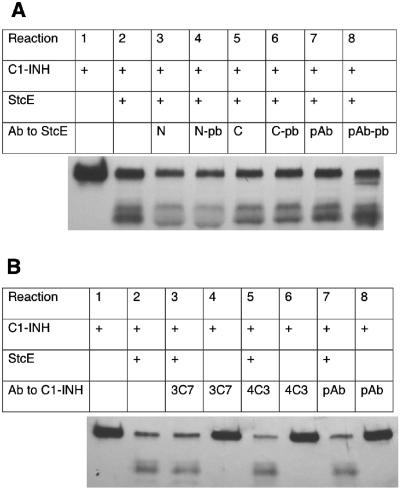
Workers in our lab demonstrated previously that StcE can be detected in the stools of EHEC-infected children (15). Another report showed not only that StcE was expressed in vivo but also that sera from convalescent-phase hemolytic-uremic syndrome patients reacted to the StcE protein (referred to as TagA) (20). Therefore, we examined whether antiserum against the StcE protein disrupts its proteolytic activity. Rabbit polyclonal antiserum against rStcE′ or against peptides derived from the protein sequence were preincubated with rStcE′. When C1-INH was added, rStcE′ was still able to cleave amounts of C1-INH similar to the amounts observed with the corresponding control sera (Fig. 4A). We then examined whether antiserum against the StcE substrate, C1-INH, inhibited the cleavage. Rabbit polyclonal antiserum and mouse monoclonal antibodies against C1-INH were preincubated with C1-INH. When rStcE′ was added to the mixture, it was able to cleave at levels comparable to the levels observed for the buffer control (Fig. 4B).
FIG. 4.
Effects of antisera and antibodies against C1-INH and rStcE′ on the ability of rStcE′ to cleave C1-INH. (A) Polyclonal antiserum (pAb) against rStcE′ (reactions 7 and 8), monospecific antiserum against the C terminus (C) of StcE (reactions 5 and 6), and monospecific antiserum against an N-terminal region (N) of StcE (reactions 3 and 4) all failed to inhibit cleavage of C1-INH by rStcE′ compared to the control sera (pAb-pb, C-pb, and N-pb [reactions 4, 7, and 8, respectively]). (B) Polyclonal antiserum (pAb) against C1-INH (reaction 7), monoclonal antibody 3C7 (reaction 3), and monoclonal antibody 4C3 (reaction 5) did not inhibit rStcE′ activity, as measured by cleaved C1-INH fragments compared to a control (reaction 2). Control (reactions 4, 6, and 8) did not show proteolytic activity in the absence of rStcE′. Ab, antibody.
StcE does not cleave MUC2 or MUC5AC.
Our previous results demonstrated that StcE cleaves heavily glycosylated, mucin-like proteins (12). Here we tested StcE activity with other mucins present in the gastrointestinal tract. Cell fractions from two mucin-producing cell lines, Caco-2 and LS174T, were treated with rStcE′ or buffer, separated by agarose gel electrophoresis, and blotted. The same immunoblot was probed with antibodies against MUC2, a major colonic mucin, and MUC5AC, a major salivary mucin. No breakdown products or differential banding was apparent in either experiment, demonstrating that rStcE′ does not cleave MUC2 or MUC5AC produced by the Caco-2 and LS174T cell lines (data not shown).
StcE does not cleave α1 acid glycoprotein.
Like C1-INH, α1 acid glycoprotein (AGP) is an acute-phase glycoprotein found in the serum that is thought to play a role as an anti-inflammatory agent and immunomodulating regulator (for a review see reference 7). AGP has a molecular mass of 41 to 43 kDa, and 45% of its molecular mass is in the form of five or six sialylated N-linked glycans (23). The physical and functional similarities of AGP and C1-INH suggested that AGP might be a substrate for StcE. AGP and C1-INH were immobilized on nitrocellulose and probed with rStcE′, and only C1-INH was bound by rStcE′ (data not shown). StcE-treated AGP was the same size as buffer-treated AGP, as determined by SDS-PAGE (data not shown). Together, these data show that StcE neither binds nor cleaves AGP.
StcE activity with MUC7 is twofold more efficient than StcE activity with C1-INH.
Proteases often exhibit different kinetics with different substrates. We examined whether StcE activity with the complement regulator C1-INH differed from its interaction with MUC7, a secreted mucin. Assays of rStcE′ activity with MUC7 and C1-INH were performed using a time course format, and the results were analyzed to determine several kinetic parameters. The Vmax revealed the maximum speed of cleavage under the conditions tested per mass of protease. The kcat/Km ratio measured the efficiency of cleavage, taking into account both binding and turnover events. From these data, the apparent Km for reactions was calculated, and the results provided insight into binding events.
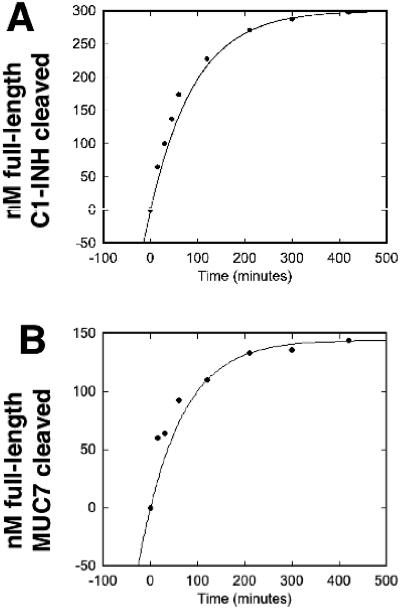
As determined using activity curves (Fig. 5 A and B), the kcat/Km for the StcE-MUC7 interaction was 2.61 ×104 M−1 s−1, while the kcat/Km for the StcE-C1-INH interaction was 1.16 ×104 M−1 s−1. The average ratio of the kcat/Km for MUC7 to the kcat/Km for C1-INH for three independent experiments was 2.5 ± 0.3, indicating that the StcE protease cleaved the MUC7 substrate more efficiently. The Vmax values for StcE with C1-INH and MUC7 were 66.8 and 70.2 nM/min/μg, respectively (Table 2), indicating that the rates of cleavage were similar. The Km was calculated to be 0.13 μM for MUC7 and 0.27 μM for C1-INH, revealing that the StcE interaction with MUC7 has a twofold-greater affinity than the StcE interaction with C1-INH.
FIG. 5.
rStcE′ interaction with C1-INH and MUC7. Samples obtained at various times during cleavage reactions of rStcE′ with C1-INH or MUC7 were separated by SDS-PAGE and stained with Pro-Q Emerald 300 glycoprotein stain. The full-length substrate bands were imaged and used to calculate progress curves for cleavage of C1-INH (A) and MUC7 (B).
TABLE 2.
Kinetic parameters for activity of StcE with C1-INH and MUC7a
| Parameter | C1-INH (rep.) | MUC7 (rep.) | Ratio (rep.) | Avg ratio (mean ± SD; n = 3)b |
|---|---|---|---|---|
| kcat/Km (M−1 s−1) | 1.16 × 104 | 2.61 × 104 | 2.25 | 2.5 ± 0.3 |
| Vmax (nM/min/μg) | 66.8 | 70.2 | 1.05 | 1.1 ± 0.3 |
The data were fitted to a modified Michaelis-Menten curve, allowing interpolation of kcat/Km and Vmax values for the representative reactions (rep.) shown in Fig. 5.
Average ratios (three independent trials) of kcat/Km for MUC7 to kcat/Km for C1-INH and of Vmax for MUC7 to Vmax for C1-INH.
DISCUSSION
We tried to further characterize the activity of the StcE protease and to test hypotheses derived from a model for the role of StcE in EHEC pathogenesis. The data show that StcE is a robust enzyme that is resistant to proteolysis by other proteases, stable for many weeks at 23°C, stable for days at 37°C, and stable after repeated freeze-thaw cycles. StcE was active in buffers at pH 6.1 to 9.0, in the presence of NaCl concentrations ranging from 0 to 600 mM, at temperatures ranging from 4°C to 55°C, and in the presence of low concentrations of detergents. Additionally, polyclonal antisera against StcE and a substrate, C1-INH, did not inhibit the cleavage of C1-INH by StcE. These data provide strong evidence that StcE could be active in various environmental or colonization conditions.
As a protein released in the oral and intestinal environments during infection, the StcE protease may be susceptible to degradation by both host and microbial proteases. Our laboratory found that StcE is present in sterile-filtered stools of EHEC-infected children, proving that StcE is present extracellularly during EHEC infection (15). Experiments in the present study demonstrated that, with the exception of the fungal protease proteinase K (22), StcE retained its protease activity after treatment with other proteases, as determined by its ability to cleave C1-INH. Figure 3 shows that StcE was resistant to a variety of proteases, even though it contained numerous cleavage sites for each of these proteases. For example, trypsin cleaves its substrates after positively charged residues, chymotrypsin cleaves after large hydrophobic residues, and elastase cleaves after small hydrophobic residues (21). Protease-cleaved StcE fragments that retain activity may prove to be useful for identifying structure/function relationships of the StcE protease.
Not only is StcE present in stools during colitis (15), but other laboratories have shown that people with EHEC infections produce antiserum against StcE (referred to as TagA by Paton and Paton [20] and by Zhang et al. [27]). The finding that polyclonal antiserum against StcE or against C1-INH did not eliminate StcE activity is intriguing. The data suggest that immunogenic regions of StcE are not present in regions of the protein involved in substrate binding or proteolysis. It would be interesting, however, to examine whether convalescent patient sera could inhibit StcE activity, since these antisera might be produced in the conditions present during normal EHEC infections.
Little is known about the three-dimensional composition of the StcE protease. Therefore, a metal analysis was performed to begin a rudimentary analysis of StcE structure. The data indicated that StcE contains one zinc atom per protein, likely carried at the histidine-rich zinc consensus sequence in the presumed active site. StcE did not contain structural calcium, which is often associated with other metalloproteases. Importantly, the proteolytically inactive mutant E435D rStcE′ contained a zinc atom. When E435D rStcE′ was treated with neutrophil elastase, the breakdown products were similar to those obtained for rStcE′ (data not shown). Like rStcE′, E435D rStcE′ was able to localize C1-INH to cell membranes (unpublished data). Therefore, the E435D mutation in the StcE protease did not affect the structural integrity and functionality in terms of zinc binding and the ability to bind substrate and associate with cells.
The characterization of StcE protease in this report places StcE in the context of other endopeptidases. StcE is a neutral zinc metalloprotease that lacks structural calcium and is active against C1-INH in the pH range tested (pH 6.1 to 9.0). Unlike other proteases, such as those in the thermolysin family (13, 22), StcE does not have a propeptide, nor is it highly thermostable. It was inactive at temperatures above 60°C. Similar to proteases used commercially in stain removers and laundry detergents (21), StcE remained active in solutions containing detergents. This finding is biologically relevant because bile secreted into the intestines acts like a detergent (24). The fact that there was StcE activity after repeated freeze-thaw cycles and extended incubation (Fig. 2) provided evidence that StcE is a stable protease. The findings that StcE activity was resistant to proteolysis, resistant to antibody interference, and active in various buffers and detergents show that StcE could be active under the physiological conditions in patients with enterohemorrhagic colitis.
The model for dual roles of StcE in EHEC infection (12) was supported by the data showing that MUC7 was a more efficient substrate for StcE than C1-INH was. The model asserts that StcE activity with mucins is important in the oral cavity to evade aggregation by first-line defense molecules, such as glycoproteins gp340 and MUC7. Also, StcE mediates the ability of EHEC to adhere intimately to intestinal epithelial cells, an essential step in colonization, by promoting penetration of EHEC through the mucin barrier lining the intestines. This activity may contribute to the low infectious dose that is a hallmark of EHEC pathogenesis. After the bacteria adhere to host cells intimately via the characteristic A/E lesions, the infection progresses. The Shiga toxins and other bacterial effectors compromise the intestinal barrier and associated endothelium, allowing complement factors carried by the blood and serum to enter the intestinal lumen. StcE then mediates binding of C1-INH to bacterial and host cell surfaces, protecting them from complement-mediated lysis. In the bovine host the mucinase activity of StcE may provide a selective advantage to EHEC by increasing the ability of the bacteria to intimately adhere to and colonize host cells within the terminal rectum.
To establish colonization, EHEC needs to rapidly evade innate immune responses, such as mucin aggregation, and needs to quickly traverse the mucus layer of the intestines to colonize the host. StcE activity against MUC7, a secreted mucin, might therefore be more efficient than the StcE interaction with C1-INH. C1-INH, which occurs at high levels in serum, likely floods the intestinal lumen during colitis. Less efficient cleavage of C1-INH may explain the previous finding that StcE localization of C1-INH to cell membranes resulted in reduced complement-mediated lysis (14). The in vitro StcE kinetic data support this model. The Vmax values for the two substrates were similar, although low, indicating that cleavage of each substrate molecule took approximately 5 min. The kcat/Km results revealed that StcE activity was more efficient against MUC7, meaning that each interaction of StcE and MUC7 was 2.5-fold more likely to produce cleavage than each StcE interaction with C1-INH. The calculated Km showed that StcE might bind MUC7 more tightly than C1-INH, although both values were surprisingly low compared to the values for other protease-substrate interactions, suggesting that StcE has a high affinity for its substrates.
These data suggest that StcE may act as an adhesin as well as a protease. This interpretation is similar to the hypothesis of Bäckhed et al. that the glycoside hydrolases and other glycan-utilizing enzymes of Bacteroides thetaiotaomicron may function as adhesive structures, since the genome of this organism lacks obvious pilin-like sequences (1). The enzymes purportedly provide effective adherence in their glycan environment.
In our initial description of the StcE protease, we found that it stably bound C1-INH that had been immobilized on a polyvinylidene difluoride membrane. The high affinity of StcE for its substrates explains why this interaction could be discovered. Since the discovery that C1-INH is an StcE substrate, we have searched for additional proteins with which StcE might interact. In this study, we identified several proteins that exhibit functional and physical similarities to known StcE substrates and examined whether they were also substrates for StcE proteolysis. For instance, antibodies against MUC2 and MUC5AC were used to examine whether StcE cleaved mucins in cell lysates, but both assays were negative for cleavage (Table 3). Mucins are produced heterogeneously, even compared to cells from the same tissue of an individual. This is due to alternative splicing, as well as varied glycosylation events (8). In addition, cancer cell lines are known to have differential glycosylation that may affect protein-protein interactions (3-5). Thus, although the mucins tested in this study were not cleaved, it is still possible that mucins produced in other types of tissue or other individuals could be susceptible to StcE cleavage. Another protein, AGP, shares physical and functional characteristics with C1-INH, but it was not cleaved by StcE (Table 3). The lack of activity with MUC2, MUC5AC, and AGP continues to support the contention that StcE is a specific protease.
TABLE 3.
Proteins tested for StcE cleavage activity
| Protein | StcE substrate | Reference |
|---|---|---|
| C1-INH | + | 15 |
| MUC7/MG2 | + | 12 |
| gp340/DMBT1/salivary agglutinin | + | 12 |
| α1 Acid glycoprotein | − | This study |
| MUC2 | − | This study |
| MUC5AC | − | This study |
| MUC5B | − | 11 |
| α1-Antitrypsin | − | 15 |
| α2-Antiplasmin | − | 15 |
| α1-Antichymotrypsin | − | 15 |
| Casein | − | 15 |
| Porcine mucin | − | 12 |
| Bovine fetuin | − | 12 |
| Bovine submaxillary mucin | − | 12 |
StcE, whose molecular mass is 95 kDa, is a large protein, unlike most metalloproteases, which generally have molecular masses of 25 to 60 kDa. One exception is lethal factor of Bacillus anthracis, which is a specific zinc metalloprotease with a molecular mass of approximately 90 kDa. The crystal structure revealed four separate domains (19). We hypothesized that StcE may contain domains other than the protease active site. The other domains could be important for substrate binding, glycan interaction, or the ability of StcE to associate with cells. Recent genome sequencing projects have revealed putative open reading frames encoding proteins with amino acid sequences similar to those of StcE. For instance, the genome of Photobacterium profundum (26) and DNA isolated from Sargasso Sea samples (25) both encode StcE-like sequences. The new StcE-like sequences are in addition to the previously identified putative homologues TagA and TagA-related protein of Vibrio cholerae and Vibrio vulnificus. It will be interesting if other members of the proposed StcE-like metalloprotease (SLiMe) family have similar activities or have diverged enough to have unique activities. In this report we characterize in vitro activities of the StcE protease and provide a reference point for characterization of other SLiMe family members.
Acknowledgments
This work was supported by NIH grant ROI AI051735. T.E.G. received support from the Biotechnology Training Program funded by NIH grant 5T32GM08349.
T.E.G. thanks J. Denu and C. Berndsen for valuable discussions.
REFERENCES
- 1.Bäckhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 2.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corfield, A. P., and C. Paraskeva. 1993. Secreted mucus glycoproteins in cell and organ culture. Methods Mol. Biol. 14:211-223. [DOI] [PubMed] [Google Scholar]
- 4.Corfield, A. P., D. Carroll, N. Myerscough, and C. S. Probert. 2001. Mucins in the gastrointestinal tract in health and disease. Front. Biosci. 6:D1321-D1357. [DOI] [PubMed] [Google Scholar]
- 5.Corfield, A. P., N. Myerscough, A. W. Einerhand, B. J. Van Klinken, J. Dekker, and C. Paraskeva. 2000. Biosynthesis of mucin cell and organ culture methods for biosynthetic study. Methods Mol. Biol. 125:219-226. [DOI] [PubMed] [Google Scholar]
- 6.de Agostini, A., M. Schapira, Y. T. Wachtfogel, R. W. Colman, and S. Carrel. 1985. Human plasma kallikrein and C1 inhibitor form a complex possessing an epitope that is not detectable on the parent molecules: demonstration using a monoclonal antibody. Proc. Natl. Acad. Sci. USA 82:5190-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fournier, T., N. Medjoubi-N, and D. Porquet. 2000. Alpha-1-acid glycoprotein. Biochim. Biophys. Acta 1482:157-171. [DOI] [PubMed] [Google Scholar]
- 8.Fowler, J., L. Vinall, and D. Swallow. 2001. Polymorphism of the human muc genes. Front. Biosci. 6:D1207-D1215. [DOI] [PubMed] [Google Scholar]
- 9.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 10.Gomis-Rüth, F. X. 2003. Structural aspects of the metzincin clan of metalloendopeptidases. Mol. Biotechnol. 24:157-202. [DOI] [PubMed] [Google Scholar]
- 11.Grys, T. E., M. J. Rock, and R. A. Welch. Unpublished data.
- 12.Grys, T. E., M. B. Siegel, W. W. Lathem, and R. A. Welch. 2005. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 73:1295-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häse, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lathem, W. W., T. Bergsbaken, and R. A. Welch. 2004. Potentiation of C1 esterase inhibitor by StcE, a metalloprotease secreted by Escherichia coli O157:H7. J. Exp. Med. 199:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lathem, W. W., T. E. Grys, S. E. Witowski, A. G. Torres, J. B. Kaper, P. I. Tarr, and R. A. Welch. 2002. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol. Microbiol. 45:277-288. [DOI] [PubMed] [Google Scholar]
- 16.Mason, A. Z., S. D. Storms, and K. D. Jenkins. 1990. Metalloprotein separation and analysis by directly coupled size exclusion high-performance liquid chromatography inductively coupled plasma mass spectroscopy. Anal. Biochem. 186:187-201. [DOI] [PubMed] [Google Scholar]
- 17.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naylor, S. W., A. J. Roe, P. Nart, K. Spears, D. G. Smith, J. C. Low, and D. L. Gally. 2005. Escherichia coli O157:H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 151:2773-2781. [DOI] [PubMed] [Google Scholar]
- 19.Pannifer, A. D., T. Y. Wong, R. Schwarzenbacher, M. Renatus, C. Petosa, J. Bienkowska, D. B. Lacy, R. J. Collier, S. Park, S. H. Leppla, P. Hanna, and R. C. Liddington. 2001. Crystal structure of the anthrax lethal factor. Nature 414:229-233. [DOI] [PubMed] [Google Scholar]
- 20.Paton, A. W., and J. C. Paton. 2002. Reactivity of convalescent-phase hemolytic-uremic syndrome patient sera with the megaplasmid-encoded TagA protein of Shiga toxigenic Escherichia coli O157. J. Clin. Microbiol. 40:1395-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao, M. B., A. M. Tanksale, M. S. Ghatge, and V. V. Deshpande. 1998. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 62:597-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings, N. D., D. P. Tolle, and A. J. Barrett. 2004. MEROPS: the peptidase database. Nucleic Acids Res. 32:D160-D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmid, K., R. B. Nimerg, A. Kimura, H. Yamaguchi, and J. P. Binette. 1977. The carbohydrate units of human plasma alpha1-acid glycoprotein. Biochim. Biophys. Acta 492:291-302. [DOI] [PubMed] [Google Scholar]
- 24.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 26.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, Z. Y., G. Zhou, J. M. Denu, L. Wu, X. Tang, O. Mondesert, P. Russell, E. Butch, and K. L. Guan. 1995. Purification and characterization of the low molecular weight protein tyrosine phosphatase, Stp1, from the fission yeast Schizosaccharomyces pombe. Biochemistry 34:10560-10568. [DOI] [PubMed] [Google Scholar]