Abstract
In the gammaproteobacteria the RpoH regulon is often equated with the stress response, as the regulon contains many of the genes that encode what have been termed heat shock proteins that deal with the presence of damaged proteins. However, the betaproteobacteria primarily utilize the HrcA repressor protein to control genes involved in the stress response. We used genome-wide transcriptional profiling to compare the RpoH regulon and stress response of Neisseria gonorrhoeae, a member of the betaproteobacteria. To identify the members of the RpoH regulon, a plasmid-borne copy of the rpoH gene was overexpressed during exponential-phase growth at 37°C. This resulted in increased expression of 12 genes, many of which encode proteins that are involved in the stress response in other species. The putative promoter regions of many of these up-regulated genes contain a consensus RpoH binding site similar to that of Escherichia coli. Thus, it appears that unlike other members of the betaproteobacteria, N. gonorrhoeae utilizes RpoH, and not an HrcA homolog, to regulate the stress response. In N. gonorrhoeae exposed to 42°C for 10 min, we observed a much broader transcriptional response involving 37 differentially expressed genes. Genes that are apparently not part of the RpoH regulon showed increased transcription during heat shock. A total of 13 genes were also down-regulated. From these results we concluded that although RpoH acts as the major regulator of protein homeostasis, N. gonorrhoeae has additional means of responding to temperature stress.
A fundamental process in all cells involves the removal of aberrant proteins, generally misfolded or aggregated protein complexes formed during cellular stress. The response to this stress involves stabilization of protein conformations, refolding of misfolded proteins, and degradation of proteins that are detrimental to the cell. These processes are performed by a variety of molecular chaperones and proteases located in different compartments of the cell. In bacteria, the main cytoplasmic components involved in protein homeostasis include the chaperones DnaK/DnaJ/GrpE, GroEL/GroES, HtpG, and ClpB, as well as the proteases ClpXP, ClpAP, HslUV, Lon, and FtsH (32). In Escherichia coli and other members of the gamma subdivision of the Proteobacteria, RpoH is the major regulator of the genes encoding these factors (32). However, members of the betaproteobacteria, such as Bordetella and Burkholderia, utilize a repressor, HrcA, which recognizes inverted repeats (CIRCE [controlling inverted repeat of chaperone expression]) in the promoter regions of many heat shock-regulated genes (29). Although Neisseria belongs to the betaproteobacteria, it is the exception in this group as in silico searches of the completed genomes of Neisseria gonorrhoeae and Neisseria meningitidis have revealed that neither a gene encoding an HrcA homolog nor CIRCE elements are present (29). An RpoH homolog, which appears to be essential for cell viability in Neisseria (24), exhibits elevated expression levels during heat stress in N. meningitidis (14) and N. gonorrhoeae (24). In N. gonorrhoeae RpoH appears to be responsible for the regulation of dnaK, dnaJ, and grpE (24), as well as the groE operon (43).
In E. coli grown at normal temperatures, low levels of RpoH (intracellular levels of less than 50 molecules per cell) are necessary for the transcription of chaperones involved in refolding of newly synthesized proteins (32). However, during heat shock rpoH transcription is rapidly increased. This, in turn, can result in increased transcription of the genes encoding the chaperones and proteases. However, this control of rpoH at the transcriptional level plays a relatively minor role in maintaining the RpoH levels in the cell (1). The regulation of RpoH is primarily at the posttranscriptional level, where translation of the protein is repressed by secondary structure in the rpoH mRNA (24, 47). Additionally, the DnaK chaperone system can influence the stability and activity of the protein (32). At normal temperatures, the small amounts of RpoH can be sequestered by DnaK/DnaJ/GrpE and targeted for degradation by FtsH, Lon, HslVU, or ClpP (1) or by GroES/EL (15). During heat shock, both the DnaK/DnaJ/GrpE and GroES/EL chaperone complexes preferentially bind misfolded proteins, releasing RpoH, which is then free to associate with RNA polymerase and direct transcription from RpoH-dependent promoters. These promoters control expression of RpoH itself, as well as the chaperones and proteases involved in protein homeostasis. Once the misfolded proteins are removed, RpoH is once again bound by the chaperone complexes, and the levels of transcription of the genes encoding the chaperones and RpoH return to normal.
In N. gonorrhoeae transcription of the chaperone genes grpE, dnaK, and dnaJ (24), as well as groESL (43), is induced from the corresponding RpoH-dependent promoters within the first 10 min of exposure to elevated temperatures. However, transcription of rpoH during heat shock is from a sigma-70-dependent promoter and increases only after 20 min of exposure to elevated temperatures. This suggests that in N. gonorrhoeae activation of preformed RpoH is sufficient to respond to a heat shock (24). In accordance with this suggestion, evaluation of the heat shock response of N. meningitidis at 45°C for 5 min has demonstrated that many genes, but not rpoH, are up-regulated (14).
The objective of this study was to investigate the RpoH regulon of N. gonorrhoeae and how this regulon overlaps with the heat shock regulon. To do this, we overexpressed RpoH from a neisserial plasmid, performed transcriptional profiling experiments to identify members of the RpoH regulon, and compared the genes with the genes induced in response to heat shock. The results suggest that RpoH acts as the major regulator of protein homeostasis during heat stress in N. gonorrhoeae, but this species also has additional means of responding to this stress.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The E. coli strain used in all cloning experiments was DH5α [F− endA1 thil-1 hsdR17 supE44 relA1 ΔlacU169 (φ80ΔlacZΔM15)]. The N. gonorrhoeae strain used was MS11-A (36). E. coli strains were routinely grown at 37°C in Luria-Bertani (LB) broth (Difco) or on LB plates supplemented with 1.5% (wt/vol) agar. When necessary, LB media were supplemented with 150 μg/ml erythromycin. Gonococcal strains were routinely cultured on GC agar base (Oxoid) or GC broth supplemented as described previously (11). When appropriate, GC agar plates were supplemented with 7 μg/ml erythromycin, 12.5 μg/ml tetracycline, or 40 μg/ml spectinomycin. Plate cultures were incubated in a 5% CO2 atmosphere, whereas GC broth was supplemented with 1% (wt/vol) NaHCO3 as a CO2 source. Gonococcal transformations and conjugations were performed as described previously (24), except that conjugations were performed by mixing 5 × 108 donor cells and 1.5 × 109 recipient cells.
Recombinant DNA techniques.
All DNA manipulations were performed by standard procedures (11). Plasmid DNA was purified using a Hi PURE plasmid isolation kit from Roche Diagnostics. The restriction endonucleases used in this study were purchased from New England Biolabs. DNA sequencing was performed with a BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems), and DNA was analyzed with an Applied Biosystems model 3730 DNA analyzer. Oligonucleotide primers (Table 1) were synthesized with an Applied Biosystems 394 oligonucleotide synthesizer.
TABLE 1.
Oligonucleotide primers used in this study
| Oligonucleotide | Sequence (5′-3′) | Usea |
|---|---|---|
| 1729 | CAGTTCGGCAATGAAGTCCG | groEL probe |
| 3260 | CACACTGGGACTGAGACATG | 16S rRNA probe |
| 3261 | CGGCAGTCTCATTAGAGTGC | 16S rRNA probe |
| 11076 | GACACCCAAATCGTCGGC | rpoH probe |
| 11077 | CGGCTATGACGGCTACGGG | rpoH probe |
| 14107 | GGACAGCGTATAACCCTTCTTC | grpE probe |
| 14108 | ATGAGCGAACAGACACAGCAG | grpE probe |
| 14109 | CTTGCAGTACGTGGATGGTTAC | dnaK probe |
| 14110 | CAAGACGTGATGGCTCTACAAC | dnaK probe |
| 14115 | GTGAGGACGTTCAAGTCGGTAT | dnaJ probe |
| 14116 | TGACACCCTTACCCTTCACG | dnaJ probe |
| 14308 | ATGCCTCGGATGTTGTTCAC | groEL probe |
| 15837 | CAGCAAACCGCGGATCCGCAGTTTC | Amplification of rpoH gene |
| 16764 | GCTGCCGTCCATTTTCATG | qRT-PCR of recA |
| 16766 | TGGCGCAAATCGAAAAAAGT | qRT-PCR of recA |
| 17456 | GACGGATAGGATCCTGTACAGCAC | Amplification of rpoH gene |
| 23047 | GTCGGCAAGCCAGTCGAT | qRT-PCR of rpoH |
| 23048 | CCGAAAAAGCCAAAATCGAA | qRT-PCR of dnaK |
| 23049 | ACCGGAAGCTATGGACAAAATC | qRT-PCR of clpB |
| 23050 | TGGTAATGTACGGCAGGTTGAT | qRT-PCR of dnaK |
| 23052 | TCGCCTTCCTGCCATTCA | qRT-PCR of marR |
| 23053 | ACCGTTTCAGGCGTATGCA | qRT-PCR of marR |
| 23054 | CGCCATCATGGCAGACAAC | qRT-PCR of rpoH |
| 23055 | TTTCAACGTGCGCCTTTTC | qRT-PCR of clpB |
| 23273 | CACGCGCCGCAAATCT | qRT-PCR of secB |
| 23274 | GGTGGAAACGCGCATATCC | qRT-PCR of secB |
qRT-PCR, quantitative real-time RT-PCR.
RNA isolation.
To prepare total RNA, 10 ml of a culture was harvested, immediately mixed with 20 ml of RNAlater RNA stabilization solution (Ambion), and incubated at room temperature for 10 min. Prior to RNA isolation cells were harvested by centrifugation (5,000 × g) for 10 min at room temperature. RNA was isolated using an RNeasy midi kit (QIAGEN) with an on-column DNase I digestion step. The quality and quantity of RNA were determined by gel electrophoresis and spectrophotometry.
RNA dot blotting.
RNA dot blotting was carried out as described previously (4) with 1 μg and 5 μg of total RNA. Probe labeling was performed using a digoxigenin nonradioactive DNA labeling kit (Roche Diagnostics) according to the manufacturer's instructions. The probes used were a 830-bp PCR product amplified from 16S rRNA and PCR products amplified from rpoH (300 bp), dnaK (570 bp), grpE (530 bp), dnaJ (590 bp), and groEL (790 bp). The oligonucleotide primers used for each PCR are described in Table 1. Hybridization of probe DNA to target RNA was detected using the method supplied with the chemiluminescent substrate CDP-Star (Roche Diagnostics).
cDNA synthesis and labeling.
For each synthesis reaction, 30 μg of total RNA was mixed with 30 μg of random hexamers, heated to 70°C for 10 min, and then rapidly chilled on ice. To this mixture 0.5 μl (20 U) of RNasin (Promega), 6 μl of Superscript II buffer (Life Technologies, Inc.), 3 μl of dithiothreitol, 1 mM dATP, 1 mM dCTP, 1 mM dGTP, 0.4 mM dTTP (Promega), 0.6 mM aminoallyl-dUTP (Sigma), and 2 μl (400 U) of Superscript II reverse transcriptase (Life Technologies, Inc) were added and incubated for 2.5 h at 42°C. The reactions were terminated, and the RNA was hydrolyzed by addition of 10 μl of 1 M NaOH and 10 μl of 0.5 M EDTA, heated to 65°C for 15 min, and neutralized by addition of 25 μl of 1 M Tris-HCl buffer (pH 7.4). Aminoallyl-modified cDNA was purified using Microcon 30 (Millipore) purification columns as follows. The cDNA mixture was diluted in 400 μl of diethyl pyrocarbonate (DEPC)-treated water, transferred to a column, and centrifuged at 12,000 × g for 7 min, and the column was washed four times with 400 μl of DEPC-treated water and centrifuged at 12,000 × g for 7 min. Purified labeled cDNA was collected according to the manufacturer's recommendations and concentrated to 12 μl, and the quantity was determined using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies). Then 2.8 μl of sodium bicarbonate (0.5 M, pH 9) was added to each cDNA preparation prior to coupling of Cy3 or Cy5 fluorophores. One-microgram portions of the dyes Cy3 and Cy5 (Amersham) were dissolved in 72 μl of dimethyl sulfoxide (Sigma), and a 4-μl aliquot of the relevant dye was added to the appropriate cDNA sample. The reaction mixtures were incubated for 1 h at room temperature in the dark and were purified using Microcon 30 columns as described above, and the eluted samples were concentrated to 10 μl. Cy3 and Cy5 dye incorporation was determined using spectrophotometry.
Microarray hybridization.
The labeled cDNA was hybridized to a pan-Neisseria microarray which contained 2,704 PCR products spotted in triplicate, corresponding to potential coding sequences from N. gonorrhoeae strain FA1090 (7), N. meningitidis strains MC58 (44) and Z2491 (28), N. gonorrhoeae strain MS11 Gonococcal Genetic Island (19), and various controls. The nucleotide sequence of the genome of N. gonorrhoeae strain FA1090 (GenBank accession number AE004969) has not been formally published, but annotations are available at the following sites: www.stdgen.lanl.gov, pathema.tigr.org/tigr-scripts/CMR/CmrHomePage.icg, and www.ncbi.nlm.nih.gov/entrez/query.fcgi. The annotation used for microarray design and the annotation submitted to GenBank can be compared at gbrowse.molbiol.ox.ac.uk/cgi-bin/gbrowse/FA1090. All gene identification numbers used in this paper correspond to the numbers in the GenBank annotation. Details of the construction of this microarray will be published elsewhere, but details and microarray slides can be obtained from the corresponding author. Prior to prehybridization, each slide was plunged into a 95°C water bath for 2 min, centrifuged for 5 min at 2,000 × g, and used immediately. Prehybridization was carried out in a 30-μl mixture containing 25% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 10 mg/ml bovine serum albumin (fraction V), and 30 μg of herring sperm DNA (Promega) under a coverslip in a humidified Corning CMT hybridization chamber (Corning) for 45 min at 42°C. The slide was rinsed in water, dried by centrifugation, and used immediately for hybridization. The labeled cDNA was added to the hybridization fluid (prehybridization solution without bovine serum albumin) in a 30-μl (total volume) mixture and denatured at 95°C for 5 min. The hybridization fluid was placed on the surface of the microarray under a coverslip. The slide was placed in the hybridization chamber and submerged in a 42°C water bath overnight. After hybridization, slides were washed once in 2× SSC-0.1% SDS for 5 min at 42°C, once in 0.1× SSC-0.1% SDS for 10 min at room temperature, and four times in 0.1× SSC for 1 min at room temperature. The slides were rinsed with water for 10 s, and excess fluid on the surface of a slide was removed by centrifugation before scanning.
Data collection and analysis.
A GMS 418 array scanner (Genetic Microsystems) was used to measure the fluorescence of the Cy3- and Cy5-labeled cDNA hybridized to the microarray. The images were combined, and quantitation of the fluorescent and background intensities for each spot was performed using the ImaGene version 5 software (BioDiscovery) as described previously (5). Data from poor spots that were manually or automatically flagged in ImaGene were not used for further analysis. The individual ImaGene data files were loaded onto a website created with BASE (33) and were converted to a common BASE format using a custom-made application. Each data set consisted of the data from two biological repeat experiments, each of which included a technical repeat with a dye swap. The spot intensities were found to be most reliable when no background correction was performed. Analysis was done using Bioconductor (9) and Limma (39). Normalization of the data to remove various biases involved two steps. First, each array was normalized independently using print tip Loess normalization (Y. H. Yang, S. Dudoit, P. Luu, and T. P. Speed, presented at SPIE BiOS 2001, San Jose, Calif., 2001). Second, diagnostic plots suggested that there was variation in scale between arrays, so the log ratios were scaled in such a way that each array had the same median for the absolute deviation. The normalized data were then used to fit a linear model (39) for each gene using generalized least squares, which took into account the correlation between replicate spots (40). The coefficient of the fitted model for each gene describes the inferred difference in RNA expression between the two strains. Empirical Bayesian methods were then used to calculate the moderated t statistics and associated P values. The P values were adjusted for multiple testing using a false discovery rate. Genes that had an absolute ratio of more than 1.5-fold and were significantly different at the 0.001 level were considered differentially expressed.
Quantitative real-time RT-PCR.
cDNA was generated from 5-μg portions of the same RNA preparations used for the microarray experiments described above, except that the concentration of each deoxynucleoside triphosphate was 1 mM and 7.8 μg of random hexamers was used. In addition, a second DNase I treatment was used prior to cDNA synthesis. Specific primer pairs were designed using the ABI PRISM Primer Express software (Applied Biosystems) and are shown in Table 1. To quantitate cDNA, the gene-specific standard curve method was employed, using serial dilutions of MS11-A genomic DNA as the templates. All assay mixtures included 12.5 μl of SYBER Green PCR master mixture (Applied Biosystems), 2 μl of each primer (final concentration, 0.5 nM), 2 μl of template (cDNA was diluted 50- to 100-fold prior to quantitative reverse transcriptase [RT] PCR), and enough DEPC-treated water so that the final volume was 25 μl. The negative controls lacked reverse transcriptase or template. The reactions were performed with an ABI 7700 sequence detection system (Applied Biosystems), and recA was used for normalization of all reactions. All RT-PCR described below amplified a single product, as determined by melting curve analysis (Applied Biosystems).
RESULTS
System for identification of genes in the RpoH regulon of N. gonorrhoeae.
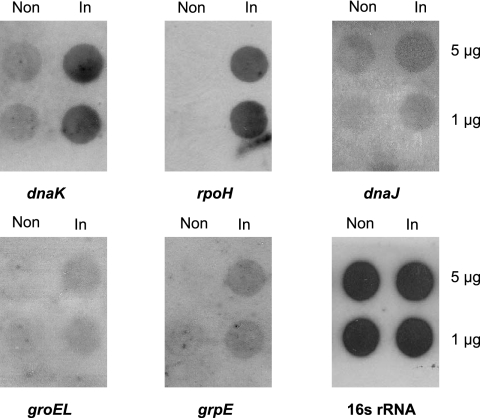
Previous work has suggested that the rpoH gene is essential for the viability of Neisseria (24). We therefore decided to overexpress RpoH by induction of a plasmid-borne copy of this gene during the exponential phase of growth at 37°C. We reasoned that an increase in the level of RpoH would lead to an increase in the level of the corresponding RpoH-associated RNA polymerase holoenzyme, which would in turn lead to an increase in the expression of the rpoH regulon. To control the expression of rpoH in the gonococcus, we cloned the entire gene into the Hermes-8 E. coli/N. gonorrhoeae shuttle plasmid (23). This shuttle vector harbors an inducible Ptrc promoter that is functional in Neisseria and DNA integration regions from the large neisserial conjugative plasmid ptetM25.2 that allows recombination between the incoming Hermes shuttle vector and a resident ptetM25.2 plasmid in Neisseria. We used oligonucleotide primers (Table 1) that allowed introduction of a modified ribosome binding site for improved translation to amplify the rpoH gene using Pfu polymerase. The PCR product was gel purified and inserted into the Hermes-8 plasmid to form plasmid pJKD2605. E. coli transformants were selected using the erm gene of Hermes, PCR screened, and sequenced to verify that no mutations had been introduced into the rpoH gene. Plasmid pJKD2605 was used to transform N. gonorrhoeae strain JKD484 (24), a derivative of strain MS11-A, containing the ptetM25.2 plasmid. A single transformant was obtained, and the recombinant plasmid was designated pJKD5067. The rpoH insert from pJKD5067 was PCR amplified and completely sequenced. The results showed that the sequence of the rpoH gene carried on pJKD5067 was identical to the sequence in N. gonorrhoeae strain MS11-A. Finally, pJKD5067 was conjugated into JKD5079, a spectinomycin-resistant mutant of strain MS11-A, to form JKD5068, which was used for transcriptional profiling. The optimal time for induction of the rpoH gene, which was under the control of the Ptrc promoter, was determined by performing RNA dot blotting and monitoring the levels of mRNA derived from rpoH and the chaperone-encoding genes dnaK, groEL, dnaJ, and grpE over a 2-h period. Gonococcal strain JKD5068 was grown to the mid-exponential phase (optical density at 560 nm, 0.6), and the culture was split between two flasks. One half of the culture was induced by addition of 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), while the other half was not induced. RNA dot blotting demonstrated that an induction time of 10 min was sufficient to obtain maximal expression of both rpoH and, subsequently, the dnaK, dnaJ, groEL, and grpE genes (Fig. 1; data not shown for later times).
FIG. 1.
RNA dot blot hybridization, showing increases in gonococcal dnaK, dnaJ, groEL, grpE, and rpoH transcripts due to overexpression of RpoH for 10 min. RNA was extracted from induced (In) and noninduced (Non) JKD5068 and transferred to the membranes as indicated on the right Filters were hybridized with probes from gonococcal 16S rRNA and from the genes indicated at the bottom.
Effect of increased RpoH levels on gene expression.
Microarrays were used to measure changes in gene expression when rpoH was overexpressed in gonococcal strain JKD5068. Genes were classified as differentially regulated if the P value using a t statistic corrected for the false discovery rate was less than 0.001 and the change in the expression ratio was 1.5-fold. A total of 11 genes were up-regulated, as determined by these criteria, when rpoH was overexpressed (Table 2). A twelfth gene (NGO1244) appeared to be up-regulated, but the P value was 0.009, largely because of a local artifact on one microarray. Quantitative real-time PCR using the same RNA preparations indicated that this gene was up-regulated 8.9-fold, and it is therefore included in Table 2. As expected, we did not identify any down-regulated genes using the same criteria. A 3.2-fold increase in rpoH transcription was detected, confirming that this gene was overexpressed in this system. Data for the same rpoH transcripts obtained by quantitative real-time PCR agreed with this overexpression, and a 10.1-fold increase was observed (Table 2).
TABLE 2.
Genes up-regulated in N. gonorrhoeae strain JKD5068 when RpoH is overexpressed
| Open reading framea | Gene | Fold change
|
Proposed functiond | |
|---|---|---|---|---|
| Microarrayb | RT-PCRc | |||
| NGO0288 | rpoH | 3.2 | 10.1 | RNA polymerase sigma factor |
| NGO1244e | marR | 2.3 | 8.9 | Transcriptional regulator MarR |
| NGO1046 | clpB | 2.3 | 5.1 | HSP, chaperone |
| NGO1429 | dnaK | 2.2 | 3.1 | HSP, chaperone |
| NGO1422 | grpE | 2.1 | HSP, chaperone | |
| NGO0775 | lon | 2.0 | ATP-dependent protease | |
| NGO1694 | folA | 2.0 | Dihydrofolate reductase | |
| NGO0116 | secB | 2.0 | 2.7 | Protein secretion |
| NGO2094 | groES | 1.9 | HSP, chaperone | |
| NGO1222 | 1.8 | OsmC-like family protein | ||
| NGO0570 | creA | 1.8 | DNA-binding protein | |
| NGO1901 | dnaJ | 1.7 | HSP, chaperone | |
| NGO1426 | 1.6 | Conserved hypothetical protein | ||
Designations from the annotation at www.ncbi.nlm.nih.gov/entrez/query.fcgi.
Average expression ratio from microarray analysis for genes that were up-regulated at least 1.5-fold with a P value of ≤0.001.
Expression ratio for genes that were analyzed by quantitative real-time RT-PCR.
Proposed functions of the genes based on sequence annotation, sequence homology, and previous investigations. HSP, heat shock protein.
P = 0.009.
Previous work with Neisseria has demonstrated that there is an increase in transcription of the dnaJ, dnaK, and grpE genes encoding the Hsp70 system upon exposure to heat shock (14, 24). As expected, dnaK, dnaJ, and grpE were up-regulated in the presence of excess RpoH (Table 2), and 3.1-fold up-regulation of dnaK was confirmed by quantitative real-time PCR (Table 2). ClpB with the assistance of DnaK has the ability to refold aggregated proteins in E. coli (8, 50) and Thermus thermophilus (26). In contrast to ClpB, other members of the Clp family are proteases involved in the removal of irreversibly damaged proteins from the intracellular pool. In E. coli, clpB together with genes encoding other members of the family have been shown to belong to the rpoH regulon (48). In Neisseria, only clpB appears to be regulated by RpoH (Table 2). An increase in transcription was also observed for clpB by quantitative real-time PCR (Table 2).
The GroE system is one of the best-characterized chaperone systems and is stress induced in many bacteria. In Neisseria groES and groEL are arranged in a bicistronic operon which is transcribed from an RpoH-dependent promoter upstream of groES under heat stress conditions (43). Transcription of groES increased in the presence of excess RpoH (Table 2). However, we did not detect a significant increase in groEL transcripts with the microarrays, although marginal induction was observed with RNA dot blots (Fig. 1). In a previous study a 12-bp inverted repeat capable of functioning as a transcriptional terminator was identified between groES and groEL in Neisseria, resulting in premature termination of some of the groESL transcripts (43). This resulted in an increased amount of groES mRNA relative to the amount of groEL and might explain our observation that there was marginal induction of groEL transcripts. A gene encoding a homolog of the chaperone SecB was identified as a member of the RpoH regulon (Table 2), and 2.7-fold up-regulation was observed for secB using quantitative real-time PCR (Table 2). In addition, lon, a gene encoding an ATP-dependent protease, also appears to be regulated by RpoH. In E. coli, the lon gene is regulated by rpoH, and the protease degrades damaged or unstable cytosolic proteins that cannot be refolded (6, 48).
An interesting observation was the up-regulation of a transcriptional regulator. NGO1244 potentially encodes a protein that belongs to the MarR family of transcriptional regulators. Two MarR family homologs have been identified in N. gonorrhoeae, and they are encoded by NGO0058 (farR) and NGO1244 (marR) (25). FarR has been shown to regulate the FarAB efflux pump of N. gonorrhoeae, which mediates resistance to antibacterial fatty acids (25). No function has been associated with MarR, and we report here that marR appears to be regulated by RpoH.
Of the remaining genes controlled by RpoH, creA (NGO0570) encodes the DNA binding protein CreA (51% identity to the CreA protein from E. coli), folA encodes a dihydrofolate reductase (2), and the final two genes encode hypothetical proteins. The conserved hypothetical protein encoded by NGO1222 exhibits similarity to the OsmC-like family of proteins. OsmC is an osmotically induced protein from E. coli (17).
Consensus sequence for neisserial RpoH-dependent promoters.
In the gammaproteobacterium E. coli, the RNA polymerase holoenzyme associated with RpoH recognizes the consensus sequence 5′-GGCTTGA(12 to 20 bp)CCCCAT-3′ with conserved motifs (underlined) located at the −35 and −10 positions relative to the transcription start points of the RpoH-regulated genes (48). The promoter consensus sequence for alphaproteobacteria is slightly different: 5′-CTTG(17 or 18 bp)CC/TTATNTNNG-3′ (37). RpoH-dependent promoter elements have previously been identified upstream of dnaK, dnaJ, and grpE (24), as well as groES (43), in N. gonorrhoeae, which belongs to the beta subdivision of the Proteobacteria.
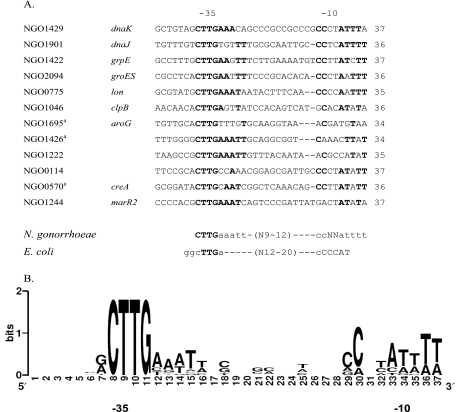
We identified a putative RpoH-dependent promoter sequence upstream of 10 of the 12 genes up-regulated in response to RpoH overexpression (Fig. 2A). No similar sequences were found upstream of the up-regulated folA and secB genes (NGO1694 and NGO0116); however, we identified the consensus sequence upstream of the adjacent upstream genes NGO1695 and NGO0114, respectively. Alignment of the nucleotide sequences of the upstream regions of these genes and putative transcriptional units allowed a comprehensive consensus sequence for RpoH-dependent promoters to be derived for N. gonorrhoeae (Fig. 2B). The −35 box of the RpoH-dependent genes shows total sequence conservation of the CTTG residues compared to the known consensus sequences of other bacteria (Fig. 2). Mutational studies have shown that the TT residues in this CTTG motif are essential for the transcription of dnaK under heat stress conditions (24). There was also some degree of sequence conservation in the spacer region with an optimum spacer distance of 9 to 12 bp. As observed previously (48), the −10 box showed some variation from the stretch of C residues found in the E. coli consensus sequence. The gonococcal consensus −10 box contains an AT-rich region (Fig. 2). Even though a previous study (24) showed that the TA residues in the −10 box of dnaK are essential for transcription under heat stress conditions, considerable variation was seen in these nucleotides when the RpoH-regulated genes were compared.
FIG. 2.
Determination of the consensus sequence for RpoH-dependent promoters. (A) The sequences upstream of the genes that were up-regulated when there was excess RpoH were aligned using ClustalW. To maximize the alignment, previously identified RpoH-dependent promoters from dnaK, dnaJ, grpE, and groES were utilized. The length of each identified promoter sequence is indicated on the right. A superscript a indicates a gene with multiple putative RpoH binding sites, and the site that best fit the consensus was used. Also shown are the E. coli and newly derived consensus sequences for RpoH-dependent promoters from N. gonorrhoeae. (B) Graphic display of the consensus RpoH-dependent promoter sequence generated by SEQUENCE LOGO (34). The height of each column reflects the nonrandom bias of a residue at the position, and the size of each letter reflects the frequency of the residue at the position.
Comparison of the rpoH regulon with the heat shock stimulon of N. gonorrhoeae.
In E. coli the alternative sigma factor RpoH regulates the transcription of many heat shock proteins (32). However, the heat shock response is a much broader stress response that involves other sigma factors and regulators and therefore multiple regulons (48). To determine whether the same is true for N. gonorrhoeae, heat shock experiments were performed using gonococcal strain MS11-A. A heat shock of 42°C for 10 min was selected since this treatment was sufficient to elicit a stress response in previous work (24) and enabled a comparison of the RpoH regulon and the heat shock response. MS11-A was grown to the mid-exponential phase, and one half of the culture was incubated at 42°C for 10 min, while the other half was left at 37°C. Two biological repeat experiments were performed on separate days, and cDNA derived from each experiment was used in two microarray hybridizations: an initial experiment and a dye swap experiment. When data from these hybridizations were analyzed, we identified 37 genes as genes that were differentially regulated in N. gonorrhoeae in response to heat stress; 24 of these genes were up-regulated (Table 3) and 13 were down-regulated (Table 4) by at least 1.5-fold, with P values of less than 0.001.
TABLE 3.
Genes up-regulated in N. gonorrhoeae strain MS11-A after heat shock at 42°C for 10 min
| Open reading framea | Gene | Fold changeb | Proposed functionc | RpoH regulond |
|---|---|---|---|---|
| Heat shock response, chaperones, and protein modification and degradation | ||||
| NGO1046 | clpB | 3.0 | HSP, chaperone | Yes |
| NGO0775 | lon | 2.4 | ATP-dependent protease | Yes |
| NGO1422 | grpE | 2.4 | HSP, chaperone | Yes |
| NGO0383 | ftsJ | 1.9 | HSP, cell division protein | |
| NGO1189 | hsp33 | 1.7 | Chaperone | |
| NGO0399 | htpX | 1.7 | HSP | |
| NGO1901 | dnaJ | 1.7 | HSP, chaperone | Yes |
| Other | ||||
| NGO2142 | 2.4 | Hypothetical protein | ||
| NGO1222 | 2.1 | OsmC-like family protein | Yes | |
| NGO1244 | marR | 2.0 | Transcriptional regulator, MarR | Yes |
| NGO0570 | creA | 2.0 | DNA-binding protein | Yes |
| NGO1426 | 1.9 | Conserved hypothetical protein | Yes | |
| NGO0848 | leuI | 1.7 | 2-Isopropylmalate synthase | |
| NGO0679 | leuC | 1.7 | 3-Isopropylmalate dehydratase | |
| NGO0114 | grxC | 1.7 | Glutaredoxin 3 | |
| NGO0568 | 1.7 | Hypothetical protein | ||
| NGO1428 | 1.7 | Hypothetical protein | ||
| NGO0999 | rpoD | 1.6 | RNA polymerase sigma factor | |
| NGO1694 | folA | 1.6 | Dihydrofolate reductase | Yes |
| NGO0678 | 1.6 | Hypothetical protein | ||
| NGO1245 | 1.5 | Hypothetical protein | ||
| NGO1425 | 1.8 | Hypothetical protein | ||
| NGO0392 | 1.6 | Conserved hypothetical protein | ||
| NGO1859 | 1.5 | Ferredoxin |
Designations from the annotation at www.ncbi.nlm.nih.gov/entrez/query.fcgi.
The fold change is the ratio of the mRNA transcript level determined by microarray analysis in heat-stressed cells to the mRNA transcript level in nonstressed cells and is the average expression ratio for genes that were up-regulated at least 1.5-fold with a P value less than 0.001 in two biological repeats.
Proposed functions of the genes based on sequence annotation, sequence homology, and previous investigations. HSP, heat shock protein
Yes indicates that the gene is up-regulated as determined by microarray analysis with RpoH overexpression (Table 1).
TABLE 4.
Genes down-regulated in N. gonorrhoeae strain MS11-A after heat shock at 42°C for 10 min
| Open reading framea | Gene | Fold changeb | Proposed functionc |
|---|---|---|---|
| NGO1235 | ilvH | 0.38 | Acetolactate synthase III, small subunit |
| NGO0410 | 0.38 | Putative transcriptional regulator CspA | |
| NGO1236 | ilvI | 0.42 | Acetolactate synthase III, large subunit |
| NGO1473 | mdaB | 0.44 | Modulator of drug activity |
| NGO1234 | 0.56 | Hypothetical protein | |
| NGO1233 | ilvC | 0.56 | Ketol-acid reductoisomerase |
| NGO0233 | nsgA | 0.57 | Outer membrane protein |
| NGO1980 | yojH | 0.61 | Malate:quinone oxidoreductase |
| NGO1415 | nqrC | 0.62 | NADH-quinone reductase, subunit C |
| NGO1449 | 0.65 | Putative l-lactate permease | |
| NGO0563 | 0.65 | Hypothetical protein | |
| NGO0904 | 0.66 | Conserved hypothetical protein | |
| NGO1406 | gcvT | 0.66 | Aminomethyl transferase |
Designations from the annotation at www.ncbi.nlm.nih.gov/entrez/query.fcgi.
The fold change is the average expression ratio for genes that were down-regulated at least 1.5 fold with a P value of <0.001.
Proposed functions of the genes based on sequence annotation, sequence homology, and previous investigations.
Nearly all of the genes previously identified as members of the RpoH regulon were also up-regulated in response to heat shock (Table 3). The three exceptions were dnaK, secB, and groES, for which the P values from the heat shock experiments were greater than 0.001 (0.002, 0.006, and 0.0013, respectively). As expected, rpoH was not identified as an up-regulated gene at this time, which is consistent with a previous finding that transcription of rpoH is only marginally elevated with 10 min of a heat shock (24). We identified marR as a gene that was both RpoH regulated and up-regulated under heat stress conditions. We also identified ftsJ as a gene that was up-regulated in response to heat shock, although not in the experiments in which RpoH was overexpressed. Situated downstream of ftsJ in the N. gonorrhoeae genome is ftsH, and these two genes appear to be transcriptionally coupled, a situation that is also observed in E. coli (21). In E. coli, ftsH is an RpoH-regulated gene, and the FtsH protein is involved in the degradation of RpoH (21).
The majority of the 13 down-regulated genes were involved in biosynthetic pathways (Table 4). The genomic arrangement of some of these genes (for example, NGO1233 to NGO1236) suggests that they could be cotranscribed. Again, a gene encoding a regulatory protein appeared to be differentially expressed. NGO0410 encodes a putative cold shock family transcriptional regulator and appeared to be down-regulated both in our analysis and in N. meningitidis subjected to heat stress (14).
DISCUSSION
Major proteins involved in the stress response are well conserved across species, but the regulation of the genes encoding these proteins tends to differ. Genome-wide transcriptional profiling has been used to document the heat shock response in bacteria such as E. coli (31, 48), N. meningitidis (14), group A Streptococcus (38), Campylobacter jejuni (41), Shewanella oneidensis (12), and Bacillus subtilis (20). The overlapping RpoH regulon has been studied in E. coli by overexpression of RpoH (48) and in several other bacterial species by analysis of rpoH mutants (22, 27, 49). The levels of RpoH and its activity are regulated at multiple levels in E. coli.
Increased expression of RpoH in N. gonorrhoeae under normal growth conditions was postulated to result in saturation of the DnaK/DnaJ/GrpE chaperone complex, allowing RpoH to associate with RNA polymerase and stimulate transcription from RpoH-dependent promoters. As expected, we identified rpoH and an additional 12 genes that appeared to be up-regulated under these conditions. The majority of the genes in the RpoH regulon of N. gonorrhoeae encode chaperones and proteases involved in protein homeostasis. The RpoH regulon that we identified in the gonococcus was significantly smaller than the more than 100 genes recently reported for E. coli (48), which is in keeping with the smaller size of the neisserial genome.
The core protease (Lon) and chaperones (DnaK, DnaJ, GroES, GrpE, and ClpB) are involved in protein homeostasis, and their genes appear to be directly regulated by RpoH during heat shock in N. gonorrhoeae. A recent proteome analysis of N. meningitidis serogroup A demonstrated that there was expression of ClpB, DnaK, and GroEL under standard physiological conditions, highlighting the involvement of these proteins in normal cellular functions (3). In addition, increased expression of groES, grpE, dnaJ, dnaK, clpB, lon, and secB was observed when N. gonorrhoeae was exposed to H2O2 (42), highlighting the fact that the general stress response can be triggered by a variety of stresses. The main role of the SecB chaperone is maintaining proteins in an export-competent state before interaction with the translocation apparatus that drives export across the cytoplasmic membrane (30). However, when SecB is overproduced in E. coli, it is able to function as a general nonspecific chaperone in a strain lacking DnaK, DnaJ, and trigger factor (45).
In E. coli, the regulatory region of rpoH consists of four promoters (P1, P3, P4, and P5) (10). At 30°C the transcription of rpoH is sigma-70 dependent and involves the P1, P4, and P5 promoters (10). When the organism is exposed to elevated temperatures, transcription is from the P3 promoter recognized by sigma-24 (35, 46). Sigma-24 belongs to the ECF sigma factor family. A separate study has shown that the ECF-type sigma factor of N. gonorrhoeae is not involved in the heat shock response of this species (16). In contrast, in N. gonorrhoeae transcription of rpoH is from a sigma-70 promoter under heat stress conditions (24). It has been suggested that preexisting levels of RpoH in the cytoplasm mediate the initial heat shock response in gonococci and that unidentified trans-acting factors are involved in the transcriptional induction of rpoH after prolonged exposure to stress (24).
Genes encoding additional heat shock proteins were identified as genes that are up-regulated during heat shock in an apparently RpoH-independent fashion. One of these proteins, Hsp33, is a unique redox-regulated chaperone which in E. coli efficiently prevents or suppresses the aggregation of folding intermediates that are derived from either chemically or thermally denatured proteins (13). FtsH plays a vital role in regulating RpoH levels in E. coli by degrading RpoH complexed with DnaJ/DnaK/GrpE (21). In E. coli, ftsH is cotranscribed from an RpoH-dependent promoter with ftsJ. ftsJ encodes a 23S rRNA methyltransferase which methylates ribosomes, improving the stability of FtsH (18). In gonococci, the ftsJ and ftsH genes have the potential to be transcriptionally coupled, but they were not RpoH regulated and there was no characteristic RpoH-dependent promoter consensus sequence upstream of ftsJ. During normal heat shock, gonococcal ftsJ, but not ftsH, was up-regulated, indicating that the promoter for ftsJ, at least, is controlled by an as-yet-unidentified stress response mechanism. From these observations we suggest that the homeostatic control mechanisms that determine the level of cytosolic RpoH in gonococci may be different from the paradigm established for gammaproteobacteria such as E. coli, which relies on coordinate expression of RpoH and FtsH. A large number of genes involved in biosynthesis functions were apparently down-regulated during heat shock in N. gonorrhoeae and N. meningitidis (14). The mode of repression after heat shock is unclear.
Our current understanding of RpoH regulons indicates that unlike other members of the betaproteobacteria, Neisseria species have retained RpoH as the master regulator of protein homeostasis. However, the mechanism for triggering the general stress response pathway via RpoH in Neisseria is distinctly different from the model advanced for the gammaproteobacteria. In addition, transcription of rpoH is not autoregulated, and the means by which a delayed increase in transcription occurs is unknown. Finally, we do not yet understand the mechanisms that result in the increase in transcription of genes that are outside the RpoH regulon or the repression of other genes during the general stress response.
Acknowledgments
This work was supported by an NHMRC program grant to the Australian Bacterial Pathogenesis Program. L.A.S.S. was supported by a Wellcome Trust project grant awarded to N.J.S. I.G. was the recipient of a Monash University postgraduate scholarship. The Gonococcal Genome Sequencing Project was supported by USPHS/NIH grant AI-38399.
The N. gonorrhoeae genome sequence was obtained from the University of Oklahoma (http://www.genome.ou.edu/gono.html).
REFERENCES
- 1.Arsene, F., T. Tomoyasu, and B. Bukau. 2000. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 55:3-9. [DOI] [PubMed] [Google Scholar]
- 2.Baccanari, D. P., R. L. Tansik, S. J. Paterson, and D. Stone. 1984. Characterization and amino acid sequence of Neisseria gonorrhoeae dihydrofolate reductase. J. Biol. Chem. 259:12291-12298. [PubMed] [Google Scholar]
- 3.Bernardini, G., G. Renzone, M. Comanducci, R. Mini, S. Arena, C. D'Ambrosio, S. Bambini, L. Trabalzini, G. Grandi, P. Martelli, M. Achtman, A. Scaloni, G. Ratti, and A. Santucci. 2004. Proteome analysis of Neisseria meningitidis serogroup A. Proteomics 4:2893-2926. [DOI] [PubMed] [Google Scholar]
- 4.Black, C. G., J. A. Fyfe, and J. K. Davies. 1998. Absence of an SOS-like system in Neisseria gonorrhoeae. Gene 208:61-66. [DOI] [PubMed] [Google Scholar]
- 5.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandu, D., and D. Nandi. 2004. Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res. Microbiol. 155:710-719. [DOI] [PubMed] [Google Scholar]
- 7.Dempsey, J. A., W. Litaker, A. Madhure, T. L. Snodgrass, and J. G. Cannon. 1991. Physical map of the chromosome of Neisseria gonorrhoeae FA1090 with locations of genetic markers, including opa and pil genes. J. Bacteriol. 173:5476-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougan, D. A., A. Mogk, and B. Bukau. 2002. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell. Mol. Life Sci. 59:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudoit, S., and Y. H. Yang. 2003. Bioconductor R packages for exploratory analysis and normalization of cDNA microarray data, p. 45-65. In G. Parmigiani, E. S. Garrett, R. A. Irizarry, and S. L. Zeger (ed.), The analysis of gene expression data: methods and software. Springer Verlag, New York, NY.
- 10.Fujita, N., and A. Ishihama. 1987. Heat-shock induction of RNA polymerase sigma-32 synthesis in Escherichia coli: transcriptional control and a multiple promoter system. Mol. Gen. Genet. 210:10-15. [DOI] [PubMed] [Google Scholar]
- 11.Fyfe, J. A., C. S. Carrick, and J. K. Davies. 1995. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a sigma 70 promoter during growth in vitro. J. Bacteriol. 177:3781-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, H., Y. Wang, X. Liu, T. Yan, L. Wu, E. Alm, A. Arkin, D. K. Thompson, and J. Zhou. 2004. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186:7796-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graf, P. C., and U. Jakob. 2002. Redox-regulated molecular chaperones. Cell. Mol. Life Sci. 59:1624-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guckenberger, M., S. Kurz, C. Aepinus, S. Theiss, S. Haller, T. Leimbach, U. Panzner, J. Weber, H. Paul, A. Unkmeir, M. Frosch, and G. Dietrich. 2002. Analysis of the heat shock response of Neisseria meningitidis with cDNA- and oligonucleotide-based DNA microarrays. J. Bacteriol. 184:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guisbert, E., C. Herman, C. Z. Lu, and C. A. Gross. 2004. A chaperone network controls the heat shock response in E. coli. Genes Dev. 18:2812-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunesekere, I. C., C. M. Kahler, C. S. Ryan, L. A. S. Snyder, N. J. Saunders, J. I. Rood, and J. K. Davies. Ecf, an alternative sigma factor from Neisseria gonorrhoeae, controls expression of msrAB, which encodes methionine sulfoxide reductase. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 17.Gutierrez, C., and J. C. Devedjian. 1991. Osmotic induction of gene osmC expression in Escherichia coli K12. J. Mol. Biol. 220:959-973. [DOI] [PubMed] [Google Scholar]
- 18.Hager, J., B. L. Staker, and U. Jakob. 2004. Substrate binding analysis of the 23S rRNA methyltransferase RrmJ. J. Bacteriol. 186:6634-6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton, H. L., N. M. Dominguez, K. J. Schwartz, K. T. Hackett, and J. P. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 20.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman, C., D. Thevenet, R. D'Ari, and P. Bouloc. 1995. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 92:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, L. H., Y. H. Tseng, and M. T. Yang. 1998. Isolation and characterization of the Xanthomonas campestris rpoH gene coding for a 32-kDa heat shock sigma factor. Biochem. Biophys. Res. Commun. 244:854-860. [DOI] [PubMed] [Google Scholar]
- 23.Kupsch, E. M., D. Aubel, C. P. Gibbs, A. F. Kahrs, T. Rudel, and T. F. Meyer. 1996. Construction of Hermes shuttle vectors: a versatile system useful for genetic complementation of transformable and non-transformable Neisseria mutants. Mol. Gen. Genet. 250:558-569. [DOI] [PubMed] [Google Scholar]
- 24.Laskos, L., C. S. Ryan, J. A. Fyfe, and J. K. Davies. 2004. The RpoH-mediated stress response in Neisseria gonorrhoeae is regulated at the level of activity. J. Bacteriol. 186:8443-8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, E. H., C. Rouquette-Loughlin, J. P. Folster, and W. M. Shafer. 2003. FarR regulates the farAB-encoded efflux pump of Neisseria gonorrhoeae via an MtrR regulatory mechanism. J. Bacteriol. 185:7145-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motohashi, K., Y. Watanabe, M. Yohda, and M. Yoshida. 1999. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. USA 96:7184-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakahigashi, K., E. Z. Ron, H. Yanagi, and T. Yura. 1999. Differential and independent roles of a σ32 homolog (RpoH) and an HrcA repressor in the heat shock response of Agrobacterium tumefaciens. J. Bacteriol. 181:7509-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 29.Permina, E. A., and M. S. Gelfand. 2003. Heat shock (sigma32 and HrcA/CIRCE) regulons in beta-, gamma- and epsilon-proteobacteria. J. Mol. Microbiol. Biotechnol. 6:174-181. [DOI] [PubMed] [Google Scholar]
- 30.Randall, L. L., and S. J. Hardy. 2002. SecB, one small chaperone in the complex milieu of the cell. Cell. Mol. Life Sci. 59:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richmond, C. S., J. E. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosen, R., and E. Z. Ron. 2002. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom. Rev. 21:244-265. [DOI] [PubMed] [Google Scholar]
- 33.Saal, L. H., C. Troein, J. Vallon-Christersson, S. Gruvberger, A. Borg, and C. Peterson. 2002. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 3:SOFTWARE0003. [DOI] [PMC free article] [PubMed]
- 34.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schurr, M. J., and V. Deretic. 1997. Microbial pathogenesis in cystic fibrosis: co-ordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 24:411-420. [DOI] [PubMed] [Google Scholar]
- 36.Segal, E., E. Billyard, M. So, S. Storzbach, and T. F. Meyer. 1985. Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell 40:293-300. [DOI] [PubMed] [Google Scholar]
- 37.Segal, G., and E. Z. Ron. 1995. The dnaKJ operon of Agrobacterium tumefaciens: transcriptional analysis and evidence for a new heat shock promoter. J. Bacteriol. 177:5952-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Applic. Genet. Mol. Biol. 3:article 3. [On line.] http://www.bepress.com/sagmb/vol3/iss1/art3. [DOI] [PubMed]
- 40.Smyth, G. K., J. Michaud, and H. S. Scott. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067-2075. [DOI] [PubMed] [Google Scholar]
- 41.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stohl, E. A., A. K. Criss, and H. S. Seifert. 2005. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tauschek, M., C. W. Hamilton, L. A. Hall, C. Chomvarin, J. A. Fyfe, and J. K. Davies. 1997. Transcriptional analysis of the groESL operon of Neisseria gonorrhoeae. Gene 189:107-112. [DOI] [PubMed] [Google Scholar]
- 44.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 45.Ullers, R. S., J. Luirink, N. Harms, F. Schwager, C. Georgopoulos, and P. Genevaux. 2004. SecB is a bona fide generalized chaperone in Escherichia coli. Proc. Natl. Acad. Sci. USA 101:7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Q. P., and J. M. Kaguni. 1989. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J. Bacteriol. 171:4248-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuzawa, H., H. Nagai, H. Mori, and T. Yura. 1993. Heat induction of sigma 32 synthesis mediated by mRNA secondary structure: a primary step of the heat shock response in Escherichia coli. Nucleic Acids Res. 21:5449-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, K., M. Liu, and R. R. Burgess. 2005. The global transcriptional response of Escherichia coli to induced sigma32 protein involves sigma32 regulon activation followed by inactivation and degradation of sigma32 in vivo. J. Biol. Chem. 280:17758-17768. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, Y. N., N. Kusukawa, J. W. Erickson, C. A. Gross, and T. Yura. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor sigma 32. J. Bacteriol. 170:3640-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zolkiewski, M. 1999. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multichaperone system from Escherichia coli. J. Biol. Chem. 274:28083-28086. [DOI] [PubMed] [Google Scholar]