Abstract
It has previously been reported that the alpha-proteobacterium Azospirillum brasilense undergoes methylation-independent chemotaxis; however, a recent study revealed cheB and cheR genes in this organism. We have constructed cheB, cheR, and cheBR mutants of A. brasilense and determined that the CheB and CheR proteins under study significantly influence chemotaxis and aerotaxis but are not essential for these behaviors to occur. First, we found that although cells lacking CheB, CheR, or both were no longer capable of responding to the addition of most chemoattractants in a temporal gradient assay, they did show a chemotactic response (albeit reduced) in a spatial gradient assay. Second, in comparison to the wild type, cheB and cheR mutants under steady-state conditions exhibited an altered swimming bias, whereas the cheBR mutant and the che operon mutant did not. Third, cheB and cheR mutants were null for aerotaxis, whereas the cheBR mutant showed reduced aerotaxis. In contrast to the swimming bias for the model organism Escherichia coli, the swimming bias in A. brasilense cells was dependent on the carbon source present and cells released methanol upon addition of some attractants and upon removal of other attractants. In comparison to the wild type, the cheB, cheR, and cheBR mutants showed various altered patterns of methanol release upon exposure to attractants. This study reveals a significant difference between the chemotaxis adaptation system of A. brasilense and that of the model organism E. coli and suggests that multiple chemotaxis systems are present and contribute to chemotaxis and aerotaxis in A. brasilense.
The ability of bacteria to detect and respond to a broad range of environmental conditions is important for their survival. The chemotaxis system allows coordination of flagellar movement with temporal sensing of the environment. Chemotaxis has been studied in great detail with the model organism Escherichia coli (27, 36). The chemotaxis system integrates environmental cues into a behavioral response by using a dedicated signal transduction pathway. This pathway is composed of chemotaxis transducers, histidine kinase protein CheA coupled to the chemotaxis transducers via docking protein CheW, response regulator CheY, and adaptation proteins CheB and CheR. Homologous chemotaxis systems have been identified for distantly related bacteria and archaea (33, 41).
Prototypical chemotaxis transducers have an N-terminal extracellular sensory domain that detects environmental cues and a cytoplasmic C-terminal signaling domain that interacts with CheA to convey the signal to the flagellar motor via the CheY protein (8, 36). In order to respond to chemical gradients, motile cells possess an adaptation mechanism that allows them to make temporal comparisons of the chemical conditions in the environment as they swim about. Adaptation in chemotaxis is due to the activity of the chemotaxis transducer-specific enzymes: the CheR methyltransferase and the CheB methylesterase (32). CheR constitutively adds methyl groups from S-adenosylmethionine to specific glutamate residues on the C-terminal domain of chemotaxis transducers. Upon activation by phosphotransfer from CheA, phosphorylated CheB removes the methyl groups as methanol, thereby resetting transducers into a “sensing mode.” The ability of CheB to remove methyl groups as methanol has been used to design a continuous-flow assay to detect CheB activity in E. coli (15). The covalent modification of chemotaxis transducers by methylation is accompanied by a return to a prestimulus swimming bias. In E. coli, CheB is activated in response to negative stimuli, such as the removal of an attractant, whereas positive stimuli, such as the addition of an attractant, suppress its activity and thus methanol release (15). These characteristics result in a single type of methanol release profile for E. coli. However, nonenteric bacteria diverge from this paradigm. As with E. coli, Bacillus subtilis possesses a single set of chemotaxis proteins, including CheR and CheB. However, with B. subtilis, methanol is released in response to both the addition and the removal of an attractant (16, 34), resulting in a single type of methanol release profile which is different from that of E. coli. Not all chemotactic responses require methylation-dependent adaptation. For example, with E. coli, aerotaxis and chemotaxis to phosphoenolpyruvate transport system sugars are methylation independent (6, 22).
The role of methylation in chemotaxis has been studied with other chemotactic organisms. The archaeon Halobacterium salinarum releases methanol in response to both positive and negative chemical and light stimuli (23, 24), showing the same type of methanol release profile as B. subtilis (16). The alpha-proteobacterium Rhodospirillum centenum releases methanol in response to a negative stimulus, a reduction in light intensity, similarly to the E. coli pattern (14). The most significant difference from the model organisms in methanol release was observed with another alpha-proteobacterium, Rhodobacter sphaeroides, where methanol release was detected only in response to positive stimuli (20). These variations suggest that there is significant diversity in the adaptation mechanisms among bacteria and archaea and that some adaptation systems are considerably more complex than that described for E. coli (33, 36).
Azospirillum brasilense is a nitrogen-fixing alpha-proteobacterium that colonizes the rhizosphere of various agronomically important grasses and cereals, promoting plant growth (5, 30). The dominant motility response of A. brasilense is a metabolism-dependent form of taxis known as energy taxis (1). It has previously been reported that A. brasilense undergoes methylation-independent taxis responses (42). A chemotaxis operon controlling motility and aerotaxis was recently identified in this species by complementation of generally nonchemotactic mutants (12). The operon contains genes encoding a CheB methylesterase and a CheR methyltransferase, suggesting a role for methylation-dependent motile behavior in A. brasilense. In this study, we investigated the role of these two proteins in A. brasilense chemotaxis and aerotaxis. Our data indicate that CheB and CheR contribute to a complex methylation-dependent and methylation-independent motility behavior and are important for chemotaxis and aerotaxis. These findings expand our current view of the diversity in bacterial chemotaxis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A wild-type A. brasilense Sp7 (ATCC 29145) strain was used throughout this study. Other bacterial strains and plasmids used are listed in Table 1. A. brasilense cells were grown at 28°C in minimal medium (MMAB) (35), supplemented with a carbon source of choice. Cells were grown aerobically at 200 rpm on a rotary shaker. The growth medium was supplemented with the antibiotics kanamycin (30 μg ml−1), tetracycline (10 μg ml−1), and gentamicin (10 μg ml−1). Kanamycin (50 μg ml−1) and gentamicin (5 μg ml−1) were added to media to grow E. coli when appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Property(ies) | Reference or source |
|---|---|---|
| Strains | ||
| A. brasilense | ||
| Sp7 | Wild-type strain | ATCC 29145 |
| GA3 | cheB mutant; cheB::gusA-km Kmr | This work |
| BS104 | cheBR mutant; Kmr | This work |
| BS109 | cheR mutant; Gmr | This work |
| BS110 | che mutant; cheAΔ::gusA-km Kmr Tetr | This work |
| E. coli | ||
| EC100 | General cloning strain | Epicentre |
| EC100D pir-116 | General cloning strain with pir-116 | Epicentre |
| JM110 | General cloning strain; dam dcm | Stratagene |
| S17-1 | thi endA recA hsdR with RP4-2Tc::Mu-Km::Tn7 integrated in chromosome | 26 |
| Plasmids | ||
| pUCBR | cheBR from pFAJ451 cloned as an AviII/EcoRI fragment into pUC18 | This work |
| pBSKBR | pBluescript II SK(+) containing cheBR from pUCBR as an EcoRI/BamHI fragment | This work |
| pBS101 | pBSKBR digested with EheI and SmaI; ΔcheBR with Kmr cassette from pHP45Ω | This work |
| pBS102 | ΔcheBR::Kmr from pBS101 cloned into pCR-Blunt | This work |
| pBS103 | ΔcheBR::Kmr cloned into pSUP202 | This work |
| pBS107 | cheR PCR fragment cloned into pCR2.1 TOPO | This work |
| pBS108 | cheR fragment from pBS107 cloned into pKNOCK-Gm | This work |
| pGA1 | pUCBR containing gusA-Kmr cassette from pWM6; ΔcheB | This work |
| pGA2 | ΔcheBR::Kmr from pGA1 cloned into pCR-Blunt | This work |
| pGA3 | ΔcheBR::Kmr from pGA2 cloned as an EcoRI fragment into pSUP202 | This work |
| pGA111 | pBluescript II SK(+) containing cheA with 545 bp upstream cloned as a HindIII/XhoI PCR fragment | This work |
| pGA112 | pGA111 with a 555-bp internal deletion of cheA | This work |
| pGA113 | pGA112 with a 1,975-bp deletion and a gusA-Kmr cassette from pWM6 | This work |
| pGA114 | pCR-Blunt containing cheAΔ::gusA-km from pGA113 | This work |
| pGA115 | cheAΔ::gusA-km from pGA114 cloned into pSUP202 | This work |
| pFAJ451 | Cosmid clone containing the A. brasilense chemotaxis operon | 12 |
| pUC18 | Cloning vector; Apr | 39 |
| pBluescript II SK(+) | Cloning vector; Apr | Stratagene |
| pCR-Blunt | Cloning vector; Kmr | Invitrogen |
| pCR2.1 TOPO | Cloning vector; Kmr | Invitrogen |
| pKNOCK-Gm | Mobilizable plasmid, suicide vector for A. brasilense; Gmr | 3 |
| pRK2013 | Helper plasmid, carries tra genes; Kmr | 10 |
| pSUP202 | Mobilizable plasmid, suicide vector for A. brasilense; Cmr Tcr Apr | 26 |
| pWM6 | Source of gusA-Kmr cassette; Apr Kmr | 21 |
| pHP45Ω-Km | Apr Kmr | 9 |
Mutant construction.
All standard cloning steps were carried out as described previously (25). The cosmid pFAJ451 was digested with AviII and EcoRI to release a fragment containing the cheB and cheR genes, which was then cloned into the SmaI and EcoRI sites of pUC18, resulting in pUCBR. pUCBR was transformed into E. coli JM110 competent cells. To create a cheB mutant, a SmaI-excised gusA-km cassette from pWM6 (21) was cloned into the SmaI and T4 Klenow fragment end-repaired BclI sites of pUCBR, yielding pGA1. pGA1 was digested completely with EcoRI and partially with BamHI, end repaired (End-it repair kit; Epicentre, Madison, WI), and cloned into pCR-Blunt (Invitrogen, Carlsbad, CA), yielding pGA2. EcoRI-digested pGA2 was cloned into the same site of pSUP202, resulting in pGA3. E. coli S17-1 competent cells were transformed with pGA3 and used as a donor in a biparental mating with A. brasilense (35). Recombinants were screened for double homologous recombination and in-frame insertion of the cassette by replica plating and testing for GusA activity (21). One such recombinant, GA3, was further characterized as a cheB mutant (Table 1).
To create a cheBR insertion-deletion mutant, an EcoRI/BamHI fragment was isolated from pUCBR and cloned into the same sites of pBluescript II SK(+), resulting in pBSKBR. A polar kanamycin cassette was excised from pHP45Ω by HindIII digestion, end repaired, and blunt cloned into the SmaI and end-repaired EheI sites of pBSKBR, yielding pBS101. An end-repaired EcoRI and NotI fragment from pBS101 containing the ΔcheBR::Kmr construct was cloned into pCR-Blunt (Invitrogen), resulting in pBS102. PCR with pBS102 as a template and the primers CheBR-F (5′-CCGGAATTCGCAAGATGAGCGGCGGGGACA) and CheBR-R (5′-CGGAATTCGCGGAAGGAGGCCATCTGGCG) was used to amplify the ΔcheBR::Kmr construct, which was then digested with EcoRI (engineered restriction sites, underlined) and cloned into EcoRI-digested pSUP202 (26), yielding pBS103. E. coli EC100 cells (Epicentre) were transformed with pBS103, which was then transferred to A. brasilense by triparental mating using pRK2013 as a helper (10), as described previously (12). Screening for double homologous recombination was performed as described above. Southern hybridization was used to verify the loss of the plasmid. One such cheBR mutant (BS104) was further characterized (Table 1).
To construct a cheR mutant, an internal fragment of cheR was amplified by PCR with the primers CheR-F1 (5′-CGACATCACGGAGGCG) and CheR-R1 (5′-CACTTGTCGCCCTGCTG) and cloned into pCR2.1 TOPO (Invitrogen), resulting in pBS107. pBS107 was digested with EcoRI and cloned into the EcoRI site of pKNOCK-Gm (3), a plasmid that does not replicate in A. brasilense, resulting in pBS108. E. coli EC100D pir-116 cells were transformed with pBS108 and introduced into A. brasilense by triparental mating. Single homologous recombination resulted in a cheR gene interrupted by the insertion of the pKNOCK-Gm vector (cheR mutant, BS109) (Table 1). Because cheR is the last gene of the chemotaxis operon (12), no polar effects were expected.
A chemotaxis operon mutant was constructed by inserting a polar cassette in cheA, the first gene of the operon (12), thereby disrupting the entire operon. First, a fragment comprising the entire cheA gene and 545 bp of DNA sequence upstream of the chemotaxis operon was amplified from pFAJ451 by using the primers cheAop-HindF (CCCAAGCTTCAGCGCGATGAACTGGTTGGACC) and cheAY-XhoR (GGGCTCGAGTCATGCGGCACCTTTCTGCTC), followed by digestion with HindIII and XhoI sites (engineered restriction sites, underlined), and cloned into the same sites of pBluescript II SK(+), yielding pGA111. An inverse-PCR-based strategy (38) using primers CheA-Del-F (5′-GAAGATCTGCGGTCGCGCGGGAATTC) and CheA-Del-R (5′-GAAGATCTGACCGCGTCCGCCGCACG) (engineered BglII sites are underlined) and pGA111 as a template generated a linear product with BglII sites at both ends of an internal 555-bp fragment to be deleted, which was precisely deleted by digestion followed by self-ligation. The construct, pGA112, was verified by sequencing. BglII and NcoI digestion followed by end repair of the restriction sites deleted an additional 1,975-bp internal fragment. A SmaI-digested gusA-km cassette from pWM6 (21) was inserted into the deleted region, yielding pGA113. The cheAΔ::gusA-km construct was released from pGA113 by digestion with XhoI, made blunt by end repair and SmaI digestion, and cloned into pCR-Blunt (Invitrogen, Carlsbad, CA), yielding pGA114. An EcoRI digestion of pGA114 released cheAΔ::gusA-km for cloning into EcoRI-digested pSUP202, yielding pGA115. pGA115 was introduced into A. brasilense by triparental mating using pRK2013 as a helper. Single homologous recombinants carrying a deletion-insertion in cheA and the inserted pSUP202 plasmid were selected. Southern hybridization and PCR were used to verify the insertion of the recombinant plasmid. One such che mutant (BS110) was further characterized (Table 1).
All final constructs were verified by sequencing (DNA Core Facility, The University of Tennessee) prior to transfer to A. brasilense for conjugation and mutant construction.
Behavioral assays.
A. brasilense cells swim by rotating a single bidirectional polar flagellum (42). A change in swimming direction (reversal) is brought about by a short backward movement along the axis of the cell due to the switch in direction of flagellar rotation coupled to a reduction in swimming speed, which randomly reorients the cells in a new direction (42, 43). Cells were grown in MMAB supplemented with a carbon source of choice (10 mM) to early exponential phase, observed with a dark-field microscope, and videotaped. Reversal frequency was measured by determining the number of reversals of a single cell during a 5-s time frame, corresponding to the average time a single cell can be tracked. Increasing the tracking time did not change the results. The reversal frequencies of at least 90 cells (from three independent cultures) were measured in each experiment. The reversal frequency of cells washed in chemotaxis buffer (10 mM phosphate buffer [pH 7.0], 1 mM EDTA) prior to analysis was similar to that described above.
Soft agar plates (so-called “swarm plates”) and temporal gradient assays for chemotaxis and aerotaxis were performed as previously described (1, 2, 42, 43). Temporal response to attractants was measured after growing cells in MMAB to an early exponential phase, washing the cells three times, and resuspending them in chemotaxis buffer. To estimate the response time to the removal of an attractant, the cells were mixed with the attractant to be tested at a final concentration of 10 mM. The cells were then stimulated by the addition of chemotaxis buffer to dilute the concentration of the attractant from 10 mM to 2.5 mM, eliciting a repellent response. The time taken for approximately 50% of the population to return to the prestimulus motility bias was measured. The temporal gradient assay for aerotaxis was performed with a microchamber ventilated with oxygen and nitrogen gases in a setup developed by Laszlo and Taylor (18) and adapted for A. brasilense (43). The spatial gradient assay for aerotaxis was performed as previously described (43) by placing motile cells in an optically flat capillary (inner dimensions, 0.1 by 2 by 50 mm; Vitro Dynamics, Inc., Rockaway, N.J.). The cells used were washed and resuspended in chemotaxis buffer supplemented with 10 mM malate.
Continuous-flow assay.
Continuous-flow assays were performed as previously described for stimulation with chemicals for E. coli (15) and with oxygen for Halobacterium salinarum (19), with some modifications. Cells were grown to late exponential phase in MMAB supplemented with 5 mM malate and 5 mM fructose. Cells were washed with chemotaxis buffer with chloramphenicol (100 μg ml−1), incubated with 50 μCi l-[methyl-3H]methionine (Amersham Biosciences) with chloramphenicol (100 μg ml−1) for 2 h, and placed on a sterile in-line filter (Acrodisc, 0.45 μm; Pall Gelman) connected to a peristaltic pump. The filter was washed at a flow rate of 3 ml/min for 35 min with chemotaxis buffer, 10 min with an attractant (10 mM) in chemotaxis buffer, and 5 min with chemotaxis buffer. Fractions were collected every 30 s. An aliquot (100 μl) from each fraction was placed in a microfuge tube in scintillation fluid (Beckman Ready Solv-HP) and subjected to vapor phase transfer for 48 h. The samples were counted for 10 min with a scintillation counter (LS 6500; Beckman Instruments). For methanol release in aerotaxis, N2 or O2 (21%) was bubbled though chemotaxis buffer or attractant and washed over the filter assembly as described previously (19).
Statistical analysis.
Microsoft Excel was used for statistical analysis of results. A t test assuming equal variances with a P value of 0.01 was used to determine whether there was a significant difference between the mutants and the wild type in behavioral assays.
RESULTS
Mutants lacking CheB and CheR are impaired in chemotaxis.
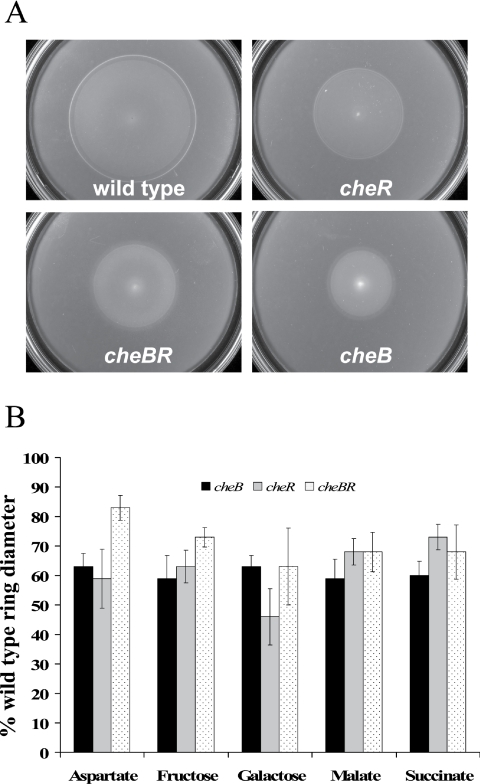
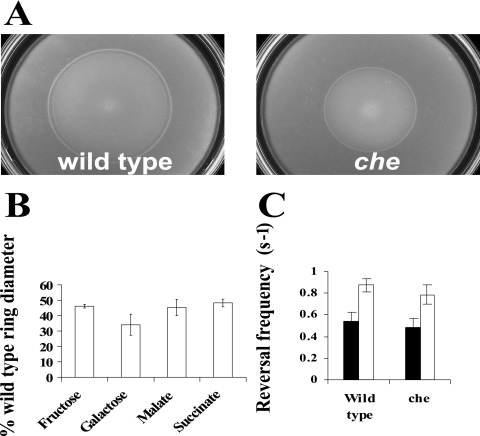
We constructed mutants defective in the cheB and/or cheR gene and characterized their motile behavior. First, we tested chemotaxis of the wild type and the cheB, cheR, and cheBR mutants to strong chemoattractants (1) by using a soft agar plate assay. We define strong and weak attractants based on their ability to trigger a response in spatial and temporal gradient assays: strong attractants elicit longer response times in a temporal gradient assay and a lower threshold in a spatial gradient assay. Malate, succinate, fructose, and oxygen are strong attractants, while galactose is a weaker attractant for A. brasilense (1). We found that the cheB, cheR, and cheBR mutants were significantly impaired in their swimming ability in this assay. The chemotactic rings formed by the mutants were significantly smaller in diameter than those formed by the wild-type strain for all substrates tested (Fig. 1). The growth rates of the mutants were measured and found to be statistically undistinguishable from that of the wild type (data not shown).
FIG. 1.
cheBR, cheB, and cheR mutants are deficient in chemotaxis. Chemotaxis of the A. brasilense wild-type strain (Sp7) and that of the mutants were compared by soft agar plate assay. Chemotactic-ring diameters were measured after incubation at 28°C for 48 h. (A) Representative soft agar plates with 10 mM malate as the sole carbon source. (B) The average chemotactic-ring diameters are expressed as percentages relative to that the of wild-type strain (defined as 100%). Error bars represent standard deviations from the means, calculated from at least three repetitions. Differences in the chemotactic-ring diameters of the wild type and the mutants were found to be statistically significant with all chemoeffectors tested.
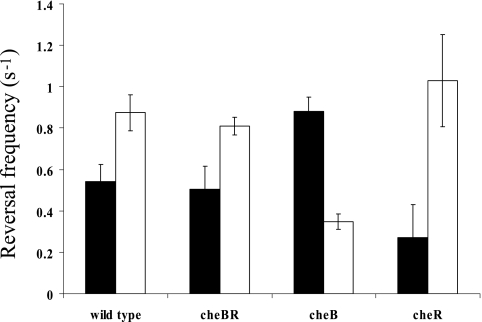
A temporal gradient assay provides a direct measurement of the frequency of flagellar rotation changes of individual cells challenged with a stimulus. Potent attractants cause free-swimming wild-type A. brasilense cells to swim smoothly and then resume the prestimulus motility pattern. We measured the response times of free-swimming cells challenged with both positive and negative stimuli. Response times of the wild-type A. brasilense strain upon addition of the strongest attractants, succinate, malate, and fructose, were similar (Table 2). Surprisingly, cells lacking CheB and/or CheR were no longer capable of responding to the addition of these attractants. On the other hand, cheB and cheBR mutants not only responded but also adapted upon addition of a weaker attractant, galactose. Upon removal of malate, succinate, fructose, and galactose, wild-type cells increased swimming reversal frequency, indicating a repellent response. The mutant cells either did not respond at all or showed no adaptation to the stimulus (Table 2). We analyzed the swimming bias of the wild-type cells in the presence of a constant concentration of an attractant (steady-state conditions). Under steady-state conditions, E. coli mutants lacking CheB and CheR have tumbly and smooth swimming biases, respectively, whereas the double mutant has a motility bias similar to that of the wild-type strain. The A. brasilense cheBR mutant had a random steady-state swimming bias similar to that of the wild type. Interestingly, the wild-type cells and the cheBR mutant had different swimming biases dependent on the substrate present: the reversal frequency was higher when cells were grown on fructose than when cells were grown on malate. Similarly, the swimming bias of cheB and cheR mutants was also dependent on the substrate present; however, it was different from that of the wild-type cells on any given substrate. On malate, the cheB mutant had increased reversal frequency, which corresponds to a tumbly bias in E. coli, and the cheR mutant had a smooth swimming bias similar to that of its E. coli counterparts. However, on fructose, the phenotype displayed by these mutants was reversed (Fig. 2).
TABLE 2.
Chemotaxis of A. brasilense wild-type strain and cheBR, cheB, and cheR mutants in a temporal gradient assay
| Strain | Response time (s) (mean ± SD)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Malatea
|
Succinate
|
Fructose
|
Galactose
|
Oxygen
|
||||||
| +b | −c | + | − | + | − | + | − | + | − | |
| Wild type | 56 ± 6 | 43 ± 3 | 52 ± 7 | 41 ± 4 | 63 ± 8 | 45 ± 3 | 34 ± 8 | 26 ± 4 | 33 ± 4 | 28 ± 2 |
| cheBR mutant | NRd | NR | NR | NR | NR | >300 | 38 ± 5 | >300 | NR | NR |
| cheB mutant | NR | NR | NR | NR | NR | NR | 82 ± 7 | NR | NR | NR |
| cheR mutant | NR | >300e | NR | NR | NR | NR | NR | NR | NR | NR |
Cells were grown with the chemical to be tested as an attractant or repellent and prepared as described in Materials and Methods.
+, addition of attractant. The chemicals were tested at a final concentration of 10 mM.
−, removal of attractant. The chemical concentration was reduced from 10 mM to 2.5 mM (final).
NR, no response, i.e., no change in the swimming bias of cells upon addition or removal of the chemoeffector tested.
Cells responded to the removal of the chemoeffector by changing the swimming bias but did not adapt within 300 s of observation.
FIG. 2.
Reversal frequencies of free-swimming cells of wild-type A. brasilense and the cheBR, cheB, and cheR mutants on different carbon sources (filled bars, malate; open bars, fructose). The reversal swimming frequency was determined as described in Materials and Methods. Standard deviations to the means are indicated.
Mutants lacking CheB and CheR are impaired in aerotaxis.
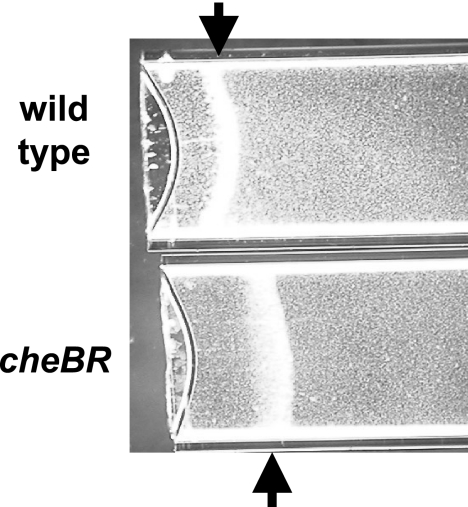
Aerotaxis is arguably the strongest behavioral response in A. brasilense (4, 43). We first used the spatial gradient assay for aerotaxis to analyze the role of CheB and CheR. In this assay, an oxygen gradient is established by oxygen diffusion through a suspension of motile cells that contain an electron donor (carbon source). Cells accumulate at the maximum energy levels generated by metabolizing the carbon source using an optimum concentration of oxygen as a terminal electron acceptor (43). We found that the cheB and cheR mutants were unable to form aerotaxis bands, although the cells were fully motile. Interestingly, the cheBR mutant did form an aerotaxis band (Fig. 3); however, it was formed with a significant delay (more than 5 min to form the band by the mutant versus less than 2 min by the wild type) and farther away from the meniscus than that formed by the wild type. Several independent studies previously showed that the time required for the formation of the aerotaxis band by A. brasilense cells in this assay is very consistent (1 to 2 min) and reflects the ability of cells to efficiently sense the oxygen gradient and navigate toward its optimum concentration (1, 4, 43). The observation that the aerotaxis band formed by the CheBR mutant is significantly delayed and formed further away from the meniscus supports the proposition that the CheBR mutant is impaired in aerotaxis.
FIG. 3.
The cheBR mutant is impaired in, but not null for, aerotaxis in a spatial gradient assay. The abilities of the A. brasilense wild-type strain (Sp7) and the cheBR mutant to form a sharp aerotaxis band in a gradient of oxygen in a capillary tube were compared. The wild-type strain (Sp7) forms a sharp aerotaxis band, while the cheBR mutant forms a diffuse band at a different position in the gradient. Malate (10 mM) was added as the sole carbon and energy source to the suspension of motile cells. The cheB and cheR mutants did not form any aerotaxis bands under similar conditions (not shown). Arrows point to the center of the aerotaxis bands.
Interestingly, none of the three mutants responded to either addition or removal of oxygen in a temporal gradient assay for aerotaxis, whereas the wild type had a positive and negative response, respectively (Table 2).
Methanol evolution upon chemostimulation.
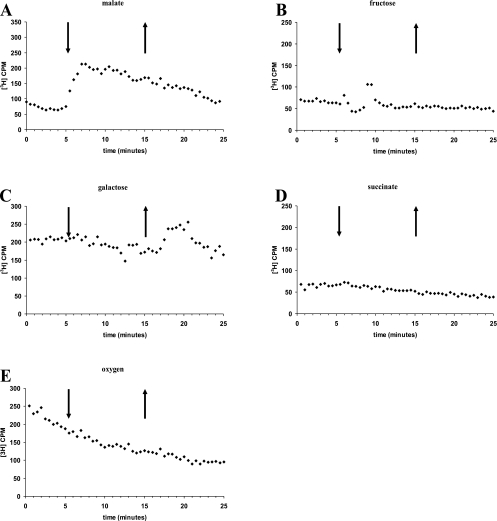
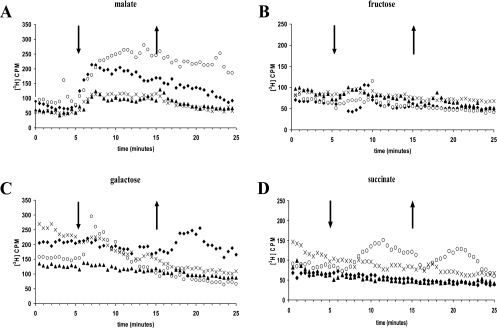
In order to determine the activities of CheB and CheR, we used a methanol release assay (15). We first determined the profile of methanol release from the wild-type cells upon stimulation with various chemoeffectors. The wild-type cells showed a release of methanol upon addition of but not upon removal of malate (Fig. 4A). Interestingly, the addition of malate resulted in continuous methanol release (rather than a peak) slowly decreasing over time. This is different from the profiles of methanol release for E. coli and B. subtilis but similar to that for R. sphaeroides. The wild-type cells also released methanol upon removal of but not upon addition of a weaker attractant, galactose (Fig. 4C). This type of profile is typical of E. coli. There was no methanol release upon the addition or removal of fructose, succinate, or oxygen (Fig. 4B, D, and E). Thus, different chemoeffectors induce different methanol release profiles in A. brasilense, and there is indication of methylation-independent taxis in response to succinate and oxygen.
FIG. 4.
Methanol release from the A. brasilense wild-type strain upon the addition (down arrows) and removal (up arrows) of chemical attractants (10 mM) and oxygen. The wild-type strain has different patterns of methanol release, represented as counts per minute (CPM), upon stimulation with malate (A), fructose (B), galactose (C), succinate (D), and oxygen (E). Representative results from at least two independent experiments are shown.
Strains deleted of cheB, cheR, or both showed a wide spectrum of methanol release profiles (Fig. 5). None of the mutants showed methanol release upon either addition or removal of fructose (Fig. 5B) or oxygen (not shown). In contrast to wild-type cells, the cheB and cheR mutants did not release methanol upon removal of galactose and they released significantly less methanol upon addition of malate (Fig. 5A and C). These results are consistent with a role for CheB and CheR in the induction of methanol release in response to these chemoeffectors.
FIG. 5.
Methanol release from the A. brasilense wild-type strain (Sp7) (⧫) and the cheBR mutant (○), cheB mutant (▴), and cheR mutant (×) upon the addition (down arrows) and removal (up arrows) of chemoeffectors. Methanol evolution, represented as counts per minute (CPM), as observed in response to malate (A), fructose (B), galactose (C), and succinate (D) is shown. Representative results from at least two independent experiments are shown.
Unexpectedly, the cheBR mutant did release methanol upon the addition of malate, succinate, and galactose as well as upon the removal of succinate (Fig. 5A, C, and D). Furthermore, the methanol release profile in response to these chemostimuli was different from that of the cheB mutant and that of the wild-type strain. The cheBR mutant released methanol upon addition of but not removal of galactose, whereas the wild type did just the opposite (Fig. 5C). Also, in response to the addition of malate, this mutant released methanol in significantly higher amounts than the cheB mutant and the wild-type strain (Fig. 5A). While succinate did not induce methanol release in the wild-type strain, the cheBR mutant released methanol upon both addition and removal of succinate (Fig. 5D). These results strongly suggest that there is a methylation/demethylation system(s) in A. brasilense in addition to the one under investigation.
A che operon mutant has modest chemotaxis defects.
The limited chemotaxis defects observed for the cheB and cheR mutants prompted us to further evaluate the function of the chemotaxis pathway in controlling motility. We constructed a chemotaxis operon insertion mutant (che mutant) and characterized its motility (Fig. 6). We found that, relative to the wild type, the che mutant was impaired in chemotaxis in the soft agar plate assay, but it did form chemotactic rings (Fig. 6A and B). These results indicated that the che mutant was not null for chemotaxis. Under steady-state conditions, the che mutant had a chemoeffector-dependent motility bias similar to that of the cheBR mutant and that of the wild-type strain (Fig. 6C). Altogether, these data suggest that this chemotaxis pathway contributes to controlling motility; however, it is probably not its primary function.
FIG. 6.
The che operon mutant is impaired in, but not null for, chemotaxis. (A) Chemotaxis toward 10 mM malate, as determined by a soft agar plate assay. Both plates were inoculated with the same number of cells and incubated at 28°C for 48 h. (B) The average chemotactic-ring diameters are expressed as percentages relative to that of the wild-type strain (defined as 100%). Error bars represent standard deviations from the means, calculated from at least three repetitions. Differences in the chemotactic-ring diameters of the wild type and the che operon mutant were found to be statistically significant with all chemoeffectors tested. (C) Reversal frequencies of free-swimming cells of the wild type and the che mutant on malate (filled bars) and fructose (open bars). The reversal swimming frequency was determined as described in Materials and Methods. Standard deviations to the means are indicated.
DISCUSSION
The identification of cheB and cheR genes (12) suggested the presence of a methylation-dependent chemotaxis pathway in A. brasilense. In this work, we used genetic and biochemical approaches to investigate the role of CheB and CheR in chemotaxis in this bacterium and found that there are both methylation-dependent and methylation-independent pathways for chemotaxis in A. brasilense. Our analysis also revealed unexpected phenotypes for the cheB, cheR, and cheBR mutants, further suggesting a more complex role for these proteins in A. brasilense than in other species.
CheB and CheR have an effect on but are not essential for chemotaxis and aerotaxis.
We have demonstrated that cheB and cheR mutants contribute to chemotaxis and aerotaxis. Remarkably, all single and double mutants did not respond to stimulation with the strong attractants malate, succinate, fructose, and oxygen in a temporal gradient assay. Addition of these attractants did not result in changes in the swimming behavior of the mutants, whereas the wild-type cells responded to all stimuli by suppressing the reversal frequency. This is the most striking departure from the behavior of the model organisms E. coli and B. subtilis, where mutations in the corresponding genes abolish adaptation but not excitation in the temporal gradient assay (31, 40). Similar behavior has been reported for R. sphaeroides, where the cheB1 and cheR2 mutants did not respond to temporal stimulation at all (20). We have found that cheB and cheR mutants in A. brasilense retained the ability (although significantly reduced) to form chemotactic rings on semisoft agar. This is again different from results for analogous mutants in E. coli (40), but similar to results for those in R. sphaeroides (20). The apparent discrepancy between the results obtained with a temporal gradient assay (lack of the response) and those obtained with a spatial gradient assay (presence of the response) was also observed for aerotaxis: the cheBR mutant formed an aerotaxis band in the spatial gradient assay but did not respond to the addition or removal of oxygen in a temporal gradient assay. It is noteworthy to mention that the chemotaxis and aerotaxis responses observed for the temporal and spatial gradient assays are not equivalent since the latter assay requires the metabolism of an attractant and cell growth (the chemotaxis assay only). Furthermore, with temporal gradient assays, cells are challenged with extremely steep step gradients of chemoeffectors (that are not physiological), while they respond to much more shallow gradients in the spatial gradient assays. Therefore, the sensitivity of the chemotaxis response tested is different for each of these assays, which could account for the discrepancies observed. Further deviations from the E. coli model were detected by analyzing the swimming motility bias of cheB and cheR mutants. Deletion of cheB and cheR in E. coli results in cells displaying tumbly and smooth-swimming phenotypes, respectively (29, 40), regardless of the substrate present. The A. brasilense cells showed a different type of behavior. The wild-type cells had different swimming biases on different substrates, and cheB and cheR mutants showed a spectrum of motility phenotypes that were dependent on the substrate present. On some substrates, the cheB mutant had a high reversal frequency, while the cheR mutant had a smooth-swimming bias, whereas on other substrates it was just the opposite. These results indicate that CheB and CheR proteins play an important role in the overall chemosensory behavior of A. brasilense that does not conform to the E. coli model. Our results also indicate that there are CheBR-independent responses in A. brasilense. For example, the cheBR mutant not only responded but also adapted to the addition of galactose in a temporal gradient assay. Thus, mutations in cheB and cheR do not affect the ability of the cells to respond to temporal stimulation by all effectors. The swimming phenotypes of cheB and cheR mutants in A. brasilense are strikingly different from those of mutants in E. coli but similar to those of mutants lacking principle chemotaxis methylesterase and methyltransferase (CheB1 and CheR2) in R. sphaeroides, which is a distant alpha-proteobacterial relative of A. brasilense. The reason(s) for these significant differences from the E. coli model is currently unknown for either R. sphaeroides (20) or A. brasilense. In contrast to chemotaxis in E. coli, metabolism-dependent chemotaxis has been proposed as a major motility behavior for both R. sphaeroides (13) and A. brasilense (1, 2, 11) and may be one of the factors contributing to the observed differences.
Evidence for additional methylation/demethylation systems contributing to chemotaxis.
We found that the profile of methanol release in A. brasilense is chemoeffector specific: some attractants caused methanol release upon addition, whereas others caused release upon removal. This behavior represents another significant difference between A. brasilense and the model organisms and has not previously been reported for any microorganism. It is important to mention that the contribution of CheB and CheR to motility and methanol evolution upon chemostimulation in A. brasilense is chemoeffector dependent. One possible explanation is that an additional methylation/demethylation system(s) in A. brasilense differentially modulates the responses observed. The strongest evidence for the presence of additional adaptation enzymes comes from the results obtained with the cheBR mutant stimulated with succinate. While the wild-type cells did not release methanol upon either addition or removal of succinate, the mutant released a significant amount of methanol upon both addition and removal of attractant. Interestingly, the same profile of methanol release has been reported for the ΔcheR1 mutant of R. sphaeroides lacking an additional methyltransferase, which did not seem to contribute to chemotaxis under conditions tested (20). Additional methylation/demethylation systems in A. brasilense might be responsible for most of the observed phenotypic differences between this bacterium and other organisms. Although the A. brasilense genome is not yet available, preliminary data from the ongoing sequencing project (http://genomics.biology.gatech.edu/research/azo) indicate that A. brasilense might possess at least three additional chemotaxis operons containing CheB and CheR pairs. Future studies will determine whether these systems contribute to chemotaxis or to other cellular functions in A. brasilense.
Complex contribution of CheB and CheR to aerotaxis.
One of the most intriguing findings of the present study is that aerotaxis in A. brasilense appears to be methylation independent. There was no methanol release from either the wild type or any of the mutants upon addition or removal of oxygen by a continuous-flow assay, whereas there was methanol release in the same experimental setup when a gradient of malate was introduced instead of oxygen. Nevertheless, the cheB and cheR mutants had a null phenotype for aerotaxis by both temporal and spatial gradient assays. Aerotaxis was shown to be methylation dependent for B. subtilis (37) and H. salinarum (19) and methylation independent for E. coli (6, 22). Normal aerotaxis in E. coli requires a CheB protein; however, the absence of the enzyme only inverts (7) but does not abolish the response in a temporal gradient assay, as it does with A. brasilense. Our results indicate a similar methylation-independent and CheBR-dependent behavior in response to the strong attractants fructose and succinate. The exact mechanism of methylation-independent sensory adaptation is currently not known for any bacterial species, including the model organism E. coli (6). However, it is conceivably a major adaptation mechanism in A. brasilense.
Model for the role of CheB and CheR.
Although this study has established an important role for the CheB and CheR proteins in chemotaxis and aerotaxis in A. brasilense, it has revealed a significant departure from the role in model organisms. The contribution of these enzymes to the overall behavior in azospirilla appears to be different from that in the model organism for chemotaxis, E. coli, and in other organisms that possess a single set of chemotaxis genes, such as B. subtilis. On the other hand, there are many similarities with chemotaxis in R. sphaeroides, which emerges as a model to study the motility behavior in organisms with multiple chemotaxis pathways (36). Most of the observed deviations from known features of the chemotactic response are likely due to the presence of multiple chemotaxis pathways in A. brasilense. We have shown that not only cheB and cheR mutants but also a mutant lacking the entire chemotaxis operon encoding the CheB and CheR proteins is impaired in, but not null for, chemotaxis. Therefore, other chemotaxis pathways contribute to the overall control of flagellar motility.
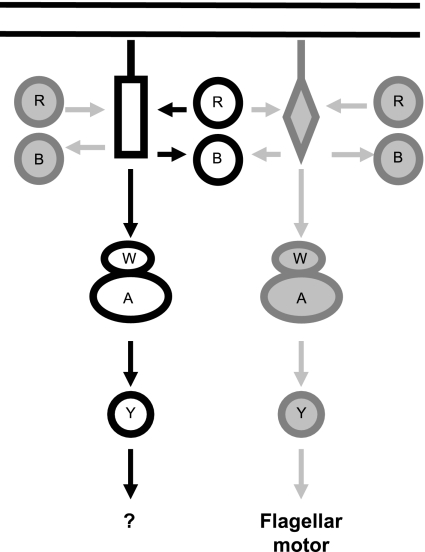
In order to reconcile the data, we propose a model in which the chemotaxis pathway under study does not primarily control motility and chemotaxis in A. brasilense. However, CheB and CheR from this pathway affect the methylation status of a chemotaxis transducer(s) that is ultimately involved in controlling chemotaxis, perhaps via cross-regulation with another chemotaxis pathway(s) at the level of chemoreceptor methylation/demethylation (Fig. 7). Our findings that multiple methylation/demethylation systems contribute to methanol release in response to a single chemostimulation are consistent with such a model. For E. coli, it was proposed that the combined activities of CheB and CheR on chemotaxis transducers modulate signal amplification and sensitivity in chemotaxis (17, 28). We argue that multiple methylation/demethylation systems contribute to the overall methylation status of chemotaxis transducers, which in turn modulates the overall sensitivity of the chemotactic response and chemosensory adaptation. We also argue that in the absence of CheB and/or CheR the overall methylation status of chemotaxis transducers is modified, altering the sensitivity of the chemotaxis response. This in turn may explain why mutants lacking CheB and/or CheR are unable to respond to essentially any stimuli in a temporal gradient assay while retaining chemotaxis in spatial gradients. Future studies aiming at the identification and characterization of other putative multiple chemotaxis homologs in A. brasilense will be critical for understanding the molecular details underlying such complex behavior and will likely lead to the modification of the working model proposed here.
FIG. 7.
Working model for the role of CheB and CheR proteins in A. brasilense chemotaxis. The CheB (B) and CheR (R) proteins under study and the che pathway are shown in solid black lines. Other putative chemotaxis homologs are shown in gray. W, A, and Y represent CheW, CheA, and CheY, respectively. Cytoplasmic C-terminal domains of putative chemotaxis transducers interacting with CheB and CheR are shown as a rectangle and a diamond. The model argues that the primary function of the chemotaxis pathway under study is probably not to control motility behavior. CheB and CheR may contribute to chemotaxis and aerotaxis because of their effect on the methylation status of the chemotaxis transducer(s) that is ultimately involved in controlling motility (see text for details).
Acknowledgments
We thank G. L. Hazelbauer for critical reading of the manuscript and many helpful suggestions and A. Lin and L. Hall for technical assistance.
This work was supported by a CAREER award from the National Science Foundation (MCB-0347218) to G.A. S.N.L. was supported by a NSF REU supplement to the CAREER award.
REFERENCES
- 1.Alexandre, G., S. E. Greer, and I. B. Zhulin. 2000. Energy taxis is the dominant behavior in Azospirillum brasilense. J. Bacteriol. 182:6042-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandre, G., S. E. Greer-Phillips, and I. B. Zhulin. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28:113-126. [DOI] [PubMed] [Google Scholar]
- 3.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-826. [DOI] [PubMed] [Google Scholar]
- 4.Barak, R., I. Nur, Y. Okon, and Y. Henis. 1982. Aerotactic response of Azospirillum brasilense. J. Bacteriol. 152:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashan, Y., G. Holguin, and L. E. de-Bashan. 2004. Azospirillum-plant relationships: physiological, molecular, agricultural, and environmental advances (1997-2003). Can. J. Microbiol. 50:521-577. [DOI] [PubMed] [Google Scholar]
- 6.Bibikov, S. I., A. C. Miller, K. K. Gosink, and J. S. Parkinson. 2004. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang, C. V., M. Niwano, J. Ryu, and B. L. Taylor. 1986. Inversion of aerotactic response in Escherichia coli deficient in CheB protein methylesterase. J. Bacteriol. 166:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 10.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greer-Phillips, S. E., B. B. Stephens, and G. Alexandre. 2004. An energy taxis transducer promotes root colonization by Azospirillum brasilense. J. Bacteriol. 186:6595-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauwaerts, D., G. Alexandre, S. K. Das, J. Vanderleyden, and I. B. Zhulin. 2002. A major chemotaxis gene cluster in Azospirillum brasilense and relationships between chemotaxis operons in alpha-proteobacteria. FEMS Microbiol. Lett. 208:61-67. [DOI] [PubMed] [Google Scholar]
- 13.Jeziore-Sassoon, Y., P. A. Hamblin, C. A. Bootle-Wilbraham, P. S. Poole, and J. P. Armitage. 1998. Metabolism is required for chemotaxis to sugars in Rhodobacter sphaeroides. Microbiology 144:229-239. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, Z. Y., and C. E. Bauer. 2001. Component of the Rhodospirillum centenum photosensory apparatus with structural and functional similarity to methyl-accepting chemotaxis protein chemoreceptors. J. Bacteriol. 183:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehry, M. R., T. G. Doak, and F. W. Dahlquist. 1984. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J. Biol. Chem. 259:11828-11835. [PubMed] [Google Scholar]
- 16.Kirby, J. R., C. J. Kristich, S. L. Feinberg, and G. W. Ordal. 1997. Methanol production during chemotaxis to amino acids in Bacillus subtilis. Mol. Microbiol. 24:869-878. [DOI] [PubMed] [Google Scholar]
- 17.Lamanna, A. C., J. E. Gestwicki, L. E. Strong, S. L. Borchardt, R. M. Owen, and L. L. Kiessling. 2002. Conserved amplification of chemotactic responses through chemoreceptor interactions. J. Bacteriol. 184:4981-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laszlo, D. J., and B. L. Taylor. 1981. Aerotaxis in Salmonella typhimurium: role of electron transport. J. Bacteriol. 145:990-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindbeck, J. C., E. A. Goulbourne, Jr., M. S. Johnson, and B. L. Taylor. 1995. Aerotaxis in Halobacterium salinarum is methylation-dependent. Microbiology 141:2945-2953. [DOI] [PubMed] [Google Scholar]
- 20.Martin, A. C., G. H. Wadhams, D. S. Shah, S. L. Porter, J. C. Mantotta, T. J. Craig, P. H. Verdult, H. Jones, and J. P. Armitage. 2001. CheR- and CheB-dependent chemosensory adaptation system of Rhodobacter sphaeroides. J. Bacteriol. 183:7135-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metcalf, W. W., and B. L. Wanner. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacements. Gene 129:17-25. [DOI] [PubMed] [Google Scholar]
- 22.Niwano, M., and B. L. Taylor. 1982. Novel sensory adaptation mechanism in bacterial chemotaxis to oxygen and phosphotransferase substrates. Proc. Natl. Acad. Sci. USA 79:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nordmann, B., M. R. Lebert, M. Alam, S. Nitz, H. Kollmannsberger, D. Oesterhelt, and G. L. Hazelbauer. 1994. Identification of volatile forms of methyl groups released by Halobacterium salinarium. J. Biol. Chem. 269:16449-16454. [PubMed] [Google Scholar]
- 24.Perazzona, B., and J. L. Spudich. 1999. Identification of methylation sites and effects of phototaxis stimuli on transducer methylation in Halobacterium salinarum. J. Bacteriol. 181:5676-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Simon, R., U. Priefer, and A. Pülher. 1983. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 27.Sourjik, V. 2004. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 12:569-576. [DOI] [PubMed] [Google Scholar]
- 28.Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 99:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Springer, W. R., and D. E. Koshland, Jr. 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. USA 74:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steenhoudt, O., and J. Vanderleyden. 2000. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 24:487-506. [DOI] [PubMed] [Google Scholar]
- 31.Stock, J. B., A. M. Maderis, and D. E. Koshland, Jr. 1981. Bacterial chemotaxis in the absence of receptor carboxyl methylation. Cell 27:37-44. [DOI] [PubMed] [Google Scholar]
- 32.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, pp. 1103-1129. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 33.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thoelke, M. S., J. R. Kirby, and G. W. Ordal. 1989. Novel methyl transfer during chemotaxis in Bacillus subtilis. Biochemistry 28:5585-5589. [DOI] [PubMed] [Google Scholar]
- 35.Vanstockem, M., K. Michiels, J. Vanderleyden, and A. P. Van Gool. 1987. Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum: physical analysis of Tn5 and Tn5-Mob insertion mutants. Appl. Environ. Microbiol. 53:410-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 37.Wong, L. S., M. S. Johnson, I. B. Zhulin, and B. L. Taylor. 1995. Role of methylation in aerotaxis in Bacillus subtilis. J. Bacteriol. 177:3985-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16:994-996. [PubMed] [Google Scholar]
- 39.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 40.Yonekawa, H., H. Hayashi, and J. S. Parkinson. 1983. Requirement of the cheB function for sensory adaptation in Escherichia coli. J. Bacteriol. 156:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhulin, I. B. 2001. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv. Microb. Physiol. 45:157-198. [DOI] [PubMed] [Google Scholar]
- 42.Zhulin, I. B., and J. P. Armitage. 1993. Motility, chemokinesis, and methylation-independent chemotaxis in Azospirillum brasilense. J. Bacteriol. 175:952-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhulin, I. B., V. A. Bespalov, M. S. Johnson, and B. L. Taylor. 1996. Oxygen taxis and proton motive force in Azospirillum brasilense. J. Bacteriol. 178:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]