Abstract
The uracil salvage pathway in Lactobacillus plantarum was demonstrated to be dependent on the upp-pyrP gene cluster. PyrP was the only high-affinity uracil transporter since a pyrP mutant no longer incorporated low concentrations of radioactively labeled uracil and had increased resistance to the toxic uracil analogue 5-fluorouracil. The upp gene encoded a uracil phosphoribosyltransferase (UPRT) enzyme catalyzing the conversion of uracil and 5-phosphoribosyl-α-1-pyrophosphate to UMP and pyrophosphate. Analysis of mutants revealed that UPRT is a major cell supplier of UMP synthesized from uracil provided by preformed nucleic acid degradation. In a mutant selection study, seven independent upp mutants were isolated and all were found to excrete low amounts of pyrimidines to the growth medium. Pyrimidine-dependent transcription regulation of the biosynthetic pyrimidine pyrR1-B-C-Aa1-Ab1-D-F-E operon was impaired in the upp mutants. Despite the fact that upp and pyrP are positioned next to each other on the chromosome, they are not cotranscribed. Whereas pyrP is expressed as a monocistronic message, the upp gene is part of the lp_2376-glyA-upp operon. The lp_2376 gene encodes a putative protein that belongs to the conserved protein family of translation modulators such as Sua5, YciO, and YrdC. The glyA gene encodes a putative hydroxymethyltransferase involved in C1 unit charging of tetrahydrofolate, which is required in the biosynthesis of thymidylate, pantothenate, and purines. Unlike upp transcription, pyrP transcription is regulated by exogenous pyrimidine availability, most likely by the same mechanism of transcription attenuation as that of the pyr operon.
Growing organisms need pyrimidine molecules to synthesize their nucleic acids (DNA and RNA). The pyrimidine needs of living cells are fulfilled either via de novo synthesis or via salvage of preformed pyrimidine bases and nucleosides provided by the surrounding medium (Fig. 1) (23). In the biosynthetic pathway, UMP is formed in six enzymatic steps. In Lactobacillus plantarum, the enzymes are encoded by the pyr operon with the gene order pyrR1-B-C-Aa1-Ab1-D-F-E (9). Uracil salvage is found in L. plantarum since a pyrD mutant impaired in de novo pyrimidine synthesis was able to use uracil as the sole pyrimidine source (26). The key enzyme in microbial pyrimidine salvage is uracil phosphoribosyltransferase (UPRT) (EC 2.4.2.9), which catalyzes the conversion of uracil and 5-phosphoribosyl-α-1-pyrophosphate (PRPP) to UMP and pyrophosphate. On the L. plantarum genome, a putative upp gene codes for a protein of 209 residues with significant sequence identity with characterized UPRT: 75% identity with Lactococcus lactis UPRT (17) and 68% identity with Bacillus subtilis UPRT (16). The upp gene was found immediately upstream of the pyrP gene (EMBL sequence accession no. AL935263). The pyrP gene encodes a putative 426-residue transmembrane protein of the conserved UraA xanthine/uracil permease family (COG2333). In L. plantarum, the upp and pyrP genes are contiguous, oriented in the same direction but separated by a 1.1-kb intergenic region with no detected open reading frames, suggesting the absence of cotranscription of the two genes. This genetic organization is different from previously studied organisms: the upp and uraA genes are unlinked in Streptomyces (GenBank accession no. NC_003888) (13) and form a bicistronic operon in Escherichia coli (1), while the pyrP gene is cotranscribed with the adjacent pyrR/pyrB genes in B. subtilis (32) and L. lactis (18). In these organisms, pyrP transcription is regulated in response to pyrimidine availability.
FIG. 1.
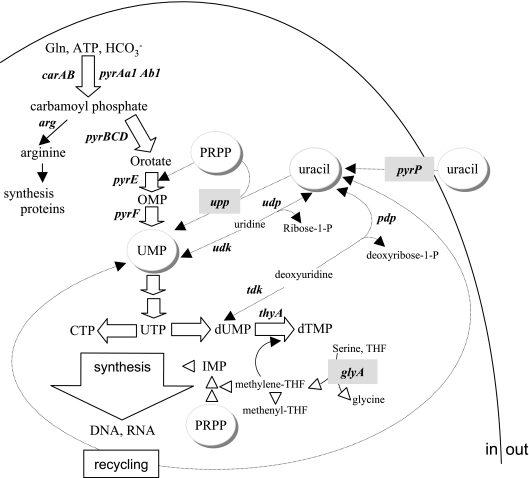
Uracil salvage and pyrimidine biosynthetic pathways. Enzymes are identified by their corresponding gene designation for the uracil salvage pathway (thin arrows), de novo pyrimidine synthesis (thick arrows), and the IMP biosynthetic pathway (arrowheads), including steps involved in the incorporation of C1 units from serine into THF. The enzymes catalyzing UMP synthesis are encoded by the pyrR1-B-C-Aa1-Ab1-D-F-E operon (9). Abbreviations for genes encoding enzymes are as follows: glyA, serine hydroxymethyltransferase (EC 2.1.2.1); ndk, nucleoside-diphospho kinase (EC 2.7.4.14); udk, uridine kinase (EC 2.7.1.48); udp, uridine phosphorylase (EC 2.4.2.3); upp, uracil phosphoribosyltransferase (EC 2.4.2.9); pdp, pyrimidine-nucleoside phosphorylase (EC 2.4.2.2); pyrP, uracil permease; tdk, thymidine kinase (EC:2.7.1.21); and thyA, thymidylate synthase (EC 2.1.1.45).
Pyrimidine-dependent repression occurs by a mechanism of transcription attenuation that has been most studied for B. subtilis (32) but that also occurs in many other gram-positive bacteria, including L. plantarum (26) and L. lactis (18). When uracil is present in the culture medium, de novo pyrimidine synthesis is inhibited due to repression of the expression of the pyrimidine biosynthetic genes encoded by the pyr operon. The first gene of the B. subtilis and L. plantarum pyr operon encodes an RNA binding regulator PyrR (called PyrR1 in L. plantarum) that represses expression of the downstream pyr genes (26). When intracellular UMP concentration is high, the repressor-UMP complex binds the target RNA hexaloop formed at the tip of the anti-antiterminator mRNA hairpin structure in the 5′ leader sequence of regulated genes. Binding of the repressor prevents formation of the antiterminator structure, allowing the terminator RNA loop to be formed and the transcription to be terminated. When the intracellular UMP is low, the repressor remains unbound, favoring formation of the antiterminator RNA loop, preventing terminator loop formation. The transcription proceeds, and the structural genes are transcribed. Evidence that in L. plantarum the pyr operon is regulated by transcription attenuation in response to exogenous uracil has been obtained using transcription studies and intracellular pyrimidine triphosphate nucleoside pool size determination for wild-type and deregulated mutants (26): (i) mutations in conserved regions of L. plantarum PyrR1 and B. subtilis PyrR (flexible loop, phosphoribosyltransferase domain, and dimer loop/mRNA binding) suggested that these proteins are functionally similar; (ii) mutations of attenuation site no. 2 are located directly downstream of the pyrR1 gene, such as in the consensus of the PyrR-binding hexaloop. Unlike B. subtilis, in L. plantarum, a second PyrR homolog encoded by the pyrR2 gene shares 62% identity at the amino acid level with PyrR1. The pyrR2 gene is not a member of the pyr operon, and its function has not been clarified yet (26).
Spontaneous uracil-deregulated mutants, including the upp-pyrP mutants characterized in this paper, were obtained in the genetic context of a strain impaired in the first step of UMP synthesis (26). This step leads to the formation of carbamoyl phosphate (CP) from glutamine, ATP, and inorganic carbon (carbon dioxide or bicarbonate) (Fig. 1). In L. plantarum, CP is synthesized by two carbamoyl phosphate synthases (CPS) (EC 6.3.5.5), the pyrimidine-repressed CPS-P encoded by the pyrAa1-pyrAb1 genes present in the pyr operon and the arginine-repressed CPS-A encoded by the carAB operon (27). The presence of two functional CPS reflects the cells' need for CP in both the pyrimidine and the arginine biosynthetic pathways (Fig. 1). Deletion of both CPS conferred pyrimidine and arginine auxotrophy (27). Strain FB335 harbors a ΔcarAB mutation so that CP synthesis relies solely on the pyrimidine-regulated CPS-P (27). In the presence of exogenous uracil believed to result in high UMP pools, strain FB335 growth was inhibited and the corresponding phenotype was designated uracil sensitive (Table 1). Uracil-resistant (UraR) spontaneous derivatives of the FB335 strain were isolated and partially characterized as previously described (26). Among the characterized genetic lesions that conferred the UraR phenotype, some mutations invalidated PyrR1 or the transcription attenuator located between pyrR1 and pyrB (26). In one UraR mutant (mutant HN38), the upp gene had an insertion of the mobile insertion sequence element ISLpl1 (25). The pyrR1 mutants were prototrophs for both arginine and pyrimidines. Conversely, the upp mutant HN38 as well as the other uncharacterized UraR mutants were high CO2-requiring (HCR) prototrophs and required pyrimidines and arginine when grown in air but not in air enriched with carbon dioxide (26). These HCR mutants harbored wild-type pyrR1 and pyrR2 genes as well as wild-type cis regulatory regions of the pyr operon (localized before pyrR1 and in the intergenic region between pyrR1 and pyrB) and wild-type structural genes pyrAa1-pyrAb1 of the CPS-P (26). In this paper, the uncharacterized deregulated mutants that displayed UraR and HCR phenotypes were shown to harbor genetic lesions in the pyrP and/or upp gene. Furthermore, the role of upp and pyrP in the pyrimidine salvage pathway in L. plantarum, their genetic organization, and their pyrimidine-dependent regulation were investigated.
TABLE 1.
Characteristics of the L. plantarum subsp. plantarum strains
| Strain | Growth phenotypeb | Pyrimidine excretion (μg/ml)c | 5FU MICd
|
Mutation(s)a | |
|---|---|---|---|---|---|
| With purines | Without purines | ||||
| CCM1904 | Prototroph | 0 | 0.03 | 0.0025 | Wild type |
| FB335 | UraS prototroph | 0 | 0.03 | 0.0025 | ΔcarAB (27) |
| AE1023 | HCR | 0 | 0.03 | NT | Attenuation site at 2 mutated between pyrR1 and pyrB, resulting in pyrimidine-independent low transcription of the pyr operon (26) |
| AE1026 | Prototroph | 4.3 | 0.03 | 0.0025 | PyrR1 D104Y (26) |
| U2 | HCR | 1.4 | 0.4 | NT | ΔcarAB Δupp-pyrP; chimeric Upp after residue S139; PyrP absent |
| U30 | HCR | 1.3 | 0.4 | 0.02 | ΔcarAB Δupp-pyrP; Upp and PyrP absent |
| U10 U35; U37 | HCR | 0.1 | 0.03 | NT | ΔcarAB Δupp; deletion of 7 residues (I12-I18) in Upp |
| U11 | HCR | 0.3 | 0.03 | NT | ΔcarAB Δupp; Upp C-terminal region truncation (S153-K209) |
| U20 | HCR | 2.0 | 0.03 | NT | ΔcarAB Δupp; Upp C-terminal region truncation (N154-K209) |
| U27 | HCR | 0.1 | 0.03 | 0.001 | ΔcarAB Δupp; Upp deletion (I55-K209) |
| U36 | HCR | 0.2 | 0.03 | NT | ΔcarAB upp; Upp G81D due to mutation G783A |
| HN38 | HCR | 0.1 | 0.03 | 0.001 | ΔcarAB upp::ISLpl1 (25) |
| U39 | HCR | 0.1 | 0.03 | NT | ΔcarAB upp; Upp truncation of the C-terminal region from Q126 to K209 due to a point mutation (C662T) |
Sequence data refer to EMBL accession number AJ012720 for the upp-pyrP locus.
CO2-dependent uracil and arginine nutritional needs were tested on the defined medium DLA that contains purines, no pyrimidines, and all amino acids but arginine. Prototrophs grew without arginine and uracil. UraS (uracil sensitive) prototroph growth was inhibited by uracil in DLA. HCR, high CO2-requiring prototroph, which required exogenous arginine and uracil only at low carbon dioxide levels but not under carbon dioxide-enriched conditions. The HCR prototrophs are uracil resistant, as defined by their growth in the presence of uracil when carbon dioxide-enriched air was provided.
μg of pyrimidine nucleotide/ml of full-grown culture supernatant.
The MIC gives the minimal concentration in μg/ml of the toxic uracil analog 5FU, which inhibited the strain growth in the absence or presence of purines (adenine, guanine, hypoxanthine, and xanthine). NT, not tested.
MATERIALS AND METHODS
Lactobacilli and physiological tests.
All tested lactobacilli derive from the pyrimidine and arginine prototroph L. plantarum subsp. plantarum CCM 1904 (Table 1). Unless specified, cultures were performed at 30°C in air enriched with 4% CO2 in a water-jacketed CH/P incubator (Forma Scientific). Pyrimidine and arginine nutritional requirements were tested in the presence of arginine (50 μg/ml) or uracil (50 μg/ml) at 30°C on DLA plates. The DLA medium contains purines: adenine (0.09 g/liter), guanine HCl (0.05 g/liter), hypoxanthine (0.03 g/liter), and xanthine (0.005 g/liter). The composition of the previously published DLA (6) was modified, with no deoxyguanosine added, Tween 80 replacing Tween 40 and oleic acid (1 g/liter). The DLP medium (26) was used for nucleotide pool size determination.
The MIC of the toxic uracil analog 5′-fluorouracil (5FU) was determined under two conditions: the presence and absence of purines. Cell suspensions (15 μl of 5.107 cells/ml) were plated on DLA agar plates supplemented with arginine and increasing amounts of 5FU and incubated for 4 days. The amount of pyrimidines present in the culture supernatant of full-grown cells cultivated in DLA media supplemented with 50 μg/ml of arginine was quantified with a bioassay using the uracil auxotroph strain HN217 (26).
DNA techniques.
The upp and pyrP genes were sequenced from strain CCM 1904 on two overlapping PCR-amplified fragments, using different primer sets (Table 2). The PCRs were performed as previously described (26).
TABLE 2.
Primer list
| Genetic locus (EMBL database accession no.) and use(s)a | PCR product size (kb) | Primer name | Primer sequence (5′→3′)b |
|---|---|---|---|
| lp_2376-glyA-upp-pyrP (AL935263) | |||
| RT-PCR primer set a | 0.3 | LPpyrP-g4 | AATCATTCACAAAAAGGGGTGCG (F) |
| LPpyrP-d2 | TCATCAGAGACTGCATCACAACG (R) | ||
| RT-PCR primer set b | 2.0 | LPupp-SG | GTTTGAGGTTTTGGATCATCCGC (F) |
| RT-PCR primer set c | 1.3 | orfph-d2 | AATTAGATTGTTGTACGTCAGCC (R) |
| RT-PCR primer set d | 0.4 | LPupp-H1 | TATCCATCGCCATGATGGCTGAACC (R) |
| RT-PCR primer set e | 0.7 | glyc-g1 | CGGTCAACAAAGAAGCGATTCCG (F) |
| RT-PCR primer set f | 2.0 | Lp2375f | TGGGCGTTATGGCAACAGATG (F) |
| RT-PCR primer set g | 0.5 | Lp2376r | CAGAGTATTTATTGGTCAGCA (R) |
| pyrP probe (LPpyrP-g4/LPpyrP-d1) | 0.6 | LPpyrP-d1 | CGGCAATGATATACCCACAGACG (R) |
| upp amplification and sequencing | 2.4 | LPupp-E2 | ATGATTCCGGCAGCTAAAGTTGG (F) |
| orfph-g1 | CGAATGGCTGACGAAGATGTACC (F) | ||
| upp cloning | 0.8 | LPupp-f | CTATCGGATCCACTAAGGAGTGTGCGTAGC (F) |
| LPupp-r | CGATATTAGTAGGCGTAAGACACTTCGAAGTAT (R) | ||
| pyrP amplification and sequencing | 1.9 | orfph-g3 | GGGTGAGAAAGTCACATTATTTC (F) |
| LPatp-d3 | CGTCAGTCCAAGGAATTTGACTG (R) | ||
| LPpyrp-g3 | CTTCAAGTGGGATGCGAATTCTG (F) | ||
| rrn (AL935263) | |||
| RT-PCR and rrn probe | 0.4 | 907r | CCGTCAATTCCTTTGAGTTT (F) |
| 530f | GTGCCAGCAGCCGCGG (F) | ||
| pyrAb1 (X99978) | |||
| RT-PCR and pyrAb1 probe | 0.4 | N58 | CAGCTTCTGGTATTGCGC (F) |
| 2005 | ATCTTCAAAGCAGCATCCCG (R) |
Primer sets are indicated in Fig. 4A. The upp probe was obtained with primers LPupp-H1/LPupp-SG.
F, forward primer; R, reverse primer.
Transcription studies.
RNAs were extracted from strains U27, FB335, and CCM 1904, which displayed similar growth rates (data not shown) in mid-exponential growth phase (an optical density at 600 nm of 0.4), using TRIzol reagent (Invitrogen) as previously described (24). Contaminant DNA was removed by incubating the RNA extracts for 20 min at 37°C in the presence of DNase I (Amersham Biosciences), which was subsequently heat inactivated. Reverse transcriptase (RT) PCR was performed using the SuperScript one-step RT-PCR with a platinum Taq kit (Invitrogen) under the conditions previously described (24). Regulation of transcription was determined using Northern slot blot analysis. The upp and pyrP probes were PCR amplified (Table 2) and digoxigenin labeled, and the concentrations were estimated using the labeled DNA control from the digoxigenin DNA labeling and detection kit (Roche). For each probe, the optimal amount of RNA to be used for quantification was tested in the range of 0.1 to 20 μg of total RNA and was found to be different from those of the upp (2 μg), pyrP (20 μg), pyrAb1 (2 μg), and rrn (0.1 μg) transcript detections. Conditions of Northern hybridization and detection of the DNA-RNA hybrids with the alkaline phosphatase chemiluminescent substrate CDP-Star (Roche) were previously specified (24). At least two independent assays were performed for each tested condition. The background level was subtracted from the measured signal quantified with Quantity One software (Bio-Rad). For each probe, the level of expression was estimated by dividing the measured probe signal by the measured rrn signal.
Cloning of L. plantarum upp gene in E. coli.
The upp gene was PCR amplified from the L. plantarum wild-type strain CCM 1904 with primer set LPupp-f/LPupp-r (Table 2), using the Promega Pfu Taq polymerase and cycles of denaturation at 94°C for 45 s, hybridization at 52°C for 45 s, and elongation at 72°C for 2 min 30 s. After 35 cycles, a postelongation at 72°C for 30 min was performed. The resulting 0.77-kb fragment was ligated in the cloning vector pUC18 (4) after digestion with the restriction enzymes BamHI and HindIII, respectively, present at unique sites in primers LPupp-f and LPupp-r. The ligation mixture was electroporated into E. coli TG2, and white transformants were selected on LB plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (32 μg/ml) and ampicillin (50 μg/ml). One of these clones harbored plasmid pLPupp, whose insert contained the L. plantarum upp gene wild-type sequence (data not shown) expressed from the vector's promoter. L. plantarum upp function has been tested by complementation in the E. coli-deficient upp udp pyrF strain BM604 (2) on minimal medium M9 supplied with either uracil or uridine at 10 or 20 μg/ml, respectively.
Uracil transport assays.
Exponentially grown cells (an optical density at 450 nm of 0.5) in DLA medium were transferred to an Eppendorf tube containing 14C-labeled uracil at a specific activity of 52 mCi/mmol. Samples were taken after 15, 30, 45, and 60 seconds using uracil concentrations ranging from 5 to 0.2 μM. The cells were trapped by filtration through a 0.22-μm filter, and the remaining medium was removed by two washes with 5 ml 0.9% NaCl and 100 μM uracil at room temperature. The radioactivity on the filters was determined in a scintillation counter. A filter with a medium sample was included in order to determine the specific activity in the individual experiments. The uptake was linear for up to 60 seconds, even at 0.5 μM. Consequently, an incubation time of 30 seconds was used to determine the kinetic constants. The classical Michaelis-Menten constants Km and Vmax were calculated by plotting the data in a double-reciprocal plot. Assuming that an optical density of 1 at 450 nm corresponds to 3.108 cells/ml, the uptake rate was determined in uracil molecules per second per cell.
Determination of the nucleotide pools.
Intracellular nucleotide pool quantification in L. plantarum has been previously described (26).
RESULTS
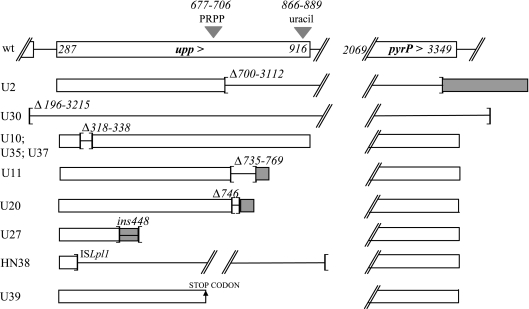
Mutations that may alter preformed uracil utilization were searched in 10 pyrimidine-deregulated derivatives of strain FB335. The phenotypes of these mutants were previously characterized as UraR, high CO2-requiring prototrophy, and excretion of low amounts of preformed pyrimidine bases and nucleosides (26). The sequence of the upp-pyrP locus in the mutants was compared to that of the wild type. All mutants harbored lesions in upp (Table 1 and Fig. 2). In addition to the upp mutation, the adjacent pyrP gene was also mutated in strains U2 and U30. The genetic lesions were point mutations in two strains (U36 and U39), deletions ranging from 1 to 2,400 nucleotides in seven strains (U2, U10, U11, U20, U30, U35, and U37), insertion of 22 nucleotides in strain U27, and insertion of ISLpl1 in the previously described strain HN38 (25). Six mutants (U30, U2, U27, U39, U11, and U20) had major truncations (25% to 100%) of the upp open reading frame (Fig. 2). The same 21-nucleotide (nt)-long deletion was found in three mutants (U10, U35, and U37) that led to the loss of the seven residues in the N-terminal part of the UPRT.
FIG. 2.
Analysis of the upp and pyrP mutations found in spontaneous uracil-resistant clones. Genes are schematized as white rectangles when the resulting protein is wild type (wt) or as gray rectangles when mutations led to chimeric proteins. Strain names are indicated on the left. PyrP was inactivated only in strains U30 and U2. Insertion and deletion events delineated by brackets are oriented outwards and inwards, respectively. In strain HN38, the mobile insertion sequence ISLpl1 inactivated upp (25). In mutant U27, the sequence 5′-TATGCCGTTGAAGGACATTGAG-3′ was inserted at nucleotide 448. Numbers refer to nucleotide positions in the EMBL database sequence accession no. AJ012720. The PRPP- and uracil-binding sites were determined using UPRT sequence alignments as determined in reference 29.
L. plantarum upp gene codes for a UPRT.
To verify that the upp gene was coding for a functional UPRT, the ability of the L. plantarum upp gene to complement a deficient upp E. coli mutant was assessed. An L. plantarum upp gene present in plasmid pLPupp was introduced by transformation into E. coli strain BM604. Strain BM604 (upp, pyrF) requires uridine in order to grow and has a pyrimidine requirement which uracil is unable to satisfy (2). Transformants were tested on minimal medium M9 in the presence of either uracil or uridine. Only transformants harboring plasmid pLPupp grew in the presence of uracil (data not shown). Thus, the L. plantarum upp gene codes for a UPRT enzyme that is functional in the heterologous E. coli genetic background.
Kinetic constants of L. plantarum uracil uptake transporter PyrP.
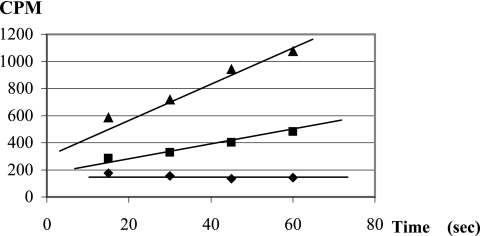
Increased resistance to the toxic uracil analog 5FU was seen only when both pyrP and upp genes were mutated as found in strains U2 and U30 (Table 1). The 5FU resistance could be explained if pyrP would encode a uracil transporter. To investigate whether pyrP encodes a uracil transporter, the uptake of 14C-labeled uracil was measured in strains U2 (ΔpyrP Δupp), U27 (Δupp), and FB335 (wild-type pyrP and wild-type upp). The U2 mutant was no longer able to transport uracil across the membrane, whereas uracil was transported across the membrane in strain U27 (Fig. 3) and in the strain FB335 (data not shown). Thus, under the tested conditions, pyrP was the only high-affinity uracil transporter in L. plantarum. The kinetic properties of uracil uptake were determined in genetic backgrounds that minimized rapid uracil utilization after cell entry. Since at least 97% of the incoming uracil was metabolized by the UPRT encoded by upp in L. lactis (17) and since UPRT is the key enzyme in the preformed uracil utilization pathway in microorganisms (22), strain U27 was used to determine the kinetic constants of uracil uptake in L. plantarum. The Km value for uracil was 0.7 ± 0.1 μM; Vmax was 3.5 × 103 ± 0.6 × 103 molecules per second per cell.
FIG. 3.
Uracil transport in the pyrP strain. Radioactively labeled uracil was added at 0.5 μM to L. plantarum U2 (upp, pyrP) (diamonds), 0.5 μM to U27 (upp) (squares), and 1.0 μM to U27 (triangles). Samples were filtered at 15-second intervals, and the radioactivity retained by the cells was measured and plotted against the incubation time.
The pyrP gene and the upp gene are not cotranscribed.
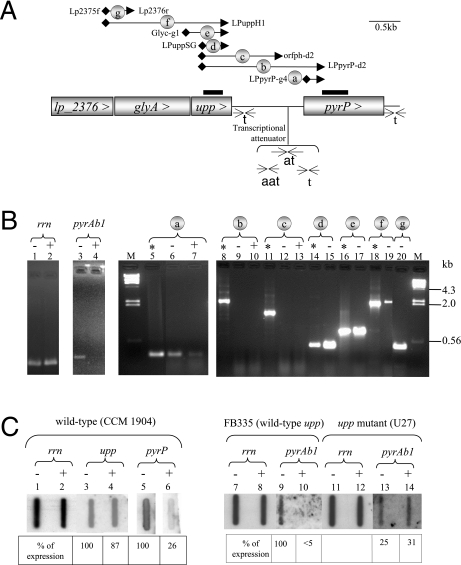
The four genes lp_2376, glyA, upp, and pyrP were contiguous on the L. plantarum genome and oriented in the same direction. The upp and pyrP 1,257-base-pair intergenic region was found to be well conserved between strains CCM 1904 and WCFS1 (less than 0.8% divergence), and no protein coding sequence was detectable in silico. To test whether these genes were cotranscribed, reverse transcriptions coupled with PCR amplifications were performed on mRNA extracts with different primer sets (Fig. 4A) and included the following controls. The specificity of hybridization of the primer sets was verified in classical PCR amplification using DNA matrices (Fig. 4B, lanes with an asterisk). The absence of contaminant DNA templates was confirmed (Fig. 4B, lanes 9, 10, 12, and 13). Transcription of genes pyrP and upp was identified using primer sets a and d, respectively (Fig. 4B, lanes 6 and 15). The upp-pyrP cotranscription was assessed with primer set b (Fig. 4B, lanes 9 and 10). Transcription from upp into the intergenic region was tested with primer set c (Fig. 4B, lanes 12 and 13). No RT-PCR products were obtained with increasing amounts (30 ng to 200 ng) of mRNA extracted from cells grown with or without uracil, indicating the absence of upp-pyrP cotranscription. Transcription arrest after upp and pyrP may occur at distinct predicted rho-independent terminators, located 11 nucleotides after the upp stop codon (nt 928 to 969; ΔG of −20.7 kcal · mol−1) and 50 nucleotides downstream of the pyrP stop codon (nt 3399 to 3440; ΔG of −14.1 kcal · mol−1) (database accession no. AJ012720).
FIG. 4.
Results of transcriptional studies of the upp-pyrP locus in L. plantarum. (A) Gene organization and transcription terminator localization. The orientation and the name of the genes are indicated in rectangles. Sequences involved in RNA hairpin structures are schematized with facing arrows and labeled t (terminator), at (antiterminator), or aat (anti-antiterminator). In circles, primer sets are named from a to g. Filled diamonds represent forward primers, and filled arrowheads represent reverse primers. Thick black lines delineate DNA probes used in Northern slot blot analysis. (B) Results of reverse transcriptase coupled with PCR amplification. The sizes of the expected bands were controlled by PCR amplification in the presence of CCM 1904 genomic DNA (lanes marked with an asterisk), and band sizes were estimated using DNA molecular marker bands (lane M). RNAs were extracted from cells grown with (+) or without (−) 50 μg/ml of uracil in defined DLA medium (for the presented experiment, cells were grown in aerobiosis but similar amplification patterns were obtained with cells grown in air enriched with 4% CO2). RT-PCR was performed with different RNA amounts: 20 ng for the 16S rrn gene (lanes 1 and 2) and the pyrAb1 gene (lanes 3 and 4), ng for the pyrP gene (lanes 6 and 7), the upp gene (lane 15), and the lp_2376-glyA-upp gene cluster (lanes 17, 19, and 20), and 200 ng for the intergenic upp-pyrP region (lanes 9 to 10 and 12 to 13). (C) Northern slot blots with probes specific to the rrn gene, the pyrP gene, the upp gene, and the pyrAb1 gene in the parental strain harboring a wild-type upp gene (lanes 1 to 10) and in its upp derivative strain U27 (lanes 11 to 14). The percentage of expression was calculated with two independent experiments. The relative amount of mRNA (signal measured with the test probe divided by the signal detected with the rrn probe) of the mutant was compared to the relative amount obtained with the wild type grown without uracil (100%).
Cotranscription of the lp_2376-glyA-upp genes was tested by RT-PCR using three primer sets, g, f, and e (Fig. 4B, lanes 20, 19, and 17, respectively). The sizes of the PCR products support the conclusion that the three genes are transcribed as a polycistronic message of approximately 3 kb. A sequence (TTGACA < 17 bp > aATAAa) with only two mismatches (shown in lowercase) compared to the L. plantarum consensus promoter sequence (TTGACA < 17 bp > TATAAT) was found 56 bp upstream of the lp_2376 initiation codon. From these transcription studies, we concluded that lp_2376-glyA-upp formed a separate transcriptional unit not including pyrP.
Pyrimidine repression of the pyrP gene and pyrimidine-independent transcription of the upp gene.
Pyrimidine-dependent expression of the pyrP and upp genes was assessed by RT-PCR and Northern slot blot analysis on mRNA extracted from cells grown with or without uracil. Two controls were performed: (i) pyrimidine repression was checked with a previously described pyrAb1-specific probe (Fig. 4C, lanes 9 and 10) (26), and (ii) pyrimidine-independent transcription of the rrn locus enabled the quantification of the relative mRNA extracted in the absence and presence of uracil (Fig. 4B [lanes 1 and 2] and C). When pyrP mRNA presence was assessed with primer set a (Fig. 4A), less RT-PCR product was obtained in mRNA extracted from cells grown in the presence of uracil than in mRNA extracted from cells grown without it (Fig. 4B, compare lanes 7 and 6). The pyrimidine-dependent repression of the pyrP gene was confirmed by Northern slot blot analysis with a pyrP probe. Only one quarter of the relative amounts of pyrP mRNA was measured in RNA extracts of cells cultivated in the presence of uracil compared to those of cells cultivated in the absence of uracil (Fig. 4C, compare lanes 6 and 5). Unlike with the pyrP probe, the relative amounts of upp mRNA detected with a specific upp probe were similar under both tested conditions (Fig. 4C, lanes 3 and 4). Thus, transcription of pyrP was regulated in response to exogenous uracil, whereas upp transcription was not.
Mutation in upp altered the uracil-dependent regulation of the biosynthetic pyr operon.
Unlike the uracil-sensitive parental strain FB335, whose growth depended on expression of the pyrimidine-repressed carbamoyl phosphate synthase (CPS-P), its upp derivatives were resistant to exogenous uracil. This ability to grow in the presence of uracil suggested the loss of pyrimidine-dependent regulation of the CPS-P-encoding genes pyrAa1-pyrAb1. Thus, the transcription levels of pyrAb1 were compared in strains harboring a wild-type and an inactive upp gene (Fig. 4C, compare lanes 7 to 10 with lanes 11 to 14). In the presence of uracil, significantly higher amounts of pyrAb1 mRNA were detected in the upp mutant U27 than in strain FB335 that harbored the wild-type upp gene (Fig. 4C, compare lanes 14 and 10). Moreover, in the absence of uracil, the amount of pyrAb1 mRNA was four times less in the upp mutant than in the wild-type strain (Fig. 4C, compare lanes 13 and 9). We concluded that in upp mutants, the pyr operon that includes the pyrAa1Ab1 genes was constitutively expressed whether or not exogenous uracil was added to the media, which explains why the upp mutants were no longer sensitive to exogenous uracil.
To investigate whether the upp mutation conferred dramatic changes in the UTP level, the nucleotide pool sizes were measured in strain U27. A mutation in the upp gene did not affect nucleotide triphosphate pool sizes, as strain U27 had the same pool size as the parental strain FB335 (data not shown).
DISCUSSION
The roles of pyrP and upp in the L. plantarum uracil salvage pathway were studied by genetic and physiological analysis. Allelic mutations in the pyrP gene conferred increased resistance to the toxic uracil analogue 5FU (Table 1) and impaired transport of radioactively labeled uracil when present in the micromolar scale (Fig. 3). Thus, the pyrP gene encodes a high-affinity uracil uptake facilitator. The upp gene codes for a functional UPRT since it complemented an E. coli upp mutant. UPRTs are members of the phosphoribosyl transferase family (Pfam accession number PF00156), defined by their protein fold and by a short sequence motif termed the “PRPP-binding site.” This motif was affected in mutant U36 by a G81D missense mutation. L. plantarum upp mutants excreted low amounts of uracil (0.1 to 0.5 μg/ml; one unexplained exception was mutant U20 that excreted 2.0 μg uracil per ml). In upp mutants of other microorganisms, such as E. coli (12) and L. lactis (17), a small increase in pyrimidine excretion has also been observed. Pyrimidine excretion can be explained by the inability of L. plantarum mutants to recover uracil arising from intracellular nucleotide turnover. We concluded that UPRT activity is required for optimal pyrimidine turnover in L. plantarum.
5-Fluorouracil metabolism in L. plantarum.
5FU is an analog of uracil, which is cytotoxic only after being converted by the pyrimidine salvage pathway enzymes to the nucleotide level. The most toxic metabolite formed inside the cell is 5-fluorodeoxyuridine monophosphate, which covalently binds to thymidylate synthase (thyA), the enzyme catalyzing the formation of dTMP from dUMP. Most microorganisms with impaired UPRT activity have increased 5FU resistance (22). However, in L. plantarum, the upp mutation had no effect on 5FU resistance in the absence of purine bases (Table 1). This is consistent with the observation made with L. lactis, where UPRT deficiency resulted only in a small effect on 5FU sensitivity. It was shown to be due to a direct conversion of 5FU to 5-fluorodeoxyuridine monophosphate through the action of pyrimidine phosphorylase utilizing deoxyribose-5-phosphate and thymidine kinase (17). A similar observation was made with B. subtilis (16). In media with purine bases, only the upp pyrP strain was more resistant towards 5FU than the parental strain. This unexpected observation could be explained by the fact that 5FU was taken up by the cell even when the uracil transporter (pyrP) was mutated, and candidates for facilitating this uptake could be purine base transporters. In the presence of exogenous purine bases, 5FU was significantly reduced due to the function of purine bases as competitive inhibitors of 5FU transport. This idea is supported by the finding that in the absence of purine bases, the pyrP mutant was 20 times more sensitive to 5FU (Table 1).
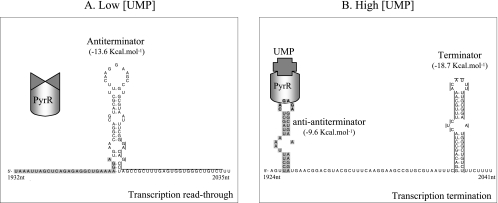
Model of pyrP pyrimidine-dependent regulation by transcription attenuation.
When wild-type L. plantarum was grown in the presence of uracil, pyrP transcription was reduced fourfold (Fig. 4B and C). PyrR-mediated transcription attenuation of the pyrP genes has been documented in other bacteria, such as B. subtilis (32), Bacillus caldolyticus (11), Enterococcus faecalis (10), and L. lactis (18). In L. plantarum, the pyrP leader mRNA harbors a good predicted transcription attenuation site (Fig. 5). The tip of the predicted anti-antiterminator stem is formed by a hexaloop CAGAGA identical to the proposed PyrR binding site (5, 26). The presence of the predicted transcription attenuation site and the measured decrease in pyrP transcription in response to exogenous uracil suggest that the pyrP gene, like the pyr operon, is regulated by PyrR-mediated transcription attenuation in L. plantarum.
FIG. 5.
Model of pyrimidine-dependent transcription attenuation. The transcription attenuation site sequence of the pyrP gene is shown (nucleotides 1924 to 2033 in EMBL database sequence accession no. AJ012720). (A) When the PyrR regulator remains unbound, the formation of the RNA loop antiterminator allows for the transcription of the downstream gene. (B) When the PyrR-UMP complex is bound to the hexaloop CAGAGA (in bold) at the tip of the anti-antiterminator hairpin, the terminator RNA loop can form and transcription stops before the downstream pyr gene. The ΔG values of the proposed RNA loops are indicated in parentheses.
Reduced CPS-P expression triggers CO2 nutritional needs.
The upp mutants displayed a high CO2 requirement for arginine and pyrimidine prototrophy. The HCR phenotype has previously been proposed to result from low intracellular CP pools under low inorganic carbon growth conditions, since CO2 is a substrate required in CP synthesis and since CP is a precursor in arginine and pyrimidine synthesis (Fig. 1). Low CP pools could be due to low CPS activity and/or to reduced amounts of CPS (7). CPS-P is the major source of CP for L. plantarum grown at low CO2 levels and can provide enough CP for both biosynthetic pathways (27). Mutant AE1023 was an HCR prototroph with low CPS-P expression due to constitutive low transcription of the pyr operon (F. Arsène-Ploetze and F. Bringel, unpublished data) (26). The upp mutants displayed the HCR phenotype and lowered CPS-P expression in the absence of uracil (Fig. 4C). These observations in different genetic contexts (AE1023 and upp mutants) raise the question of whether low CPS-P expression implies higher CO2 nutritional needs under conditions where arginine and pyrimidines are not provided. Previous studies have suggested that intracellular CO2 levels may be naturally low in L. plantarum grown under laboratory conditions since the two CO2-producing steps of the Krebs cycle (isocitrate dehydrogenase and 2-oxoglutarate dehydrogenase) are inoperative in L. plantarum (20). Low CO2 levels would be a limiting factor of CPS-P activity in L. plantarum grown without CO2 enrichment. In the wild-type strain, the CPS-P amount is sufficient to compensate for CO2 substrate shortage. However, in the case of reduced CPS expression (as in AE1023 and upp mutants) and substrate shortage (as when grown in ordinary air), CP synthesis would be too low to sustain arginine and pyrimidine prototrophy. By increasing the CO2 supply, CPS-P activity would increase, enough CP would be synthesized, and arginine and pyrimidine prototrophy would be restored in the mutants. We concluded that reduced CPS-P expression in the absence of uracil in L. plantarum explains its higher CO2 nutritional needs.
Can the UMP pools explain deregulated expression of the pyr operon in the upp mutants?
Compared to a strain harboring wild-type upp, in the upp mutant, transcription of the pyr operon was reduced fourfold when uracil was absent (Fig. 4C, compare lanes 9 and 13). Therefore, a functional upp is required for maximal expression in the absence of exogenous uracil. On the other hand, in the presence of uracil, higher expression of the pyr operon was observed in the upp mutant than in the wild type, where no expression was detected (Fig. 4C, compare lanes 14 and 10). Thus, a functional upp gene would be required in pyrimidine-dependent repression of the pyr operon. Inactivation of the upp gene resulted in the loss of the pyrimidine-dependent regulation so that the pyr operon expression levels were similar whether or not exogenous uracil was present (Fig. 4C, compare lanes 13 and 14). The L. plantarum repressor PyrR1 mediates pyrimidine-dependent transcription attenuation of the pyr operon (26). Mutations in the UMP/PRPP coregulator binding sites of L. plantarum PyrR1 reduced its repressor function. So, as demonstrated for B. subtilis PyrR (31), the ratio of UMP to PRPP is likely to regulate L. plantarum PyrR1 activity. This would imply that in the upp mutants, deregulation of the pyr operon may result from different PyrR1 expression levels or different UMP/PRPP ratios compared to those of the wild-type strain. Concerning the UMP/PRPP ratios, at least in the absence of uracil, the intracellular PRPP levels were similar in the wild-type and upp strains (data not shown), suggesting that the PRPP pool was not responsible for the observed deregulation. Since we have demonstrated that upp mutants were impaired in exogenous uracil assimilation and recycling of uracil derived from RNA degradation, the change in UMP concentration would infer deregulation. As a conclusion, we propose that the deregulation of the pyr operon observed in the upp strains would be the result of altered control of the activity or expression of the repressor PyrR1 by impaired intracellular pools of its effectors. Since PyrR1 was demonstrated not to be entirely responsible for pyr operon repression (26), another mechanism may also be involved.
lp_2376-glyA-upp operon.
The upp gene is cotranscribed with two upstream genes: lp_2376 and glyA. Gene lp_2376 codes for a putative 334-amino-acid-long protein that belongs to a conserved protein family with the C-terminal conserved domain (154 residues) of the sua5-yciO-yrdC domain (Pfam01300, Swiss-Prot/TrEMBL database), whose members can be found in prokaryotes and eucaryotes. Suggested members of this family are YciO and HypF (E. coli), YwlC (B. subtilis), Sua5 (Saccharomyces cerevisiae), and YrdC (Drosophila melanogaster and humans). Sua5 has been proposed to be involved in the reinitiation of translation (21). The C-terminal domain of E. coli YrdC has been shown to preferentially bind to double-stranded RNA and has been proposed as a putative ribosome maturation factor (14, 30). The implication of lp_2376 in translation has not been tested so far in L. plantarum.
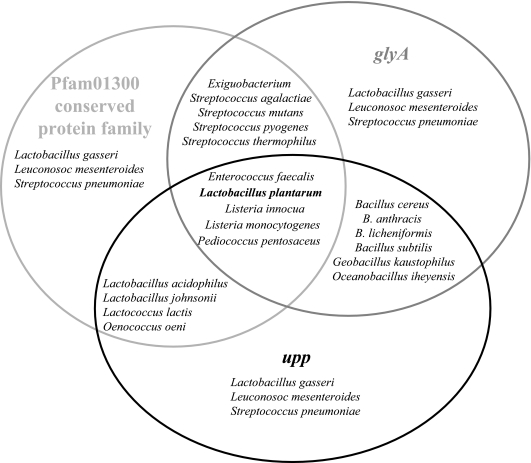
To evaluate whether other gram-positive bacteria might also harbor such an operon, the genetic linkage and respective gene orientation of the three genes were analyzed in sequenced genomes (Fig. 6). The gene order lp_2376-glyA-upp found in L. plantarum was conserved in the two Listeria species, E. faecalis, and Pediococcus pentosaceus. In other gram-positive bacteria, only two of the three genes were contiguous or interspaced by fewer than two genes. The genes were always oriented in the same direction within a gene cluster. This genetic linkage was not found in Lactobacillus gasseri, Leuconostoc mesenteroides, and Streptococcus pneumoniae. The fact that at least two genes were genetically linked in 16 gram-positive bacteria (Fig. 6) prompted us to search for metabolic links between them. Whereas the function of lp_2376 is unknown, the glyA gene is well characterized. The corresponding enzyme is responsible for the incorporation of C1 units from serine into tetrahydrofolate (THF) (Fig. 1). The C1 units of formyl-THF and methenyl-THF are the C1 donors in pantothenate (vitamin B5) biosynthesis in reactions catalyzed by PanB (EC 2.1.2.11), as well as in the purine biosynthetic pathway in reactions catalyzed by the PurN and PurH transformylases. Furthermore, one-carbon units are also required for the synthesis of the unique nucleotide component of DNA, thymidylate. The conversion of dUMP to thymidylate involves a methylation at C-5 of the uracil ring, with one carbon unit donated by methylene-THF (19). Thus, a gene involved in pyrimidine metabolism is linked and coexpressed with a gene involved in purine metabolism. The glyA gene is a member of the PurR regulon and is induced under general purine starvation in L. lactis (3) and B. subtilis (28). PRPP is the effector molecule in PurR-mediated regulation and functions as an inducer of pur genes by binding to the PurR protein. In B. subtilis, PurR acts as a repressor, preventing transcription initiation by binding in the promoter region. PRPP induces transcription since the PRPP-PurR complex no longer binds to the DNA (8). In L. lactis, the PRPP-PurR complex is an activator of transcription (15).
FIG. 6.
Gene linkage of lp_2376-glyA-upp genes in gram-positive bacteria. Black, gray, and light-gray circles refer to the distributions of the upp, glyA, and lp_2376 genes, respectively. Names of bacteria in which a genetic linkage of two genes was observed (cluster) are found in the circle intersection. In the intersection of the three circles are bacteria names with linked lp_2376-glyA-upp genes.
Under conditions of high intracellular uracil and PRPP pools, UPRT would catalyze UMP synthesis from uracil, while GlyA would provide C1 units for PRPP-PurR-induced IMP synthesis and thymidylate synthesis. Thus, under conditions of high PRPP and low UMP pools, GlyA and UPRT would be coexpressed and would contribute to balance purine and pyrimidine metabolisms in these microorganisms. This model will be tested by proteomic and transcriptional studies of L. plantarum under conditions of purine depletion or deregulation.
REFERENCES
- 1.Andersen, P. S., D. Frees, R. Fast, and B. Mygind. 1995. Uracil uptake in Escherichia coli K-12: isolation of uraA mutants and cloning of the gene. J. Bacteriol. 77:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, P. S., J. M. Smith, and B. Mygind. 1992. Characterization of the upp gene encoding uracil phosphoribosyltransferase of Escherichia coli K12. Eur. J. Biochem. 204:51-56. [DOI] [PubMed] [Google Scholar]
- 3.Beyer, N. H., P. Roepstorff, K. Hammer, and M. Kilstrup. 2003. Proteome analysis of the purine stimulon from Lactococcus lactis. Proteomics 3:786-797. [DOI] [PubMed] [Google Scholar]
- 4.Bolivar, F., R. L. Rodriguez, M. C. Betlach, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene 2:75-93. [DOI] [PubMed] [Google Scholar]
- 5.Bonner, E. R., J. N. D'Elia, B. K. Billips, and R. L. Switzer. 2001. Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR. Nucleic Acids Res. 29:4851-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bringel, F., L. Frey, S. Boivin, and J. C. Hubert. 1997. Arginine biosynthesis and regulation in Lactobacillus plantarum: the carA gene and the argCJBDF cluster are divergently transcribed. J. Bacteriol. 179:2697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bringel, F., and J.-C. Hubert. 2003. Extent of genetic lesions of the arginine and pyrimidine biosynthetic pathways in Lactobacillus plantarum, L. paraplantarum, L. pentosus, and L. casei: prevalence of CO2-dependent auxotrophs and characterization of deficient arg genes in L. plantarum. Appl. Environ. Microbiol. 69:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebbole, D. J., and H. Zalkin. 1989. Bacillus subtilis pur operon expression and regulation. J. Bacteriol. 171:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elagöz, A., A. Abdi, J. C. Hubert, and B. Kammerer. 1996. Structure and organisation of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: a PCR strategy for sequencing without cloning. Gene 182:37-43. [DOI] [PubMed] [Google Scholar]
- 10.Ghim, S. Y., C. C. Kim, E. R. Bonner, J. N. D'Elia, G. K. Grabner, and R. L. Switzer. 1999. The Enterococcus faecalis pyr operon is regulated by autogenous transcriptional attenuation at a single site in the 5′ leader. J. Bacteriol. 181:1324-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghim, S. Y., and J. Neuhard. 1994. The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J. Bacteriol. 176:3698-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer-Jespersen, K., and A. Munch-Petersen. 1973. Mutants of Escherichia coli unable to metabolize cytidine: isolation and characterization. Mol. Gen. Genet. 126:177-186. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, L. E., D. A. Beck, and G. A. O'Donovan. 2005. Pathways of pyrimidine salvage in Streptomyces. Curr. Microbiol. 50:8-10. [DOI] [PubMed] [Google Scholar]
- 14.Kaczanowska, M., and M. Ryden-Aulin. 2005. The YrdC protein—a putative ribosome maturation factor. Biochim. Biophys. Acta 1727:87-96. [DOI] [PubMed] [Google Scholar]
- 15.Kilstrup, M., and J. Martinussen. 1998. A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. J. Bacteriol. 180:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinussen, J., P. Glaser, P. S. Andersen, and H. H. Saxild. 1995. Two genes encoding uracil phosphoribosyltransferase are present in Bacillus subtilis. J. Bacteriol. 177:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinussen, J., and K. Hammer. 1994. Cloning and characterization of upp, a gene encoding uracil phosphoribosyltransferase from Lactococcus lactis. J. Bacteriol. 176:6457-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinussen, J., J. Schallert, B. Andersen, and K. Hammer. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 183:2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews, R. G. 1996. One-carbon metabolism, p. 600-611. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 20.Morishita, T., and M. Yajima. 1995. Incomplete operation of biosynthesis and bioenergetic functions of the citric acid cycle in multiple auxotrophic lactobacilli. Biosci. Biotechnol. Biochem. 59:251-255. [Google Scholar]
- 21.Na, J. G., I. Pinto, and M. Hampsey. 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuhard, J. 1983. Utilization of preformed pyrimidine bases and nucleosides, p. 95-148. In A. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press, London, United Kingdom.
- 23.Neuhard, J., and R. A. Kelln. 1996. Biosynthesis and conversion of pyrimidines, p. 580-599. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 24.Nicoloff, H., F. Arsène-Ploetze, C. Malandain, M. Kleerebezem, and F. Bringel. 2004. Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J. Bacteriol. 186:6059-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicoloff, H., and F. Bringel. 2003. ISLpl1 is a functional IS30-related insertion element in Lactobacillus plantarum that is also found in other lactic acid bacteria. Appl. Environ. Microbiol. 69:5832-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicoloff, H., A. Elagöz, F. Arsène-Ploetze, B. Kammerer, J. Martinussen, and F. Bringel. 2005. Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels. J. Bacteriol. 187:2093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicoloff, H., J.-C. Hubert, and F. Bringel. 2000. In Lactobacillus plantarum, carbamoyl phosphate is synthesized by two carbamoyl-phosphate synthetases (CPS): carbon dioxide differentiates the arginine-repressed from the pyrimidine-regulated CPS. J. Bacteriol. 182:3416-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxild, H. H., K. Brunstedt, K. I. Nielsen, H. Jarmer, and P. Nygaard. 2001. Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO. J. Bacteriol. 183:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher, M. A., D. Carter, D. M. Scott, D. S. Roos, B. Ullman, and R. G. Brennan. 1998. Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding. EMBO J. 17:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teplova, M., V. Tereshko, R. Sanishvili, A. Joachimiak, T. Bushueva, W. F. Anderson, and M. Egli. 2000. The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci. 9:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomchick, D. R., R. J. Turner, R. L. Switzer, and J. L. Smith. 1998. Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure 6:337-350. [DOI] [PubMed] [Google Scholar]
- 32.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 176:3708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]