Abstract
Shewanella oneidensis MR-1 is a facultatively anaerobic bacterium capable of using soluble and insoluble forms of manganese [Mn(III/IV)] and iron [Fe(III)] as terminal electron acceptors during anaerobic respiration. To assess the structural association of two outer membrane-associated c-type decaheme cytochromes (i.e., OmcA [SO1779] and MtrC [SO1778]) and their ability to reduce soluble Fe(III)-nitrilotriacetic acid (NTA), we expressed these proteins with a C-terminal tag in wild-type S. oneidensis and a mutant deficient in these genes (i.e., ΔomcA mtrC). Endogenous MtrC copurified with tagged OmcA in wild-type Shewanella, suggesting a direct association. To further evaluate their possible interaction, both proteins were purified to near homogeneity following the independent expression of OmcA and MtrC in the ΔomcA mtrC mutant. Each purified cytochrome was confirmed to contain 10 hemes and exhibited Fe(III)-NTA reductase activity. To measure binding, MtrC was labeled with the multiuse affinity probe 4′,5′-bis(1,3,2-dithioarsolan-2-yl)fluorescein (1,2-ethanedithiol)2, which specifically associates with a tetracysteine motif engineered at the C terminus of MtrC. Upon titration with OmcA, there was a marked increase in fluorescence polarization indicating the formation of a high-affinity protein complex (Kd < 500 nM) between MtrC and OmcA whose binding was sensitive to changes in ionic strength. Following association, the OmcA-MtrC complex was observed to have enhanced Fe(III)-NTA reductase specific activity relative to either protein alone, demonstrating that OmcA and MtrC can interact directly with each other to form a stable complex that is consistent with their role in the electron transport pathway of S. oneidensis MR-1.
As a facultatively anaerobic gram-negative bacterium, Shewanella oneidensis MR-1 possesses one of the more versatile respiration capabilities that have been demonstrated to date by a single microorganism. In addition to using O2 as a terminal electron acceptor during respiration, it uses a variety of alternative terminal electron acceptors, such as nitrate, fumarate, trimethylamine N-oxide, dimethyl sulfoxide, thiosulfate, and soluble and insoluble forms of iron [Fe(III)] and manganese [Mn(III/IV)] during anaerobic respiration (12). To maintain its diverse respiration capabilities, S. oneidensis MR-1 has developed a sophisticated electron transport network that has been hypothesized to be responsible for its metabolic flexibility. This network is believed to be highly branched so that different terminal electron acceptors can be used under various growth conditions. Furthermore, to reduce insoluble metals such as Fe(III) and Mn(III/IV) at neutral pH in oxidizing environments, this network must be able to transfer electrons from the cytoplasmic membrane where electrons are generated to the extracellular surface of the outer membrane, where reduction is thought to occur (27). Identification of the proteins and their functions in mediating metal reduction is of considerable importance both from a standpoint of understanding bacterial physiology and in order to facilitate bioremediation of contaminated sites, since these metal-reducing activities may also function in the reduction and immobilization of toxic metals, including U(VI), Tc(VII), and Cr(VI) (25, 36-40).
c-type cytochromes are the major components of the electron transport network of S. oneidensis MR-1, whose genome contains up to 42 open reading frames that encode putative c-type cytochromes. Most of them (33 out of 42) possess more than one heme-containing binding site. In addition, 5 out of 15 membrane-bound c-type cytochromes of S. oneidensis MR-1 are predicted to be localized to the outer membrane. This is in contrast to the membrane c-type cytochromes found in most other bacteria, which are commonly associated with the inner cytoplasmic membrane (12, 18). Because of their location, these outer membrane cytochromes are believed to be directly involved in reduction of Fe(III) or Mn(III/IV) oxides via either direct contact with these metal oxides or other components of the electron transport network (15). Indeed, two of these outer membrane c-type cytochromes, OmcA (SO1779) and MtrC (SO1778; also known as OmcB), were previously demonstrated to play an important role in the reduction of Fe(III) and Mn(III/IV) (4, 21, 23).
OmcA and MtrC are c-type cytochromes with 10 putative heme-binding sites and a lipoprotein consensus sequence (4, 22). Although omcA and mtrC are adjacent to each other on the genome of S. oneidensis MR-1, they are not in the same operon (23). Disruption of the genes encoding OmcA or MtrC has no effect on the ability of S. oneidensis MR-1 to reduce many electron acceptors, including nitrate, nitrite, or anthraquinone-2,6-disulfonate. However, in comparison to wild-type cells, mutants defective in either omcA or mtrC have reduced rates of MnO2 reduction (4, 21, 23). Also, mutation of mtrC reduces the capacity of S. oneidensis MR-1 to reduce Fe(III) citrate and Fe(III) oxide (4, 21). These latter results suggest that MtrC plays a critical role in iron reduction through either a direct mechanism involving electron transport or in conjunction with OmcA. In this respect, prior measurements have suggested that MtrC is involved in the proper localization of OmcA to the outer membrane (23).
While OmcA and MtrC have been implicated in the reduction of Fe(III) or Mn(III/IV) oxide (4, 21, 23), it is unclear whether either protein can directly reduce metals, or if they interact with other heme proteins or redox-active metabolites to reduce metal oxides. To clarify the roles of OmcA and MtrC, we have expressed and purified these membrane-associated multiheme cytochromes, which for the first time are demonstrated to independently reduce Fe(III)-nitrilotriacetic acid (NTA). Upon expression in wild-type S. oneidensis MR-1, OmcA and MtrC copurify with one another. Consistent with this observation, the purified proteins are demonstrated to associate with submicromolar affinity. Following association, purified OmcA and MtrC have a higher Fe(III)-NTA reductase specific activity than either protein alone, demonstrating that OmcA and MtrC interact directly with each other to form a stable complex that has the capacity to function as part of the electron transport pathway of S. oneidensis MR-1.
MATERIALS AND METHODS
Standard procedures.
Protein concentrations were measured with a bicinchoninic acid protein assay kit from Pierce (Rockford, IL). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblot assays were conducted according the instructions from Invitrogen (Carlsbad, CA). To visualize proteins directly, gels were stained with GelCode blue stain from Pierce. Heme staining was carried out according the protocol described by Thomas and coworkers (35). Antibodies against MtrC permitted the visualization of this protein by immunoblot assays (3).
Gene cloning.
Genes encoding membrane-associated decaheme cytochrome OmcA (SO1779) and MtrC (SO1778) were identified from the annotated genome of S. oneidensis MR-1 (7, 12) and were cloned and expressed by a previously developed strategy for soluble c-type cytochromes with the PCR primers listed in Table 1 (32). SO1779C4ER and SO1778C4ER, two reverse primers, contain the sequence encoding a tetracysteine tag (i.e., AACCPGCCK) that binds to the multiuse affinity probe 4′,5′-bis(1,3,2-dithioarsolan-2-yl)fluorescein(1,2-ethanedithiol)2 (FlAsH-EDT2) (1, 11). The reverse primer of SO1779EKER contains a coding sequence for the cut site of enterokinase (DDDDKL). SO1778EF and SO1778C4ER were used to amplify an MtrC-encoding gene to create plasmid pLS123. Similarly, SO1779EF paired with SO1779C4ER or SO1779EKER to generate plasmids pLS127 and pLS132, respectively. All resulting plasmids were sequenced for verification before they were used to transform wild-type S. oneidensis MR-1 and a knockout mutant lacking expressed OmcA and MtrC (ΔomcA mtrC) (A. S. Beliaev, unpublished data). Upon incorporation into the cloning vector pBAD202/D-TOPO, a V5 epitope and a His6 sequence are included at the C termini of recombinant OmcA and MtrC to facilitate subsequent protein detection and purification.
TABLE 1.
Oligonucleotide primers for gene cloninga
| Name | Sequence |
|---|---|
| SO1779EF | CACCTAAGAAGGAGATATACATCCCATGATGAAACGGTTCAATTTC |
| SO1779C4ER | CTTGCAACAACCAGGGCAACATGCAGCGTTACCGTGTGCTTCCATC |
| SO1779EKER | AAGCTTGTCATCGTCATCGTTACCGTGTGCTTCCATCAATTG |
| SO1778EF | CACCTAAGAAGGAGATATACATCCCATGATGAACGCACAAAAATC |
| SO1778C4ER | CTTGCAACAACCAGGGCAACATGCAGCCATTTTCACTTTAGTGTGATCTG |
The sequences are from 5′ to 3′. The forward primers (EF) contain a stop codon (in italics) and a ribosome binding site (in boldface). The reverse primers (ER) contain a coding sequence for AACCPGCCK (C4) or DDDDKL (EK) (underlined).
Protein purification.
To purify recombinant OmcA with Ni2+-NTA agarose, S. oneidensis strain MR-1 containing pLS127 or pLS132 was cultured in 10 ml of LB plus 50 μg/ml kanamycin at 30°C overnight. Five milliliters of an overnight culture was used to inoculate 1 liter of LB plus 50 μg/ml kanamycin. After the culture was grown at 30°C until its optical density at 600 nm reached 0.6, l-arabinose was added to a final concentration of 1 mM. The cells were grown for another 17 h and harvested by centrifugation at 6,000 × g and 4°C for 15 min. The harvested cells were washed once with 30 ml of ice-cold buffer A (20 mM HEPES [pH 7], 150 mM NaCl) and stored at −20°C. To isolate recombinant OmcA, the cell pellets were resuspended with 30 ml of ice-cold buffer B (20 mM HEPES [pH 7], 5 mM β-mercaptoethanol), to which protease inhibitor (Roche Diagnostic, Indianapolis, IN) was added in accordance with the manufacturer's instructions, and lysed by passage through a French press three times at 8,000 lb/in2. The unbroken cells and debris were removed by centrifugation at 15,000 × g and 4°C for 30 min. The supernatant was transferred to a 60-ml ultracentrifugation tube and centrifuged at 150,000 × g for 1 h. The pellet (i.e., the membrane fraction that was dark red) was resuspended with 50 ml of ice-cold buffer C (buffer B plus 1% [wt/vol] n-octyl-β-d-glucopyranoside [OGP]), transferred to a 50-ml centrifugation tube, and solubilized by rotating the tube at 4°C for at least 1 h. The unsolubilized proteins were removed by centrifugation at 15,000 × g and 4°C for 30 min. The supernatant was loaded onto a column (1 by 5 cm) of Ni2+-NTA agarose preequilibrated with buffer C. The column was washed with 25 ml of the following ice-cold buffers in sequential order: buffer C, buffer D (buffer C plus 150 mM NaCl+ and 10% [vol/vol] glycerol), and buffer E (buffer D plus 40 mM imidazole). OmcA was eluted with 25 ml of buffer F (buffer D plus 80 mM imidazole), aliquoted, and stored at −20°C. Before measurement of Fe(III)-NTA reductase activity, aliquoted OmcA was thawed and changed to buffer G (20 mM HEPES [pH 7], 5 mM β-mercaptoethanol, protease inhibitor, 150 mM NaCl, 1% [wt/vol] OGP, 10% [vol/vol] glycerol) with a spin column of Microcon YM-10 (molecular mass cutoff of 10 kDa) from Millipore (Billerica, MA), and its protein concentration was determined.
The procedures for purifying recombinant MtrC and OmcA by anion-exchange chromatography were identical. After membrane-bound MtrC or OmcA was solubilized as described above, the supernatant was loaded onto a 5-ml Econo-Pack High Q cartridge (Bio-Rad, Hercules, CA) that was equilibrated with buffer C. The cartridge was washed with 50 ml of the following ice-cold buffers in sequential order: buffer C, buffer H (buffer B plus 0.1% [wt/vol] OGP and 10% [vol/vol] glycerol), buffer I (buffer H plus 150 mM NaCl), buffer J (buffer H plus 500 mM NaCl), buffer K (buffer H plus 1,000 mM NaCl), and buffer H. Under these conditions, recombinant MtrC and OmcA bound to the cartridge even in the presence of 5 M NaCl. Recombinant MtrC or OmcA was eluted with at least 50 ml of ice-cold buffer L (20 mM HEPES [pH 7], 5 mM β-mercaptoethanol, protease inhibitor, 150 mM NaCl, 1% [wt/vol] OGP, 10% [vol/vol] glycerol). After determination of their concentrations, the proteins were aliquoted and stored at −20°C. The yield was about 5 mg each for OmcA and MtrC. The identities of isolated heme-containing OmcA and MtrC were confirmed by Western blot analysis with anti-V5 antibody (Invitrogen) and heme staining, and their purities were judged by staining with GelCode blue stain reagent after SDS-PAGE. Cloning, expression, and purification of MtrA (SO1777), a soluble decaheme c-type cytochrome of S. oneidensis MR-1, and determination of the absorption spectra and heme contents of purified OmcA and MtrC were carried out as described before (2, 32).
Membrane enrichment.
To enrich the membrane of wild-type S. oneidensis MR-1, 100 ml of S. oneidensis MR-1 culture was grown and harvested in the same way as described in the protein purification section. After cells were resuspended in 30 ml of buffer B and lysed by passing the suspension through a French press three times at 8,000 lb/in2, the cell lysates were centrifuged at 15,000 × g and 4°C for 30 min to remove unbroken cells. The supernatant was then further centrifuged at 150,000 × g and 4°C for 1 h. The pellets containing enriched membrane were resuspended with 10 ml of buffer C to solubilize proteins. After protein concentration determination, solubilized proteins were used for SDS-PAGE analysis.
Enrichment of the proteins copurified with MtrC.
To identify the proteins copurified with MtrC, the MtrC isolated by anion-exchange chromatography was loaded on a 10-cm-long tube containing 10% (wt/vol) acrylamide gel and further separated from other proteins by preparative SDS-PAGE with a Mini Prep Cell from Bio-Rad (Hercules, CA) according to the manufacturer's protocol. A total of 10 mg of MtrC isolated by anion-exchange chromatography was used for several runs of preparative SDS-PAGE, and fractions (2 ml) were collected. The fractions without MtrC, which lacked visible color, were pooled, concentrated to about 1 ml by an Amicon Ultra Centrifugal Filter Device from Millipore, and used for mass spectrometry (MS) analysis.
Identification of the proteins copurified with MtrC by MS analysis.
The enriched proteins were analyzed by two MS methods by following either a previously described protocol (8) or the instructions from the manufacturer. Briefly, for liquid chromatography in conjunction with Fourier transform ion cyclotron resonance (LC-FTICR) MS analysis, 50 μg of total protein was digested with trypsin (Promega, Madison, WI). Following clean-up on a strong cation-exchange SPE column (SUPELCO, Bellefonte, PA), the resulting peptides were identified with a 9.4 T FTICR mass spectrometer (Bruker Daltonics, Billerica, MA). The FTICR data were processed, and peptide sequences were determined following analysis in triplicate by the accurate mass and time tag approach developed at the Pacific Northwest National Laboratory (8, 28).
For matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS analysis, 10 μg of enriched proteins was separated by SDS-PAGE and the proteins were stained with GelCode blue. Individual bands of stained proteins were cut from the polyacrylamide gel. Each gel piece was then ground in a single 0.5-ml tube. The pieces were destained and dehydrated with 50% (vol/vol) acetonitrile-25 mM ammonium bicarbonate and 100% acetonitrile, respectively. After drying, each gel piece was covered with 15 μl of trypsin solution (20 ng/μl trypsin) and incubated at 37°C for 2 h. Peptides were then collected and resuspended in 10 μl of 25 mM ammonium bicarbonate in 50% (vol/vol) acetonitrile. Peptides were crystallized in α-cyanohydroxycinnamic acid and analyzed with an Autoflex II MALDI-TOF mass spectrometer (Bruker Daltonics) operated in reflector mode. The top 15 parent ions detected were selected for tandem MS, where 1,000 shots were collected for each corresponding fragment ion spectrum. A combination of peptide mass fingerprinting and peptide tandem MS data was submitted for identification with the Mascot tool. Multiple pieces of data were acquired, including combinations of peptide mass fingerprint and tandem MS data (Mowse scores with P < 0.05) or multiple tandem MS peptide matches.
Reductase activity of purified OmcA and MtrC.
Transient kinetic measurements of absorbance changes at 552 nm were used to determine the oxidation rate of either OmcA or MtrC that were chemically reduced with sodium dithionate by addition of electron acceptors, essentially as described previously (9, 16, 27, 41). Briefly, an SFA-20 stopped-flow apparatus (Hi-Tech Ltd., Salisbury, United Kingdom) is integrated with an Agilent 8453 UV/Vis diode array spectrometer (Agilent, Palo Alto, CA) in an anaerobic chamber. The stopped-flow system has a minimum dead time of about 20 ms and is computer assisted to synchronize stop flow and data collection. All reactions were carried out in a solution containing 5 μM cytochrome, 150 μM electron acceptors [such as Fe(III)-NTA, NaNO2, and NaNO3], 100 mM HEPES (pH 7), 1% (wt/vol) OGP, and 10% (vol/vol) glycerol. Alternatively, NADH-dependent reductase activity toward Fe(III)-NTA was assayed by a variation of a previously described procedure (6, 10, 16). Briefly, the procedure was modified for use in 96-well plates. The reaction solution, whose final volume was 210 μl in either 100 mM PIPES (pH 6.2 to 6.8) or HEPES (pH 6.8 to 8.2), contained 1.0 mM 3-(2-pyridyl)-5,6-bis(4-phenylsulfonic acid)-1,2,4-triazine monosodium salt (Ferrozine), 40 mM MgCl2, 0.2 mM NADH, 10 mM Fe(III)-NTA, 1% (wt/vol) OGP, 10% (vol/vol) glycerol, and 3 μg of purified MtrC or OmcA (or a combination of equal amounts of both proteins). In control solutions, either NADH, purified proteins, or Fe(III)-NTA was omitted. The reaction was prepared on ice, carried out at room temperature for 10 min, and terminated by placing the reaction plates on ice. The formation of an Fe(II)-3-(2-pyridyl)-5,6-bis(4-phenylsulfonic acid)-1,2,4-triazine monosodium salt complex was measured spectrophotometrically at 562 nm essentially as described previously (6), with a SpectraMax UV/Vis spectrophotometer from Molecular Devices (Sunnyvale, CA). To determine whether NADH could reduce purified cytochromes, 1 mM NADH was added to an anaerobic glass cell (STARNA, Atascadero, CA) containing purified OmcA (80 μg), MtrC (80 μg), or MtrA (40 μg) in 1 ml of 100 mM HEPES (pH 7), 1% OGP, and 10% glycerol. The absorption spectrum of OmcA, MtrC, or MtrA was then recorded by a spectrometer (UV-2101 PC; Shimadzu, Japan). Controls included omission of NADH and replacement of NADH with 1 mM NADPH, 1 mM lactate, or H2. H2 was added by purging the solution with pure H2 for 30 min. All solutions were purged with 80% N2 and 20% CO2 (room temperature, 2 h) before use, and all reactions were performed in an anaerobic chamber (6, 10).
Labeling of MtrC with FlAsH-EDT2 and fluorescence polarization measurements.
FlAsH-EDT2 was synthesized according to previously published protocols (1, 17). Before addition of 30 μM FlAsH-EDT2, 1 ml of MtrC (15 μM) in buffer P (50 mM HEPES [pH 7.5], 140 mM KCl, 5 mM β-mercaptoethanol, 0.1% [wt/vol] OGP) was incubated with 1.0 mM tris-(carboxyethyl)-phosphate for 1 h at room temperature to reduce any disulfide bonds (5). The labeling reaction with FlAsH-EDT2 was carried out at 4°C overnight. After labeling, free FlAsH-EDT2 was removed by dialyzing the reaction solution in a dialysis cassette (molecular mass cutoff of 10 kDa; Pierce) against 1 liter of buffer P twice at 4°C overnight. The fluorescence emission spectra of FlAsH bound to MtrC were recorded by a FluoroMax-2 fluorometer (SPEX, Edison, NJ) with excitation and emission slits of 5 nm and excitation at 500 nm. Fluorescent polarization (P) was measured as follows: P = (Ivv − Ivh)/(Ivv + Ivh).
Fluorescence intensities (I) were measured with excitation polarizers in the vertical position and emission polarizers in the vertical (Ivv) or horizontal (Ivh) position (13).
Determination of binding affinity.
Increases in the polarization of FlAsH-labeled MtrC were measured upon increasing amounts of OmcA, permitting detection of the formation of the MtrC-OmcA complex (i.e., [OmcA]bound) and calculation of the amount of unbound OmcA (i.e., [OmcA]free) as follows: [OmcA]bound = [(P − PO)/(Pmax − PO)] × [MtrC]total and [OmcA]free = [OmcA]total − [OmcA]bound.
The binding isotherm was fitted to the Hill equation as follows: [OmcA]bound = {[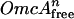 /(Kd + [
/(Kd + [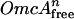 )]} × Max + Min.
)]} × Max + Min.
Kd is the sum of all equilibrium dissociation constants for the complex between MtrC and OmcA, n is the Hill coefficient associated with cooperative binding, and Min and Max are fitting parameters associated with the initial value and full range of the bound complex.
RESULTS
Expression and purification of OmcA and MtrC.
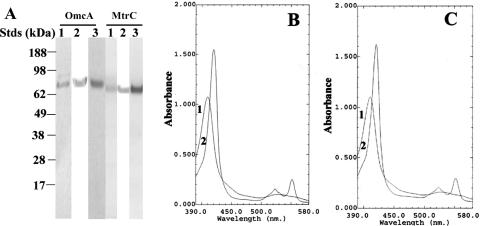
Following induction, recombinant proteins OmcA and MtrC were expressed in the ΔomcA mtrC double mutant. After cell lysis and ultracentrifugation, OmcA and MtrC are principally found in the membrane fraction (data not shown). To facilitate purification, a screen with various detergents, including Triton X-100 and Tween-20, was implemented. Optimal solubilization of OmcA and MtrC from the membrane pellet was obtained with 1% (wt/vol) OGP (data not shown). Under these conditions, solubilized OmcA binds to Ni2+-NTA columns, permitting its purification by immobilized metal ion affinity chromatography (IMAC). Following elution, purified OmcA migrates as a single band on SDS-PAGE with an apparent mass of approximately 75 kDa (Fig. 1A). Similar results were obtained for OmcA expressed without a tetracysteine tag. The two purified recombinant OmcA proteins are nearly electrophoretically homogeneous and exhibit identical absorption spectra.
FIG. 1.
Purified OmcA and MtrC. (A) SDS-PAGE (8 to 12% [wt/vol] acrylamide gradient) demonstrating electrophoretic purity of purified (1.0 μg) OmcA (left) and MtrC (right) following visualization with GelCode blue stain (lanes 1), immunoblot assays with antibodies against the V5 epitope tag (lanes 2), and heme staining (lane 3). Migration positions of protein standard (Stds) are indicated at the left. (B and C) Absorption spectra of oxidized proteins (80 μg/ml) (curve 1) and following reduction with dithionite (curve 2) for either OmcA (B) or MtrC (C) in 50 mM HEPES (pH 7.5), 150 mM NaCl, and 1% (wt/vol) OGP. Following purification of 10 mg of MtrC, nine additional proteins (<0.1 mg total) were identified by LC-FTICR and MALDI-TOF. In order of peptide abundance, they were the ATP synthase F1 beta subunit (SO4747), a putative outer membrane porin (SO3896), the ATP synthase F1 alpha subunit (SO4749), the MotA/TolQ/ExbB proton channel (SO1825), putative long-chain fatty acid transport protein (SO3099), MSHA pilin MshA (SO4105), putative outer membrane protein OmpK (SO1215), a conserved hypothetical protein (SO3343), and outer membrane protein A (SOA0114).
In contrast to OmcA, recombinant MtrC was refractory to IMAC purification following solubilization with variable amounts of OGP or other detergents. For this reason, anion-exchange chromatography was employed to purify MtrC solubilized in 1% (wt/vol) OGP. After exhaustive washing to release nonspecific binding proteins, MtrC was eluted in a buffer containing 150 mM NaCl and 1% (wt/vol) OGP. The eluted MtrC protein was close to electrophoretically homogeneous as judged by staining with GelCode blue (Fig. 1A), which revealed a predominant band of approximately 71 kDa. In all cases, the purified heme-containing proteins migrate as a single band on SDS-PAGE with electrophoretic mobilities similar to those previously reported for these heme-containing proteins (9, 27). Western immunoblot assays and heme staining confirm the identities of OmcA and MtrC (lanes 2 and 3, Fig. 1A). Contaminating proteins were identified by LC-FTICR and MALDI-TOF MS analyses following their separation (<0.1 mg) from 10 mg of purified protein (legend to Fig. 1). No other protein is present in sufficient quantity to perturb the absorption spectra.
The absorption peaks of oxidized OmcA (Fig. 1B) and MtrC (Fig. 1C) were maximal at 408 nm (i.e., Soret [γ] peak) (curve 1, Fig. 1B and C). When reduced with sodium dithionite, the Soret peak maximum shifted to 419 nm and characteristic β and α Soret absorption peaks became prominent at 523 and 552 nm, respectively (curve 2, Fig. 1B and C). These redox-dependent changes in the absorption spectra of purified OmcA and MtrC are typical for c-type cytochromes and indicate that these purified proteins contain functional hemes. On the basis of the difference absorption coefficient of 11,300 M−1 cm−1 at 550 nm (9), the average heme contents of purified OmcA and MtrC were calculated to be 10.2 and 10.4, respectively, consistent with the prediction of 10 heme-binding sites identified in each polypeptide (4, 22).
Functional activity of purified decaheme cytochromes.
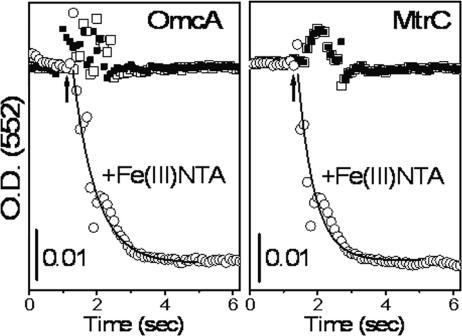
The functional activities of the purified OmcA and MtrC proteins were measured, permitting an assessment of whether these membrane proteins are properly folded with all bound cofactors following their expression. After reduction by dithionite, OmcA or MtrC was rapidly mixed with Fe(III)-NTA and transient changes in optical density associated with the α Soret absorption peak at 552 nm were measured (Fig. 2). There was a rapid decrease in absorbance consistent with the oxidation of OmcA or MtrC; fits to the transient decay indicate apparent rate constants of 1.5 ± 0.1 and 2.0 ± 0.2 s−1 for OmcA and MtrC, respectively. In contrast, no change in the absorption spectrum of either OmcA or MtrC was observed upon rapid mixing with another electron acceptor (e.g., NaNO2 or NaNO3). These results indicate that the purified proteins are functional reductases with activity toward Fe(III)-NTA and are not readily oxidized by nitrate or nitrite, a finding that is consistent with prior in vivo measurements that mutation of either omcA or mtrC only affects the metal-reducing activity of Shewanella and does not affect its ability to reduce nitrate or nitrite (4, 22).
FIG. 2.
Reduction of Fe(III)-NTA by purified OmcA and MtrC. Time-dependent changes in the absorption band at 552 nm permitted the measurement of the oxidation of dithionite-reduced OmcA or MtrC (5 μM) upon rapid mixing with 150 μM Fe(III)-NTA (○), NaNO2 (□), or NaNO3 (▪), as indicated by an arrow. Transient decreases upon addition of Fe(III)-NTA were fitted to a model by assuming apparent rate constants of 1.5 ± 0.1 and 2.0 ± 0.2 s−1 for OmcA and MtrC, respectively. The experimental conditions involved pH 7.0 and 25°C, as described in Materials and Methods. O.D. (552), optical density at 552 nm.
NADH-dependent Fe(III)-NTA reductase activity.
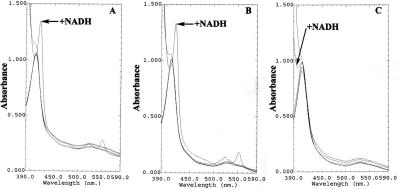
The ability of NADH to reduce OmcA or MtrC was considered to explore alternative assays of measuring OmcA- or MtrC-mediated Fe(III)-NTA reduction, as NADH does not directly reduce this Fe(III) complex. Addition of NADH resulted in the reduction of MtrC or OmcA, as evidenced by characteristic changes in the absorption spectra associated with a shift of the Soret peak maximum to 419 nm and characteristic β and α Soret absorption peaks that became prominent at 523 and 552 nm, respectively. NADH-mediated reduction of OmcA and MtrC was specific, as neither glycerol nor an alternate electron donor such as NADPH, lactate, or hydrogen was able to reduce the oxidized proteins. Furthermore, NADH failed to reduce MtrA, a soluble decaheme c-type cytochrome (Fig. 3). With NADH as the electron donor, the Fe(III)-NTA reductase activity of OmcA and MtrC was measured. Purified recombinant OmcA and MtrC displayed Fe(III)-NTA reductase activity, with similar specific activities of 1.5 and 1.6 μmol min−1 mg−1 (Fig. 4C), indicating that under these conditions each enzyme has an apparent turnover value of about 2 s−1. Virtually identical turnover values were measured with NADH as an electron donor relative to that measured following mixing of the enzymes reduced by sodium dithionite with Fe(III)-NTA (Fig. 2), indicating that the NADH-dependent reduction of these enzymes is fast in comparison to the rate-limiting reduction of Fe(III)-NTA. Thus, the reduction of Fe(III)-NTA is directly mediated by OmcA and/or MtrC because (i) OmcA and MtrC are unable to reduce Fe(III)-NTA when NADH is omitted, (ii) NADH does not reduce Fe(III)-NTA when OmcA and MtrC are absent, (iii) neither OmcA nor MtrC reacts directly with 3-(2-pyridyl)-5,6-bis(4-phenylsulfonic acid)-1,2,4-triazine monosodium salt when Fe(III)-NTA is omitted, and (iv) purified MtrA exhibits no ferric reductase activity toward Fe(III)-NTA when NADH is used as an electron donor.
FIG. 3.
NADH-mediated reduction of OmcA and MtrC. Change in the absorption spectrum of purified OmcA (A) or MtrC (B) upon addition of NADH (indicated by an arrow) in comparison to the spectrum obtained in the presence of glycerol, NADPH, lactate, or H2, as described in Materials and Methods. Purified MtrA (C) was used as a control.
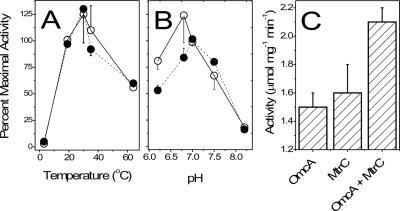
FIG. 4.
NADH-dependent reductase activities of OmcA and MtrC. Temperature (A) and pH (B) dependence of Fe(III) reductase activities [micromoles of Fe(III)-NTA reduced per minute per milligram of protein] of OmcA (○) and MtrC (•) alone or following their co-reconstitution (C) at pH 7.0 and 25°C, as described in Materials and Methods. The values reported are the means and standard deviations of triplicate measurements. For points with no error bar, the error was smaller than the symbol.
The optimal Fe(III)-NTA reductase activity of OmcA and/or MtrC occurred at temperatures between 20 and 37°C. Outside of this range, the activity was reduced significantly; no activity was found at 4°C, while about 25 to 60% of the original activity remained at 65°C (Fig. 4A). Both proteins were active over a pH range of 6.2 to 8.2. The maximal activity of OmcA was at pH 6.8, and that of MtrC was at pH 7.0 (Fig. 4B). OmcA purified from ΔomcA mtrC mutant cells by anion-exchange chromatography also displayed Fe(III)-NTA reductase activity with a specific activity similar to that of OmcA purified by IMAC (data not shown), clearly demonstrating that Fe(III)-NTA reductase activity is associated with the isolated OmcA polypeptide. When equimolar amounts of OmcA and MtrC were combined and assayed, the Fe(III)-NTA reductase specific activity increased by approximately 40% in comparison to that obtained by assaying either protein alone, demonstrating a synergistic reduction activity (Fig. 4C). The enhanced activity following the addition of equimolar amounts of OmcA and MtrC suggests a possible intermolecular interaction between OmcA and MtrC, which indicates that these proteins may function as a protein complex.
Copurification of OmcA and MtrC.
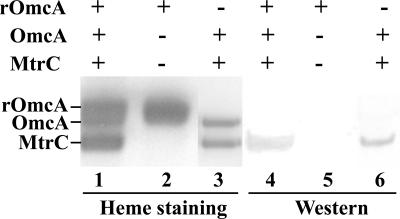
To further assess possible interactions between OmcA and MtrC, OmcA was expressed and purified from wild-type S. oneidensis MR-1 cells. Following purification of tagged OmcA, two heme staining bands are apparent (lane 1 in Fig. 5). The upper band, with a molecular mass of approximately 75 kDa, is OmcA, as evidenced by immunoblot assays against the V5 tag (data not shown). The lower band, with an apparent mass of approximately 68 kDa, is MtrC, as indicated by Western immunoblot assays with an antibody against MtrC (lanes 4 and 6 in Fig. 5) (3). The mobilities of these proteins are, furthermore, similar to that of endogenous OmcA and MtrC present in enriched vesicles prepared from wild-type cells (lane 3 in Fig. 5); the broader band associated with OmcA following purification is consistent with the higher molecular mass of tagged OmcA, which contains a tetracysteine binding motif in addition to the V5 epitope and His6 tags. Additional confirmation of the identities of expressed OmcA is possible with a ΔomcA mtrC double mutant, where it is apparent that following the purification of tagged OmcA, the lower band, with an apparent molecular mass of approximately 68 kDa, no longer copurified with OmcA (lane 2 in Fig. 5). Under these conditions, there is also a loss of density near the bottom of the 75-kDa band, consistent with the disruption of omcA. These results indicate that OmcA and MtrC associate and form part of a functional Fe(III)-NTA reductase complex. The ability of the tagged OmcA protein to copurify with endogenous OmcA suggests that this is a multimeric complex involving both tagged and endogenous OmcA in association with MtrC.
FIG. 5.
Copurification of MtrC with OmcA. Heme staining (lanes 1 to 3) and immunoblot assays against MtrC (lanes 4 to 6) following separation by SDS-PAGE of purified recombinant OmcA (rOmcA) isolated from either wild-type (lanes 1 and 4) or ΔomcA mtrC double-mutant (lanes 2 and 5) S. oneidensis MR-1 cells. The enriched membrane of wild-type S. oneidensis MR-1 cells (lanes 3 and 6) was used as a control.
Direct binding between OmcA and MtrC.
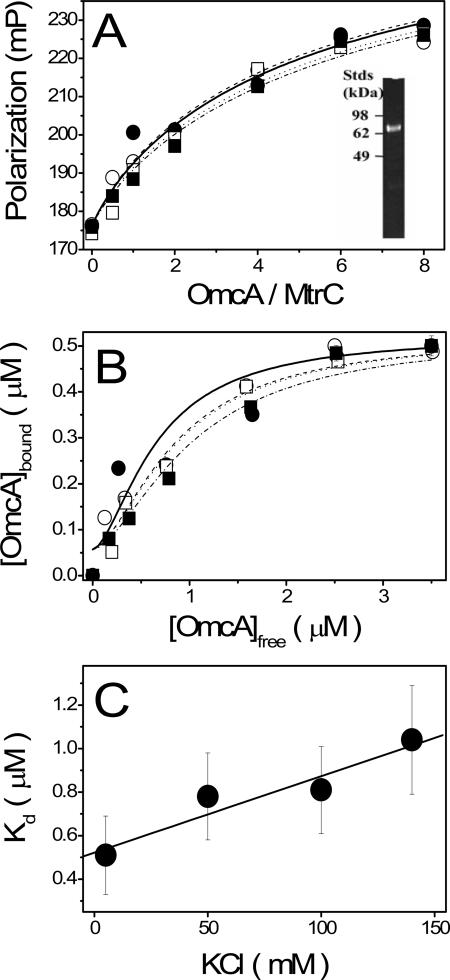
The failure of recombinant MtrC to bind to Ni2+-NTA agarose prevented direct determination of whether OmcA could be copurified with recombinant MtrC under identical conditions. To validate the association between MtrC and OmcA, we selectively labeled the tetracysteine tag encoded on purified MtrC with FlAsH-EDT2 (Fig. 6A, insert), permitting the use of fluorescence spectroscopy to monitor binding through changes in polarization. This assay takes advantage of the rigid association between the tetracoordinate FlAsH labels and protein molecules, which prevents free rotation of the fluorophore (5, 17). Thus, changes in the overall rotational dynamics of MtrC can be measured by fluorescence polarization, which is sensitive to the overall size of the protein complex.
FIG. 6.
High-affinity association between OmcA and MtrC. Fluorescence polarization changes (A) and associated binding isotherms (B) for purified FlAsH-labeled MtrC (0.5 μM) (see inset showing fluorescence in panel A) following titration with variable amounts of purified OmcA in 50 mM HEPES (pH 7.5) and 2% OGP and increasing amounts of KCl, which varied from 5 mM (▪) to 50 mM (□) to 100 mM (•) to 140 mM (○), and associated measured dissociation constants (C) were determined as described in Materials and Methods. Values are the averages and standard deviations of three independent measurements. mP, milli-polarization units.
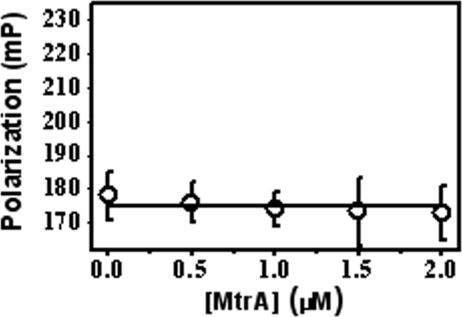
In the absence of OmcA, the steady-state polarization of FlAsH-labeled MtrC is approximately 0.175. Upon addition of OmcA, there is a progressive increase in the steady-state polarization, with a half point at an [OmcA]/[FlAsH-MtrC] ratio of close to 2, that approaches a limiting value of approximately 0.23 (Fig. 6A). In contrast, addition of purified MtrA, which is also implicated in the Fe(III) electron transport pathway (3, 27, 32), did not affect the measured polarization values for FlAsH-labeled MtrC (Fig. 7), emphasizing the specificity of the interaction. Furthermore, binding was reduced by addition of salt, resulting in a shift in the OmcA-dependent increase in polarization to higher ratios of OmcA relative to MtrC (Fig. 6A), suggesting that association is mediated, in part, by electrostatic interactions. These results indicate that MtrC and OmcA directly associate as part of a high-affinity complex in S. oneidensis MR-1.
FIG. 7.
Lack of association between MtrA and MtrC. Fluorescence polarization measured for FlAsH-labeled MtrC upon addition of indicated amounts of MtrA in the presence of 50 mM HEPES (pH 7.5), 140 mM KCl, and 2% OGP. Excitation was centered at 500 nm, and emission was centered at 528 nm. mP, milli-polarization units.
Determination of binding affinities.
Binding affinities were calculated for the formation of the MtrC-OmcA complex with the Hill equation (the last equation in Materials and Methods). All binding isotherms were simultaneously fitted by assuming a common mechanism (i.e., linkage between the Hill coefficients) in which binding affinities were dependent upon the salt concentration (Fig. 6B). Calculated fits indicate that the apparent dissociation constant (Kd) varies between 0.5 and 1.0 μM, depending on the salt concentration (Fig. 6C). Given that both MtrC and OmcA are membrane proteins that are constrained in a two-dimensional lattice, these results suggest the formation of a stable oligomeric complex. Furthermore, the Hill coefficient (n) was 1.6 ± 0.2, indicating positive cooperativity of binding. These latter results suggest that two OmcA molecules associate with each MtrC molecule, consistent with the results of the pull-down affinity purification measurements where the intensity of heme staining of both endogenous and recombinant OmcA bands is approximately twice that of MtrC (lane 1 in Fig. 5). These results strongly suggest that more than one OmcA may associate in complex with MtrC.
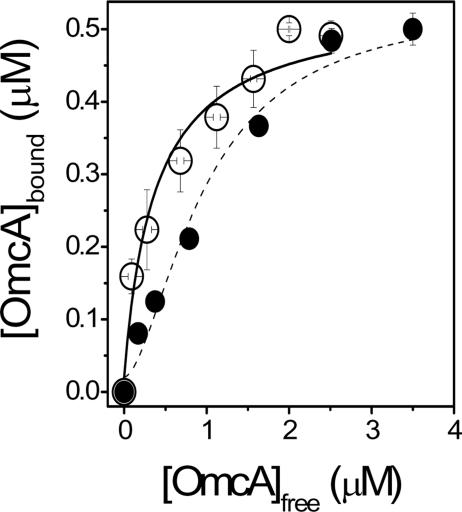
To further investigate the possible association between multiple OmcA protein molecules with each MtrC molecule to form an oligomeric protein complex, we took advantage of the fact that OmcA with a tetracysteine tag engineered at the C terminus forms a stable dimer under nonreducing conditions. In comparison to the binding isotherms for untagged monomeric OmcA, one observes that binding of dimeric OmcA associates with an apparent affinity that is substantially higher (i.e., Kd = 0.4 ± 0.1 μM) than that of the monomeric protein (i.e., Kd = 1.0 ± 0.1 μM) (Fig. 8). Furthermore, in comparison to that of the monomeric protein (n = 1.6 ± 0.2), the Hill coefficient is 1.0 ± 0.1, consistent with the direct binding of the dimeric complex. These latter results suggest that multiple OmcA molecules cooperatively associate with MtrC to form a multimeric protein complex. If one assumes that the equilibrium constants associated with OmcA binding to MtrC are equal, these results suggest that the actual Kd for the association between monomeric OmcA and MtrC is 0.5 ± 0.1 μM, in agreement with results obtained for the dimeric OmcA construct.
FIG. 8.
Cooperative association between OmcA and MtrC. Binding isotherms are shown for association between OmcA and FlAsH-labeled MtrC, using OmcA without a tetracysteine tag (•) or with a tetracysteine tag (○) that promotes dimer formation, i.e., (OmcA)2. Curves represent respective fits to the Hill equation (see the last equation in Materials and Methods). For monomeric OmcA, Kd = 1.0 ± 0.1 μM and n = 1.7 ± 0.2. Upon dimerization, Kd = 0.4 ± 0.1 μM and n = 1.0 ± 0.1. In all cases, experimental conditions involved 0.5 μM FlAsH-labeled MtrC in 50 mM HEPES (pH 7.5), 140 mM KCl, and 2% OGP.
DISCUSSION
We have cloned, expressed, and purified the outer membrane-associated decaheme cytochromes OmcA and MtrC to electrophoretic homogeneity, permitting, for the first time, direct measurements of their metal reductase activity and functional association. Both purified cytochromes are functional Fe(III)-NTA reductases that lack activity toward nitrite or nitrate. Independently of whether dithionite or NADH is used to reduce OmcA or MtrC, virtually identical rates of reduction of Fe(III)-NTA are observed (2 s−1) (Fig. 2 and 4). Thus, the reduction of OmcA or MtrC by NADH is fast in comparison to the reduction of Fe(III)-NTA, which is rate limiting (2 s−1). OmcA cooperatively associates with MtrC (Kd < 500 nM) and forms an oligomeric protein complex involving multiple OmcA subunits with enhanced Fe(III)-NTA reductase activity. The high-affinity association between OmcA and MtrC is consistent with the copurification of these proteins. In contrast, another decaheme cytochrome (i.e., MtrA) does not bind with this complex, consistent with the absence of other heme-containing proteins following its purification (32). These results suggest that prior observations of MtrA in promoting electron transfer between the inner and outer membranes of S. oneidensis MR-1 would involve transient low-affinity associations (27).
Prior measurements have identified the presence of OmcA, MtrC, and other c-type cytochromes in the outer membranes of Shewanella that function to couple conserved electron transfer chains present in the inner membrane to mediate the reduction of insoluble metals (20, 23). The involvement of the outer membrane decaheme cytochrome MtrC in facilitating the reduction of iron oxides in Shewanella was first identified following gene disruption, although residual metal-reducing activity remains (4). Likewise, OmcA has been reported to participate in the reduction of metal oxides [i.e., Mn(IV) and Fe(III)], although it does not appear to be required for the reduction of chelated soluble iron (9, 23). Our measurements, demonstrating a functional interaction between purified OmcA and MtrC, which facilitates the Fe(III)-NTA reductase activity of these enzymes, is consistent with the fact that neither OmcA nor MtrC is absolutely required for growth, as both enzymes have some capacity to reduce Fe(III)-NTA. Since OmcA and MtrC reduce Fe(III)-NTA independently, this functional redundancy may be the reason that the ΔomcA mutant is still capable of reducing soluble iron(III) complexes (9, 23). An OmcA homolog from Shewanella frigidimarina that is 55% identical to OmcA of S. oneidensis MR-1 was previously purified free from other heme-containing proteins, and this preparation was reported to reduce soluble Fe(III) complexes. Further, OmcA of S. frigidimarina was found to contain 10 hemes, consistent with sequence-based predictions (9). Our present results are consistent with these prior measurements and sequence-based predictions that MtrC contains 10 hemes.
The reduction of OmcA and MtrC by NADH is specific, as other electron donors, such as NADPH, lactate, and H2, failed to reduce OmcA or MtrC. In addition, NADH does not reduce MtrA, and thus MtrA is not capable of any NADH-dependent Fe(III)-NTA reductase activity. In contrast, following reduction by sodium dithionite, MtrA is known to reduce soluble Fe(III)-EDTA as well as Fe(III)-maltol (27). Thus, differences in assay conditions underlie differences in the observed ability of MtrA to reduce soluble Fe(III) complexes. NADH dehydrogenase is not involved in the observed NADH-mediated reduction of either OmcA or MtrC, since both enzymes are reduced by NADH following their purification to near electrophoretic homogeneity. Furthermore, contaminating proteins (<1% of the total protein) were identified by MS analysis and none of the subunits associated with predicted NADH dehydrogenase from MR-1 were found to copurify with MtrC by two different MS analyses. The failure to identify the subunits of NADH dehydrogenase from our sample is not caused by the limitation of MS analyses as some subunits of NADH dehydrogenase were previously identified from the membrane fraction used in our purification (J. K. Fredrickson, unpublished data).
The ability of NADH to directly reduce either OmcA or MtrC is puzzling, as physiological concentrations of NADH are not expected to be present in the periplasm. Furthermore, the polypeptide sequence of neither OmcA nor MtrC is predicted to contain a Rossman dinucleotide binding fold (commonly associated with NADH binding) (30). Thus, the mechanism and physiological significance associated with the ability of NADH to reduce either OmcA or MtrC remain unclear. Nevertheless, the ability of NADH to reduce periplasmic c-type cytochromes purified from Geobacter sulfurreducens has been previously observed (31). While the physiological significance of the NADH-dependent reduction of periplasmic and outer membrane-associated proteins remains unclear, from a practical point of view the specificity of NADH in the reduction of either OmcA or MtrC has important experimental advantages in the measurement of their kinetic properties. This latter advantage is highlighted by the fact that similar turnover values (i.e., 2 s−1) are observed irrespective of whether NADH is coupled to enzyme turnover or rapid mixing is used to measure the transient oxidation of the dithionite-reduced enzyme by Fe(III)-NTA (Fig. 2 and 4). These latter results indicate that the association and reduction of the highly soluble Fe(III)-NTA complex (where diffusional barriers would be expected to be minimal) represent the rate-limiting step of the reaction (Fig. 9). Taken together, all of these results clearly demonstrate that both purified OmcA and MtrC have Fe(III)-NTA reductase activity and ensure that the reported measurements of binding between purified OmcA and MtrC are physiologically significant.
FIG. 9.
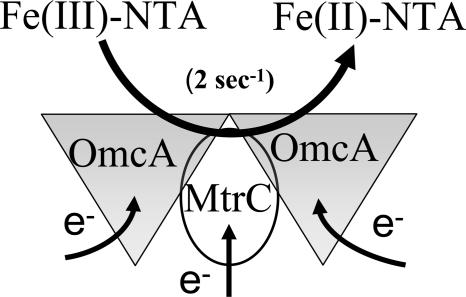
Model of the protein complex between OmcA and MtrC. An oligomeric complex involving outer membrane decaheme cytochromes OmcA and MtrC permits coupling between dehydrogenases in the inner membrane that couple the oxidation of NADH and a range of different substrates through classical oxidative pathways involving quinol intermediates and periplasmic proteins and the reduction of Fe(III)-NTA by either OmcA or MtrC. Rates of Fe(III)-NTA reduction are rate limiting (2 s−1), as evidenced by the virtually identical rate constants obtained from direct measurements of OmcA or MtrC oxidation (Fig. 2) relative to that associated with the NADH-dependent reduction of Fe(III)-NTA (Fig. 4).
The Fe(III)-NTA-reducing activities of OmcA and MtrC are 1.5 and 1.6 μmol min−1 mg−1, respectively, which are the highest Fe(III)-NTA reductase activities reported to date (10, 16). Synergistic reduction activity of OmcA and MtrC toward Fe(III)-NTA further supports the direct interaction between OmcA and MtrC. The measured affinity suggests that these proteins exist as a stable complex, and their affinity is comparable to that of other proteins that form stable complexes, including the soluble proteins ferredoxin I and pyruvate-ferredoxin oxidoreductase from Desulfovibrio africanus (Kd = 38.5 μM), soluble cytochrome c and membrane-bound complex III of beef mitochondria (Kd = 0.2 μM), subunits of the photosynthetic reaction center of Rhodobacter sphaeroides (Kd = 4 μM), and the integral membrane proteins phospholamban and Ca2+-ATPase in cardiac muscle (Kd = 140 μM), in which approximately 70% of the Ca-ATPase is in complex with phospholamban (14, 26, 33, 34). In the case of membrane proteins, the stability of the complex is considerably enhanced by the diffusional constraints associated with the membrane environment. In addition, polarization measurements of binding between purified proteins indicate that OmcA and MtrC form a heterotrimer in which two OmcA protein molecules interact with one MtrC protein molecule. Consistent with these binding measurements, endogenous OmcA was copurified with recombinant and tagged OmcA along with endogenous MtrC at an apparent ratio of approximately two OmcA protein molecules per MtrC molecule. Lane 3 in Fig. 5 represents the relative abundance of OmcA and MtrC in wild-type cells, which is unrelated to the stoichiometry of the isolated complex (Fig. 5).
The formation of a stable complex between MtrC and OmcA is consistent with prior dynamic models in which dehydrogenases in the inner membrane couple the oxidation of NADH and a range of different substrates through classical oxidative pathways involving quinol intermediates that function to reduce the inner membrane protein CymA that are then coupled to the outer membrane through soluble periplasmic proteins (e.g., MtrA and Cct) that function as intermediate electron carriers (4, 19, 24, 27, 29). These results are, furthermore, consistent with the ability of MtrA to couple electron transport between spatially distant reductases following its expression in Escherichia coli (27). In this model, the integral membrane protein MtrB, previously demonstrated to play a critical role in Fe(III)-NTA reduction, is suggested to represent a protein scaffold that permits the transient association between reduced periplasmic proteins and the OmcA-MtrC protein complex. Thus, depending on the metabolic needs of the organism, different redox partners can couple with either OmcA or MtrC as a means to maintain critical metabolic pathways important to the energy metabolism of the organism. The ability to couple a range of different metabolic pathways to mediate metal reduction through a versatile electron transport chain involving OmcA and MtrC underlies the ability of Shewanella to colonize and populate anaerobic environments and represents an important understanding of how metal reductase activities can function in the reductive immobilization of toxic metals in contaminated sites. Future experiments should measure the possible association and function of a reconstituted OmcA-MtrC complex against solid metals to determine whether binding and metal reduction involve a direct association with individual proteins.
Acknowledgments
This research was supported by the U.S. Department of Energy Office of Biological and Environmental Research under the Genomics-Genomes to Life Program and an EMSL Scientific Grand Challenge project at the W. R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy Office of Biological and Environmental Research program located at Pacific Northwest National Laboratory. The Pacific Northwest National Laboratory is operated for the Department of Energy by the Battelle Memorial Institute under contract DE-AC05-76RLO1830.
REFERENCES
- 1.Adams, S. R., R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis, and R. Y. Tsien. 2002. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124:6063-6076. [DOI] [PubMed] [Google Scholar]
- 2.Bartsch, R. G. 1971. Cytochromes: bacterial. Methods Enzymol. 34:344-363. [Google Scholar]
- 3.Beliaev, A. S., and D. A. Saffarini. 1998. Shewanella putrefaciens mtrB encodes an outer membrane protein required for Fe(III) and Mn(IV) reduction. J. Bacteriol. 180:6292-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. [DOI] [PubMed] [Google Scholar]
- 5.Chen, B., M. U. Mayer, and T. C. Squier. 2005. Structural uncoupling between opposing domains of oxidized calmodulin underlies the enhanced binding affinity and inhibition of the plasma membrane Ca-ATPase. Biochemistry 44:4737-4747. [DOI] [PubMed] [Google Scholar]
- 6.Cowart, R. E., F. L. Singleton, and J. S. Hind. 1993. A comparison of bathophenanthrolinedisulfonic acid and Ferrozine as chelators of iron(II) in reduction reactions. Anal. Biochem. 211:151-155. [DOI] [PubMed] [Google Scholar]
- 7.Daraselia, N., D. Dernovoy, Y. Tian, M. Borodovsky, R. Tatusov, and T. Tatusova. 2003. Reannotation of Shewanella oneidensis genome. Omics 7:171-175. [DOI] [PubMed] [Google Scholar]
- 8.Elias, D. A., M. E. Monroe, M. J. Marshall, M. F. Romine, A. S. Belieav, J. K. Fredrickson, G. A. Anderson, R. D. Smith, and M. S. Lipton. 2005. Global detection and characterization of hypothetical proteins in Shewanella oneidensis MR-1 using LC-MS based proteomics. Proteomics 5:3120-3130. [DOI] [PubMed] [Google Scholar]
- 9.Field, S. J., P. S. Dobbin, M. R. Cheesman, N. J. Watmough, A. J. Thomson, and D. J. Richardson. 2000. Purification and magneto-optical spectroscopic characterization of cytoplasmic membrane and outer membrane multiheme c-type cytochromes from Shewanella frigidimarina NCIMB400. J. Biol. Chem. 275:8515-8522. [DOI] [PubMed] [Google Scholar]
- 10.Gaspard, S., F. Vazquez, and C. Holliger. 1998. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl. Environ. Microbiol. 64:3188-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin, B. A., S. R. Adams, and R. Y. Tsien. 1998. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281:269-272. [DOI] [PubMed] [Google Scholar]
- 12.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 13.Lakowicz, J. R. 1999. Principles of fluorescence spectroscopy. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 14.Larson, J. W., and C. A. Wraight. 2000. Preferential binding of equine ferricytochrome c to the bacterial photosynthetic reaction center from Rhodobacter sphaeroides. Biochemistry 39:14822-14830. [DOI] [PubMed] [Google Scholar]
- 15.Lloyd, J. R. 2003. Microbial reduction of metals and radionuclides. FEMS Microbiol. Rev. 27:411-425. [DOI] [PubMed] [Google Scholar]
- 16.Magnuson, T. S., A. L. Hodges-Myerson, and D. R. Lovley. 2000. Characterization of a membrane-bound NADH-dependent Fe3+ reductase from the dissimilatory Fe3+-reducing bacterium Geobacter sulfurreducens. FEMS Microbiol. Lett. 85:205-211. [DOI] [PubMed] [Google Scholar]
- 17.Mayer, M. U., L. Shi, and T. C. Squier. 2005. One-step, non-denaturing isolation of an RNA polymerase enzyme complex using an improved multi-use affinity probe resin. Mol. BioSystems 1:53-56. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, T. E., A. I. Tsapin, I. Vandenberghe, L. de Smet, D. Frishman, K. H. Nealson, M. A. Cusanovich, and J. J. van Beeumen. 2004. Identification of 42 possible cytochrome C genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. Omics 8:57-77. [DOI] [PubMed] [Google Scholar]
- 19.Myers, C. R., and J. M. Myers. 1992. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174:3429-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers, C. R., and J. M. Myers. 1997. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim. Biophys. Acta 1326:307-318. [DOI] [PubMed] [Google Scholar]
- 21.Myers, J. M., and C. R. Myers. 2002. Genetic complementation of an outer membrane cytochrome omcB mutant of Shewanella putrefaciens MR-1 requires omcB plus downstream DNA. Appl. Environ. Microbiol. 68:2781-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers, J. M., and C. R. Myers. 1998. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S. putrefaciens. Biochim. Biophys. Acta 1373:237-251. [DOI] [PubMed] [Google Scholar]
- 23.Myers, J. M., and C. R. Myers. 2001. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl. Environ. Microbiol. 67:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers, J. M., and C. R. Myers. 2000. Role of the tetraheme cytochrome CymA in anaerobic electron transport in cells of Shewanella putrefaciens MR-1 with normal levels of menaquinone. J. Bacteriol. 182:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nealson, K. H., and D. Saffarini. 1994. Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48:311-343. [DOI] [PubMed] [Google Scholar]
- 26.Pieulle, L., M. Nouailler, X. Morelli, C. Cavazza, P. Gallice, S. Blanchet, P. Bianco, F. Guerlesquin, and E. C. Hatchikian. 2004. Multiple orientations in a physiological complex: the pyruvate-ferredoxin oxidoreductase-ferredoxin system. Biochemistry 43:15480-15493. [DOI] [PubMed] [Google Scholar]
- 27.Pitts, K. E., P. S. Dobbin, F. Reyes-Ramirez, A. J. Thomson, D. J. Richardson, and H. E. Seward. 2003. Characterization of the Shewanella oneidensis MR-1 decaheme cytochrome MtrA: expression in Escherichia coli confers the ability to reduce soluble Fe(III) chelates. J. Biol. Chem. 278:27758-27765. [DOI] [PubMed] [Google Scholar]
- 28.Qian, W. J., D. G. Camp II, and R. D. Smith. 2004. High-throughput proteomics using Fourier transform ion cyclotron resonance mass spectrometry. Expert Rev. Proteomics 1:87-95. [DOI] [PubMed] [Google Scholar]
- 29.Richardson, D. J. 2000. Bacterial respiration: a flexible process for a changing environment. Microbiology 146(Pt. 3):551-571. [DOI] [PubMed] [Google Scholar]
- 30.Rossmann, M. G., D. Moras, and K. W. Olsen. 1974. Chemical and biological evolution of nucleotide-binding protein. Nature 250:194-199. [DOI] [PubMed] [Google Scholar]
- 31.Seeliger, S., R. Cord-Ruwisch, and B. Schink. 1998. A periplasmic and extracellular c-type cytochrome of Geobacter sulfurreducens acts as a ferric iron reductase and as an electron carrier to other acceptors or to partner bacteria. J. Bacteriol. 180:3686-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi, L., J. T. Lin, L. M. Markillie, T. C. Squier, and B. S. Hooker. 2005. Overexpression of multi-heme c-type cytochromes. BioTechniques 38:297-299. [DOI] [PubMed] [Google Scholar]
- 33.Speck, S. H., and E. Margoliash. 1984. Characterization of the interaction of cytochrome c and mitochondrial ubiquinol-cytochrome c reductase. J. Biol. Chem. 259:1064-1072. [PubMed] [Google Scholar]
- 34.Tatulian, S. A., B. Chen, J. Li, S. Negash, C. R. Middaugh, D. J. Bigelow, and T. C. Squier. 2002. The inhibitory action of phospholamban involves stabilization of alpha-helices within the Ca-ATPase. Biochemistry 41:741-751. [DOI] [PubMed] [Google Scholar]
- 35.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 36.Tiedje, J. M. 2002. Shewanella—the environmentally versatile genome. Nat. Biotechnol. 20:1093-1094. [DOI] [PubMed] [Google Scholar]
- 37.Viamajala, S., B. M. Peyton, W. A. Apel, and J. N. Petersen. 2002. Chromate reduction in Shewanella oneidensis MR-1 is an inducible process associated with anaerobic growth. Biotechnol. Prog. 18:290-295. [DOI] [PubMed] [Google Scholar]
- 38.Viamajala, S., B. M. Peyton, W. A. Apel, and J. N. Petersen. 2002. Chromate/nitrite interactions in Shewanella oneidensis MR-1: evidence for multiple hexavalent chromium [Cr(VI)] reduction mechanisms dependent on physiological growth conditions. Biotechnol. Bioeng. 78:770-778. [DOI] [PubMed] [Google Scholar]
- 39.Viamajala, S., B. M. Peyton, and J. N. Petersen. 2003. Modeling chromate reduction in Shewanella oneidensis MR-1: development of a novel dual-enzyme kinetic model. Biotechnol. Bioeng. 83:790-797. [DOI] [PubMed] [Google Scholar]
- 40.Wade, R., and T. J. DiChristina. 2000. Isolation of U(VI) reduction-deficient mutants of Shewanella putrefaciens. FEMS Microbiol. Lett. 184:143-148. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Z., C. C. Ainsworth, D. M. Friedrich, P. L. Gassman, and A. G. Joly. 2000. Kinetics and mechanism of surface reaction of salicylate on alumina in colloidal aqueous suspension. Geochem. Cosmochim. 64:1159-1172. [Google Scholar]