Abstract
Osteoclasts are essential for bone dynamics and calcium homeostasis. The cells form a tight seal on the bone surface, onto which they secrete acid and proteases to resorb bone. The seal is associated with a ring of actin filaments. Cortactin, a c-Src substrate known to promote Arp2/3-mediated actin assembly in vitro, is expressed in osteoclasts and localizes to the sealing ring. To address the role of cortactin and actin assembly in osteoclasts, we depleted cortactin by RNA interference. Cortactin-depleted osteoclasts displayed a complete loss of bone resorption with no formation of sealing zones. On nonosteoid surfaces, osteoclasts flatten with a dynamic, actin-rich peripheral edge that contains podosomes, filopodia, and lamellipodia. Cortactin depletion led to a specific loss of podosomes, revealing a tight spatial compartmentalization of actin assembly. Podosome formation was restored in cortactin-depleted cells by expression of wild-type cortactin or a Src homology 3 point mutant of cortactin. In contrast, expression of a cortactin mutant lacking tyrosine residues phosphorylated by Src did not restore podosome formation. Cortactin was found to be an early component of the nascent podosome belt, along with dynamin, supporting a role for cortactin in actin assembly.
INTRODUCTION
Cortactin is a prominent Src substrate (Maa et al., 1992) expressed in many vertebrate cell types. Hematopoietic cells generally express the cortactin homologue hematopoietic cell-specific 1 (HS1) instead of cortactin. In cultured fibroblasts, cortactin localizes to sites of dynamic actin assembly, including lamellipodia (Weed et al., 2000). Biochemical studies have shown that cortactin can bind Arp2/3 complex and actin filaments, which enhances Arp2/3-induced nucleation and stabilizes actin filament branches (Weaver et al., 2001). Wiskott-Aldrich syndrome protein (WASp) family members also bind Arp2/3 complex, but they bind actin monomers, which makes them more efficient at nucleation. Cortactin and WASp can be synergistic in vitro because they are able to bind to Arp2/3 complex at the same time (Weaver et al., 2002).
Cortactin has a Src homology (SH)3 domain and a proline-rich region, which are not required for F-actin and Arp2/3 interaction (Figure 1A). These regions may allow cortactin to function as an adaptor to amplify cellular signaling pathways. The signaling and actin assembly functions of cortactin may both exist, working in a positive feedback loop.
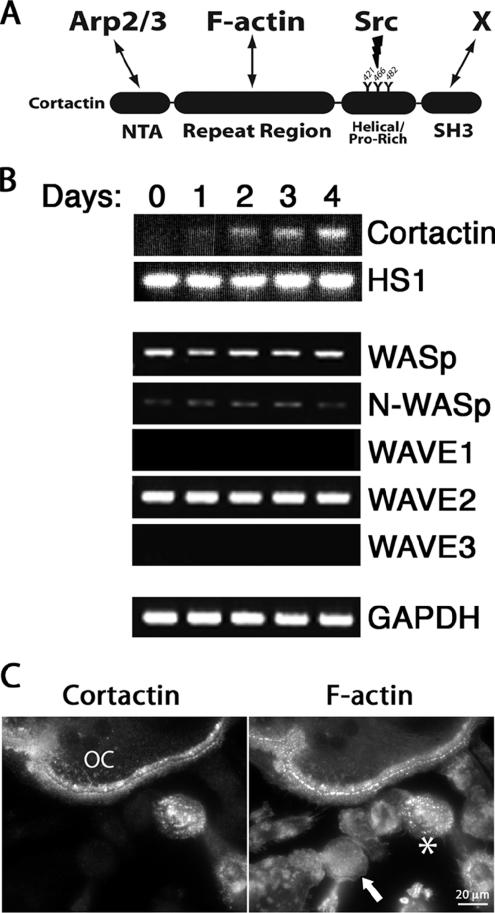
Figure 1.
Expression of cortactin and other Arp2/3 activators during osteoclast differentiation. (A) Schematic representation of mouse cortactin and major interacting partners. Cortactin’s N-terminal acidic (NTA) domain binds Arp2/3 complex and the repeat region binds actin filaments. The helical/proline-rich domain contains three tyrosines (421, 466, and 482) that are targeted for phosphorylation by active Src. The SH3 domain of cortactin binds a number of proline-rich–containing proteins (denoted by X), including N-WASp and dynamin. (B) Expression of cortactin and other Arp2/3 complex activator genes by RT-PCR of primary macrophages cultured in the presence of RANKL for the indicated number of days. GAPDH is a control. (C) Anti-cortactin and rhodamine-phalloidin staining of cells differentiated for 4 d. Cortactin is present in multinucleated osteoclasts (OC) but not in macrophages (arrow). An asterisk marks a mononucleate cortactin-positive cell, which is presumably a maturing osteoclast before fusion.
We chose osteoclasts as a system to study the cellular function of cortactin for several reasons. First, osteoclasts are available as primary cells whose physiological function, namely, resorption of bone, can be assayed directly. Second, osteoclasts express cortactin, but they are derived from monocytic precursors that lack cortactin. Finally, cortactin is a target of c-Src, which is critical for osteoclast function (Soriano et al., 1991).
In vertebrates, bone remodeling and turnover requires osteoclasts to resorb bone. Osteoclasts form an integrin-based contact with the bone surface (Vaananen et al., 2000), which in association with actin forms a ring-shaped sealing zone (Kanehisa et al., 1990; Saltel et al., 2004). Osteoclasts secrete acid and proteases inside the sealing zone, which degrades the bone.
On nonosteoid surfaces (Saltel et al., 2004), actin in osteoclasts forms podosomes, filopodia, and lamellipodia at the cell edge. Podosomes are small columns of F-actin, surrounded by a ring of proteins characteristic of integrin-based focal adhesions. Podosomes are also a prominent feature of the actin cytoskeleton of dendritic cells (Burns et al., 2001), macrophages (Linder et al., 1999), smooth muscle cells (Gimona et al., 2003), and Src-transformed fibroblasts (Mizutani et al., 2002). The F-actin core of podosomes contains the actin nucleator Arp2/3 complex and its activators cortactin, WASp, and neuronal-WASp (N-WASp) (Linder and Aepfelbacher, 2003). Arp2/3 complex depletion results in fewer podosomes and no sealing zone formation in an osteoclast-like cell line (Hurst et al., 2004).
In vivo evidence about the importance of cortactin-based actin assembly in osteoclast function and podosome formation is lacking. We depleted cortactin in primary mouse osteoclasts using short hairpin RNA (shRNA) delivered with a lentiviral system (Rubinson et al., 2003). We found that cortactin was absolutely necessary for bone resorption, sealing zone formation, and podosome formation in osteoclasts but that cell differentiation and actin-based motility were otherwise remarkably normal. In addition, rescue studies of cortactin knockdown cells with exogenous cortactin and cortactin mutants suggest a critical role for cortactin’s tyrosine phosphorylation downstream of Src.
MATERIALS AND METHODS
Antibodies and Reagents
Antibodies were raised in chickens against a N-terminal fragment of mouse cortactin (C90) and used for immunoblots. Anti-Arp3 antibody (R43) was used as described previously (Welch et al., 1997). The 3F2 monoclonal antibody (mAb) against capping protein β2 was used for immunoblots as described previously (Schafer et al., 1994). The 4F11 mAb against cortactin was a gift from Dr. Scott Weed (West Virginia University, Health Sciences Center, Morgantown, WV) and was used in immunofluorescence staining (Kanner et al., 1990). The monoclonal anti-c-Src antibody was a gift from Dr. Steven Teitelbaum (Department of Pathology, Washington University School of Medicine, St. Louis, MO). The monoclonal anti-actin antibody C4 was a gift from Dr. James Lessard (Department of Pediatrics, University of Cincinnati, Cincinnati, OH) (Lessard, 1988). Anti-c-Fos, anti-nuclear factor of activated T-cells c1 (NFATc1), anti-N-WASp, and anti-WASp rabbit polyclonal antibodies and anti-dynamin2 goat polyclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-β3 integrin rabbit polyclonal antibody was purchased from Cell Signaling Technology (Beverly, MA). Anti-Pyk2 rabbit polyclonal antibody was purchased from BioSource International (Camarillo, CA). Anti-FLAG mAb M2 and rhodamine- and coumarin-labeled phalloidin were purchased from Sigma-Aldrich (St. Louis, MO). Horseradish peroxidase (HRP)-conjugated anti-chicken, anti-mouse, and anti-rabbit second antibodies (Pierce Chemical, Rockford, IL) were used in immunoblots. Alexa-conjugated anti-mouse, anti-rabbit, and anti-goat second antibodies (Invitrogen, Carlsbad, CA) were used for immunofluorescence. Glutathione S-transferase (GST)-receptor activator of nuclear factor κB ligand (RANKL) was purified as described previously (McHugh et al., 2000).
Mouse Osteoclast Culture
Bone marrow macrophages were isolated from 4- to 8-wk-old mice, cultured overnight in α-minimal essential medium (MEM) containing 10% heat-inactivated fetal bovine serum (FBS), and subjected to Ficoll-Hypaque gradient purification as described previously (Feng et al., 2001). Cells (5 × 106) collected from the gradient interface were plated in a P100 suspension culture dish (Corning Costar, New York, NY) in the presence of 1:10 CMG14-12 culture supernatant (Takeshita et al., 2000), containing the equivalent of 120 μg/ml recombinant macrophage colony-stimulating factor (MCSF). The cells were infected with lentivirus at 3 d after harvest. For in vitro generation of osteoclasts, macrophages were cultured in α-MEM containing 10% heat-inactivated FBS in the presence of 100 ng/ml RANKL and 100 μg/ml MCSF for 5 d. In culture, lentivirus-infected and uninfected osteoclasts were viable and looked morphologically healthy for at least 10 d in the presence of MCSF and RANKL. Experiments on glass were performed after 5 d in culture, and experiments on bone were performed after 8 d in culture.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was collected from macrophages after 0–4 d of RANKL treatment using the RNeasy kit (QIAGEN, Valencia, CA). Reverse transcription from equal amounts of total RNA was performed using the SuperScript system (Invitrogen), followed by PCR with oligonucleotides specific for cortactin, HS1, WASp, N-WASp, WAVE1, WAVE2, WAVE3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Lentivirus and RNA Interference (RNAi)
The pLL3.7 lentiviral-based shRNA expression system (kindly provided by Dr. Frank Gertler, Massachusetts Institute of Technology, Cambridge, MA) was used as described previously (Rubinson et al., 2003). Short hairpin RNA sequences were selected to target the coding region of mouse cortactin. Sequence 1 and 2 targeted the 5′-ggattcggcgggaagtatg-3′ coding region and the 5′-gactgagaagcatgcctcc-3′ coding region of the cortactin gene, respectively. As a control, we scrambled sequence 1 to give 5′-ggacaggtaggttcgtgag-3′, which has no significant similarity to known genes as assessed by National Center for Biotechnology Information BLASTn searches. 293T cells were cotransfected with pLL3.7 and packaging plasmids pHR’CVM8.2ΔR and pCMV-VSV-G (gifts from Dr. Sheila Stewart, Washington University School of Medicine) using FuGENE (Roche Diagnostics, Indianapolis, IN). Optimal viral particle numbers for infection were based on infection efficiency, determined from the percentage of target cells with green fluorescent protein (GFP), which is expressed independently from the RNAi sequence. Concentrated viral titers for multiplicity of infection (MOI) studies were obtained by centrifugation of viral supernatants at 83,000 × g for 90 min. Based on MOI studies, viral titers of ∼3 × 106 viral particles were used for optimal infection.
To infect macrophages, the virus-containing supernatant was collected 3 d after transfection of packaging cells and added to bone marrow macrophages plated on a P100 culture dish along with 8 μg/ml protamine sulfate. After 4 h, the viral supernatant was replaced with fresh macrophage media. Cells were cultured 2–3 d in macrophage medium, lifted using trypsin/EDTA, and resuspended in Hank’s balanced salt solution supplemented with 1% FBS for sorting. Infectivity efficiencies were determined by observing GFP expression in infected cells as a function of lentiviral titer. Cells from the highest infectivity titer where then sorted for GFP expression using fluorescence-activated cell sorter (FACS) to enrich for infected cell populations. Cells were sorted for GFP expression in the Siteman Cancer Center High Speed Sorter Core Facility using a MoFlo high-speed flow cytometer (Dako Colorado, Fort Collins, CO). GFP-positive cells were plated on glass or dentine slices in osteoclast differentiation medium.
Retrovirus and Cortactin Mutants
The pBABE murine leukemia virus retroviral-based expression system was kindly provided by Dr. Sheila Stewart (Washington University School of Medicine). FLAG-tagged cortactin wild-type and mutant (3YF and W525K) cDNAs were expanded by PCR from previously described plasmids (Head et al., 2003; gifts from Dr. Scott Weed, West Virginia University, Health Sciences Center, Morgantown, WV) and cloned into the pBABE expression plasmid. RNAi-resistance was conferred by mutating three nucleotides in the region targeted by RNAi sequence 1 from 5′-ggattcggcgggaagtatg-3′ to 5′-ggcttcggagggaaatatg-3′ with the QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA). To generate retrovirus, 293T cells were cotransfected with pBABE constructs and packaging plasmids pUMVC3 and pCMV-VSV-G (gifts from Dr. Sheila Stewart, Washington University School of Medicine) using FuGENE (Roche Diagnostics). Viral titers of ∼3 × 106 viral particles were used for optimal infection. This viral titer had no cytotoxic effects on target cells. The virus-containing supernatant was collected 3 d after transfection and added to bone marrow macrophages plated on a acid-washed glass coverslips in culture dishes along with 8 μg/ml protamine sulfate. After 4 h, the viral supernatant was replaced with fresh macrophage medium. Cells were cultured 2–3 d and then treated with 2 μg/ml puromycin in osteoclast medium containing RANKL, as described above.
Western Blot Analysis
Cells were lysed in SDS sample buffer, boiled, and electrophoresed on 10% SDS-polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes, which were blocked with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) containing 0.1% Tween 20. Membranes were incubated with the appropriate antibody for 1 h in blocking solution, followed by HRP-conjugated second antibody incubation for 1 h in blocking solution. Bands were visualized with an ECL kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), followed by exposure to autoradiography film.
Immunofluorescence and Time-Lapse Movies
For immunofluorescence, cells on coverslips or dentine were fixed with 2% paraformaldehyde in PBS at room temperature for 15 min, permeabilized with 0.5% Triton X-100 in PBS for 1 min, followed by blocking with 8% BSA in PBS for 1 h. For anti-Arp3 staining, cells were treated with an additional methanol rinse and costained for actin using monoclonal anti-actin (C4) antibodies. All other actin staining was done with fluorescent-labeled phalloidin. Cells were incubated in primary antibody solutions containing 2% BSA in PBS for 1 h, followed by Alexa-conjugated second antibody solution containing 2% BSA in PBS for 1 h. Coverslips were mounted on glass slides, and dentine slices were mounted on coverslips. Images were collected with an Olympus IX70 inverted microscope (Olympus, Melville, NY) equipped with a 10× Plan 0.25 Ph1 phase objective, a 100× PlanApo 1.40 oil objective (immunofluorescence microscopy), and a 60× PlanApo 1.40 oil Ph3 phase objective (immunofluorescence and time-lapse microscopy). Images were captured with a Coolsnap HQ camera (Photometrics, Tucson, AZ) and processed using QED In Vivo software (Media Cybernetics, Silver Spring, MD). For time-lapse movies, cells on glass coverslips were mounted on glass slides and bathed in 10% FBS in DMEM containing 25 mM HEPES buffer without phenol red (Invitrogen). Images were captured every 10 s for ∼5 min. The sample temperature was maintained at 37°C.
Scanning Electron Microscopy (SEM)
Cells on glass coverslips were fixed for 2 h with 2% phosphate-buffered glutaraldehyde, postfixed with 1% aqueous osmium tetroxide for 1 h, dehydrated in an ethyl alcohol series, and critical point dried from liquid CO2. Mounted coverslips were sputter-coated with 30 nm of gold and examined by Dr. Michael Veith (Washington University, St. Louis, MO) in a Hitachi S-450 scanning electron microscope (Hitachi America, Brisbane, CA) at 20-KV accelerating voltage.
Tartrate-resistant Acid Phosphatase (TRAP) Staining and Bone Resorption
Bone resorption was assayed by culturing macrophages on dentine slices in the presence of RANKL and MCSF for 8 d. Cells were fixed with 2% paraformaldehyde in PBS and stained for TRAP to confirm osteoclastogenesis using a kit (Sigma-Aldrich, St. Louis, MO). For resorption pit visualization, cells were removed by brief treatment with 2 N NaOH followed by 0.5% toluidine blue solution staining.
RESULTS
Cortactin Is Expressed during Osteoclast Differentiation
Cortactin is expressed in all vertebrate cells and tissues, except in cells of hematopoietic origin, which generally express only the cortactin homologue HS1 (Kitamura et al., 1995). Because osteoclasts are hematopoietic-derived cells and are known to express cortactin (Hiura et al., 1995), we asked whether cortactin would be expressed during differentiation of osteoclasts in culture, from monocyte precursors. RT-PCR was performed on primary bone marrow macrophages induced to differentiate into osteoclasts with RANKL and MCSF (Yasuda et al., 1998). After 5 d, ∼90% of macrophages differentiated into osteoclasts. Cortactin was undetectable initially, and expression was induced during differentiation (Figure 1B). Immunofluorescence staining confirmed that osteoclasts and their mononuclear precursors express cortactin, whereas the few macrophages that did not differentiate were negative for cortactin (Figure 1C). HS1 was expressed, at a constant level, during the time course of differentiation (Figure 1B).
In vitro, cortactin activates Arp2/3 complex in a manner synergistic with WASp family members (Weaver et al., 2002). We asked whether the expression of any other Arp2/3 activators changed during osteoclast differentiation. The levels of WASp, N-WASp, and WAVE2, assessed by RT-PCR, did not change during osteoclast differentiation (Figure 1B). WAVE1 expression was low, detected only with additional PCR cycles, and its level did not change (our unpublished data). WAVE3 expression was not detected, even with additional cycles.
Cortactin Depletion in Osteoclasts Using Lentiviral RNAi Expression
To determine the role of cortactin in actin assembly and the function of osteoclasts, we depleted cortactin using RNAi. A lentiviral system was used. Lentiviruses integrate into the host cell genome, so the cells maintained a constitutive expression of RNAi after infection. The lentiviral vector expressed GFP as an independent marker, allowing us to sort and collect a homogenous population of infected primary macrophages, which we then induced to differentiate into osteoclasts.
First, we assessed the efficiency of the infection and the health of the cells as a function of viral titer. To assess infection efficiencies, the percentage of cells expressing GFP was measured by FACS, as a function of viral titer (Figure 2A). The highest infectivity rates were achieved with 3 × 106 viral particles for control and knockdown lentivirus, with lower infectivity rates at lower and higher viral titers. This optimal viral titer was used for most subsequent experiments, followed by sorting of GFP-positive cells to enrich for infected cells. To test for adverse effects of viral infection on the cells, we assessed the viability of macrophages, as a function of viral titer. Cells were infected, sorted by FACS, and plated on glass. The number of cells that were alive and adherent was followed by phase-contrast microscopy for 2–5 d, which is the number of days required for in vitro differentiation of macrophages into osteoclasts. Control and knockdown infected macrophages remained fully viable and adherent over time regardless of the viral titer used for infection (Figure 2B).
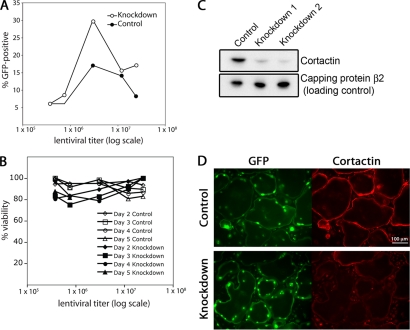
Figure 2.
Knockdown of cortactin in osteoclasts. (A) Infectivity rate, based on GFP expression, as a function of viral titer. Macrophages were infected with lentivirus expressing a cortactin RNAi knockdown sequence (1) or a control scrambled RNAi sequence. The RNAi plasmid expressed GFP independently. (B) FACS-sorted macrophages were cultured on glass in osteoclast medium. Viability was assessed by phase-contrast morphology and by substrate attachment. (C) Level of cortactin in knockdown cells, by immunoblot. Infected and FACS-sorted macrophages were differentiated into osteoclasts for 5 d in culture. The samples are the control scrambled RNAi sequence, the cortactin knockdown RNAi sequence 1, and the cortactin knockdown RNAi sequence 2. Capping protein β2 is a loading control. (D) Anti-cortactin immunofluorescence of cortactin in osteoclasts expressing control RNAi or knockdown RNAi sequence 1, plated on glass. The level of fluorescence in the knockdown cells was similar to that seen for controls without first antibody. GFP accumulates in the nuclei of infected cells.
We assessed the efficiency of cortactin knockdown with immunoblots and immunofluorescence. Two different RNAi sequences targeting cortactin (1 and 2) and a scrambled RNAi control sequence were used. The level of cortactin knockdown with RNAi sequence 1 or 2 in osteoclasts was >90% by immunoblot analysis (Figure 2C). Cortactin immunofluorescence staining was diminished to background levels in cells expressing RNAi sequence 1 or 2, with no change in cells expressing control RNAi (Figure 2D).
Cortactin-depleted Osteoclasts Fail to Form Sealing Rings and Resorb Bone
We assessed the ability of cortactin-depleted osteoclasts to resorb bone. Macrophages expressing RNAi were plated on dentine and differentiated into osteoclasts. The number of mature osteoclasts on dentine was the same for cortactin-depleted and control cells, revealed by counting cells (Figure 3A) and staining for TRAP, an osteoclast marker (Figure 3B). Large, multinucleated cells were observed in control and knockdown cells, indicating normal fusion during differentiation (Figure 3B). Osteoclasts expressing scrambled RNAi produced numerous bone resorption pits, assessed by toluidine blue staining (Figure 3C). In contrast, cells expressing cortactin RNAi did not produce any resorption pits at all. Cortactin RNAi sequence 1 and 2 gave the same result. Control cells expressing scrambled RNAi formed normal sealing rings that contained F-actin and cortactin (Figure 3D). Cells depleted for cortactin did not form actin-rich sealing rings. Their morphology resembled that of macrophages, as assessed by rhodamine-phalloidin (Figure 3D).
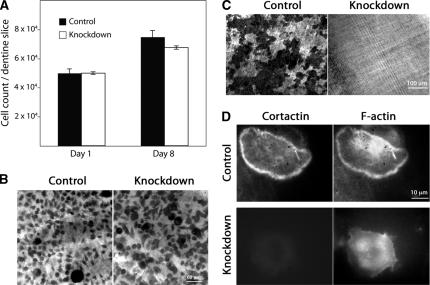
Figure 3.
Cortactin-depleted osteoclasts are viable and differentiate on bone, but they fail to resorb bone or form sealing rings. (A) Cell viability. Macrophages expressing scrambled or cortactin RNAi sequence 1 were plated on dentine slices and differentiated into osteoclasts for 8 d. Cells were counted on day 1 and day 8. (B) Osteoclast differentiation. Cells on dentine as in A were stained for an osteoclast-specific marker, TRAP. RNAi control and knockdown sequence 1 samples are shown. The number of cells and the degree of TRAP staining is similar. (C) Bone resorption. After removal of osteoclasts, dentine slices were stained with toluidine blue solution to reveal resorption pits. Pits seem dark by transmitted light microscopy. Control cells produced pits, and knockdown cells showed no evidence of resorption. (D) Actin sealing ring formation. Osteoclasts on dentine were stained with rhodamine-phalloidin and anti-cortactin antibody. Control osteoclasts formed actin rings, and knockdown cells did not. Representative examples are shown.
The Role of Cortactin in Osteoclast Actin Assembly on Nonosteoid Surfaces
On glass and other nonosteoid surfaces, the most prominent feature of the actin cytoskeleton of osteoclasts is a belt of podosomes just behind the cell edge, with lamellipodia and filopodia at the cell edge. Podosomes have a central core of F-actin and actin-binding proteins, surrounded by a ring of proteins found in focal adhesions (Linder and Aepfelbacher, 2003). We observed cortactin staining in podosomes in all mature osteoclasts (Figure 4A), along with F-actin and Arp2/3 complex, assessed by rhodamine-phalloidin and anti-Arp3 staining, respectively (Figure 4, A and B).
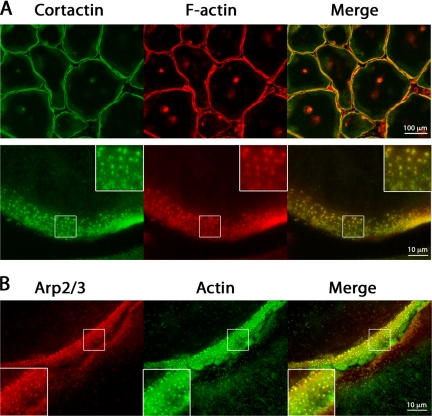
Figure 4.
Cortactin, actin, and Arp2/3 complex localization to podosomes. Osteoclasts on glass were costained for cortactin and F-actin, or Arp3 and actin. (A) Low- and high-magnification fluorescence images of osteoclasts stained with anti-cortactin antibody and rhodamine-phalloidin. (B) Higher magnification images of the periphery of two adjacent osteoclasts stained with anti-Arp3 and anti-actin antibodies. In all images, podosomes look like punctate, fluorescent structures. Diffuse staining for actin, cortactin, and Arp3 is observed between podosomes.
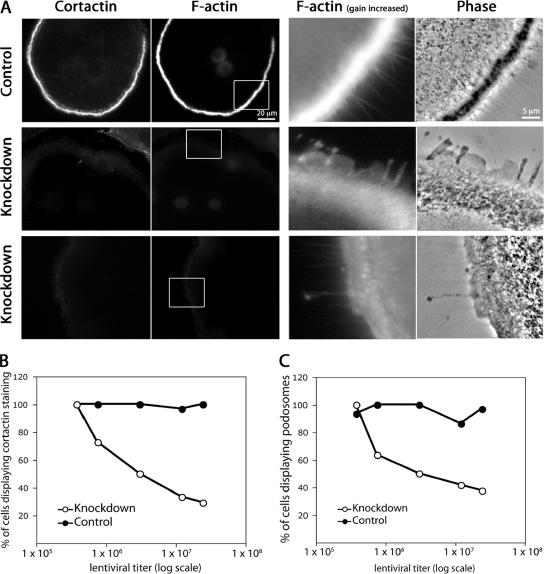
Depletion of cortactin caused an essentially complete loss of podosomes, as revealed by rhodamine-phalloidin fluorescence and phase-contrast microscopy (Figure 5A). The area where podosomes were lost showed weak diffuse F-actin staining, when the gain was increased, and the phase-contrast image had a ground-glass appearance. These features are consistent with the presence of an actin filament network that excludes vesicles. This phenotype was rescued by expression of RNAi-resistant wild-type cortactin expression, as described below, which confirms the specificity of the knockdown effects.
Figure 5.
Cortactin depletion results in loss of podosomes in osteoclasts. (A) Fluorescence and phase-contrast images of osteoclasts stained with anti-cortactin antibodies and rhodamine-phalloidin after infection with lentivirus expressing a control scrambled RNAi (top) or cortactin RNAi sequence 1 or 2 (middle and bottom, respectively). The level of fluorescence in the anti-cortactin images of cells depleted for cortactin was the same as that seen in cells stained with second antibody alone (our unpublished data). The F-actin images on the left were captured at a gain setting optimal for observing podosomes. The F-actin images on the right are high magnifications of the boxed areas, collected with a higher gain setting, which reveals that the knockdown cells have a persisting presence of F-actin in the region that would be occupied by podosomes, along with lamellipodia- and filopodia-like structures at the cell edge. (B) The percentage of cells displaying immunofluorescence staining for cortactin as a function of lentiviral titer for control and knockdown cell samples. Samples were assessed visually using fluorescence microscopy. Cells displaying any detectable cortactin staining above background were included as positive. (C) The percentage of cells displaying podosomes as a function of lentiviral titer for control and knockdown cell samples. Samples were assessed visually using rhodamine-phalloidin staining and fluorescence microscopy. Cells displaying any number of podosomes were included as positive, as assessed by rhodamine-phalloidin staining.
We asked whether cortactin-depleted cells had short-lived podosomes by performing time-lapse phase-contrast movies (Supplemental Movies 1 and 2). In control cells, podosomes looked like a belt of phase-dark puncta a short distance from the cell edge. Characteristic of podosomes, the puncta appeared and disappeared on a time scale of minutes, and they did not move relative to the substrate (Supplemental Movie 1). Cortactin-depleted cells had no podosomes, and the area had a ground-glass appearance (Figure 5A and Supplemental Movie 2, respectively).
We asked how well the presence of cortactin correlated with the presence of podosomes, by infecting cells with different amounts of virus. As the viral titer increased, the percentage of cells displaying cortactin staining decreased. The percentage of cells with podosomes also decreased in a similar manner, indicating a correlation between the presence of cortactin and podosomes (Figure 5, B and C).
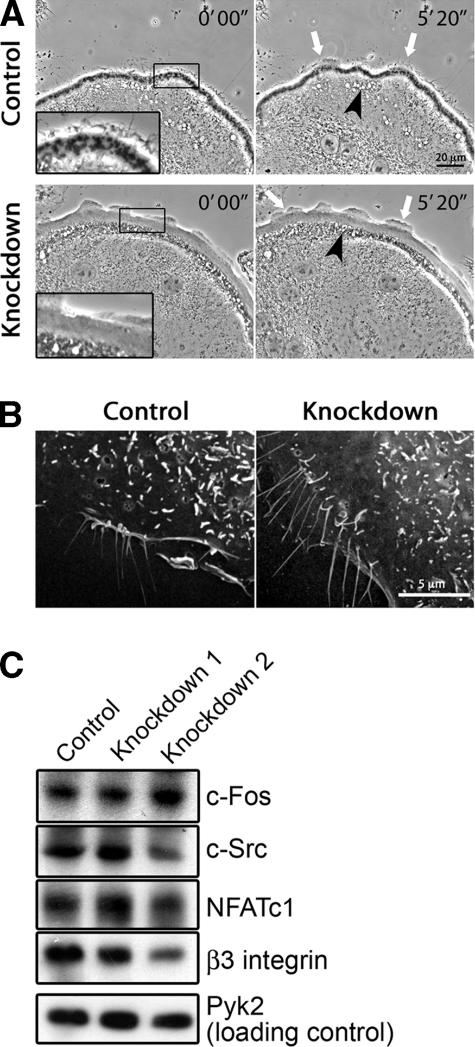
We asked whether other aspects of actin-based motility were affected by cortactin depletion. Lamellipodial and filopodial structures were seen at the cell edge of cortactin-depleted cells by rhodamine-phalloidin staining and phase-contrast imaging of fixed preparations (Figure 5A) and by time-lapse phase-contrast imaging of live cells (Supplemental Movies 1 and 2 and Figure 6A). Compared with control cells, cortactin-depleted cells showed a slight increase in the ratio of lamellipodia to filopodia in the phase-contrast movies. The movies also revealed phase-light and phase-dark vesicles undergoing rapid saltatory motion in a region just behind the podosome area, with no obvious effect from cortactin depletion (Figure 6A). The overall shape and size of the osteoclasts on glass were not changed by cortactin depletion, based on phase-contrast light microscopy and SEM images. SEM showed filopodia-like structures and membrane protrusions on the dorsal surface of knockdown and control cells (Figure 6B). Overall, control and knockdown cells were relatively similar in these other manifestations of actin assembly and actin-based motility.
Figure 6.
Effect of cortactin depletion on podosomes, membrane protrusions, and osteoclast maturation. (A) Membrane and cell edge dynamics in time-lapse phase-contrast microscopy movies of osteoclasts expressing scrambled or cortactin RNAi sequence 2. These images are frames from Supplemental Movies 1 and 2. Podosomes are seen as phase-dark puncta just behind the cell edge in control cells. Cortactin-depleted cells show no podosomes. Control and cortactin-depleted cells exhibit dynamic membrane protrusions, including lamellipodia (white arrows). Rapidly moving phase-light and phase-dark vesicles are seen farther back from the cell edge in control and knockdown cells (black arrowheads). These vesicles may be endocytic. (B) Scanning electron microscopy of osteoclasts expressing scrambled or cortactin RNAi sequence 1. Control and cortactin-depleted cells show filopodia-like structures and dorsal membrane protrusions. (C) Immunoblot of osteoclast markers in cells expressing control scrambled RNAi, knockdown sequences 1 or knockdown sequence 2. Pyk2 is a loading control. All four osteoclast markers tested are present in control and cortactin-depleted cell samples.
We asked whether RANKL-induced osteoclast differentiation occurred normally in cells depleted for cortactin. RANKL signaling results in the expression of a number of osteoclast-specific proteins. Control and cortactin-depleted cells expressed c-Fos, c-Src, NFATc1 and β-3 integrin upon RANKL treatment, all of which are markers of osteoclast maturation not found in macrophages (Figure 6C). Cell fusion also occurred, which is characteristic of osteoclast differentiation, with no apparent difference in the average number of nuclei per cell between control and knockdown cells (Figure 2D; our unpublished data).
Cortactin Is an Early Component for Podosome Belt Formation
To understand how cortactin depletion affects podosome dynamics, we examined the assembly and disassembly of podosomes. When a cell edge was advancing, a single belt of podosomes was generally observed, in the area just behind the cell edge (Supplemental Movie 1). The belt was several podosomes wide. New podosomes formed at the peripheral, outer, edge of the belt, whereas podosomes disappeared from the central, inner, edge of the belt. The lifetime of a podosome was ∼2 min. These findings are consistent with previous studies (Destaing et al., 2003).
In contrast, when a cell was stationary, we often observed two belts of podosomes separated by 1–2 μm (Figure 7A and Supplemental Movie 3). Alone, each belt resembled a single belt described above, in terms of the number, arrangement, and behavior of the individual podosomes. Each belt moved outward, in the manner of single belts. The key difference was in their life cycle. At some point in time, the outer belt disappeared, because all its podosomes disassembled, and a new inner belt appeared, with podosomes assembling de novo. The inner belt would move out, and then the cycle would repeat. Thus, when two belts of podosomes were present, the inner belt was newer than the outer one, in every case.
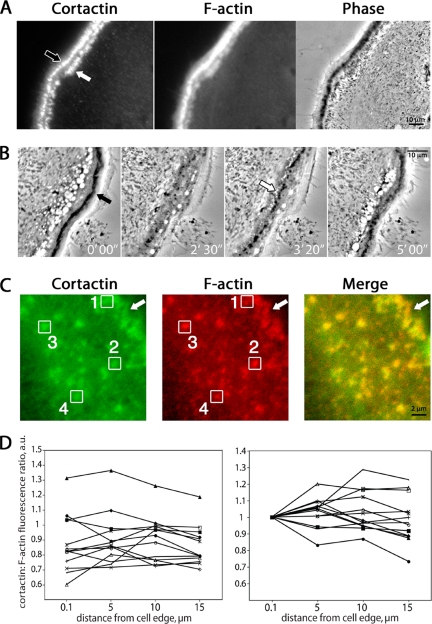
Figure 7.
Cortactin and F-actin in podosome belt maturation. (A) An example of anti-cortactin and F-actin (rhodamine-phalloidin) staining of a cell region with two podosome belts. Cortactin fluorescence intensity is similar in the two belts, whereas the F-actin fluorescence and phase-contrast darkness are more pronounced in the outer belt (black arrow) than in the inner belt (white arrow). (B) Podosome belt formation in time-lapse phase-contrast microscopy. Images are frames from Supplemental Movie 3. These movies reveal that the outer belt (black arrow) is older, and the inner belt (white arrow) is younger. (C) Quantitation of anti-cortactin and F-actin (rhodamine-phalloidin) fluorescence of individual podosomes within a single belt. Boxes were placed around podosomes, and pixel fluorescence values were determined. Boxes 1–4 are examples of podosomes ∼0.1, 5, 10, and 15 μm from the cell edge (arrow). (D) Fluorescence intensities for multiple podosomes within a single belt were averaged and plotted as a function of distance from the cell edge. Each plot of the 12 curves represents the averaged podosome intensity ratios from a single osteoclast. The graph on the left plots the actual ratios. For the graph on the right, the curves were adjusted up or down to begin at the same point.
In fixed cell preparations, costained for cortactin and F-actin, the inner, newer belt had less intense rhodamine-phalloidin staining than did the outer belt, and this difference was reflected in the darkness of the podosomes in the phase-contrast images (Figure 7A). In contrast, cortactin stained both belts equally well, suggesting that cortactin occurs early in the life of a podosome belt.
We asked whether a similar trend in cortactin:F-actin ratio could be observed by comparing individual podosomes within a single belt. In this case, new podosomes are those on the outer edge of the belt. We measured the fluorescence intensities of cortactin and F-actin for individual podosomes in double-stained images (Figure 7B). We calculated the ratio of cortactin:F-actin fluorescence for each podosome, measured its distance from the cell edge and plotted the average ratio versus the distance (Figure 7, C and D). Overall, the curves showed no consistent trend up or down, even when the curves were aligned at the starting point (Figure 7D). That a difference is seen when comparing two belts, and not podosomes within one belt, may reflect the spatial regulation of the assembly process or the magnitude of the temporal differences in age.
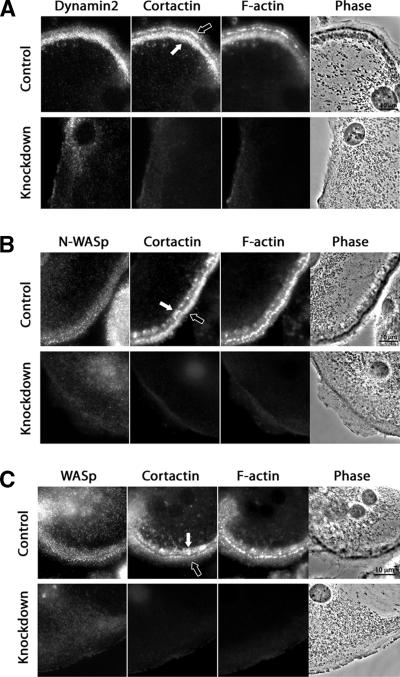
Because cortactin localized to nascent podosome belts and cortactin depletion resulted in the loss of podosomes, we asked whether other podosome components were able to localize in a podosome-like pattern despite the absence of cortactin and F-actin. Dynamin2 has been found in podosomes of osteoclasts and RSV-transformed cells (Ochoa et al., 2000), and the SH3 domain of cortactin can bind directly to the proline-rich region of dynamin (Weed and Parsons, 2001). N-WASp localizes to podosomes of Src-transformed fibroblasts (Mizutani et al., 2002), and cortactin can bind and activate N-WASp in vitro (Kowalski et al., 2005). WASp is important for podosome formation in macrophages (Linder et al., 1999) and osteoclasts (Calle et al., 2004). Here, dynamin2, N-WASp and WASp localized in a punctate pattern with podosomes in control osteoclasts (Figure 8). Dynamin2 showed preferential staining of the inner podosome belt, when two belts were present (Figure 8A). On cortactin depletion, the distribution of all three proteins was diffuse in the cytoplasm, without any appreciable punctate podosome pattern (Figure 8, A–C). Thus, assembly of dynamin, N-WASp, and WASp at the podosome either depends on or, more likely, is cooperative with that of cortactin and F-actin.
Figure 8.
Localization of dynamin2, N-WASp, and WASp in control and cortactin-depleted osteoclasts. (A) Dynamin2 localized predominantly to the inner podosome belt in osteoclasts expressing control scrambled RNAi. Dynamin2 was not observed at the cell edge in cells expressing cortactin RNAi sequence 1. N-WASp (B) and WASp (C) localized equally well to the inner and outer podosome belts in control osteoclasts expressing scrambled RNAi. This localization was lost, and the staining was diffuse after cortactin depletion (cortactin RNAi sequence 2 in B and cortactin RNAi sequence 1 in C). Black arrows point to the older outer belts, and white arrows point to the new inner belts.
Expression of Cortactin Mutants in Control and Knockdown Cells
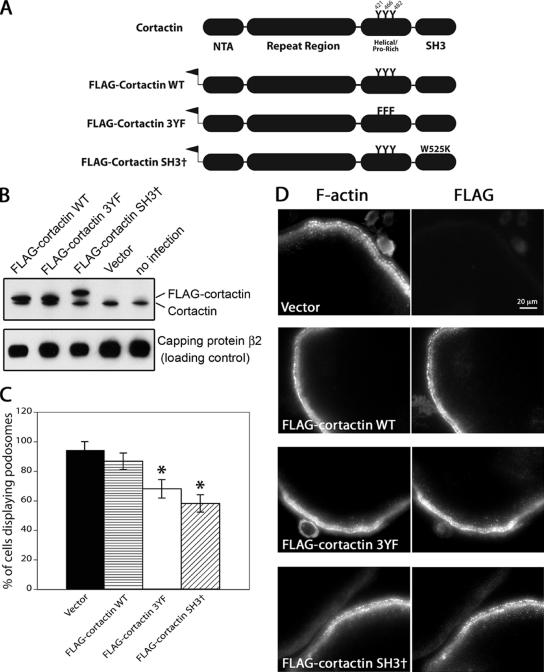
To investigate the biochemical basis of cortactin’s function in osteoclasts, we expressed cortactin mutants using an N-terminal FLAG tag and a retroviral system (Figure 9A). The 3YF mutant of cortactin contained 3 tyrosine-to-phenylalanine mutations at residues Y421, Y466, and Y482, which are known to be phosphorylated downstream of Src in cells and to account for essentially all of the Src-induced tyrosine phosphorylation of cortactin in cells (Head et al., 2003). The SH3 mutant of cortactin contained a tryptophan-to-lysine mutation at W525, which causes loss of binding to proline-rich ligands (Kinley et al., 2003).
Figure 9.
Effects of overexpression of cortactin mutants on podosome formation. (A) Diagram illustrating the nature of the cortactin mutants. All constructs included an N-terminal FLAG tag. 3YF contains three tyrosine to phenylalanine mutations (residues 421, 466, and 482), and SH3† contains the mutation W525K in the SH3 domain. (B) Anti-cortactin immunoblot of osteoclasts expressing the cortactin mutants. The FLAG-tagged cortactins run higher than endogenous cortactin, and the SH3 W525K mutant is anomalously high in SDS gels, as seen in previous studies. Capping protein β2 is a loading control. (C) Percentage of osteoclasts with podosomes. Osteoclasts expressing the cortactin mutant rescue constructs were fixed, stained with rhodamine-phalloidin and viewed by fluorescence microscopy to visualize podosomes. Error bars are SE of proportion, and asterisks indicate statistical difference (p < 0.01) compared with vector and wild type. Statistical significance was determined by the binomial probability distribution function. The number of cells was >35 in each sample. (D) Osteoclasts expressing cortactin mutants were fixed and costained with rhodamine-phalloidin and anti-FLAG.
First, we expressed the cortactin mutants in osteoclasts that had normal levels of endogenous cortactin. The level of exogenously expressed FLAG-tagged cortactin was two- to threefold higher than that of endogenous cortactin, based on anti-cortactin immunoblots (Figure 9B). The SH3 point mutant of cortactin migrated slower than wild-type cortactin in the SDS gel, as seen previously (Kowalski et al., 2005).
Overexpression of wild-type cortactin in osteoclasts did not significantly alter the percentage of cells containing podosomes, and the cells looked normal by phase-contrast microscopy and phalloidin staining (Figure 9, C and D). Expression of either the SH3 mutant or the 3YF mutant of cortactin resulted in an ∼20–30% statistically significant decrease in the number of osteoclasts with podosomes (Figure 9C). In cells with podosomes, the FLAG-tagged SH3 and 3YF mutant cortactins localized normally to podosomes, as did FLAG-tagged wild-type cortactin (Figure 9D). Biochemically, cortactin is a monomer (Weaver et al., 2002), so the ability of the FLAG-tagged mutants to localize normally is probably not due to association with endogenous wild-type cortactin. Thus, the SH3 and 3YF cortactin mutants caused small but statistically significant dominant negative effects on podosome assembly.
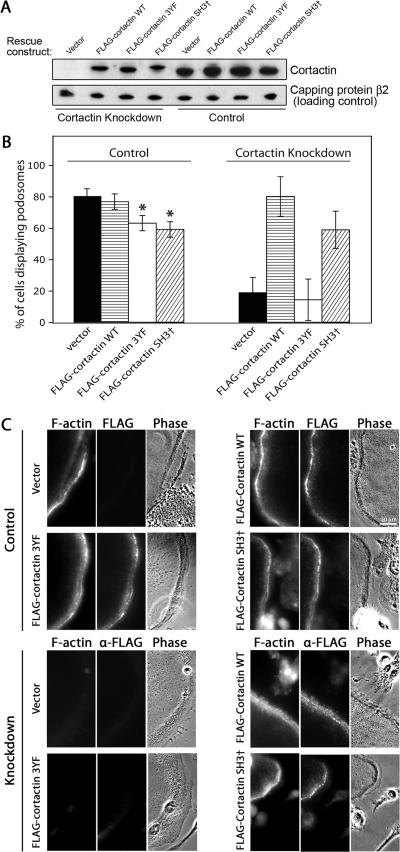
Next, we tested the ability of the mutants to rescue podosome assembly in cells depleted of cortactin by knockdown. These FLAG-tagged cortactin rescue expression constructs were rendered resistant to the cortactin RNAi knockdown sequence 1 by three codon-neutral nucleotide changes. Expressed in 293T cells, the protein levels for the mutants and wild-type cortactin rescue constructs were similar to each other and near that of endogenous cortactin, based on anti-cortactin immunoblots (Figure 10A). Primary macrophages were infected with control and cortactin knockdown lentivirus and sorted for GFP expression as described above. Those cells were then infected with retroviral cortactin expression constructs. The retroviral constructs contained a puromycin resistance marker, which allowed us to select for infected cells. Puromycin-resistant cells were differentiated into osteoclasts, and the presence of podosomes was assessed.
Figure 10.
Rescue of cortactin knockdown phenotype with cortactin mutants. (A) Expression of RNAi-resistant FLAG-tagged cortactin constructs in 293T cells expressing knockdown or control RNAi. Anti-cortactin antibody was used, revealing absence of cortactin in cells expressing knockdown RNAi, and restoration of cortactin in knockdown cells expressing FLAG-tagged wild-type and mutant cortactin constructs. (B) Presence of podosomes in cells expressing rescue constructs. Cells expressing control scrambled RNAi (left) or cortactin knockdown RNAi sequence 1 (right) were infected with RNAi-resistant, FLAG-tagged cortactin rescue constructs. The percentage of cells with podosomes is plotted, based on rhodamine-phalloidin staining of the differentiated osteoclasts. Error bars are SE of proportion, and asterisks indicate statistical difference (p < 0.01) compared with vector and wild type. Statistical significance was determined by the binomial probability distribution function. (C) Fluorescence and phase-contrast images of osteoclasts expressing RNAi and cortactin mutants, fixed, and stained with rhodamine-phalloidin and anti-FLAG. The top half is cells expressing control scrambled RNAi, and the bottom half is cells expressing cortactin knockdown RNAi sequence 1.
In cells expressing control scrambled RNAi, the 3YF and SH3 cortactin mutants had small dominant negative effects (Figure 10B, left) similar to those described above for cells without RNAi (Figure 9C). Cortactin knockdown cells infected with an empty rescue expression construct had a greatly decreased percentage of osteoclasts with podosomes, as expected, and expression of wild-type cortactin restored that value to near normal levels (Figure 10B, right). The rescued cells appeared normal in phase-contrast and rhodamine-phalloidin fluorescence images (Figure 10C). That wild-type cortactin rescues confirms that the cortactin knockdown phenotype is not a nonspecific effect of the RNAi sequence.
In contrast, the 3YF cortactin mutant did not rescue the cortactin knockdown phenotype, producing results similar to those for empty vector (Figure 10B, right). The SH3 mutant of cortactin was able to rescue, at a level less than but not significantly different from that of wild type (Figure 10B, right). We asked how the mutant cortactins localized, by anti-FLAG staining. In cells expressing control scrambled RNAi, the cortactin mutants localized to podosomes (Figure 10C), as seen for cells not expressing RNAi (Figure 9D). In cortactin knockdown cells, 3YF-cortactin was localized diffusely, whereas wild-type and SH3 mutant cortactin localized to podosomes (Figure 10C).
DISCUSSION
We discovered that cortactin is necessary in osteoclasts for actin ring assembly, bone resorption, and podosome assembly. The knockdown phenotypes were specific, strong, and highly penetrant. A great deal is known about cortactin’s biochemical properties to promote actin assembly via Arp2/3 complex and to act as a scaffold for signaling molecules in vitro (Weed et al., 2000; Weaver et al., 2001, 2002). However, the function of cortactin in cells has remained somewhat elusive. The only study of loss of cortactin function in an intact organism is in Drosophila, where complete loss-of-function mutations of the sole cortactin gene had only minor effects, producing smaller F-actin ring canals of germline cells and impaired migration of border cells in oogenesis (Somogyi and Rorth, 2004). In cultured cells, a variety of studies point to a role for cortactin in actin-based processes such as pathogen invasion (Lambotin et al., 2005; Selbach and Backert, 2005), endocytosis (Merrifield et al., 2005; Sauvonnet et al., 2005), post-Golgi transport (Cao et al., 2005), cell adhesion (El Sayegh et al., 2004; Helwani et al., 2004), and lamellipodial persistence (Bryce et al., 2005). In these studies, cortactin was generally found to associate with sites of actin assembly, and, in some cases, expression of cortactin mutants or knockdown of cortactin produced partial loss of actin-based motility or actin cytoskeleton assembly.
We hypothesized that cortactin might have a more crucial role in osteoclasts because cortactin is expressed in osteoclasts (Hiura et al., 1995), and osteoclasts are derived from hematopoietic cell precursors that express the cortactin homologue HS1 but not cortactin itself (Miglarese et al., 1994). Here, we found that cortactin expression was indeed induced upon osteoclast differentiation. In contrast, the expression of HS1 and other Arp2/3 activators, including WASp-family members, did not change with osteoclast differentiation.
To test our hypothesis, we depleted cortactin from osteoclasts using RNAi delivered with lentivirus. Cortactin-depleted osteoclasts plated on bone did not form actin-based sealing rings and were completely deficient in bone resorption. We suspect that the actin sealing ring defect is the cause of the failure in bone resorption, but we cannot rule out the possibility that secretion of acid and proteases may have been decreased. On nonosteoid surfaces, osteoclasts form actin-based podosomes, and we found that cortactin depletion led to the absence of podosomes, based on rhodamine-phalloidin staining of fixed cells and phase-contrast imaging of live cells. Thus, cortactin is necessary for podosome assembly in this setting. Recently, cortactin expression was also shown to be necessary for the podosome formation that results from phorbol ester treatment of the A7r5 line of vascular smooth muscle cells (Zhou et al., 2006).
Whether the mechanisms for actin assembly in sealing rings and podosomes are similar is an open question in osteoclast biology. Early studies suggested the hypothesis that podosomes might represent precursors of sealing rings, based on the localization of podosome-like structures around resorption pits in fixed cells and on certain similarities in molecular composition (Kanehisa et al., 1990; Lakkakorpi and Vaananen, 1991). In contrast, in a recent study of living osteoclasts on mineralized substratum, actin sealing rings were observed by GFP-actin imaging, but podosomes were not seen (Saltel et al., 2004). Inhibition of Rho did convert the sealing ring into a belt of podosomes (Saltel et al., 2004), suggesting that podosomes might be rapidly and efficiently incorporated into the sealing ring by the action of Rho. Our results here show that the sealing ring and podosomes both require cortactin to form, providing further evidence for their relationship.
Perhaps the most remarkable finding from our study was the specificity of the actin assembly defect caused by the loss of cortactin in osteoclasts on glass. The cell edge was highly dynamic in cortactin-depleted cells, in phase-contrast movies. Lamellipodia and filopodia were both present and dynamic. The cortactin-depleted cells showed a slight increase in the amount of lamellipodial activity, relative to filopodia. In a detailed study of lamellipodia in the HT1080 fibrosarcoma cell line, cortactin depletion decreased the persistence time of lamellipodia by half, with no effect on protrusion speed, protrusion distance, or retraction speed (Bryce et al., 2005). Given the proximity of podosomes and lamellipodia at the cell edge of the osteoclast, and that both seem to be nucleated by Arp2/3 complex, we find it remarkable that podosomes are completely absent, whereas lamellipodia are relatively normal. Therefore, different regulators of Arp2/3 complex may have distinct functions at close locations within one cell.
A priori, cortactin might promote podosome actin assembly either by increasing actin filament polymerization or by decreasing depolymerization. In vitro, cortactin activates Arp2/3 complex for actin nucleation, which increases the rate of polymerization, but cortactin also stabilizes actin filament branches formed by Arp2/3 complex, which decreases the rate of depolymerization (Weaver et al., 2001). Cortactin appears before filamentous actin when podosomes assemble in phorbol ester-stimulated A7r5 smooth muscle cells (Webb et al., 2006), suggesting that cortactin promotes actin polymerization. Two results from our study also support this idea. First, in time-lapse movies of cortactin-depleted cells, podosomes were not observed. If the primary role of cortactin was to stabilize actin filaments in podosomes, then one might have expected to see podosomes form at normal rates but disassemble rapidly. Second, in cases where cells had two belts of podosomes at their periphery, cortactin accumulation seemed to precede that of F-actin, based on differential staining of the newer, inner belt. In contrast, when we compared old and new podosomes within one belt, in cases where cells only had one belt, the ratio of cortactin to F-actin was the same. Perhaps the age differences between old and new podosomes are greater when a second belt is created.
A number of other molecular components, in addition to cortactin, are known to play important roles in the assembly of podosomes and the function of osteoclasts (reviewed by Linder and Aepfelbacher, 2003). Integrins, signaling proteins, and actin cytoskeleton components have all been implicated in various studies. For example, gelsolin is an actin filament severing protein found in the podosome, and osteoclasts from gelsolin-deficient mice have no podosomes (Chellaiah et al., 2000). In addition, WASp knockout mice have moderate deficits in the number of podosome-forming osteoclasts (Calle et al., 2004). In this study, WASp-deficient osteoclasts on bone showed a decrease in the area of bone surface resorbed, whereas other parameters, such as volume and depth of resorption pits, were unaffected. In addition, calpain, which can proteolyze the podosome proteins talin, Pyk2 and filamin, has been found to be important for osteoclast activity (Marzia et al., 2006).
Dynamin localizes to osteoclast podosomes, and expression of dynamin mutants can inhibit osteoclast activity (Bruzzaniti et al., 2005). In vitro, dynamin and cortactin can bind directly to each other, and their interaction can cause actin assembly on lipid vesicles, in the presence of Arp2/3 (Schafer et al., 2002). Here, we observed dynamin staining of podosomes, with increased staining of the inner younger belt, when cells had two belts, consistent with dynamin being important for podosome assembly.
Dynamin binds to the SH3 domain of cortactin (Schafer et al., 2002), as do several other proteins, including N-WASp (Kowalski et al., 2005), faciogenital dysplasia (Kim et al., 2004), WASp-interacting protein (Kinley et al., 2003), and CD2AP/CIN85 (Lynch et al., 2003). In osteoclasts, the physiologically relevant binding partners of cortactin’s SH3 domain have yet to be defined. Overexpression of a C-terminal fragment of cortactin, which included the SH3 domain, inhibited podosome formation in smooth muscle cells (Webb et al., 2006), presumably by causing nonproductive interactions with cortactin binding partners. Here, we tested an SH3 point mutation (W525K) in full-length cortactin for the ability to rescue the osteoclast knockdown phenotype. The mutant was able to rescue, at a value less than, but not significantly different from, that of wild-type cortactin. When expressed in control cells with normal levels of endogenous cortactin, this SH3 mutant of cortactin had a modest dominant negative effect. Therefore, the SH3 domain may have a minor role in cortactin function. The cortactin SH3 mutant localized to podosomes, so the SH3 domain does not target cortactin to podosomes. Instead, the SH3 domain may recruit other proteins. This is consistent with studies in fibroblasts, in which the N-terminal acidic Arp2/3-binding domain and the F-actin binding domain of cortactin are each necessary and together sufficient for cortactin localization to lamellipodia (Weed et al., 2000).
The role of cortactin’s tyrosine phosphorylation, downstream of Src, has been an important question. Here, we found that the three tyrosines targeted for phosphorylation by Src are necessary for podosome formation in osteoclasts, based on failure of a 3YF mutant to suppress the knockdown phenotypes. When expressed in cells with endogenous cortactin, this mutant had only a small dominant negative effect, and the mutant protein localized to podosomes, suggesting that the tyrosine residues have a primary or direct role in cortactin function. Consistent with our results, the 3YF cortactin mutant failed to rescue a cortactin knockdown defect in endothelial cells assayed for the ability to support leukocyte transmigration (Yang et al., 2006).
In conclusion, cortactin-depleted osteoclasts fail to form actin-based sealing rings and fail to resorb bone, showing that cortactin-based actin assembly is necessary for the sealing ring to function. On glass, actin-based podosomes do not form, which supports the notion that podosomes and sealing rings have molecular mechanisms in common. In other respects, cortactin-depleted osteoclasts are remarkably normal in terms of actin assembly at the cell edge. A 3YF mutant of cortactin localizes normally but fails to restore podosome formation in cortactin-depleted cells, demonstrating the importance of cortactin’s tyrosine phosphorylation downstream of Src. In contrast, the SH3 domain of cortactin is largely dispensable in this context.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Ken Blumer and Sheila Stewart for helpful insight and discussion and to Dr. Stewart for assistance with lentivirus. We thank Dr. Frank Gertler for the lentivirus vector. Dr. Scott Weed kindly provided cortactin antibodies. This work was supported in part by National Research Service Award, Medical Scientist, GM-07200 (to S.T.); National Institutes of Health Grant GM-38542 (to J.A.C.); and National Institutes of Health Grants AR-48812 and AR-46852 (to F.P.R.). The Siteman Cancer Center is supported in part by National Cancer Institute Cancer Center Support Grant P30 CA91842.
Abbreviations used:
- FACS
fluorescence activated cell sorter
- HS1
hematopoietic cell line-specific protein
- MCSF
macrophage colony-stimulating factor
- MOI
multiplicity of infection
- NFATc1
nuclear factor of activated T-cells c1
- N-WASp
neuronal-WASp
- RANKL
receptor activator of nuclear factor κB ligand
- RNAi
RNA interference
- SEM
scanning electron microscopy
- TRAP
tartrate-resistant acid phosphatase
- WASp
Wiskott-Aldrich syndrome protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0187) on April 12, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bruzzaniti A., Neff L., Sanjay A., Horne W. C., De Camilli P., Baron R. Dynamin forms a Src kinase-sensitive complex with Cbl and regulates podosomes and osteoclast activity. Mol. Biol. Cell. 2005;16:3301–3313. doi: 10.1091/mbc.E04-12-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce N. S., Clark E. S., Leysath J. L., Currie J. D., Webb D. J., Weaver A. M. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Burns S., Thrasher A. J., Blundell M. P., Machesky L., Jones G. E. Configuration of human dendritic cell cytoskeleton by Rho GTPases, the WAS protein, and differentiation. Blood. 2001;98:1142–1149. doi: 10.1182/blood.v98.4.1142. [DOI] [PubMed] [Google Scholar]
- Calle Y., Jones G. E., Jagger C., Fuller K., Blundell M. P., Chow J., Chambers T., Thrasher A. J. WASp deficiency in mice results in failure to form osteoclast sealing zones and defects in bone resorption. Blood. 2004;103:3552–3561. doi: 10.1182/blood-2003-04-1259. [DOI] [PubMed] [Google Scholar]
- Cao H., Weller S., Orth J. D., Chen J., Huang B., Chen J. L., Stamnes M., McNiven M. A. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat. Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- Chellaiah M., Kizer N., Silva M., Alvarez U., Kwiatkowski D., Hruska K. A. Gelsolin deficiency blocks podosome assembly and produces increased bone mass and strength. J. Cell Biol. 2000;148:665–678. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O., Saltel F., Geminard J. C., Jurdic P., Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol. Biol. Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sayegh T. Y., Arora P. D., Laschinger C. A., Lee W., Morrison C., Overall C. M., Kapus A., McCulloch C. A. Cortactin associates with N-cadherin adhesions and mediates intercellular adhesion strengthening in fibroblasts. J. Cell Sci. 2004;117:5117–5131. doi: 10.1242/jcs.01385. [DOI] [PubMed] [Google Scholar]
- Feng X., Novack D. V., Faccio R., Ory D. S., Aya K., Boyer M. I., McHugh K. P., Ross F. P., Teitelbaum S. L. A Glanzmann’s mutation in beta 3 integrin specifically impairs osteoclast function. J. Clin. Investig. 2001;107:1137–1144. doi: 10.1172/JCI12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M., Kaverina I., Resch G. P., Vignal E., Burgstaller G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol. Biol. Cell. 2003;14:2482–2491. doi: 10.1091/mbc.E02-11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head J. A., Jiang D., Li M., Zorn L. J., Schaefer E. M., Parsons J. T., Weed S. A. Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol. Biol. Cell. 2003;14:3216–3229. doi: 10.1091/mbc.E02-11-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwani F. M., Kovacs E. M., Paterson A. D., Verma S., Ali R. G., Fanning A. S., Weed S. A., Yap A. S. Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization. J. Cell Biol. 2004;164:899–910. doi: 10.1083/jcb.200309034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura K., Lim S. S., Little S. P., Lin S., Sato M. Differentiation dependent expression of tensin and cortactin in chicken osteoclasts. Cell Motil. Cytoskeleton. 1995;30:272–284. doi: 10.1002/cm.970300405. [DOI] [PubMed] [Google Scholar]
- Hurst I. R., Zuo J., Jiang J., Holliday L. S. Actin-related protein 2/3 complex is required for actin ring formation. J. Bone Miner. Res. 2004;19:499–506. doi: 10.1359/JBMR.0301238. [DOI] [PubMed] [Google Scholar]
- Kanehisa J., Yamanaka T., Doi S., Turksen K., Heersche J. N., Aubin J. E., Takeuchi H. A band of F-actin containing podosomes is involved in bone resorption by osteoclasts. Bone. 1990;11:287–293. doi: 10.1016/8756-3282(90)90082-a. [DOI] [PubMed] [Google Scholar]
- Kanner S. B., Reynolds A. B., Vines R. R., Parsons J. T. Monoclonal antibodies to individual tyrosine-phosphorylated protein substrates of oncogene-encoded tyrosine kinases. Proc. Natl. Acad. Sci. USA. 1990;87:3328–3332. doi: 10.1073/pnas.87.9.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Hou P., Gorski J. L., Cooper J. A. Effect of Fgd1 on cortactin in Arp2/3 complex-mediated actin assembly. Biochemistry. 2004;43:2422–2427. doi: 10.1021/bi036173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinley A. W., Weed S. A., Weaver A. M., Karginov A. V., Bissonette E., Cooper J. A., Parsons J. T. Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr. Biol. 2003;13:384–393. doi: 10.1016/s0960-9822(03)00107-6. [DOI] [PubMed] [Google Scholar]
- Kitamura D., Kaneko H., Taniuchi I., Akagi K., Yamamura K., Watanabe T. Molecular cloning and characterization of mouse HS1. Biochem. Biophys. Res. Commun. 1995;208:1137–1146. doi: 10.1006/bbrc.1995.1452. [DOI] [PubMed] [Google Scholar]
- Kowalski J. R., Egile C., Gil S., Snapper S. B., Li R., Thomas S. M. Cortactin regulates cell migration through activation of N-WASP. J. Cell Sci. 2005;118:79–87. doi: 10.1242/jcs.01586. [DOI] [PubMed] [Google Scholar]
- Lakkakorpi P. T., Vaananen H. K. Kinetics of the osteoclast cytoskeleton during the resorption cycle in vitro. J. Bone Miner. Res. 1991;6:817–826. doi: 10.1002/jbmr.5650060806. [DOI] [PubMed] [Google Scholar]
- Lambotin M., Hoffmann I., Laran-Chich M. P., Nassif X., Couraud P. O., Bourdoulous S. Invasion of endothelial cells by Neisseria meningitidis requires cortactin recruitment by a phosphoinositide-3-kinase/Rac1 signaling pathway triggered by the lipo-oligosaccharide. J. Cell Sci. 2005;118:3805–3816. doi: 10.1242/jcs.02514. [DOI] [PubMed] [Google Scholar]
- Lessard J. L. Two monoclonal antibodies to actin: one muscle selective and one generally reactive. Cell Motil. Cytoskeleton. 1988;10:349–362. doi: 10.1002/cm.970100302. [DOI] [PubMed] [Google Scholar]
- Linder S., Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- Linder S., Nelson D., Weiss M., Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc. Natl. Acad. Sci. USA. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D. K., Winata S. C., Lyons R. J., Hughes W. E., Lehrbach G. M., Wasinger V., Corthals G., Cordwell S., Daly R. J. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 2003;278:21805–21813. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- Maa M. C., Wilson L. K., Moyers J. S., Vines R. R., Parsons J. T., Parsons S. J. Identification and characterization of a cytoskeleton-associated, epidermal growth factor sensitive pp60c-src substrate. Oncogene. 1992;7:2429–2438. [PubMed] [Google Scholar]
- Marzia M., Chiusaroli R., Neff L., Kim N. Y., Chishti A. H., Baron R., Horne W. C. Calpain is required for normal osteoclast function and is downregulated by calcitonin. J. Biol. Chem. 2006;281:9745–9754. doi: 10.1074/jbc.M513516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh K. P., Hodivala-Dilke K., Zheng M. H., Namba N., Lam J., Novack D., Feng X., Ross F. P., Hynes R. O., Teitelbaum S. L. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J. Clin. Investig. 2000;105:433–440. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield C. J., Perrais D., Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Miglarese M. R., Mannion-Henderson J., Wu H., Parsons J. T., Bender T. P. The protein tyrosine kinase substrate cortactin is differentially expressed in murine B lymphoid tumors. Oncogene. 1994;9:1989–1997. [PubMed] [Google Scholar]
- Mizutani K., Miki H., He H., Maruta H., Takenawa T. Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 2002;62:669–674. [PubMed] [Google Scholar]
- Ochoa G. C. A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 2000;150:377–389. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson D. A. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Saltel F., Destaing O., Bard F., Eichert D., Jurdic P. Apatite-mediated actin dynamics in resorbing osteoclasts. Mol. Biol. Cell. 2004;15:5231–5241. doi: 10.1091/mbc.E04-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N., Dujeancourt A., Dautry-Varsat A. Cortactin and dynamin are required for the clathrin-independent endocytosis of gammac cytokine receptor. J. Cell Biol. 2005;168:155–163. doi: 10.1083/jcb.200406174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. A., Korshunova Y. O., Schroer T. A., Cooper J. A. Differential localization and sequence analysis of capping protein beta-subunit isoforms of vertebrates. J. Cell Biol. 1994;127:453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. A., Weed S. A., Binns D., Karginov A. V., Parsons J. T., Cooper J. A. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr. Biol. 2002;12:1852–1857. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Backert S. Cortactin: an Achilles’ heel of the actin cytoskeleton targeted by pathogens. Trends Microbiol. 2005;13:181–189. doi: 10.1016/j.tim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Somogyi K., Rorth P. Cortactin modulates cell migration and ring canal morphogenesis during Drosophila oogenesis. Mech. Dev. 2004;121:57–64. doi: 10.1016/j.mod.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Soriano P., Montgomery C., Geske R., Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- Takeshita S., Kaji K., Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J. Bone Miner. Res. 2000;15:1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- Vaananen H. K., Zhao H., Mulari M., Halleen J. M. The cell biology of osteoclast function. J. Cell Sci. 2000;113:377–381. doi: 10.1242/jcs.113.3.377. [DOI] [PubMed] [Google Scholar]
- Weaver A. M., Heuser J. E., Karginov A. V., Lee W. L., Parsons J. T., Cooper J. A. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr. Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- Weaver A. M., Karginov A. V., Kinley A. W., Weed S. A., Li Y., Parsons J. T., Cooper J. A. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- Webb B. A., Eves R., Mak A. S. Cortactin regulates podosome formation: roles of the protein interaction domains. Exp. Cell Res. 2006;312:760–769. doi: 10.1016/j.yexcr.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Weed S. A., Parsons J. T. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 2001;20:6418–6434. doi: 10.1038/sj.onc.1204783. [DOI] [PubMed] [Google Scholar]
- Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M. D., DePace A. H., Verma S., Iwamatsu A., Mitchison T. J. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Kowalski J. R., Zhan X., Thomas S. M., Luscinskas F. W. Endothelial cell cortactin phosphorylation by Src contributes to polymorphonuclear leukocyte transmigration in vitro. Circ. Res. 2006;98:394–402. doi: 10.1161/01.RES.0000201958.59020.1a. [DOI] [PubMed] [Google Scholar]
- Yasuda H. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Webb B. A., Eves R., Mak A. S. Effects of tyrosine phosphorylation of cortactin on podosome formation in A7r5 vascular smooth muscle cells. Am. J. Physiol. 2006;290:C463–C471. doi: 10.1152/ajpcell.00350.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.