Abstract
Although we have detailed information on the alterations occurring in steady-state levels of all cellular mRNAs during differentiation, we still know little about more global changes. Therefore, we investigated the numbers of molecules of RNA polymerase II that are active—and the way those molecules are organized—as two mouse cells (aneuploid F9 teratocarcinoma, and euploid and totipotent embryonic stem cells) differentiate into parietal endoderm. Quantitative immunoblotting shows the number of active molecules roughly halves. Transcription sites (detected by light and electron microscopy after allowing engaged polymerases to extend nascent transcripts in bromouridine-triphosphate) are uniformly distributed throughout the nucleoplasm. The numbers of such sites fall during differentiation as nuclei become smaller, but site density and diameter remain roughly constant. Similar site densities and diameters are found in salamander (amphibian) cells with 11-fold larger genomes, and in aneuploid HeLa cells. We conclude that active polymerases and their nascent transcripts are concentrated in a limited number of discrete nucleoplasmic sites or factories, and we speculate that the organization of transcription is conserved during both differentiation and evolution to a high C value.
INTRODUCTION
Differentiation involves the repression of some genes and the activation of others. For example, during erythropoiesis nuclei become smaller and heterochromatic as many genes are silenced; concurrently, globin genes become active by associating with specific nuclear sites known as “factories” or “active chromatin hubs” (Cook, 1999, 2002; de Laat and Grosveld, 2003; Osborne et al., 2004). Although microarrays provide detailed information on changes in steady-state mRNA levels during differentiation (Kelly and Rizzino, 2000; Harris and Childs, 2002; Tanaka et al., 2002; Sperger et al., 2003), there is little information on how primary rates of transcription change. For example, we do not know by how much global rates vary as any mammalian cell differentiates. Eukaryotic genomes also differ in size more than 200,000-fold in a way unrelated to organismal complexity; there is no consensus as to the basis of this C-value enigma (Hartl, 2000; Gregory, 2001), or on how transcription might be organized in such differently sized genomes.
Against this background, we investigated the changes that occur in the rates and organization of transcription as two mouse cells differentiate. Mouse F9 teratocarcinoma cells are a well-studied aneuploid line that can be induced to differentiate into parietal endoderm, which is characterized by flattened cells that secrete tissue-type plasminogen activator, laminin, and type IV collagen (Strickland and Mahdavi, 1978; Strickland et al., 1980; Hogan et al., 1983). ESF/48–1 cells are totipotent embryonic stem (ES) cells with a normal karyotype; when transferred into host blastocysts, they contribute to all tissues (including germ lines) of the resulting chimeras (Brook and Gardner, 1997), and they can be induced to differentiate into parietal endoderm much like F9 cells. We find the numbers of active molecules of RNA polymerase II roughly halve as mouse F9 and ES cells differentiate. The number of nucleoplasmic transcription sites also falls as nuclei shrink, but site density and diameter remain much the same. This constancy of site density and diameter in the face of a changing nucleoplasmic volume prompted us to examine a larger nucleus. A1 cells are derived from the limb of the red-spotted salamander (newt), which has an 11-fold larger genome (http://www.genomesize.com/); they can differentiate into multinucleated myotubes (Ferretti and Brockes, 1988; Lo et al., 1993). They have similar site densities and diameters, despite their larger nuclei. We conclude that active polymerases are concentrated in a limited number of discrete nucleoplasmic sites or factories, and we speculate that the organization of transcription is conserved during both differentiation and evolution to a high C value.
MATERIALS AND METHODS
Cells and Buffers
F9 cells (Bernstine et al., 1973) were grown in DMEM + 15% fetal calf serum (FCS) in flasks coated with 0.1% gelatin (all coated plastics/coverslips were BioCoat cellware; BD Biosciences, Franklin Lakes, NJ); few cells differentiate spontaneously, but differentiation was induced by adding 10−6 M retinoic acid, 10−4 M dibutyryl cyclical AMP, and 10−4 M isobutylmethyl xanthine (Mason et al., 1986). A clone of ES cells (ESF 48/1) from a male 129 mouse was kindly provided by F. A. Brook and R. L. Gardner (Department of Zoology, University of Oxford, Oxford, United Kingdom). They are the cells derived from the single-cell suspension of a fifth-day epiblast that are described in Table 3 of Brook and Gardner (1997), although the name ESF 48/1 was not used in this article. They have a normal karyotype and are totipotent. They were cultured in the same medium plus leukemia inhibitory factor (LIF) in flasks coated with 0.1% gelatin (Robertson, 1997) and plated on to coverslips coated with collagen I; this coat minimizes detachment during permeabilization and does not induce differentiation (because Oct-4 levels remain constant; Figure 2). ESF 48/1 was induced to differentiate like F9, except LIF was removed as drugs were added. A1 cells (Ferretti and Brockes, 1988) were grown in minimal essential medium + 10% FCS + 24% water + 100 μg/ml insulin on collagen-coated coverslips.
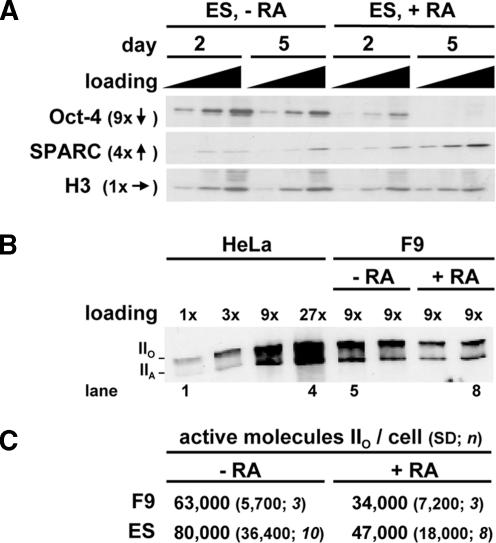
Figure 2.
Amounts of Oct-4, SPARC, and the largest (catalytic) subunit of RNA polymerase II (RPB1) in differentiating F9 and ES cells. (A) Blots illustrating changes in Oct-4, SPARC, and histone H3 (a loading control) induced by RA. Cells were grown ± RA for 2 or 5 d; 1, 3, or 9 μg of protein was resolved on gels, proteins were detected by immunoblotting, and selected regions of blots were imaged. Relative changes in band intensities (averages from 3 blots) on day 2 (−RA) and day 5 (+RA) are indicated by arrows. (B) Blot illustrating loss of forms IIO and IIA of RPB1 induced by RA in F9 cells. Cells were grown ± RA for 1 (−RA) or 5 d (+RA), proteins from known numbers of cells were resolved on a gel, forms IIO and IIA were detected by immunoblotting, and a selected region of the blot was imaged. HeLa proteins are included for reference; a 1× loading represents protein from 103 cells. (C) Numbers of active molecules of form IIO were calculated from blots like that in B, assuming a HeLa cell contains 65,000 molecules. Undifferentiated and differentiated populations were significantly different; the Kolmogorov–Smirnov test applied to the log data showed a normal distribution, and p values (determined using the two-sample Welch t test) were 0.047 (F9) and 0.034 (ES). Values for differentiated populations necessarily contain SPARC +ve and −ve cells.
“Physiological” buffer (PB; pH 7.4; Pombo et al., 1999a) contains 100 mM potassium acetate, 30 mM KCl, 10 mM Na2HPO4, 1 mM MgCl2, 1 mM Na2ATP, and 1 mM dithiothreitol (the original buffer also contains phenylmethylsulfonyl fluoride and RNase inhibitor). PBB and PBF contain PB plus 100 mg/ml bovine serum albumin (BSA) and 50 mg/ml Ficoll PM400 (GE Healthcare; Little Chalfont, Buckinghamshire, United Kingdom), respectively. Phosphate-buffered saline (PBS)-Tween is PBS plus 0.1% Tween 20. PBS+, pH 8.0, is PBS plus 10 mg/ml BSA and 0.2% gelatin from cold water fish skin (Sigma Chemical, Poole, Dorset, United Kingdom). Transcription buffer is PBF plus either 100 μM ATP, CTP, GTP, and bromouridine-triphosphate (Br-UTP) plus MgCl2 to a final concentration of 1.4 mM, or just 100 μM Br-UTP plus MgCl2 to a final concentration of 1.1 mM (the additional MgCl2 ensures Mg2+ ions are equimolar to nucleotide triphosphates). The buffer with all triphosphates supports “run-on” transcription at high rates (Iborra and Cook., unpublished data).
Immunoblotting
Cells were lysed (10 min; 4°C) in 10 mM Tris-HCl, pH 8.0, 0.5 M NaCl, and 1% Triton X-100; proteins were resolved on SDS-polyacrylamide gels (5–15% for Figure 2A, 6% for Figure 2B; Sambrook et al., 1989) and blotted onto membranes; antigens were detected (Kimura et al., 1999) using an enhanced chemiluminescence kit (GE Healthcare); digital images were collected; and band intensities were measured using Photoshop (Adobe Systems, Mountain View, CA). Primary antibodies were 1) mouse monoclonal anti-Oct-4 (2 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA), 2) goat anti-secreted protein which is acidic and rich in cysteines (SPARC) (2 μg/ml; R&D Systems, Minneapolis, MN), 3) rabbit anti-histone H3 (COOH-terminal 130–135 peptide; 1/1000–1/3000; Stemmer et al., 1997), and 4) mouse monoclonal anti-polymerase (7C2; 1/10,000 dilution; Besse et al., 1995). Secondary antibodies were 1) donkey anti-mouse IgG (heavy and light chains) conjugated with horseradish peroxidase (HRP; 0.16 μg/ml; Jackson ImmunoResearch Laboratories, West Grove, PA; used with anti-polymerase), 2) sheep anti-mouse IgG conjugated with HRP (1/4000; GE Healthcare; used with anti-Oct-4), 3) donkey anti-goat IgG (heavy and light chains) conjugated with HRP (0.16 μg/ml; Jackson ImmunoResearch Laboratories), and 4) donkey anti-rabbit IgG (heavy and light chains) conjugated with HRP (0.16 μg/ml; Jackson ImmunoResearch Laboratories).
Run-On Transcription in Br-UTP
Transcription was monitored by permeabilizing cells and allowing engaged polymerases to extend their transcripts in Br-UTP, before levels of the resulting (nascent) Br-RNA were monitored by immunofluorescence (Jackson et al., 1993). Various detergents and conditions were tested to ensure that permeabilization was efficient (>95% cells became permeable judged from the fraction of cells with Br-RNA signal) and endogenous triphosphate pools were removed (judged from a low signal when three nucleotide triphosphates were omitted; Figure 4B), without reducing polymerizing activity (judged by a reduction in signal intensity per nucleus) due either to “overpermeabilization” (Pombo et al., 1999a) or cell detachment. Cells on coverslips were washed once in PBF (4°C), permeabilized by incubation (5 min; 4°C) with occasional agitation in saponin in PBF, and washed in PBF (4°C). The amount of saponin and number of washes after permeabilization varied with cell type and differentiation status. Undifferentiated F9 and ES cells, and differentiated F9 and ES cells were permeabilized in 150, 180, 170, and 275 μg/ml saponin, respectively; undifferentiated and differentiated cells were washed eight times and 13 times over 20 min, respectively. A1 cells were permeabilized using 140 μg/ml saponin and washed eight times over 20 min. Permeabilized cells were now incubated (25°C) in transcription buffer, and reactions were terminated at different times by replacing the buffer with 4% paraformaldehyde in 250 mM HEPES, pH 7.4, and incubation (15 min) at 4 or 20°C when labeling cryosections or whole cells, respectively. Note that 1) the permeabilized cells analyzed here were all more fragile than the HeLa cells studied previously (Pombo et al., 1999a), and 2) we were unable to develop conditions that enabled us to perform similar experiments on both undifferentiated cells and their differentiated counterparts isolated directly from animals.
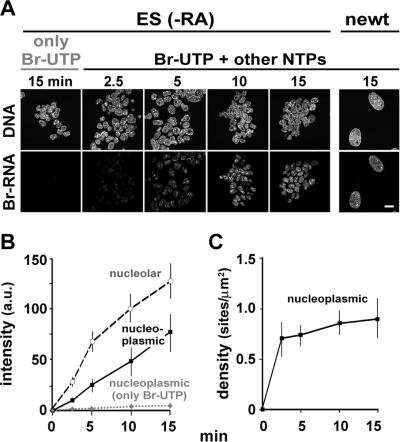
Figure 4.
Most transcription sites are detected. Cells were grown, permeabilized, incubated for various times in either Br-UTP alone or in Br-UTP + ATP + CTP + GTP, and fixed. (A) Nascent Br-RNA in ES (−RA) or (larger) newt cells was indirectly immunolabeled with Cy3, DNA counterstained with TO-PRO-3, and images were collected on a confocal microscope. Br-RNA signal increases progressively on incubation in Br-UTP and the other triphosphates (but not in Br-UTP alone). Bar, 20 μm. (B) Signal intensity due to Br-RNA (arbitrary units [au] ± SD; n = 30–60 cells for each data point) was measured in nucleoplasmic and nucleolar areas of ES cells (−RA) using images like those in A. Intensities given by cells incubated in all four triphosphates increase with time, whereas those incubated only in Br-UTP barely do so. Essentially similar curves were found with the other cell types. (C) After incubation in all four triphosphates, fixed cells (ES, −RA) were cryosectioned, Br-RNA was indirectly immunolabeled, confocal images of individual cryosections were collected, and the density of Br-RNA foci in nucleoplasmic areas of ≥35 nuclei were determined. Site density does not increase significantly between 10 and 15 min (p = 0.08, determined using the two-sample Welch t test), suggesting all sites are detected (if some sites initially remained undetected, the sixfold increase in incorporation seen in whole cells would lead to a corresponding increase in density). Similar curves were seen with all cell types; thus, densities after 2.5 and 15 min were 0.7 ± 0.2 and 1.0 ± 0.2 (F9, −RA), 0.7 ± 0.2 and 1.0 ± 0.3 (F9, +RA), 1.0 ± 0.2 and 1.4 ± 0.3 (ES, +RA, SPARC −ve), 0.9 ± 0.1 and 1.4 ± 0.3 (ES, +RA, SPARC +ve), and 1 ± 0.2 and 1.1 ± 0.3 (newt).
Cryosections
Cells were further fixed (20°C; 1 h) in 8% paraformaldehyde in 250 mM HEPES, pH 7.4, and gold (120-nm) and pink/yellow (150-nm) cryosections were prepared (Tokuyasu, 1980; Pombo et al., 1999a) with the modification that cell pellets were immersed (16 h; 4°C) in 2.1 M sucrose in PBS. Sections were placed on coverslips or carbon-coated grids for light (LM) and electron microscopy (EM), respectively.
Immunofluorescence
After fixation, cells were washed three times in PBS, incubated (20 min) to quench aldehydes in 25 mM glycine in PBS, permeabilized (10 min) in 1% Triton X-100 in PBS to maximize antibody access, and washed three times over 30 min in PBS-0.1% Tween. Unspecific binding sites were blocked (20 min) in PBS+, and antigens were indirectly immunolabeled (Pombo et al., 1999a). All washes were in PBS-0.1% Tween, and antibody incubations were in PBS+ at room temperature unless stated otherwise. For single labeling, cells were incubated (1.5–2 h) with primary antibody, washed (4–6 times over 0.5–1.5 h), incubated (1–1.5 h) with secondary antibody, washed (3 times over 30 min), and rewashed (10 min) in PBS. Nucleic acids were counterstained (dyes from Invitrogen, Carlsbad, CA) by incubation (10 min) in 0.67 μM TO-PRO-3, 0.4 μM SYTO 16, or 1 μg/ml 4′,6′-diamino-2-phenylindole (DAPI), and washed (2 times over 20 min) in PBS. Finally, coverslips were mounted using Vectashield (Vector Laboratories, Burlingame, CA). Cryosections were treated as described above except for 1) permeabilization (2 min) in 0.1% Triton X-100 in PBS followed by washing five times over 30 min in PBS, 2) all subsequent washes and antibody incubations were in PBS+ (20°C) unless stated otherwise, and 3) counterstaining was accomplished with 20 μM TO-PRO-3.
For double labeling, cells or cryosections were incubated simultaneously with both primary antibodies and then with both secondary antibodies. The following antibody pairs were used for indirect immunolabeling. For Figure 1, primary mouse monoclonal anti-Oct-4 (2 μg/ml; Santa Cruz Biotechnology) with secondary donkey anti-mouse IgG (heavy and light chains) conjugated with Cy3 (6 μg/ml; Jackson ImmunoResearch Laboratories), and primary goat anti-SPARC (2 μg/ml; R&D Systems) with secondary donkey anti-goat IgG (heavy and light chains) conjugated with Alexa 488 (6.5 μg/ml; Alexa dye from Invitrogen was conjugated with donkey anti-goat IgG antibody from Jackson ImmunoResearch Laboratories according to the manufacturer’s instructions). For Figures 3 and 4, primary mouse monoclonal anti-5-bromo-2′-deoxyuridine BrdU (used to detect Br-RNA; 1/100; Caltag Laboratories, Burlingame, CA) with donkey anti-mouse IgG conjugated with Cy3 (6 μg/ml). For Figure 5, primary goat anti-SPARC plus mouse monoclonal anti-BrdU with secondary donkey anti-goat IgG conjugated with Alexa 488 plus donkey anti-mouse IgG conjugated with Cy3 (all as described above).
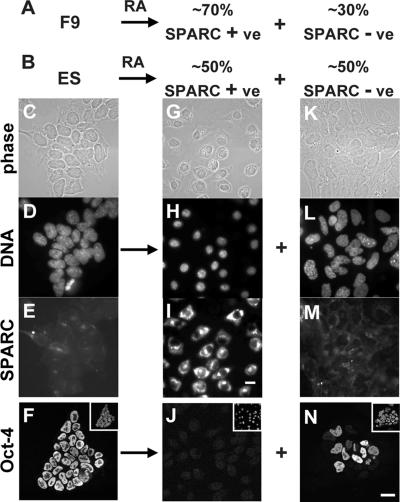
Figure 1.
Differentiation of F9 and ES cells. (A and B) F9 and ES cells differentiate into SPARC +ve and −ve populations, respectively. (C–N) ES cells were grown ± RA for 5 d, SPARC or Oct-4 was indirectly immunolabeled with Alexa 488, DNA was stained with DAPI, and single equatorial images were collected using a phase contrast or fluorescence microscope. SPARC −ve, Oct-4 +ve, cells differentiate into two populations—one population with small Oct-4 −ve nuclei and large amounts of cytoplasmic SPARC, and the other population with large irregularly shaped nuclei (some SPARC +ve; others SPARC −ve). Insets, DAPI-stained view of fields. Bar, 20 μm.
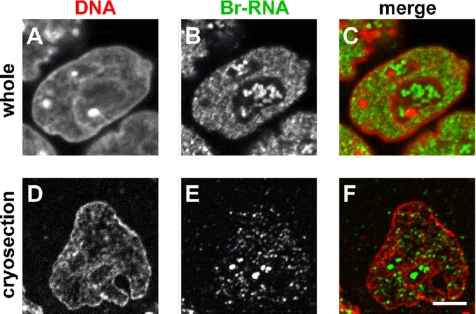
Figure 3.
Individual Br-RNA sites in F9 cells can only be resolved in cryosections. Cells were permeabilized, engaged polymerases were allowed to extend nascent transcripts in Br-UTP and fixed, and in some cases (D–F) cryosectioned. After indirectly immunolabeling Br-RNA with Cy3, and counterstaining DNA with TO-PRO-3, single equatorial images were collected on a confocal microscope. Bar, 5 μm. (A–C) Whole cells. Individual sites containing Br-RNA cannot be resolved (B) because too many are contained in the optical section (≥700 nm). (D–F) Cryosections. Individual sites are easily resolved and counted (E) in the thin section (120 nm).
Figure 5.
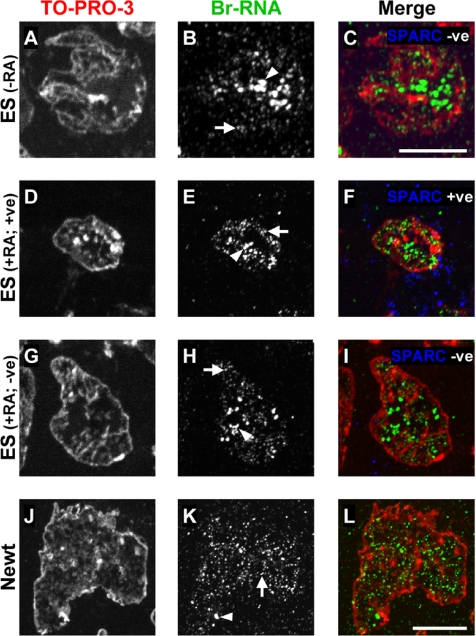
Imaging transcription sites in cryosections by light microscopy. Cells were grown ± RA and permeabilized, and engaged polymerases were allowed to extend nascent transcripts in Br-UTP plus the other triphosphates for 15 min before they were fixed and cryosectioned. Br-RNA was indirectly immunolabeled with Cy3, DNA was counterstained with TO-PRO-3, and fluorescence images were collected and merged. The pseudocolors used are indicated. Bright nucleolar (Br-RNA) foci are marked by arrowheads, and fainter nucleoplasmic foci by arrows. Bars, 10 μm. (A–I) ES cells. SPARC was also indirectly immunolabeled with Alexa 488. In all cells, Br-RNA is concentrated in nucleoplasmic sites present at roughly the same density. (J–L) A1 cells. The density of nucleoplasmic Br-RNA foci is much as in B, E, and H.
Images were collected using a Radiance 2000 MP confocal microscope controlled by LaserSharp 2000 (Bio-Rad, Hemel Hempstead, Hertsfordshire, United Kingdom) attached to an inverted microscope (Nikon TE300; Nikon UK Limited, Surrey, United Kingdom) and a 60× PlanApo oil immersion objective (numerical aperture 1.4). Kalman-filtered images were collected using a low iris aperture (0.7–1.2 mm) with minimum laser power to fill the whole gray scale in low scan/low signal mode. For multiple labeling, sequential series were obtained by alternating the different channels to minimize z-axis changes during collection; no “bleedthrough” was detected between channels.
Nuclear and nucleoplasmic volumes were measured in fixed whole cells after counterstaining with TO-PRO-3; sequential equatorial sections were collected using the confocal microscope at 0.5-μm intervals from bottom of the cell to the top, three-dimensional reconstructions were prepared, and volumes were determined using Imaris software (Bitplane, St. Paul, MN). The estimated average volume of 10 fluorescent microspheres (6 and 15 μm in diameter; Molecular Probes) lay within 1% of the real volume. Volumes can also be determined from diameters seen in sections using stereological procedures if it is assumed that the structures analyzed are spheres or ovoids (Iborra et al., 1998); this approach was not used here, because some nuclei were shaped so irregularly.
Immunogold Labeling
Br-RNA in cryosections on grids was incubated with the primary anti-BrdU, washed (all as described above) and incubated with either 1) goat anti-mouse IgG conjugated with 5-nm gold particles (1 μg/ml; British Biocell International, Cardiff, United Kingdom) for 3–4 h or 2) rabbit anti-mouse IgG (13 μg/ml; MP Biomedicals, Irvine, CA) for 1 h followed by goat anti-rabbit IgG conjugated with 5-nm particles (1 μg/ml; British Biocell International) for 3–4 h. Because double and triple sandwiches gave similar site densities and diameters, results were pooled. After the final antibody, cryosections were washed (4 times over 40 min) in PBS+, rewashed (16 h; 4°C) in PBS and then in water (5 drops; 1 h), and stained (10 min; 4°C) with 0.3% uranyl acetate in 2% methylcellulose. Images were collected on a Zeiss EM 912 Omega (Carl Zeiss, Oberkochen, Germany) transmission electron microscope equipped with a charge-coupled device camera (2048 × 2048 pixels; Proscan, Cincinnati, OH) running under AnalySIS EasiVision software (Soft Imaging Systems, Münster, Germany).
Image Analysis
For Figure 4B, the intensity of Br-RNA signal in nucleolar and nucleoplasmic regions was measured using MetaMorph software (Molecular Devices, Sunnyvale, CA), and background over noncellular regions was subtracted. The number of foci per unit area of a fluorescent image like that in Figure 4C can be measured automatically, but then we are confronted with the problem of how to define a focus. This requires arbitrary decisions on how many contiguous pixels are required to constitute a focus, where to draw the threshold between signal and noise, and whether two peaks of intensity are part of one extended focus. In practice, human eyes and brains are well adapted to this kind of complex image analysis (Iborra et al., 1996), and we found they gave reproducible results. For example, site numbers in images like those in Figure 5H counted (blind) by three untutored individuals lay within 10% of that determined by an expert. Numbers of sites in electron micrographs (Figure 6) were estimated more rigorously, where lone gold particles represent background and clusters the sites. A cluster of particles marking a site was defined as three or more particles in which each particle in the cluster lies within 40 nm of another. The background cluster density obtained by run-on only in Br-UTP was subtracted. Standard stereological procedures were then used to calculate site densities and diameters (Williams, 1977; Iborra and Cook, 1998; Pombo et al., 1999b). The density of foci (LM) or clusters (EM) per unit volume (nv) was obtained from the density per unit area of image (ns; Weibel, 1979) using the formula of DeHoff and Rhines, corrected by Abercrombie to take into account the “Holmes” effect. The diameter (d) of a cluster (EM) was determined after measurement of major (2a) and minor (2b) orthogonal axes and by treating the underlying objects as spheres; then, the average diameter (D) of the population was calculated using Fullman’s formula. Diameters were determined using a two-layer immunolabeling sandwich to minimize errors introduced by probe size (Iborra and Cook, 1998), and corrected for the Holmes effect. The minimum sample size required was estimated using the progressive mean; ≥77% of the last values analyzed lay within 10% of the mean.
Figure 6.
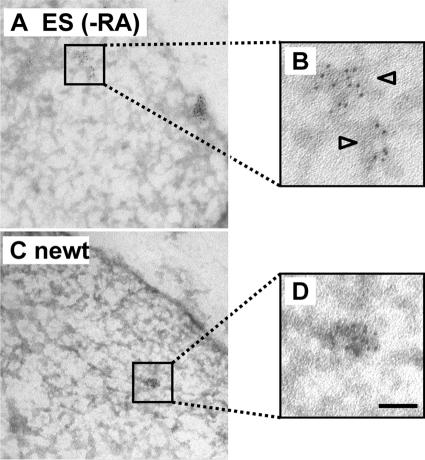
Imaging transcription sites in cryosections by electron microscopy. After preparing cryosections as in Figure 5, Br-RNA was indirectly immunolabeled with 5-nm gold particles, sections were stained with uranyl acetate, and micrographs were collected. Bars, 50 nm. (A and B) ES cells. Clusters of gold particles mark Br-RNA. (C and D) A1 cells. Clusters are of similar size to those in B.
The percentage of sites seen in the peripheral 20% of the nucleoplasm was determined using images like those in Figure 5; the nucleoplasmic area in the image (which reflects the volume of the cryosection) was selected, eroded pixel by pixel from the periphery using MetaMorph to leave 80% area, and the fraction of sites in the eroded (peripheral) 20% was determined. A one-sample parametric t test was used to determine whether means were significantly different from the expected mean. Changes in TO-PRO-3 intensity occurring during differentiation were analyzed using images of undifferentiated and differentiated (SPARC +ve) ES cells like those in Figure 5, A and D. We assume TO-PRO-3 intensity reflects chromatin density. Intensities were first stretched to fill the gray scale.
Data were analyzed to see whether they were normally distributed using the Kolmogorov–Smirnov test (applied to log data in Figures 2C and 4C, plus nucleoplasmic volumes, two-dimensional (2D) LM site densities in Table 1, the number of sites in the peripheral 20% of the nucleoplasm, and changes in heterochromatin measured using TO-PRO-3 intensity). If normally distributed, a Welch two-sample parametric t test was used to determine whether differences in means were significant. If not normally distributed (for 2D EM site density in F9 and for nucleoplasmic volume in differentiated SPARC −ve ES cells in Table 1), a two-sample Wilcoxon test was used.
Table 1.
Nucleoplasmic and nucleolar volumes, plus transcription site densities, diameters, and numbers in the nucleoplasm of various types of cells
| F9 |
ES |
|||
|---|---|---|---|---|
| LM | EMa | LM | EMa | |
| Undifferentiated | ||||
| 1. Nucleoplasmic volume (μm3) | 810 ± 300 (12) | 840 ± 240 (14) | ||
| 2. Nucleolar volume (μm3) | 260 ± 86 (12) | 480 ± 134 (14) | ||
| 3. 2D site density, ns (sites/μm2) | 1.0 ± 0.2 (79)b | 1.6 (41) | 1.1 ± 0.3 (50)b | 1.4c (47) |
| 4. 3D site density, nv (sites/μm3) | 6 | 7.9 | 6.1 | 6.8 |
| 5. Site diameter [range] (nm) | 49 [20–60] (96) | 52 [22–68] (139) | ||
| 6. Site no. | 4800 | 6400 | 5100 | 5700 |
| Differentiated (SPARC +ve) | ||||
| 7. Nucleoplasmic volume (μm3) | 460 ± 160 (11)d | 400 ± 140 (14)d | ||
| 8. Nucleolar volume (μm3) | 200 ± 66 (11) | 240 ± 140 (14)d | ||
| 9. 2D site density, ns (sites/μm2) | 1.0 ± 0.3 (39)b | 1.4 ± 0.3 (19)b | 2.0e,f (45) | |
| 10. 3D site density, nv (sites/μm3) | 6.1 | 8.3 | 9.9e | |
| 11. Site diameter [range] (nm) | 54e [20–60] (171) | |||
| 12. Site no. | 2800 | 3300 | 3900e | |
| Differentiated (SPARC −ve) | ||||
| 13. Nucleoplasmic volume (μm3) | 1500 ± 760 (12) | |||
| 14. Nucleolar volume (μm3) | 750 ± 390 (12) | |||
| 15. 2D site density, ns (sites/μm2) | 1.4 ± 0.3 (38)b | 2.0e,f (45) | ||
| 16. 3D site density, nv (sites/μm3) | 8.2 | 9.9e | ||
| 17. Site diameter [range] (nm) | 54e [20–60] (171) | |||
| 18. Site no. | 12,300 | 15,000e | ||
| A1 |
HeLag |
|||
|---|---|---|---|---|
| LM | EM | LM | EM | |
| 19. Nucleoplasmic volume (μm3) | 5400 ± 2200 (10) | 660 | ||
| 20. Nucleolar volume (μm3) | 2400 ± 910 (10) | |||
| 21. 2D site density, ns (sites/μm2) | 1.1 ± 0.2 (39)b | 2 | ||
| 22. 3D site density, nv (sites/μm3) | 6.1 | 14 | ||
| 23. Site diameter [range] (nm) | 58 [24–75] (81) | 46 | ||
| 24. Site no. | 33,000 | 9700 | ||
Data were obtained from images like those in Figures 5 and 6. Because cryosections of 90 (HeLa), 120 (all other cells; LM), and 150 nm (all other cells; EM) were used, three-dimensional (3D) site densities (not 2D densities) should be compared. Values ± SD; n (number of images/sites analyzed) is given in parentheses. Site diameters (range in square brackets) are corrected for the Holmes effect (sectioning can leave a “polar cap” with a diameter less than that of uncut structure).
a Area of ≥47 μm2 analyzed for each sample.
b Data sets were normally distributed (Kolmogorov–Smirnov test applied to log data); mean values for undifferentiated F9, differentiated F9, undifferentiated ES, and A1 were not significantly different (determined using two-sample Welch t test; p > 0.05), whereas those for SPARC +ve and −ve ES cells were significantly different from others (p < 0.05) but not from each other.
c Not significantly different from F9 (determined using Wilcoxon text; p = 0.13).
d Significantly smaller (p < 10−3) than undifferentiated counterpart (determined using two-sample Welch t test).
e Value for mixed population of SPARC +ve and −ve cells.
f Not significantly different from undifferentiated ES (determined using two-sample Welch t test; p = 0.05).
g Values from Pombo et al. (1999a).
RESULTS
Monitoring Differentiation of F9 and ES Cells
Differentiation was monitored using two markers. Oct-4 (also known as Oct-3/4) is a marker for pluripotency and a member of the Pit1-Oct1-Unc86 family of transcription factors; it is required for stem cell self-renewal and is lost when ES cells differentiate (Niwa et al., 2000). Conversely, SPARC is a calcium-binding matricellular glycoprotein and a specific marker for parietal endoderm (Mason et al., 1986; Nomura et al., 1988; Yiu et al., 2001).
F9 cells differentiate on addition of retinoic acid, dibutyryl cyclical AMP, and 3-isobutyl-1-methylxanthine (hereafter this treatment is called RA; Strickland and Mahdavi, 1978; Strickland et al., 1980; Gardner, 1983; Hogan et al., 1983). About 70% of the population become individual and migratory, losing Oct-4 and gaining SPARC (Figure 1A; our unpublished data); this is typical of parietal endoderm. The remainder remains SPARC −ve (Figure 1A). ES cells differentiate into two distinct populations (Figure 1B) on withdrawing LIF and adding the three inducers (also called RA; Smith et al., 1988; Williams et al., 1988; Mummery et al., 1990; Smith, 2001). Generally, all cells become more migratory, growth slows, and cell death increases. About 50% have small round nuclei (Figure 1H) with increased SPARC (Figure 1I) but less Oct-4 (Figure 1J). The second population is also clearly different from the starting population; cells generally have large irregular nuclei (Figure 1L) with little SPARC (Figure 1M), and some retain Oct-4 (Figure 1N). We conclude the first population contains parietal endoderm, whereas the second has differentiated along other pathways. These two differentiated populations serve as excellent controls for each other because they are present side by side in the same culture (and so in one image in the microscope). Quantitative immunoblotting confirms that RA treatment increases SPARC and reduces Oct-4 in both F9 and ES cells (Figure 2A; our unpublished data). These results show that at least half of both populations differentiate into parietal endoderm.
Numbers of Active Molecules of RNA Polymerase II Halve on Differentiation
RNA polymerase II is a multisubunit enzyme responsible for transcribing most genes in the nucleoplasm, and the C-terminal domain of its catalytic subunit becomes hyperphosphorylated during elongation. As a result, such hyperphosphorylation is widely used as a marker for the active enzyme (Dahmus, 1996; Buratowski, 2003). The changes in polymerase numbers occurring on differentiation were monitored by quantitative immunoblotting, using an antibody that recognizes both the (active) hyper- and (inactive) hypophosphorylated forms of the polymerase (i.e., IIO and IIA; Besse et al., 1995). Band intensities given by two samples of differentiated F9 cells (inevitably a mixed population; Figure 1A) were roughly one-half those given by two samples of undifferentiated cells (Figure 2B; compare lanes 5 and 6 with 7 and 8). To convert relative numbers to absolute numbers, band intensities were compared with those given by HeLa cells (Figure 2B, lanes 1–4), which are known to contain ∼65,000 elongating molecules of polymerase II (Kimura et al., 1999). Quantitative analysis of several blots showed the number of active molecules in F9 and ES cells fell by 40–50% as they differentiate (Figure 2C).
Detecting Transcription Sites in Cryosections
Nucleoplasmic transcription sites can be detected with high sensitivity and resolution after permeabilizing cells and allowing engaged polymerases to extend their transcripts in Br-UTP. After immunolabeling with fluors (or gold particles), sites containing the resulting nascent Br-RNA are imaged in cryosections by light (or electron) microscopy (Jackson et al., 1993). Br-UTP is incorporated into the same transcripts as [3H]UTP, and the Br-RNA (in which every U is replaced by Br-U) does not leave synthetic sites (Iborra et al., 1998). The use of cryosections offers several advantages (Pombo et al., 1999a,b). Nucleoplasmic sites are so numerous they are impossible to resolve using conventional confocal microscopy, which has a resolution (at best) of ∼200 nm in xy and ∼700 nm in z (Figure 3, A–C). However, most sites can be resolved by confocal imaging of 120-nm cryosections, where resolution in z is defined by section thickness; they are generally found in DNA-poor regions (Figure 3, D–F). Nucleolar sites are typically larger and brighter than nucleoplasmic ones (Figure 3, D–F). Antibodies also have better access to their targets because they must penetrate ≤120 nm (compared with the many micrometers in whole cells). Background is also lower, because there is no out-of-focus flare.
We next established that most nucleoplasmic sites are detected. We reasoned that if many less-active sites escaped detection, then increased incorporation of Br-UTP should raise more above the detection threshold; if all sites are detected, then increased incorporation should increase site intensity but not site number (Iborra et al., 1996; Pombo et al., 1999a). Incorporation of Br-UTP in the absence of ATP, CTP, or GTP served as a control; essentially no Br-RNA was detected because essentially no RNA synthesis is possible because endogenous pools are washed away (Figure 4A, left, and B, only Br-UTP). In contrast, extension in Br-UTP plus ATP, CTP, and GTP progressively increases signal in both nucleoplasm and nucleoli (Figure 4A, right). [See Materials and Methods and Pombo et al. (1999a) for other labeling controls.] Quantitative analysis confirms the Br-RNA signal increases with time (Figure 4B). In contrast, site density increases and then remains essentially constant (Figure 4C). This is consistent with faint sites initially going undetected, but by 10 min most sites contain sufficient Br-RNA to be detected. Similar results are obtained with all cells examined (Figure 4C, legend). The method proves so sensitive because each site contains many Br-RNA molecules each containing many target epitopes (rather than the one found in most proteins).
Relative Amounts of Nucleolar and Nucleoplasmic Synthesis
As bacterial growth slows, rRNA synthesis declines more than mRNA synthesis (Bremer and Dennis, 1996); therefore, it is of interest to see whether this occurs during eukaryotic differentiation. However, no obvious switch could be detected. Thus, the ratio of nucleolar (i.e., rRNA) to nucleoplasmic (i.e., mRNA) synthesis in the various cells analyzed (determined using curves like those in Figure 4B) is 1.8 (F9, −RA), 1.2 (F9, +RA), 1.5 (ES, −RA), 1.5 (ES, +RA, SPARC +ve), and 3.9 (newt).
Density and Intensity of Nucleoplasmic Sites
Quantitative analysis showed the density of nucleoplasmic foci to be similar in undifferentiated and differentiated F9 cells, and in undifferentiated ES cells (Table 1, LM data, rows 3, 9, and 15); however, the density in differentiated ES cells was marginally higher (Figure 5, A–I, and Table 1, LM data, rows 9 and 15).
Small SPARC +ve and large SPARC −ve ES cells are found side by side in the same cryosection; therefore, intensities of individual foci in two different differentiated populations can be compared directly. Individual nucleoplasmic foci in SPARC +ve cells had a mean ± SD fluorescence intensity of 33 ± 20 arbitrary units (n = 369), compared with 32 ± 21 (n = 730) in the SPARC −ve cells, an insignificant difference (p = 0.79; two-sample t test). The mean intensity per unit area of nucleoplasm in SPARC +ve cells (i.e., 12 ± 7 arbitrary units; n = 20) was also similar to that in SPARC −ve cells (i.e., 10 ± 5; n = 28; p = 0.20). This is to be expected, because densities and intensities of individual foci are similar. Given this result, if follows that the total amount of nucleoplasmic transcription depends on nucleoplasmic volume. We conclude that 1) both the amount of nascent RNA in one focus and the amount of transcription per unit volume of nucleoplasm is similar in cells that differentiate along two different pathways, and 2) cells with increased or decreased global levels of transcription have similar site intensities and densities. (This second conclusion is not inconsistent with results in Figure 2C, which were obtained using a mixed population of SPARC +ve and −ve cells.)
Similar Site Densities in the Interior and at the Periphery
Because heterochromatin, which is assumed to be inactive (but see Discussion), is often found at the periphery, and because active genes are often silenced when translocated there (Andrulis et al., 1998), it was of interest to determine whether there were fewer sites at the periphery. Therefore, we determined the percentage of sites seen in the peripheral 20% of the nucleoplasm (see Materials and Methods). In undifferentiated ES cells, 21 ± 6% sites (mean ± SD; n = 24) were peripheral, an insignificant difference from the 20% expected of a uniform distribution (p = 0.38; one-sample t test). In differentiated SPARC +ve cells, 21 ± 7% (n = 19) were peripheral, again an insignificant difference (p = 0.73). However, in the heterogeneous SPARC −ve population, 16 ± 6% (n = 39) were peripheral, a small, but significant, reduction from 20% (p = 0.0004). A different set of comparisons confirmed these similarities and slight difference; the value for undifferentiated ES cells was no different from that for differentiated SPARC +ve cells (p = 0.37) but marginally different from that for the SPARC −ve population (p = 0.0002). We conclude there is no consistent pattern and that sites are distributed randomly in the SPARC +ve ES nuclei—the most compact differentiated nuclei analyzed.
Changes in Nucleoplasmic and Nucleolar Volumes on Differentiation
Nucleoplasmic volumes were calculated by subtracting nucleolar from nuclear volumes determined from reconstructions of whole cells prepared from serial confocal sections of cells stained with TO-PRO-3. Nucleoplasmic volume halves as F9 and ES cells differentiate into (SPARC +ve) parietal endoderm (Table 1, rows 1 and 7). However, when ES cells differentiate into SPARC −ve progeny, volume doubles (Table 1, compare rows 7 and 13), consistent with results illustrated in Figure 1, K–N. Moreover, this population yields a higher SD (Table 1, rows 7 and 13), again consistent with the variability seen above. Clearly, the nucleoplasmic volume of an ES cell can double or halve depending on differentiation pathway. Nucleolar volumes also became larger or smaller roughly in concert with the changes in nucleoplasmic volumes (Table 1, rows 2, 8, and 14).
Changes in Chromatin Compaction Occurring during Differentiation
We next investigated the changes in chromatin compaction accompanying differentiation of ES cells into their smaller SPARC +ve derivatives (see Materials and Methods). We assume the TO-PRO-3 intensity seen in cryosections reflects DNA density, but note the dye binds to RNA to give a small amount of fluorescence (e.g., see the RNA-rich nucleolar regions in Figure 5). Inspection of intensity profiles shows a continuous range of nucleoplasmic intensities from faint to bright with no obvious step that could be used to distinguish euchromatin from heterochromatin (our unpublished data). Analysis of living cells also shows this to be so, with the local nucleosome concentration varying less than twofold (Weidemann et al., 2003). Therefore, we analyzed a range of different thresholds (after stretching image intensities to fill the gray scale from 0 to a maximum of 255 arbitrary units). (Tagging with the green fluorescent protein shows ∼75% nucleosomes in HeLa and ES cells are relatively immobile—and so presumably heterochromatic—and this provides one estimate of the heterochromactic fraction; Kimura and Cook, 2001; Meshorer et al., 2006.) Comparison of undifferentiated and differentiated ES cells shows nucleoplasmic intensity in the former to be fainter than in the latter (our unpublished data); this is expected because nucleoplasmic volume is lower (see above). Differentiated nucleoplasm also yields relatively more brighter pixels and fewer fainter pixels—the change expected of heterochromatinization. For example, 75% of the total nucleoplasmic intensity in undifferentiated cells was contained in the brightest pixels with intensities of 20–255 arbitrary units; this compares with the 26–255 arbitrary units seen in differentiated cells. Corresponding values for 50, 25, and 10% of the total intensity were 30–255, 44–255, and 62–255 (undifferentiated) compared with 33–255, 45–255, and 61–255 (differentiated), respectively. These results are consistent with 1) no obvious demarcation between euchromatin and heterochromatin (Weidemann et al., 2003), and 2) this type of differentiation being accompanied by incremental changes in compaction across the whole range of all but the densest chromatin (also see Meshorer et al., 2006).
Site Densities and Diameters Determined by EM
We next repeated the analysis using the higher resolution provided by the electron microscope. Br-RNA in cryosections was indirectly immunolabeled with 5-nm gold particles; then, clusters of gold particles—defined as three or more particles in which each particle in a cluster lies within 40 nm of another (Figure 6, A and B)—are seen against a background of a few lone particles. As is so often the case, the approach used is a compromise; here, we maximize sensitivity of detection (validated as described above) at the expense of preservation of ultrastructure and ease of detection of gold particles. Various controls indicate the clusters mark sites containing Br-RNA, and lone particles represent background. Thus, only lone particles are seen when transcripts are extended in Br-UTP without the other triphosphates, or when the primary antibody is omitted; in addition, the number of lone particles remains constant on incubation with Br-UTP plus the other NTPs (our unpublished data; Pombo et al., 1999a). In contrast, the number of particles in clusters increases with time on incubation in all four triphosphates, and by ∼5 min most sites became saturated (i.e., the number of particles/clusters becomes constant and is essentially similar in all cell types examined; our unpublished data; Pombo et al., 1999a). Most sites are again seen; longer incubations in Br-UTP lead to more particles per cluster (up to the plateau) without affecting cluster density (our unpublished data; Pombo et al., 1999a), and labeling sites in the same section with both fluors and gold particles shows that clusters mark the same structures as the foci seen by LM (Pombo et al., 1999b). Here, differentiated ES cells were not subdivided into SPARC −ve and +ve populations because 1) high-magnification views are needed to resolve gold particles of different sizes needed to mark Br-RNA and SPARC in nuclei and cytoplasm respectively, and then tracking from one compartment to the other is laborious; and 2) LM had shown sites were present at similar densities in SPARC −ve and +ve cells. This EM analysis confirms that site densities seen in the two-dimensional images of the different cells are all roughly similar (Table 1, EM data, rows 3, 9, and 15). It did not confirm the marginal difference in density seen by LM in undifferentiated and differentiated ES cells (Table 1, EM data, rows 3, 9, and 15).
Site diameters can only be measured by EM because sites lie below the resolution of the LM. They were similar in all cells examined (Table 1, rows 5, 11, and 17), which suggests that all sites contain the same numbers of nascent transcripts. This is consistent with the results found by LM, where the intensity of foci in SPARC +ve and −ve ES cells lying side by side in the same cryosection are similar (see above).
Numbers of Nucleoplasmic Sites and Active Polymerases per Site
The total number of nucleoplasmic sites—imaged as fluorescent foci (LM) or clusters of gold particles (EM)—was now calculated using standard stereological procedures (Table 1). Thus, the density of sites seen in the two dimensions of an image (ns; e.g., row 3) was converted into the density in the three dimensions of the section (nv; e.g., row 4) using the known section thickness (after correction for the Holmes effect; see Materials and Methods); finally, total site number (e.g., row 6) was calculated using the nucleoplasmic volume (e.g., row 1).
Site densities (nv)—and derived site numbers—seen by LM and EM are reassuringly similar, with one estimate lending credibility to the other (Table 1). (ns determined by EM is also higher, but this is partly due to the use of thicker sections; Table 1, legend.) If some sites went undetected, the two methods, which might be expected to have different thresholds of detection, would probably give different results. Moreover, the EM estimates of nv are 11–26% higher than those seen by LM (Table 1), which is to be expected. Thus, we have previously used probes labeled with both fluorochromes and gold particles to image exactly the same sites in both microscopes. About 20% more sites are seen in the EM, because some LM foci are resolved into two or more clusters of gold particles (Pombo et al., 1999b); the excess seen here is in the same range.
The three-dimensional site density (nv) remains roughly similar during differentiation, despite the halving of the number of active polymerases (Figure 2C) and despite a halving or doubling of nucleoplasmic volume (Table 1).
Newt Cells with 11-Fold Higher C Value
The above-mentioned results indicate that changes in nuclear volume have little effect on site density and diameter. Therefore, we analyzed salamander (newt) nuclei, which are much larger and contain 11-fold larger genomes (Figure 4A). Nucleoplasmic volume is 6- to 13-fold larger than that of the other cells examined (Table 1, row 19). This increase is associated with an increased number of active polymerases; thus, quantitative immunoblotting showed there are 151,000 ± 58,000 molecules of IIO per cell (average of 9 experiments like those in Figure 2B; our unpublished data). However, site densities and diameters are roughly similar to the values found in the other cells (Figures 5, J–L, and 6, C and D, and Table 1, rows 21–23). As with the differentiated SPARC −ve population of ES cells, marginally fewer sites are found at the periphery of newt cells, with 16 ± 4% (n = 18) of the sites in the peripheral 20% nucleoplasmic volume—a slight, but significant reduction (p = 0.0012; value determined as described above).
DISCUSSION
Differentiation involves a complicated set of interconnected changes as some genes become more active, and others less so. Although we have detailed information on steady-state levels of each mRNA in many differentiating cells (see Introduction), we know surprisingly little about more global changes. For example, does the number of active polymerases rise or fall, and by how much? Therefore, we investigated the changes that occur in the rates and organization of transcription as mouse F9 teratocarcinoma and totipotent ES cells differentiate in vitro. The ES cells prove particularly useful, because they differentiate into two distinct populations with roughly one-half and double the nucleoplasmic volume. We also studied the organization in newt A1 cells with an 11-fold larger genome.
Changes in Numbers of Active Molecules of RNA Polymerase II
The number of active molecules of the catalytic subunit of RNA polymerase II (i.e., form IIO) was estimated by quantitative immunoblotting by reference to the known number in a HeLa cell; numbers almost halved as F9 and ES cells differentiate (Figure 2, B and C). We can be fairly confident of numbers in HeLa, because three independent methods (immunoblotting, run-on analysis of nascent [32P]RNA, and imaging of nascent Br-RNA) gave similar results (i.e., 55,000–65,000 molecules; Jackson et al., 1998). Because F9 and ES both give similar values as they differentiate along the same pathway, we are also reasonably confident of these estimates. The larger newt cells also had two- to fourfold more active molecules of the enzyme (see Results). We speculate that as cells evolve to a higher C value—presumably by duplicating large genomic regions—more polymerases become active on the “extra” DNA. Note that it is often assumed that the extra DNA will all be heterochromatic and so transcriptionally inactive, but it is now known that many sequences within even the “deepest” heterochromatin are transcribed (Abranches et al., 1998; Claverie, 2005), that the density of transcription sites in euchromatin and “deep” heterochromatin is similar (Abranches et al., 1998), and that centromeric heterochromatin even depends on continuing transcription for compaction (Cheng and Gartenberg, 2000; Martienssen et al., 2005). Therefore, genome duplication may increase genome size with only some—but certainly not all—of the extra DNA becoming heterochromatic.
Constancy of Nucleoplasmic Site Density and Diameter
How confident can we be of site number? Previous work has shown that essentially all sites can be detected (Iborra et al., 1996; Pombo et al., 1999b), and we confirm this here. Thus, if many less-active sites went undetected, increased Br-UTP incorporation should raise more above the detection threshold, but it does not; rather, the same numbers become labeled more intensely (Figure 4). Moreover, sectioning cuts through some sites to leave “polar” caps, and we can estimate how small such caps must be before they go undetected. It turns out that sites containing 1/20th the nascent Br-RNA of the average can be detected by EM, so any undetected sites would contain only a tiny fraction of the total (Pombo et al., 1999b). Finally, both LM and EM, which might be expected to have different detection thresholds, give roughly similar results (Table 1), so it is unlikely that many sites can be missed. We believe EM gives the most accurate estimate, because it allows resolution of some sites seen by LM into two or more sites (see Results). Note that Osborne et al. (2004) recently observed only hundreds of sites containing RNA polymerase II in various primary mouse cells. However, their results are not necessarily inconsistent with ours; they did not demonstrate that all sites could be detected, so many sites could have been missed. Nevertheless, it remains possible—although we believe unlikely—that the organization of transcription in primary cells differs from that in the cells cultured in vitro that we study.
Site diameter and density are surprisingly similar in all cells examined; diameters lie between 46 and 54 nm, whereas densities seen in the EM are between 6.8 and 14 sites/μm3 (Table 1; data obtained previously for aneuploid HeLa are included). The identical densities and intensities of foci seen by LM in two different cell types lying side by side in a cryosection (i.e., SPARC +ve and −ve ES cells) provide a striking example of how little differentiation affects these aspects of the organization. The constancy in diameter coupled to a twofold variation in density (at most) should be compared with the 11-fold range seen in C value (ES, A1), an approximately fourfold variation in nucleoplasmic volume as ES cells differentiate into either smaller (SPARC +ve) or larger (SPARC −ve) cells, and the ∼13-fold range in nucleoplasmic volume in different cell types (SPARC +ve ES, A1). The slight variations in diameter and density are also seen despite cells being derived from different classes (amphibian A1, mammalian HeLa, F9, and ES), whether or not they are diploid (F9 and ES), and irrespective of differentiation state (undifferentiated ES and SPARC +ve/−ve derivatives).
Number of Active Polymerases per Site
Previous studies on HeLa indicate that approximately active molecules of RNA polymerase II (and their nascent transcripts) are contained in a typical nucleoplasmic site (Cook, 1999). Roughly similar values are found here: ∼63,000 active molecules in an undifferentiated F9 cell are concentrated in ∼6400 sites (Figure 2C and Table 1), and if ∼80% sites contain polymerase II and the other ∼20% polymerase III (as in HeLa; Pombo et al., 1999a), it follows there would be ∼12 molecules per site. Based on the same assumptions, sites in the other cells would contain 6–17 (calculated using polymerase numbers in Figure 2C; see Results and data in Table 1).
How many active transcription units are there per site? We can imagine a range of possibilities, from each site containing one unit (with 6–17 engaged polymerases) to 6–17 units (each with one engaged enzyme). The latter is consistent with our previous results (Cook, 1999; 2002), and with observations that few eukaryotic units are associated with more than one engaged polymerase at any moment (reviewed by Jackson et al., 2000). Moreover, microarray analyses reveal only 73 of the several thousand active open reading frames in yeast are typically associated with one or more engaged polymerases (Bon et al., 2006b). In addition, recent results using “chromosome conformation capture” provide good evidence that sites contain at least two active units (reviewed by Cook, 2003); thus, the mouse Hbb-b1 globin gene and its locus control region are found in the same factory in erythroid nuclei (where both are transcribed), but not in brain nuclei (where globin is not expressed; Osborne et al., 2004).
Speculations on Genome Reorganization during Differentiation and Evolution
We assume these results are representative of other differentiating cell types found in higher eukaryotes, and we discuss them in the context of a general model for genome organization (Cook, 2002). When genes strung along a template are transcribed, we suggest polymerases and transcription factors bound to DNA cluster to loop the intervening DNA; the result is a “cloud” of loops around the cluster. We call such a cluster a factory because it contains many polymerizing machines (Figure 7A). Then, polymerases act both as enzymes and the critical molecular ties maintaining loops. Our results fit comfortably with this model (Table 1).
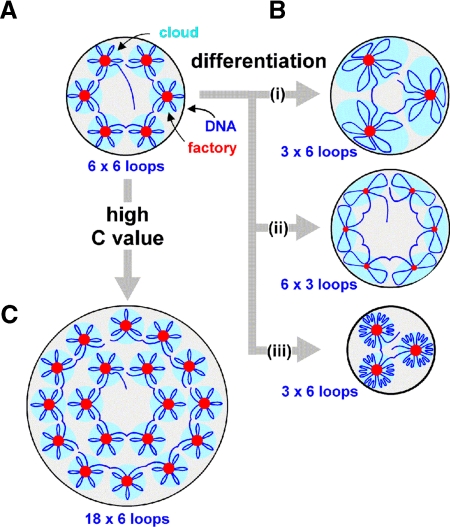
Figure 7.
Models for genome reorganization during differentiation and evolution. (A) Thirty-six transcription units strung along the chromatin fiber are attached to polymerases and transcription factors in six factories to give six surrounding clouds (each with 6 loops). (B) During differentiation, one-half the polymerases and transcription units become inactive, so one-half the factory volume (which reflects activity) and one-half the ties attaching loops are lost (giving 18 longer loops). Three ways of adjusting to this are shown. (i) Factory density halves, while factory diameter and nucleoplasmic volume remain constant; each cloud has twice the DNA in double the volume. (ii) Factory density and nucleoplasmic volume remain the same, while factory diameter and number of loops per factory both fall. The same amount of DNA is associated with each factory so cloud volume remains unchanged. (iii) Factory density and diameter remain unchanged, as factory number and nucleoplasmic volume both fall; this is the result found. Nucleoplasmic compaction is likely to occur spontaneously; doubling loop length increases the radius, r, of the volume occupied by one randomly diffusing loop ∼1.5-fold, and r will be even closer to 1 as the extra DNA is far from a factory and likely to be tightly packed into heterochromatin (Bon et al., 2006a). Then the system is self-regulatory, with increased loop length having little effect on site density or cloud volume (although nucleoplasmic volume falls and chromatin density increases). (C) During evolution, partial genome duplications increase C value (3-fold in this case); site density and diameter remain constant, and nucleoplasmic volume increases. However, the numbers of active polymerases and nucleoplasmic volume increase less than predicted by this model. A simple modification (our unpublished data) provides a good fit to all our data. Because we find fewer active polymerases than expected, some loops must be longer; this gives fewer clouds and a smaller nucleoplasmic volume (as in B, iii).
How might this organization change as F9 and ES cells differentiate into parietal endoderm? As the number of active polymerases halves, there will be fewer ties to maintain loops, so loop length doubles. This increase is in accord with old results showing that loop length progressively increases as transcriptionally active chicken erythroblasts differentiate into inert erythrocytes (Cook and Brazell, 1976). Three scenarios for the changes that occur are illustrated in Figure 7B, but only the third is consistent with results (Table 1).
How might the organization change during evolution to a higher C value (which we assume occurs through successive and partial genome duplications)? Only the third possibility is shown in Figure 7C, because constancy of site density and diameter is maintained (Table 1). However, numbers of active polymerases and nucleoplasmic volume increase less than predicted by this model. A simple modification (our unpublished data) involving the same changes occurring during differentiation accommodates all the observed changes. Because there are fewer active polymerases than predicted, some loops will be longer; this gives fewer clouds and a smaller nucleoplasmic volume (as in Figure 7B, iii).
CONCLUSIONS
In summary, we find nascent transcripts made by RNA polymerase II are concentrated in a limited number of discrete sites or factories that are generally distributed uniformly throughout the nucleoplasm. The density and diameter (and so activity) of these sites are roughly similar in newt, mouse, and human cells growing in tissue culture. Moreover, the same densities and diameters are found in the face of ∼13-fold variations in nucleoplasmic volume and approximately fourfold differences in numbers of active polymerases. Strikingly, the density and intensity (and so activity) of sites in small SPARC +ve ES cells growing next to their larger SPARC −ve counterparts in the same culture are very similar. Because the same constancy in site diameter and density is found in cells from different classes (amphibian A1, mammalian HeLa, F9, and ES), whether or not they are diploid (ES) or aneuploid (F9 and HeLa), and irrespective of differentiation state (undifferentiated ES and SPARC +ve/−ve derivatives), it is attractive to suppose that the same general organization is found in the tissues of all higher eukaryotes. Then, global rates of transcription would change by varying the total number of sites (through changes in volume) as site density and the activity of individual sites remain the same.
ACKNOWLEDGMENTS
We thank J. Brockes, F. A. Brook, R. L. Gardner, R. Hollinshead, A. Kumar, S. Muller, M. Shaw, and M. Vigneron for help and reagents and the Foundation for Science and Technology (Portugal) plus the European Social Fund in the context of the III Community Support Panel for support.
Abbreviations used:
- ES
embryonic stem(s)
- LIF
leukemia inhibitory factor
- RA
retinoic acid
- SPARC
secreted protein which is acidic and rich in cysteines.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1024) on April 19, 2006.
REFERENCES
- Abranches R., Beven A. F., Aragon-Alcaide L., Shaw P. J. Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 1998;143:5–12. doi: 10.1083/jcb.143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E. D., Neiman A. M., Zappulla D. C., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Bernstine E. G., Hooper M. L., Grandchamp S., Ephrussi B. Alkaline phosphatase activity in mouse teratoma. Proc. Nat. Acad. Sci. USA. 1973;70:3899–3903. doi: 10.1073/pnas.70.12.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse S., Vigneron M., Pichard E., Puvion-Dutilleul F. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expr. 1995;4:143–161. [PMC free article] [PubMed] [Google Scholar]
- Bon M., Marenduzzo D., Cook P. R. Modeling a self-avoiding chromatin loop: relation to the packing problem, action-at-a-distance, and nuclear context. Structure. 2006a;14:197–204. doi: 10.1016/j.str.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Bon M., McGowan S., Cook P. R. Many expressed genes in bacteria and yeast are transcribed only once per cell cycle. FASEB J. 2006b doi: 10.1096/fj.06-6087fje. in press. [DOI] [PubMed] [Google Scholar]
- Bremer H., Dennis P. P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt . C., Curtiss R. III, Ingraham J. L., Lin E.C.C., Brooks Low K., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: American Society for Microbiology Press; 1996. pp. 1553–1569. [Google Scholar]
- Brook F. A., Gardner R. L. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat. Struct. Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Cheng T. H., Gartenberg M. R. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 2000;14:452–463. [PMC free article] [PubMed] [Google Scholar]
- Claverie J. M. Fewer genes, more non-coding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Conformational constraints in nuclear DNA. J. Cell Sci. 1976;22:287–302. doi: 10.1242/jcs.22.2.287. [DOI] [PubMed] [Google Scholar]
- Cook P. R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Cook P. R. Predicting three-dimensional genome structure from transcriptional activity. Nature Genetics. 2002;32:347–352. doi: 10.1038/ng1102-347. [DOI] [PubMed] [Google Scholar]
- Cook P. R. Nongenic transcription, gene regulation and action at a distance. J. Cell Sci. 2003;116:4483–4491. doi: 10.1242/jcs.00819. [DOI] [PubMed] [Google Scholar]
- Dahmus M. E. Reversible phosphorylation of the C–terminal domain of RNA polymerase II. J. Biol. Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- de Laat W., Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Ferretti P., Brockes J. P. Culture of newt cells from different tissues and their expression of a regeneration-associated antigen. J. Exp. Zool. 1988;247:77–91. doi: 10.1002/jez.1402470111. [DOI] [PubMed] [Google Scholar]
- Gardner R. L. Origin and differentiation of extraembryonic tissues in the mouse. Int. Rev. Exp. Path. 1983;24:63–133. [PubMed] [Google Scholar]
- Gregory T. R. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Harris T. M., Childs G. Global gene expression patterns during differentiation of F9 embryonal carcinoma cells into parietal endoderm. Funct. Integr. Genomics. 2002;2:105–119. doi: 10.1007/s10142-002-0062-6. [DOI] [PubMed] [Google Scholar]
- Hartl D. L. Molecular melodies in high and low C. Nat. Rev. Genet. 2000;1:145–149. doi: 10.1038/35038580. [DOI] [PubMed] [Google Scholar]
- Hogan B.L.M., Barlow D. P., Tilly R. F9 teratocarcinoma cells as a model for the differentiation of parietal and visceral endoderm in the mouse embryo. Cancer Surveys. 1983;2:115–140. [Google Scholar]
- Iborra F. J., Cook P. R. The size of sites containing SR proteins in human nuclei: problems associated with characterizing small structures by immunogold labelling. J. Histochem. Cytochem. 1998;46:985–992. doi: 10.1177/002215549804600901. [DOI] [PubMed] [Google Scholar]
- Iborra F. J., Jackson D. A., Cook P. R. The path of transcripts from extra-nucleolar synthetic sites to nuclear pores: transcripts in transit are concentrated in discrete structures containing SR proteins. J. Cell Sci. 1998;111:2269–2282. doi: 10.1242/jcs.111.15.2269. [DOI] [PubMed] [Google Scholar]
- Iborra F. J., Pombo A., Jackson D. A., Cook P. R. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J. Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Hassan A. B., Errington R. J., Cook P. R. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Iborra F. J., Manders E.M.M., Cook P. R. Numbers and organization of RNA polymerases, nascent transcripts and transcription units in HeLa nuclei. Mol. Biol. Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. A., Pombo A., Iborra F. J. The balance sheet for transcription: an analysis of nuclear RNA metabolism in mammalian cells. FASEB J. 2000;14:242–254. [PubMed] [Google Scholar]
- Kelly D. L., Rizzino A. DNA microarray analyses of genes regulated during the differentiation of embryonic stem cells. Mol. Reprod. Dev. 2000;56:113–123. doi: 10.1002/(SICI)1098-2795(200006)56:2<113::AID-MRD1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Kimura H., Cook P. R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Tao Y., Roeder R. G., Cook P. R. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell Biol. 1999;19:5383–5392. doi: 10.1128/mcb.19.8.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo D. C., Allen F., Brockes J. P. Reversal of muscle differentiation during urodele limb regeneration. Proc. Natl. Acad. Sci. USA. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R. A., Zaratiegui M., Goto D. B. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Mason I. J., Murphy D., Munke M., Francke U., Elliott R. W., Hogan B. L. Developmental and transformation-sensitive expression of the Sparc gene on mouse chromosome 11. EMBO J. 1986;5:1831–1837. doi: 10.1002/j.1460-2075.1986.tb04434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery C. L., Feyen A., Freund E., Shen S. Characteristics of embryonic stem cell differentiation: a comparison with two embryonal carcinoma cell lines. Cell Differ. Dev. 1990;30:195–206. doi: 10.1016/0922-3371(90)90139-n. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Nomura S., Wills A. J., Edwards D. R., Heath J. K., Hogan B. L. Developmental expression of 2ar (osteopontin) and SPARC (osteonectin) RNA as revealed by in situ hybridization. J. Cell Biol. 1988;106:441–450. doi: 10.1083/jcb.106.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C. S., Chakalova C., Brown K. E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J. A., Lopes S., Reik W., Fraser P. Active genes dynamically co-localize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pombo A., Hollinshead M., Cook P. R. Bridging the resolution gap: imaging the same transcription factories in cryosections by light and electron microscopy. J. Histochem. Cytochem. 1999b;47:471–480. doi: 10.1177/002215549904700405. [DOI] [PubMed] [Google Scholar]
- Pombo A., Jackson D. A., Hollinshead M., Wang Z., Roeder R. G., Cook P. R. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999a;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson E. J. Derivation and maintenance of embryonic stem cell cultures. Methods Mol. Biol. 1997;75:173–184. doi: 10.1385/0-89603-441-0:173. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Volume 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith A. G. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Sperger J. M., Chen X., Draper J. S., Antosiewicz J. E., Chon C. H., Jones S. B., Brooks J. D., Andrews P. W., Brown P. O., Thomson J. A. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer C., Briand J. P., Muller S. Mapping of linear histone regions exposed at the surface of the nucleosome in solution. J. Mol. Biol. 1997;273:52–60. doi: 10.1006/jmbi.1997.1270. [DOI] [PubMed] [Google Scholar]
- Strickland S., Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978;15:393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- Strickland S., Smith K. K., Marotti K. R. Hormonal induction of differentiation in teratocarcinoma stem cells: generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell. 1980;21:347–355. doi: 10.1016/0092-8674(80)90471-7. [DOI] [PubMed] [Google Scholar]
- Tanaka T. S., Kunath T., Kimber W. L., Jaradat S. A., Stagg C. A., Usuda M., Yokota T., Niwa H., Rossant J., Ko M. S. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–1928. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K. T. Immunochemistry on ultrathin frozen sections. Histochem. J. 1980;12:381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Weibel E. R. London: Academic Press; 1979. Stereological Methods: Practical Methods for Biological Morphometry. [Google Scholar]
- Weidemann T., Wachsmuth M., Knoch T. A., Muller. G., Waldeck W., Langowski J. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J. Mol. Biol. 2003;334:229–240. doi: 10.1016/j.jmb.2003.08.063. [DOI] [PubMed] [Google Scholar]
- Williams M. A. Quantitative methods in biology. In: Glauert A. M., editor. Practical Methods in Electron Microscopy. Amsterdam: North-Holland Publishing Company; 1977. [Google Scholar]
- Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Yiu G. K., Chan W. Y., Ng S. W., Chan P. S., Cheung K. K., Berkowitz R. S., Mok S. C. SPARC (secreted protein acidic and rich in cysteine) induces apoptosis in ovarian cancer cells. Am. J. Pathol. 2001;159:609–622. doi: 10.1016/S0002-9440(10)61732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]