Abstract
Protein stability of the c-jun-like yeast bZIP transcriptional activator Gcn4p is exclusively controlled in the yeast nucleus. Phosphorylation by the nuclear Pho85p cyclin-dependent protein kinase, a functional homolog of mammalian Cdk5, initiates the Gcn4p degradation pathway in complex with the cyclin Pcl5p. We show that the initial step in Gcn4p stabilization is the dissociation of the Pho85p/Pcl5p complex. Pcl7p, another nuclear and constantly present cyclin, is required for Gcn4p stabilization and is able to associate to Pho85p independently of the activity of the Gcn4p degradation pathway. In addition, the nuclear cyclin-dependent Pho85p kinase inhibitor Pho81p is required for Gcn4p stabilization. Pho81p only interacts with Pcl5p when Gcn4p is rapidly degraded but constitutively interacts with Pcl7p. Our data suggest that Pcl7p and Pho81p are antagonists of the Pho85p/Pcl5p complex formation in a yet unknown way, which are specifically required for Gcn4p stabilization. We suggest that dissociation of the Pho85p/Pcl5p complex as initial step in Gcn4p stabilization is a prerequisite for a shift of equilibrium to an increased amount of the Pho85p/Pcl7p complexes and subsequently results in decreased Gcn4p phosphorylation and therefore increased stability of the transcription factor.
INTRODUCTION
Cyclin-dependent kinases (CDKs) play a crucial role in the regulation of eukaryotic cell cycle progression (Morgan, 1997), gene transcription, and various cellular processes including subcellular localization and trafficking or interaction with other proteins, respectively. Activation of the kinases requires specific cyclin subunits, which mediate the specificity for targeting the kinase to the respective substrates (Jeffrey et al., 1995; Huang et al., 1998; Wilson et al., 1999). Deregulation of CDKs as Cdk5 in humans are assumed to promote neurodegenerative processes, e.g., in Alzheimer’s disease (Patrick et al., 1999). The activities of numerous kinases are modulated by various CDK inhibitors (CKIs), which are able to interact with the kinase–cyclin complexes (Mendenhall, 1998).
In the eukaryotic model Saccharomyces cerevisiae there are six different CDKs of which Pho85p is the functional homolog of the mammalian Cdk5 cyclin-dependent protein kinase (Huang et al., 1999) and capable of interacting with 10 different cyclin partners (Measday et al., 1997). Therefore, a deletion of PHO85 results in a pleiotropic phenotype (Lenburg and O’Shea, 1996; Tennyson et al., 1998).
The more complex filamentous fungus Aspergillus nidulans carries the three CDKs, NIMXcdc2, PHOA, and PHOB, and among them PHOA and PHOB are highly related to the S. cerevisiae Pho85p (Bussink and Osmani, 1998; Dou et al., 2003). Manual annotation and genome analysis of different Aspergilli revealed the presence of homologues of 10 different yeast Pho85p cyclins exhibiting relatively low similarities (Galagan et al., 2005).
The cyclins that bind and activate the S. cerevisiae kinase Pho85p to perform different functions have been divided into two subfamilies according to their sequence homology and functional relationship (Measday et al., 1997). Members of the Pcl1,2 subfamily as Pcl1p, Pcl2p, Pcl5p, Pcl9p, or Clg1p are involved in association with Pho85p in cell cycle control (Measday et al., 1997; Tennyson et al., 1998) as well as in the regulation of cell wall maintenance (Andrews and Measday, 1998).
The Pho80 subfamily consisting of Pho80p, Pcl6p, Pcl7p, Pcl8p, and Pcl10p is functionally involved in distinct metabolic pathways (Andrews and Measday, 1998). The two cyclins Pcl6p and Pcl7p participate in carbon source utilization (Lee et al., 2000; Wang et al., 2001), whereas Pcl7p-dependent Pho85p activity is regulated by the CKI Pho81p (Measday et al., 1997; Lee et al., 2000). Besides Sic1p and Far1p, Pho81p is one of the three CKIs identified in S. cerevisiae (Mendenhall, 1998).
The Pho85p/Pho80p kinase phosphorylates the basic transcription factor Pho4p in phosphate-rich environment, resulting in its reduced activity (O’Neill et al., 1996). In response to phosphate starvation, phosphorylation of Pho4p is inhibited and Pho4p is able to increase the transcription of its target genes (Kaffman et al., 1998). The reduced kinase activity of Pho85p/Pho80p in low phosphate is mediated by the CKI Pho81p (Huang et al., 2001) that binds stably to Pho85p/Pho80p under both high and low phosphate conditions, yet it only inhibits when cells are starved for phosphate (Schneider et al., 1994).
The S. cerevisiae inhibitor domain of Pho81p (Huang et al., 2001) is conserved in the mammalian protein C42, a neuronal regulator protein that displays an inhibitory effect on Cdk5 kinase activity (Ching et al., 2002). An increased Cdk5 activity has been implicated in Alzheimer’s and Parkinson’s disease (Lau and Ahlijanian, 2003; Smith et al., 2003). In addition, the Neurospora crassa and A. nidulans CKIs Nuc-2 and AN4310 show high sequence homology to yeast Pho81p (Poleg et al., 1996; Galagan et al., 2005).
The S. cerevisiae Pho85p cyclin Pcl5p is specifically required for phosphorylation of the transcription factor Gcn4p in sated cells (Shemer et al., 2002). The c-jun homolog Gcn4p is a global key regulator of the appropriate cellular response to starvation of amino acids, purines, or various drugs as rapamycin (Hinnebusch and Fink, 1983). This genetic network is known as the “general control” (GC) system of amino acid biosynthesis (Natarajan et al., 2001) and is conserved from yeast to man. In mammalian cells, the Gcn4p like ATF4 is the central activator of the GC, which functions also to guide food selection, learning, and memory (Costa-Mattioli et al., 2005; Hao et al., 2005). Activity of the yeast GCN4 gene product is regulated via control of protein synthesis in the cytoplasm and control of protein degradation in the nucleus. Starvation for amino acids results in an increased GCN4 mRNA translation, mediated by phosphorylation of the general translation initiation factor eIF2α by the kinase Gcn2p (Hinnebusch, 1984; Dever et al., 1992). In addition, protein stability of the highly unstable Gcn4p increases in response to amino acid starvation (Kornitzer et al., 1994). Two CDKs are involved in Gcn4p degradation: Pho85p and Srb10p. Srb10p phosphorylation of Gcn4p occurs constitutively and independently of the availability of amino acids. As a component of the RNA polymerase II holoenzyme, Srb10p might be part of promoter clearing after activation of transcription and is also required for the activation function of Gcn4p (Chi et al., 2001).
The initial and committing step of the Gcn4p degradation pathway is its phosphorylation at the specific residue Thr165 by the kinase Pho85p/Pcl5p (Meimoun et al., 2000). This step is regulated and depends on the presence or absence of amino acids. Phosphorylated Gcn4p is then polyubiquitinated by the E2 ubiquitin–conjugating enzyme Cdc34 together with the E3 SCFCDC4 RING ubiquitin ligase (Kornitzer et al., 1994; Meimoun et al., 2000). Gcn4p stability regulation depends on its phosphorylation and occurs exclusively in the yeast nucleus (Pries et al., 2002). Nuclear import of Gcn4p is triggered by the α-importin Srp1p and the β-importin Kap95p (Pries et al., 2004).
In this work, we elucidated the molecular mechanisms of the amino acid–dependent Gcn4p stability regulation. Our data present the first evidence of a novel regulatory pathway within the fine tuned network of Gcn4p stability regulation, namely the dissociation and disassembly of the Pho85p/Pcl5p complex in response to amino acid starvation. The CKI Pho81p and the cyclin Pcl7p are identified to be specifically required for Gcn4p stabilization. We propose a molecular mechanism for the stabilization of Gcn4p, where the Pho81p or Pho85p association to Pcl5p is disrupted and replaced by Pho81p/Pcl7p and Pho85p/Pcl7p complexes.
MATERIALS AND METHODS
S. cerevisiae Strains and Growth Conditions
All yeast strains used in this study are listed in Table 1. They are either congenic to S. cerevisiae S288c (RH1168) or the W303 genetic background. Standard methods for genetic crosses and transformation were used as described (Ito et al., 1983). Yeast strains RH3237 and RH3238 were obtained by replacing the mutant his3-11 allele of yeast strains KY346 and KY826 by a wild-type HIS3 allele using BamHI linearized plasmid B1683 (Table 2).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| RH1168 | MATa, leu2-3, ura3-52, gal2 | Our collection |
| RH2711 | MATa, ade2, trp1, leu2-3, his3, ura3-52 | O’Neill et al. (1996) |
| RH2712 | MATa, ade2, trp1, leu2-3, his3, ura3-52, pho81Δ::TRP1 | O’Neill et al. (1996) |
| RH3306 | MATa, ade2, trp1, leu2-3, his3, ura3-52, pho81Δ::TRP1, GCN4-9Myc-natNT2 | This study |
| RH3307 | MATa, ade2, trp1, leu2-3, his3, ura3-52, pho81Δ::TRP1, kanMX4-GALLprom-GCN4-9Myc-natNT2 | This study |
| RH2977 | MATa, ura3-1, can1-100, leu2-3, trp1-1, PCL7–9Myc-k1TRP1 | This study |
| RH3237 | MATa, ura3-1, can1-100, leu2-3, trp1-1 | This study |
| RH3238 | MATa, ura3-1, can1-100, leu2-3, trp1-1, pcl5::hisG | This study |
| RH3241 | MATa, ade2, trp1, leu2-3, his3, ura3-52, pho81Δ::HIS3 | Ogawa et al. (1995) |
| RH3255 | MATa, ura3-1, can1-100, leu2-3, trp1-1, pcl7Δ::kanMX4 | This study |
| KY346 | MATa, ura3-1, can1-100, leu2-3, trp1-1, his3-11 | Kornitzer, pers. comm. |
| KY826 | MATa, ura3-1, can1-100, leu2-3, trp1-1, his3-11; pcl5::hisG | Kornitzer, pers. comm. |
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pBKSII | 2.96-kb vector, AmpR (bla), lacZ, ori | Stratagene (La Jolla, CA) |
| pRS424 | TRP, 2μm, AmpR (bla), ori | Sikorski and Hieter (1989) |
| pRS425 | LEU2, 2μm, AmpR (bla), ori | Sikorski and Hieter (1989) |
| pRS426 | URA, 2μm, AmpR (bla), ori | Sikorski and Hieter (1989) |
| pYM6 | 9Myc-k1TRP1 module | Knop et al. (1999) |
| pYM21 | 9Myc-natNT2module | Janke et al. (2004) |
| pYM-N27 | kanMX4-GALL promoter module | Janke et al. (2004) |
| p424MET25 | pRS424 containing MET25 promoter, CYC1 terminator | Mumberg et al. (1994) |
| p425GAL1 | pRS425 containing GAL1 promoter, CYC1 terminator | Mumberg et al. (1994) |
| p426MET25 | pRS426 containing MET25 promoter, CYC1 terminator | Mumberg et al. (1994) |
| pYGEX-2T | GAL1-10prom-GST-CYC1term, URA3, 2μm | Pries et al. (2002) |
| KB294 | GAL1-10prom-myc3-GCN4 fusion in URA3-marked 2μm vector | Pries et al. (2002) |
| pME2228 | MET25prom-GFP-PHO81 fusion in p426MET25 | This study |
| pME2230 | MET25prom-GFP-PCL7 fusion in p426MET25 | This study |
| pME2564 | MET25prom-GFP- fusion in p426MET25 | This study |
| pME2933 | MET25prom-PCL7 fusion in p424MET25 | This study |
| pME2863 | MET25prom-PHO81 fusion in p424MET25 | This study |
| pME2865 | GAL1-10prom-myc9-PCL5 fusion in p425GAL1-10 | This study |
| pME2866 | GAL1-10prom-GST-PHO85 fusion in pYGEX-2T | This study |
| pME2867 | GAL1-10prom-GST-PHO81fusion in pYGEX-2T | This study |
| B1683 | 1720-bp HIS3 gene in pBKSII | Hill, pers. comm. |
Yeast strain RH3306 was obtained by PCR-based C-terminal tagging of chromosomal GCN4-ORF (open reading frame; Janke et al., 2004). Primers were designed for amplification of the 9Myc-natNT2-module from plasmid pYM21. Yeast strain RH2712 was transformed with the PCR product and transformants were selected on YEPD with 100 mg/l natNT2, nourseothricin (ClonNAT, Werner BioAgents, Jena-Cospeda, Germany). Transformants were replica-plated onto the same medium, and the correct integration of the 9Myc-tag was confirmed by Western hybridization. Yeast strain RH3307 was created by PCR-based N-terminal promoter exchange of GCN4 (Janke et al., 2004). The kanMX4-GALL module was amplified from plasmid pYM-N27 using designed primers with homologous sequences to the GCN4-ORF. RH3306 was transformed with the PCR product and plated onto rich medium supplemented with 200 μg/ml G418 (Geneticin, Invitrogen, Karlsruhe, Germany). Transformants were replica-plated and the correct integration was confirmed by Western hybridization.
Yeast strain RH3255 (pcl7Δ::kanMX4) was constructed by PCR-mediated gene replacement (Longtine et al., 1998). Primers were designed specifically for amplification of pcl7Δ::kanMX4 with chromosomal DNA of the Euroscarf strain EY1443 (pcl7Δ::kanMX4; Brachmann et al., 1998). The PCR product was transformed into strain RH3237 and plated on rich medium supplemented with 200 μg/ml G418 (Geneticin, Invitrogen). Transformants were replica-plated and deletions were confirmed by Southern hybridization.
The yeast strain RH2977 was obtained by PCR-based C-terminal tagging of chromosomal PCL7-ORF (Knop et al., 1999). Primers were designed for PCR amplification of the 9Myc-k1TRP1 module from plasmid pYM6. The PCR product was transformed into the yeast strain RH3237 to be introduced at the desired chromosomal location via homologous recombination. Tryptophan auxotrophic cells were plated on medium without tryptophan. Transformants were replica-plated onto the same medium, and the correct integration of the 9Myc-tag was confirmed by Southern hybridization.
The strains were grown in standard yeast extract-peptone-dextrose (YPD: 1% yeast extract, 2% peptone, 2% dextrose) and minimal yeast nitrogen base media (YNB: 1.5 g/l yeast nitrogen base lacking amino acids, 5 g/l ammonium sulfate, 2% dextrose or galactose and supplemented with the appropriate amino acids).
Plasmid Constructions
All plasmids used in this study are listed in Table 2. Construction of plasmid KB294 is described in Pries et al. (2002). Plasmid pME2228 expressing GFP-PHO81 was obtained by amplifying the 750-base pair GFP-ORF with Pfu-Polymerase from plasmid pBAD-GFP (Clontech, Heidelberg, Germany). The GFP-ORF was introduced as a BglII fragment into BamHI-restricted p426MET25. The PHO81-ORF was amplified with Pfu-Polymerase and ligated as a ClaI fragment behind the GFP-ORF. Plasmid pME2230 expressing GFP-PCL7 was constructed by amplifying the PCL7-ORF followed by introduction via BamHI/HindIII into p426MET25. In front of the PCL7 coding region, GFP was introduced as a BglII-fragment into the BamHI-restricted plasmid. pME2564, pME2933, and pME2863 expressing GFP, PCL7, and PHO81 were constructed by amplifying the GFP-ORF, the PCL7-ORF, and the PHO81-ORF with Pfu-Polymerase and introducing them as a SmaI/ClaI fragment (GFP) into p426MET25 (pME2564), as BamHI/blunt-SmaI/ (PCL7), or ClaI fragments (PHO81) into p424MET25 (pME2933, pME2863). Plasmid pME2865 expressing a ninefold epitope-tagged version of PCL5 was obtained by amplifying PCL5 with Pfu-Polymerase and inserting it into p425GAL1 as SmaI//HindIII fragment. A 360-base pair BglII-fragment carrying myc9 was introduced into a BglII restriction site in front of the third amino acid of Pcl5p. Plasmids pME2866 and pME2867 expressing GST-PHO85 or GST-PHO81 were constructed by amplifying the PHO85-ORF or PHO81-ORF with Pfu-Polymerase. The ORFs were inserted via SpeI/SmaI/ (pME2866) or SpeI/SalI (pME2867) into pYGEX-2T.
Protein Analysis
Shut-off Western Procedure.
Yeast cells were pregrown in selective minimal medium with glucose as the carbon source. Cells were collected by centrifugation and incubated in minimal medium containing 2% galactose to express myc3-GCN4 from the GAL1 promoter or GCN4-myc9 from the GALL promoter. After 3 h, the cells were collected via centrifugation, and half of these leu2-deficient cells were starved for leucine by shifting them to minimal medium lacking leucine. Two percent glucose was added to shut off the promoter after half an hour of leucine starvation. In case of GFP-Pho81p or GFP-Pcl7p, leucine auxotrophic cells were collected after pregrowing in selective minimal medium, and half of them were shifted to a medium lacking leucine to induce the “general control of amino acid biosynthesis.” After half an hour of starvation, 1 mM methionine was added to reduce the MET25 promoter activity to 10% of the induced level. Samples were analyzed at the indicated time points after promoter shut-off (0-min time point).
Purification of GST-Fusions.
Yeast strains expressing GST, GST-PHO85 or GST-PHO81 together with an myc-tagged version of PCL5 or PCL7 were pregrown in selective minimal medium containing raffinose as the carbon source. Two percent galactose was added to induce the expression of the GAL1-driven fusions. After 3 h of induction, a bigger part of the cells was collected by centrifugation and shifted to minimal medium lacking tryptophan for half an hour to stabilize Gcn4p. Protein extracts were prepared exactly as previously described (Roberts et al., 1997). Extracts were incubated with glutathione-agarose overnight at 4°C, and the beads were repeatedly washed and collected to purify GST-fusions and any associated proteins. Samples were denatured by heating at 65°C for 15 min in SDS loading dye and equal amounts of each sample were analyzed by Western hybridization.
Whole-Cell Extracts of S. cerevisiae.
Extracts were prepared from yeast cultures grown to exponential phase. Cells were washed in ice-cold buffer B (50 mM Tris-HCl, pH 7, 5, 1 mM EDTA, 50 mM dithiothreitol), lysed with glass beads in 200 μl of buffer B + PIM (1 mM each phenylmethylsulfonyl fluoride, tosyl-l-lysine-chloromethylketone, tosyl-l-phenylalanine-chloromethylketone, p-aminobenzamidine–HCl and o-phenanthroline) + 3% Triton X-100 + 0.8% SDS at 4°C and spun at 3500 rpm for 15 min to remove glass beads and large cell debris. Extracts (10 μl) were removed to determine total protein concentration using a protein assay kit from Bio-Rad (München, Germany). Proteins were denatured in SDS loading dye by heating at 65°C for 15 min and were subjected to SDS-PAGE followed by transfer to nitrocellulose membranes. GFP, GST, and the myc-fusion proteins, Cdc28p, eIF2p, and eIF2α-Pp were detected using ECL technology (Amersham, Amersham, United Kingdom). For the first incubation, monoclonal mouse anti-GFP (Clontech), polyclonal rabbit anti-GST (Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal mouse anti-myc (9E10), polyclonal rabbit anti-Cdc28p, polyclonal rabbit anti-eIF2p, or anti-eIF2α-Pp (Biosource, Nivelles, Belgium) antibodies were used. Peroxidase-coupled goat anti-rabbit or goat anti-mouse IgG were used as secondary antibodies (Dianova, Hamburg, Germany). Gcn4p protein bands were quantified using the Kodak 1D Image Analysis Software (Eastman Kodak, Rochester, NY).
GFP Fluorescence Microscopy
Yeast strains harboring plasmids encoding GFP-Pho81p or GFP-Pcl7p were grown to early log phase and analyzed under sated and starved conditions. Leucine or tryptophan starvation was induced by transferring these leu2-deficient yeast cells from minimal medium containing leucine to minimal medium lacking leucine for 1 h. Cells from 1 ml of the cultures were harvested by centrifugation and immediately viewed in vivo on a Zeiss Axiovert microscope (Oberkochen, Germany) by either differential interference contrast microscopy (DIC) or fluorescence microscopy using a GFP filter set (AHF Analysentechnik AG, Tübingen, Germany) or in case of 4′,6-diamidino-2-phenylindole (DAPI) staining, a standard DAPI filter set. DAPI staining was used for visualization of nuclei. Cells were photographed using a Hamamatsu-Orca-ER digital camera (Bridgewater, NJ) and the Improvision Openlab software (Improvision, Coventry, United Kingdom).
RESULTS
The CKI Pho81p Is Involved in the Control of Yeast Gcn4p Stabilization
In sated yeast cells, rapid Gcn4p decay is initiated by phosphorylation at residue Thr165 by the kinase/cyclin complex Pho85p/Pcl5p (Shemer et al., 2002). This marks the transcription factor for ubiquitination by the SCFCDC4-mediated pathway and guarantees subsequent Gcn4p degradation at the 26S proteasome. Amino acid–dependent Gcn4p degradation is restricted to the yeast nucleus (Pries et al., 2002). In response to amino acid starvation, Gcn4p is stabilized from a half-life of 5 min up to 20 min (Irniger and Braus, 2003). Our aim was to analyze the stability regulation of Gcn4p with respect to the molecular mechanisms and interactions of the involved proteins.
Stabilization of Yeast Gcn4p Requires the CKI Pho81p.
Several inhibitors of CDKs are described. In case of the kinase Pho85p, the CKI Pho81p is known to inhibit the Pho85p activity in response to phosphate starvation when this kinase associates with another cyclin, Pho80p (Kaffman et al., 1998). We asked whether Pho81p fulfils a similar function for the Pho85p/Pcl5p activity in an amino acid–limiting environment resulting in an altered Gcn4p stability.
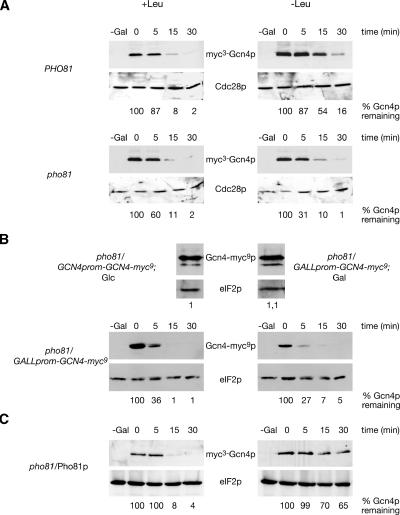
Therefore, we analyzed the requirement of Pho81p for Gcn4p stabilization during amino acid starvation. Gcn4p stabilization was induced in a PHO81 wild-type strain and compared with the corresponding pho81 mutant strain. High copies of myc3-GCN4 driven from an inducible GAL1 promoter were expressed, and the amounts of myc3-Gcn4p were analyzed after GAL1 promoter shut-off in sated and amino acid–starved cells. Our data show that the pho81 mutant strain is impaired in stabilizing Gcn4p in starved cells when compared with the wild-type PHO81 strain, suggesting that Pho81p is required for tuning down the protein degradation pathway (Figure 1A). These data were confirmed with endogenous GCN4-myc9 expressed from the less active GALL promoter. Before, it was shown that expression of GCN4-myc9 from the authentic GCN4 promoter in glucose-containing medium is similar to the level of GCN4-myc9 driven from the GALL promoter in galactose containing medium (Figure 1B).
Figure 1.
A pho81 mutation leads to unstable Gcn4p in amino acid–starved yeast cells. (A) The isogenic S. cerevisiae strains RH2711 (PHO81) and RH2712 (pho81) were transformed to express the GAL1-driven myc3-GCN4 on plasmid KB294. (B) The pho81 mutant strains RH3306 and RH3307 express endogenous GCN4-myc9 from the authentic GCN4 promoter under glucose conditions or from the GALL promoter in galactose containing medium. (C) In addition, GCN4 was expressed from the GAL1 promoter (KB294) together with MET25 driven PHO81 (pME2863) in the pho81 mutant strain RH3241 (pho81/Pho81p). Protein levels of myc3-Gcn4p, Gcn4-myc9p and Cdc28p or eIF2p as loading control were determined in sated (+Leu) and amino acid–starved (−Leu) cells after the GAL promoter shut-off. A twofold protein amount was loaded for overexpressed PHO81 to obtain similar amounts of Gcn4p at time point 0. Numbers given below indicate remaining Gcn4p percentages when compared with Cdc28p or eIF2p as internal standard quantified by image station of the gel shown.
We next asked whether an overexpression of PHO81 affects the stability of Gcn4p in sated and amino acid–starved cells. Therefore, GCN4 was expressed from the GAL1 promoter together with MET25-driven PHO81 in leucine auxotrophic pho81 cells. A promoter shut-off experiment was performed as described above. Figure 1C shows that Gcn4p is rapidly degraded in sated cells resulting in a half-life of only a few minutes similar to PHO81 wild-type cells. Furthermore, the stabilization of Gcn4p under leucine starvation conditions indicates the functional complementation of the pho81 mutation by Pho81p.
In summary, our data underline a novel and specific role for the CKI Pho81p, which is required for the stabilization of the short-lived transcription factor Gcn4p. Therefore, this inhibitor might be able to modulate the Pho85p/Pcl5p activity dependent on the presence or absence of amino acids. We compared the subcellular localization, the stability, and the protein–protein interaction of Pho81p under conditions where Gcn4p is either unstable or stabilized to establish the mechanism by which amino acid availability regulates Gcn4p stability.
Pho81p Is a Nuclear Protein in S. cerevisiae Independent of the Stability of Gcn4p.
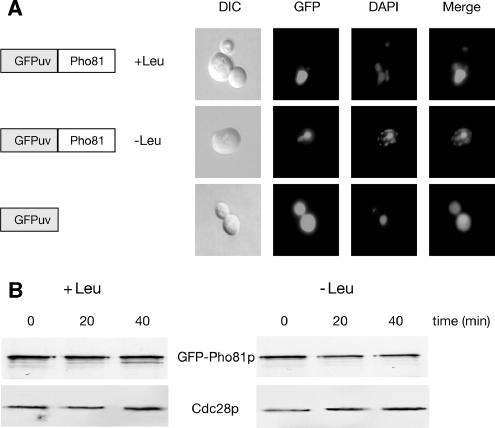
Pho81p is required for stabilization of Gcn4p. Because the regulation of Gcn4p degradation occurs exclusively in the yeast nucleus (Pries et al., 2002), we investigated the subcellular localization of Pho81p as a putative tool for Gcn4p stability regulation. Therefore, a pho81 mutant strain was transformed to express chimeric GFP-PHO81 from the efficient MET25 promoter instead of the weak native PHO81 promoter. Localization of GFP-Pho81p was monitored in living yeast cells under leucine starvation or nonstarvation conditions by fluorescence microscopy. Figure 2A illustrates that Pho81p is enriched in the nucleus under conditions when Gcn4p is unstable because of a sufficient supply of amino acids as well as under amino acid starvation conditions when Gcn4p is stabilized. DAPI staining confirms the nuclear enrichment of Pho81p.
Figure 2.
Nuclear localization and stability of Pho81p are unaffected by the availability of amino acids. (A) Nuclear import of Pho81p is independent of the presence or absence of amino acids. Yeast pho81 mutant strain RH2712 was transformed to express GFP-PHO81 driven from the MET25 promoter in high amounts (pME2228). Cells were analyzed under sated (+Leu) and starved (−Leu) conditions. On the left, DIC microscopy is shown and on the right, GFP and DAPI signals are merged (Merge). (B) Amino acid starvation results in unchanged GFP-Pho81p protein stability. The pho81-deficient yeast strain RH2712 was transformed to express GFP-PHO81 from the repressible MET25 promoter on a 2-μm plasmid (pME2228). Protein levels of GFP-Pho81p and Cdc28p as loading controls were determined in sated (+Leu) and amino acid–starved (−Leu) cells after the MET25 promoter shut-off.
In summary, we have shown that Pho81p is necessary for Gcn4p stabilization within the yeast nucleus and that Pho81p is a nuclear protein under both Gcn4p-degrading and -stabilizing conditions. Accordingly, Pho81p-dependent regulation of Gcn4p stabilization most likely also takes place in the yeast nucleus.
Pho81p Is a Stable Protein in S. cerevisiae Independent of the Stability of Gcn4p.
The requirement of Pho81p for the stabilization of Gcn4p leads to the question whether the stability of this inhibitor is affected by amino acid starvation, conditions where Pho85p/Pcl5p activity is reduced. We used the MET25 promoter, which can be repressed by methionine to analyze the stability of Pho81p. GFP-PHO81 was expressed in exponentially growing yeast cells and subsequently the MET25 promoter was shut off by the addition of methionine resulting in a halted Pho81p expression. Because Gcn4p stabilization occurs within a time window of ∼30 min (Kornitzer et al., 1994), samples were collected 20 and 40 min after the promoter shut-off. Figure 2B demonstrates that no significant differences in the amount of the chimeric GFP-Pho81 protein and therefore in the rate of Pho81p degradation were observed under conditions when Gcn4p is rapidly degraded or stabilized. This indicates that amino acid starvation does not affect the stability of Pho81p.
Interaction of Pcl5p with Pho81p and Pho85p Is Disrupted When Gcn4p Is Stabilized
The Gcn4p degradation pathway is initiated by the kinase activity of Pho85p/Pcl5p. The activity of CDKs is predominantly regulated by the presence or absence of specific cyclin subunits mediating the specificity for targeting the kinase to the respective substrate (Jeffrey et al., 1995; Huang et al., 1998; Wilson et al., 1999). We wanted to know whether additional mechanisms are essential for the regulation of Gcn4p degradation and therefore analyzed the association of Pho81p with the Pho85p/Pcl5p complex under conditions when Gcn4p is either unstable or is stabilized.
The Pho81p/Pcl5p Complex Dissociates under Conditions When Gcn4p is Stabilized.
Pho81p forms a stable ternary complex with Pho85p/Pho80p independently of the kinase activity of Pho85p. This occurs by the recognition and binding of Pho81p to the Pho80p cyclin subunit (Schneider et al., 1994), which leads to decreased Pho85p activity in low phosphate. Because the inhibitor, Pho81p, is involved in Gcn4p stabilization, we asked whether Pho81p is able to interact with the Pho85p/Pcl5p complex by binding to the unstable cyclin Pcl5p.
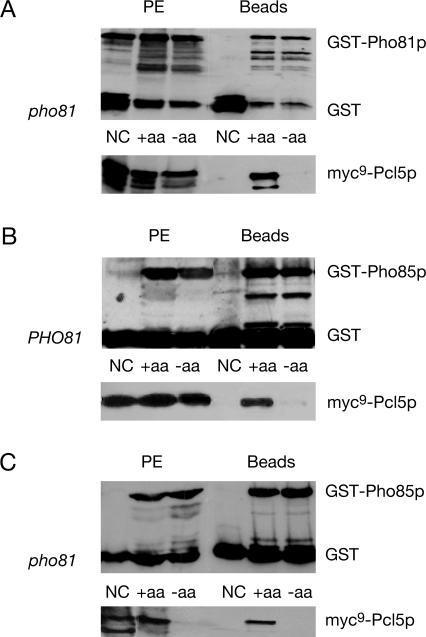
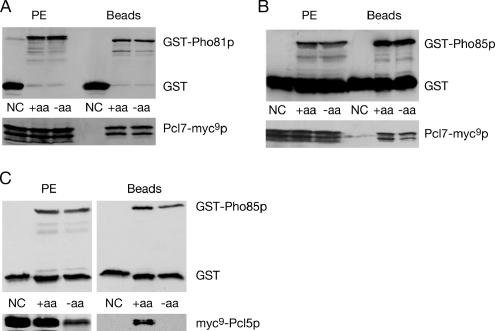
Pho81p/Pcl5p interaction was investigated by an in vivo coprecipitation assay under conditions when cellular Gcn4p is unstable because of a sufficient supply of amino acids. Functional versions of GST-PHO81 and myc9-PCL5 were expressed from the GAL1 promoter. The protein fusions were induced and purified with glutathione beads to isolate the GST-fusion and its associated proteins. Figure 3A shows that myc9-Pcl5p copurifies with GST-Pho81p under conditions when Gcn4p is rapidly degraded.
Figure 3.
Protein–protein interaction of Pcl5p with Pho81p and Pho85p are disrupted in S. cerevisiae when Gcn4p is stabilized. The strain RH3241 (pho81) (A) was transformed to express either myc9-PCL5 (pME2865) with glutathione S transferase (GST on pYGEX-2T) as negative control (NC), or myc9-PCL5 (pME2865) together with GST-PHO81 (pME2867). In addition, yeast strains RH3238 (PHO81) (B) and RH3241 (pho81) (C) were transformed to express either myc9-PCL5 (pME2865) with glutathione S-transferase (GST on pYGEX-2T) as negative control, or myc9-PCL5 (pME2865) together with GST-PHO85 (pME2866). Protein levels of the fusion proteins were determined in sated (+aa) and amino acid–starved (−aa) cells. The left part represents the GST, GST-Pho81p, GST-Pho85p, and myc9-Pcl5p before GST-agarose incubation to ensure that the initial protein extracts (PE) contain similar amounts of the fusion proteins. On the right, the elutions of the glutathione beads are shown (Beads). The twofold amount of protein extract and elution of the glutathione beads of amino acid–starved cells were loaded in case of the myc-antibody reaction in the pho81 mutant strain (A and C) to obtain similar amounts of myc9-Pcl5p in the initial protein extract (A).
Next, we were interested whether there is any difference in the association of Pho81p with Pcl5p, when Gcn4p is stabilized. The half-life of Gcn4p is increased from 3 to 5 min up to 20 min when cells are starved for tryptophan (unpublished data). Under conditions when Gcn4p is stabilized, both Pho81p and Pcl5p are clearly detectable in the protein extract, but myc9-Pcl5p does not copurify with Pho81p (Figure 3A).
Our data show that Pho81p interacts with the Pho85p/Pcl5p complex by binding to the cyclin under conditions when Gcn4p is rapidly degraded, whereas dissociation occurs in response to stabilization of Gcn4p in tryptophan starved cells.
Pho85p and Pcl5p Physically Interact, and This Interaction Is Impaired When Gcn4p Is Stabilized.
An interaction between Pho85p and Pcl5p had only been shown genetically by the yeast two-hybrid system (Measday et al., 1997). Because we successfully monitored the in vivo interaction between Pho81p and the unstable cyclin Pcl5p, we focused on the in vivo physical interaction between kinase Pho85p and the cyclin Pcl5p. A fusion between glutathione S transferase (GST) and PHO85 was constructed (as described in Huang et al., 2001; Measday et al., 1997) and expressed from the GAL1 promoter together with myc9-PCL5. Fusion proteins were induced in sated cells under conditions when Gcn4p is unstable. Protein extracts with physiologically activated Pho85p/Pcl5p complexes were prepared. We analyzed the interaction of coexpressed Pho85p and Pcl5p and found that myc9-Pcl5p copurifies with GST-Pho85p under conditions when Gcn4p is rapidly degraded, i.e., in the presence of sufficient amino acids (Figure 3B).
Under amino acid starvation, i.e., conditions that stabilize Gcn4p, we found that the Pho81p/Pcl5p interaction is disrupted. Therefore, we wanted to know whether the kinase Pho85p and the cyclin Pcl5p are still interacting under these conditions. Gcn4p stabilization was induced by tryptophan starvation, and both Pho85p and Pcl5p were expressed and detectable in protein extracts prepared under these conditions. However, myc9-Pcl5p was not copurified together with GST-Pho85p, when Gcn4p is stabilized and therefore this interaction was disrupted.
Stabilization of Gcn4p was also induced using the histidine analogue 3AT to corroborate the Pho85p/Pcl5p-dissociation effect under conditions when Gcn4p degradation is decreased. The use of high concentrations of 3AT (100 mM) also resulted in the stabilization of Gcn4p (unpublished data) and a decreased interaction between GST-Pho85p and myc9-Pcl5p compared with sated conditions (unpublished data).
These findings show that Pho85p interacts with Pcl5p when the Gcn4p degradation pathway is initiated. Under conditions when this transcription factor is required, the Pho85p/Pcl5p complex is disassembled and therefore allowing Gcn4p stabilization.
Pcl5p Is Not Detectable in a pho81 Mutant under Amino Acid Starvation Conditions.
We have shown that the protein–protein interaction of Pcl5p with Pho81p and Pho85p is disrupted under conditions when Gcn4p is stabilized, i.e., in response to amino acid starvation. Furthermore Pho81p is required for this Gcn4p stabilization. Now we asked whether Pho81p is required for disruption of the Pho85p/Pcl5p complex under amino acid starvation conditions leading to a stabilized Gcn4p. To test this, expression of GST-tagged PHO85 and myc9 epitope-tagged PCL5 was induced in sated and amino acid–starved pho81 mutant cells. Induction of amino acid starvation and protein purification and detection were accomplished as described before. We found that myc9-Pcl5p copurifies with GST-Pho85p in sated cells when Gcn4p is instable. Under amino acid starvation conditions a reduced amount of the cyclin Pcl5p in a pho81 mutant strain is detectable in the protein extract. Therefore, myc9-Pcl5p did not purify with GST-Pho85p under these conditions compared with sated pho81 mutant cells (Figure 3C). These data suggest that although no interactions between the CKI Pho81p and Pcl5p were recognized under amino acid starvation conditions, this inhibitor is required to obtain a certain Pcl5p-level under these conditions.
Cyclin Pcl7p Participates in Gcn4p Stability Regulation
Yeast Gcn4p Stabilization Requires the Cyclin Pcl7p.
The CDK Pho85p is known to bind at least 10 different cyclins. One of these cyclins is Pcl7p, which is involved in the regulation of glycogen biosynthesis and catabolism. Additionally, it is known that besides Pho85p, the CKI Pho81p is also able to interact with the cyclin Pcl7p (Measday et al., 1997; Lee et al., 2000). Furthermore, depending on phosphate availability, Pho81p is suggested to regulate the activity of both Pho85p/Pho80p and Pho85p/Pcl7p complexes (Lee et al., 2000). We have shown here that Pho81p is required for Gcn4p stabilization under amino acid starvation conditions. The disassembly of the Pho81p/Pcl5p and Pho85p/Pcl5p complexes during Gcn4p stabilization prompted us to analyze whether other cyclins such as Pcl7p are also involved in the control of Gcn4p stability.
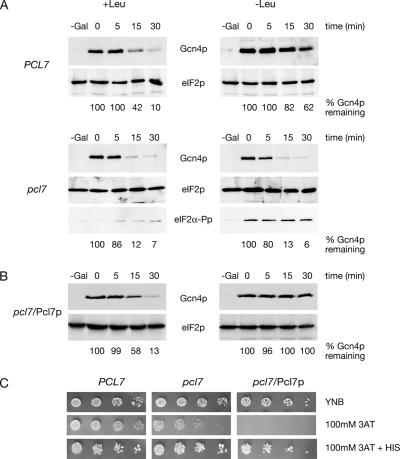
Therefore, we analyzed stability regulation of Gcn4p in a pcl7 mutant strain and the corresponding wild-type strain expressing myc3-GCN4 driven from the inducible GAL1 promoter. Protein levels of myc3-Gcn4p were analyzed after GAL1 promoter shut-off in sated and amino acid–starved cells. Western hybridization analysis revealed that a pcl7 mutant strain is unable to stabilize Gcn4p in contrast to wild-type cells (Figure 4A).
Figure 4.
A pcl7 mutation leads to unstable Gcn4p in amino acid–starved yeast cells. (A) The isogenic yeast strains RH3237 (PCL7) and RH3255 (pcl7) were transformed to express myc3-GCN4 from the GAL1 promoter from the high copy number plasmid KB294. (B) In addition GCN4 was expressed from the GAL1 promoter (KB294) together with MET25 driven PCL7 (pME2933) in the pcl7 mutant strain RH3255 (pcl7/Pcl7p). Protein levels of myc3-Gcn4p and Cdc28p or eIF2p as loading control were determined in sated (+Leu) and amino acid–starved (−Leu) cells after the GAL1 promoter shut-off. In addition, phosphorylated eIF2α-Pp confirms amino acid starvation. Numbers given below indicate remaining Gcn4p-percentages when compared with eIF2p as internal standard quantified by image station of the gel shown. (C) Overexpression of PCL7 results in sensitivity toward amino acid starvation. Yeast strains RH3237 (PCL7), RH3255 (pcl7), and pcl7 mutant cells expressing PCL7 (pME2933) from the MET25 promoter on 2-μm plasmids (pcl7/Pcl7p) were spotted in fivefold dilution on minimal medium (YNB), YNB with 100 mM 3AT, and YNB with 100 mM 3AT and histidine. Plates were incubated at 30°C for 3 d.
Amino acid starvation induces within the cell the phosphorylation of the translation initiation factor eIF2αp. The presence of eIF2α-Pp was verified as an evidence that the general control signal transduction pathway controlling Gcn4p synthesis is not impaired. Therefore, these data strongly support for the first time that the cyclin Pcl7p is required for Gcn4p stabilization under amino acid starvation (Figure 4A).
The PCL7 overexpression phenotype was determined by expressing GCN4 from the GAL1 promoter together with MET25 driven PCL7 in pcl7 cells. Promoter shut-off experiments were carried out indicating that Gcn4p is rapidly degraded in sated cells with a half-life of only a few minutes as in PCL7 wild-type cells. A strong stabilization of Gcn4p under leucine starvation conditions indicates proper functioning of expressed Pcl7p protein (Figure 4B).
For further characterization studies, wild-type (PCL7), pcl7 mutant cells and cells overexpressing PCL7 were spotted in fivefold dilutions on minimal medium (YNB), YNB with 100 mM 3AT and YNB with 100 mM 3AT and histidine. Plates were incubated at 30°C for 3 d. As shown in Figure 4C, yeast cells overexpressing PCL7 are unable to grow in the presence of 100 mM 3AT, whereas growth can be restored by the addition of histidine. Growth of pcl7 mutant is slightly reduced on media containing 100 mM 3AT. In the case of 10 mM 3AT all strains are able to grow like the wild-type control (unpublished data). These data indicate that a pcl7 deletion as well as an overexpression of PCL7 leads to sensitivity against amino acid starvation further corroborating the importance of Pcl7p in the control of Gcn4p degradation.
Pcl7p Is a Predominantly Nuclear Protein.
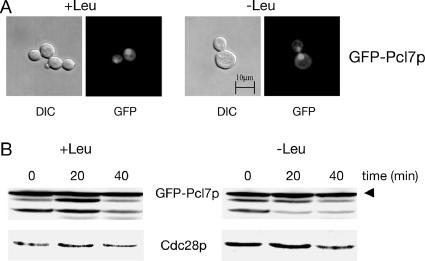
Stability regulation of yeast Gcn4p is restricted to the nucleus (Pries et al., 2002) and Pcl7p is involved in this process. Therefore, the localization of Pcl7p was monitored in vivo by expressing GFP-PCL7 from the repressible MET25 promoter. Localization of GFP-Pcl7p was examined by fluorescence microscopy in pcl7 mutant cells grown in sated and amino acid–starved cells. GFP-Pcl7p was localized mainly in the yeast nucleus under both conditions with an additional weak cytoplasmic localization (Figure 5A), indicating that the Gcn4p stability regulation is independent of the subcellular localization of this cyclin.
Figure 5.
Nuclear localization and stability of cyclin Pcl7p are unaffected by the availability of amino acids. (A) The GFP-Pcl7p protein fusion is predominantly enriched in the yeast nucleus in sated and amino acid–starved cells. pcl7 mutant cells (RH3255) expressing GFP-PCL7 from the MET25 promoter (pME2230) were analyzed under sated (+Leu) and starved (−Leu) conditions by DIC microscopy and fluorescence microscopy (GFP). (B) GFP-Pcl7p is a stable yeast protein independently of the availability of amino acids. The yeast strain RH1168 was transformed to express GFP-PCL7 (pME2230). Protein levels of GFP-Pcl7p and Cdc28p as loading control were determined in sated (+Leu) and amino acid–starved (−Leu) cells after the MET25 promoter shut-off. The GFP antibody recognizes additional bands of different sizes, which might be products of premature translation termination or protein degradation.
Pcl7p Protein Stability Is Not Influenced by Amino Acid Starvation in Yeast Cells.
Because Gcn4p could hardly be stabilized in pcl7 mutant cells under amino acid starvation conditions, we asked whether varying amounts of Pcl7p might be the cause for rapid Gcn4p turnover. Most cyclins are known to undergo rapid synthesis and turnover processes according to the cellular requirements. The cyclin Pcl5p is affected by the availability of amino acids on two levels. Although, PCL5 is transcriptionally induced by Gcn4p in response to amino acid starvation (Jia et al., 2000), the levels of the constitutively unstable protein Pcl5p are decreased in amino acid–starved cells (Shemer et al., 2002). To analyze whether Pcl7p is affected by the amino acid availability, the stability of Pcl7p was investigated.
Promoter shut-off experiments were carried out to analyze the relative turnover rates of GFP-Pcl7p in sated and amino-starved cells. A functional GFP-PCL7-fusion was expressed from the repressible MET25 promoter in exponentially growing yeast cells. After MET25 promoter shut-off, analysis of GFP-Pcl7p showed similar protein levels of Pcl7p under both conditions and a highly stable protein independent of the availability of amino acids (Figure 5B). These data indicate a tentative difference between Pcl7p and Pcl5p, with Pcl7p being not regulated at its protein level. Therefore, the function of Pcl7p in the Gcn4p stability control seems to be not dependent on regulating the amount of Pcl7p within the cell.
Pcl7p Interacts with Pho81p and Pho85p Independently of Gcn4p Stability.
We investigated the complex formation of both Pho81p/Pcl7p and Pho85p/Pcl7p under conditions when Gcn4p is rapidly degraded in sated yeast cells and when Gcn4p is stabilized under amino acid starvation conditions. Therefore, GST-PHO81 or GST-PHO85 were expressed from the GAL1 promoter and transformed into a strain containing a genomic fusion of PCL7-myc9. Fusion proteins were expressed in sated and amino acid–starved cells, whereas amino acid starvation was induced as described above. The GST-fusion protein with its associated proteins were purified by glutathione agarose beads and analyzed by Western hybridization. Figure 6A shows that the interaction of Pcl7p with Pho81p significantly differs from the interaction of the other relevant cyclin, Pcl5p. Although Pcl5p is unable to assemble with Pho81p in starved cells, a similar amount of Pcl7-myc9p is coprecipitated with the GST-Pho81p in sated and starved cells. Similarly, Pcl7p interacts constitutively with Pho85p, as opposed to Pcl5p, which only interacts with Pho85p under sated conditions (Figure 6B). These results verify the in vivo interaction between the cyclin Pcl7p with the CKI Pho81p and the kinase Pho85p but hint that Pcl7p plays an auxiliary role during Gcn4p stability control. Pcl7p interactions are independent of the amino acid concentration in the medium, whereas the Pcl5p interactions correlate to the amino acid availability. Therefore, both Pho81p and Pho85p are able to distinguish between different cyclins in response to amino acid starvation.
Figure 6.
Pcl7p interacts with Pho81p and Pho85p independently of the stability of Gcn4p. The S. cerevisiae strain RH2977 containing PCL7-myc9 behind its endogenous promoter expresses either PCL7-myc9 with glutathione S-transferase (GST on pYGEX-2T) as negative control (NC), PCL7-myc9 together with GST-PHO81 (pME2867) (A) or PCL7-myc9 together with GST-PHO85 (pME2866) (B). In addition, yeast strain RH3255 (pcl7) was transformed to express either myc9-PCL5 (pME2865) with glutathione S-transferase (GST on pYGEX-2T) as negative control, or myc9-PCL5 (pME2865) together with GST-PHO85 (pME2866) (C). Protein levels of the fusion proteins were determined in sated (+aa) and amino acid–starved (−aa) cells. Left, GST, GST-Pho81p, GST-Pho85p and Pcl7-myc9p before GST-agarose incubation (PE). Right, the elutions of the glutathione beads (Beads).
In addition, we analyzed the complex formation of Pho85p/Pcl5p in a pcl7 mutant strain to examine whether Pcl7p is involved in the dissociation of Pho85p/Pcl5p in low amino acids. Complex formation was not significantly affected compared with the analyzed pcl5 mutant strain (Figure 3B). Pho85p and Pcl5p are not interacting in amino acid–starved pcl7 mutant cells (Figure 6C), indicating that Pcl7p is not required for direct disruption of the Pho85p/Pcl5p interaction in amino acid–starved cells.
DISCUSSION
It is generally assumed that the presence or absence of specific cyclins is a major prerequisite for controlling the activity of a cyclin-dependent protein kinase in a eukaryotic cell. Accordingly, decreasing levels of the highly unstable cyclin Pcl5p were assumed to be the reason for reduced phosphorylation of Gcn4p by the kinase Pho85p during amino acid starvation (Shemer et al., 2002; Irniger and Braus, 2003). Rapid Gcn4p decay in the nucleus (Pries et al., 2002) is initiated in sated yeast cells by phosphorylation of Thr165 by the kinase cyclin complex Pho85p/Pcl5p (Shemer et al., 2002), whereas in response to a reduced amino acid supply Gcn4p is stabilized. In this work, we present evidence that a new mechanism including additional proteins is essential to control amino acid–dependent Gcn4p stabilization. The Pho85p/Pcl5p complex dissociates when Gcn4p is required under amino acid starvation, and the Gcn4p degradation pathway therefore has to be inhibited. The cyclin Pcl7p is able to interact constitutively with Pho85p and Pho81p independently of the presence or absence of amino acids. The CKI Pho81p behaves like Pho85p and is therefore only able to associate with Pcl5p in sated yeast cells. Our findings suggest that there has to be a yet unknown molecular mechanism by which Pho81p and Pcl7p affect the Pho85p/Pcl5p activity and therefore Gcn4p stability.
Pho81p and Gcn4p Stability Regulation
Previous studies have shown that Pho81p inhibits another Pho85p complex, the Pho85p/Pho80p activity, by binding to the Pho80p cyclin subunit. Pho81p is only activated as inhibitor under phosphate starvation but forms a stable complex with Pho85p/Pho80p under both high and low phosphate conditions (Schneider et al., 1994). Huang et al. (2001) proposed an increased or altered affinity of the Pho81p/Pho80p interaction in low phosphate, leading to an inhibited kinase activity. We show here that binding and release seems to be an issue for the Pho81p/Pcl5p interaction, because Pho81p interacts with Pcl5p only in sated cells. The fact that the interaction of Pho81p with Pcl5p is lost precisely under conditions where Pho85p/Pcl5p becomes inactive contradicts an inhibitory role of Pho81p in the process. When we analyzed the possibility whether Pho81p and Pcl5p interact only as part of a complex with Pho85p, we found that Pho81p/Pcl5p interaction still occurs in cells impaired in PHO85 (unpublished data), indicating that Pho85p is not required for binding of Pho81p to Pcl5p. Therefore, we can exclude a putative model where Pho81p/Pcl5p interaction can only occur in a ternary complex with Pho85p, which is disrupted in response to amino acid starvation leading to separated Pho81p and Pcl5p.
We have excluded as possible function of Pho81p in the regulation of Gcn4p stability that Pho81p is required to disrupt directly the Pho85p/Pcl5p complex in amino acid–starved cells (Figure 3C). When we analyzed the role of Pho81p in the dissociation of Pho85p/Pcl5p in low amino acids, we found surprisingly that myc9-Pcl5p is no more detectable in amino acid–starved cells impaired in PHO81 (Figure 3C). The N-terminal myc9-tagged version of Pcl5p resembles native instable Pcl5p concerning its half-life of only a few minutes. In contrast, a GFP at the C-terminus of Pcl5p leads to a more stabilized cyclin. Only this stabilized Pcl5-GFP-fusion can be expressed in sated as well as amino acid–starved pho81 mutant cells (unpublished data). Therefore, we assume that Pho81p has an important function in the regulation of Pcl5p stability.
We have shown that Gcn4p is constitutively degraded in pho81 or pcl7 mutant cells, even under amino acid limitation conditions (Figures 1A and 4A), where Pcl5p is hardly detectable (Figures 3C and 6C). This suggests that even very low levels of Pcl5p might be sufficient to trigger Gcn4p degradation. To rule out the alternative that there might be an additional Pho85p- (Meimoun et al., 2000) or Srb10p- (Chi et al., 2001) independent Gcn4p degradation pathway, which is uncovered in a pho81 mutant background, we measured Gcn4p half-lives in pho81/pho85 and pho81/srb10 double knockouts. Under sated and amino acid–starved conditions we did not observe any difference in Gcn4p stability compared with pho85 and srb10 single mutants (unpublished data). This suggests that in the absence of PHO81 the Gcn4p degradation pathway still requires the kinases Pho85p and Srb10p. We assume that the small amounts of Pcl5p in starved pho81 mutant cells are still sufficient to mediate phosphorylation of Gcn4p and therefore to initiate turnover of this transcription factor.
Another important question is how Pho81p itself is regulated in response to the availability of amino acids. Activation might include phosphorylation because it was shown that Pho81p phosphorylation is required for the inhibition of Pho85p kinase activity (Knight et al., 2004). It was proposed that Pho81p inhibitor activity is regulated by Pho85p-mediated phosphorylation of Pho81p (Waters et al., 2004). The Pho85p/Pcl5p complex differs from the Pho85p/Pho80p complex because Pho81p binds to the cyclin of Pho85p/Pcl5p only in sated cells. This interaction is not required for the degradation of Gcn4p because under sated conditions Gcn4p is similarly degraded in the presence or absence of functional Pho81p (Figure 1A). It is an attractive model that under amino acid starvation, Pho81p is posttranslationally modified, e.g., by phosphorylation or dephosphorylation resulting in an activated Pho81p. This leads to the question, why are Pho81p and Pcl5p interacting under conditions when Gcn4p is rapidly degraded, i.e., in the presence of sufficient amino acids. One possibility is that Pho81p binds to Pcl5p in sated cells as prerequisite or support for a strong binding between Pho85p and Pcl5p.
Pcl7p and Gcn4p Stability Regulation
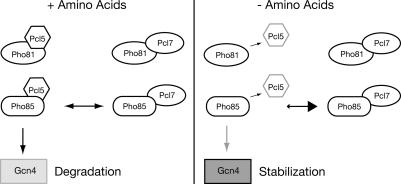
We show that a second cyclin, Pcl7p, is involved in the control of Gcn4p stability besides the cyclin Pcl5p. Pcl7p is required for the stabilization of Gcn4p in low amino acids and is able to interact constitutively with Pho85p or Pho81p. A possible mechanism of Pcl7p function in Gcn4p stability control is antagonistic binding to the kinase Pho85p and therefore a competition for binding to Pho85p between Pcl7p and Pcl5p. Amino acid starvation induces the disruption of Pho85p/Pcl5p complexes and therefore a shift in the equilibrium toward an increased number of Pho85p/Pcl7p complexes. Because Pcl7p is also more stable than Pcl5p, Gcn4p remains preferentially unphosphorylated and stable under these conditions (Figure 7). Analyses of the Pho85p-Pcl5p complex formation in pcl7 mutant cells revealed that Pcl7p is not required to disrupt directly the Pho85p/Pcl5p interaction in low amino acids (Figure 6C).
Figure 7.
Model for Gcn4p stability regulation. In sated cells, when amino acids are available (+Amino Acids), both cyclins, Pcl5p, and Pcl7p compete for binding to the kinase Pho85p. This results in a possible equilibrium of both complexes with sufficient amounts of Pho85p/Pcl5p for Gcn4p phosphorylation and therefore rapid protein degradation. In response to amino acid starvation (−Amino Acids), Gcn4p is stabilized because the Pho85p/Pcl5p complex is dissociated, and Pcl5p is replaced by Pcl7p, which is present in high amounts. The decrease of Pho85p/Pcl5p complexes results in less phosphorylation of Gcn4p and subsequently in its stabilization.
A more detailed analysis indicated that a pcl7 deletion as well as an overexpression of PCL7 leads to sensitivity against amino acid starvation induced by 100 mM 3AT (Figure 4C). A gcn4 mutant is hypersensitive to 10 mM 3AT (Hinnebusch, 1992). In contrast, a pcl5 strain is more able to deal with an induced general control, indicating the negative role of Pcl5p on Gcn4p activity (Shemer et al., 2002). Based on the constitutive Gcn4p degradation in pcl7 cells together with the fact that Gcn4p is required for 3AT resistance, the decreased growth of pcl7 cells on 100 mM 3AT suggests a positive effect of Pcl7p on Gcn4p activity and therefore an antagonistic role to Pcl5p. An overexpression of PCL7 leads to a strong stabilization of Gcn4p under amino acid starvation and also to a hypersensitivity to high amounts of 3AT (Figure 4, B and C). One possible explanation for the 3AT-sensitive phenotype of overexpressed PCL7 is a strong stabilized Gcn4p under 100 mM 3AT with a decreased activity that is not more able to mediate full 3AT resistance.
In summary, our data illustrate that the stability of the global transcriptional activator Gcn4p is highly controlled in yeast by a complex network of various regulatory proteins including two cyclins, Pcl5p and Pcl7p, the CDK Pho85p, and the inhibitor Pho81p. The Gcn4p stability network is only part of a still larger number of proteins, which are orchestrating the modulation of Gcn4p activity on additional levels such as protein synthesis or interaction with the transcriptional machinery to secure proper Gcn4p function within the “general control of amino acid biosynthesis,” a control mechanism that is conserved from yeast to man (Costa-Mattioli et al., 2005; Hao et al., 2005).
ACKNOWLEDGMENTS
We are grateful to Malte Kleinschmidt, Claudia Fischer, Oliver Valerius, Oliver Draht, and Nirmala Padmanabhan for helpful comments of the manuscript and critical reading and especially thank D. Kornitzer, E. M. O’Neill, and A. G. Hinnebusch for yeast strains, plasmids, and antibodies. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Lower-Saxony-Israeli and the Volkswagen Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-10-0975) on April 12, 2006.
REFERENCES
- Andrews B., Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bussink H. J., Osmani S. A. A cyclin-dependent kinase family member (PHOA) is required to link developmental fate to environmental conditions in Aspergillus nidulans. EMBO J. 1998;17:3990–4003. doi: 10.1093/emboj/17.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y., Huddleston M. J., Zhang X., Young R. A., Annan R. S., Carr S. A., Deshaies R. J. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching Y. P., Pang A. S., Lam W. H., Qi R. Z., Wang J. H. Identification of a neuronal Cdk5 activator-binding protein as Cdk5 inhibitor. J. Biol. Chem. 2002;277:15237–15240. doi: 10.1074/jbc.C200032200. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M., et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2α kinase GCN2. Nature. 2005;436:1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2α by protein kinase Gcn2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Dou X., Wu D., An W., Davies J., Hashmi S. B., Ukil L., Osmani S. A. The PHOA and PHOB cyclin-dependent kinases perform an essential function in Aspergillus nidulans. Genetics. 2003;165:1105–1115. doi: 10.1093/genetics/165.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Hao S., et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1983;80:5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Broach J. R., Jones E. W., Pringle J. R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 319–414. [Google Scholar]
- Huang D., Moffat J., Wilson W. A., Moore L., Cheng C., Roach P. J., Andrews B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol. Cell. Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Patrick G., Moffat J., Tsai L. H., Andrews B. Mammalian Cdk5 is a functional homolog of the budding yeast Pho85 cyclin-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 1999;96:14445–14450. doi: 10.1073/pnas.96.25.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Jeffery D. A., Anthony M. D., O’Shea E. K. Functional analysis of the cyclin-dependent kinase inhibitor Pho81 identifies a novel inhibitory domain. Mol. Cell. Biol. 2001;21:6695–6705. doi: 10.1128/MCB.21.19.6695-6705.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Braus G. H. Controlling transcription by destruction: the regulation of yeast Gcn4p stability. Curr. Genet. 2003;44:8–18. doi: 10.1007/s00294-003-0422-3. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jeffrey P. D., Russo A. A., Polyak K., Gibbs E., Hurwitz J., Massague J., Pavletich N. P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Jia M. H., Larossa R. A., Lee J. M., Rafalski A., Derose E., Gonye G., Xue Z. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics. 2000;3:83–92. doi: 10.1152/physiolgenomics.2000.3.2.83. [DOI] [PubMed] [Google Scholar]
- Kaffman A., Rank N. M., O’Neill E. M., Huang L. S., O’Shea E. K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Knight J. P., Daly T. M., Bergman L. W. Regulation by phosphorylation of Pho81p, a cyclin-dependent kinase inhibitor in Saccharomyces cerevisiae. Curr. Genet. 2004;46:10–19. doi: 10.1007/s00294-004-0502-z. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kornitzer D., Raboy B., Kulka R. G., Fink G. R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Ahlijanian M. K. Role of cdk5 in the pathogenesis of Alzheimer’s disease. Neurosignals. 2003;12:209–214. doi: 10.1159/000074622. [DOI] [PubMed] [Google Scholar]
- Lee M., O’Regan S., Moreau J. L., Johnson A. L., Johnston L. H., Goding C. R. Regulation of the Pcl7-Pho85 cyclin-cdk complex by Pho81. Mol. Microbiol. 2000;38:411–422. doi: 10.1046/j.1365-2958.2000.02140.x. [DOI] [PubMed] [Google Scholar]
- Lenburg M. E., O’Shea E. K. Signaling phosphate starvation. Trends Biochem. Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Measday V., Moore L., Retnakaran R., Lee J., Donoviel M., Neiman A. M., Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol. Cell. Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimoun A., Holtzman T., Weissman Z., McBride H. J., Stillman D. J., Fink G. R., Kornitzer D. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin-ligase complex. Mol. Biol. Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall M. D. Cyclin-dependent kinase inhibitors of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Curr. Top. Microbiol. Immunol. 1998;227:1–24. doi: 10.1007/978-3-642-71941-7_1. [DOI] [PubMed] [Google Scholar]
- Morgan D. O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell. Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K., Meyer M. R., Jackson B. M., Slade D., Roberts C., Hinnebusch A. G., Marton M. J. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E. M., Kaffman A., Jolly E. R., O’Shea E. K. Regulation of Pho4 nuclear localization by the Pho80-Pho85 cyclin-cdk complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Ogawa N., Noguchi K., Sawai H., Yamashita Y., Yompakdee C., Oshima Y. Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick G. N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L. H. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- Poleg Y., Aramayo R., Kang S., Hall J. G., Metzenberg R. L. NUC-2, a component of the phosphate-regulated signal transduction pathway in Neurospora crassa, is an ankyrin repeat protein. Mol. Gen. Genet. 1996;252:709–716. doi: 10.1007/BF02173977. [DOI] [PubMed] [Google Scholar]
- Pries R., Bömeke K., Irniger S., Grundmann O., Braus G. H. Amino acid-dependent Gcn4p stability regulation occurs exclusively in the yeast nucleus. Eukaryot. Cell. 2002;1:663–672. doi: 10.1128/EC.1.5.663-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pries R., Bömeke K., Draht O., Künzler M., Braus G. H. Nuclear import of yeast Gcn4p requires karyopherins Srp1p and Kap95p. Mol. Genet. Genomics. 2004;271:257–266. doi: 10.1007/s00438-003-0955-7. [DOI] [PubMed] [Google Scholar]
- Roberts R. L., Mösch H.-U., Fink G. R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- Schneider K. R., Smith R. L., O’Shea E. K. Phosphate-regulated inactivation of the kinase Pho80-Pho85 by the CDK inhibitor Pho81. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- Shemer R., Meimoun A., Holtzman T., Kornitzer D. Regulation of the transcription factor Gcn4 by Pho85 cyclin Pcl5. Mol. Cell. Biol. 2002;22:5395–5404. doi: 10.1128/MCB.22.15.5395-5404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. D., et al. Cyclin-dependent kinase 5 is a mediator of dopaminergic neuron loss in a mouse model of Parkinson’s disease. Proc. Natl. Acad. Sci. USA. 2003;100:13650–13655. doi: 10.1073/pnas.2232515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson C. N., Lee J., Andrews B. J. A role for the Pcl9-Pho85 cyclin-cdk complex at the M/G1 boundary in Saccharomyces cerevisiae. Mol. Microbiol. 1998;28:69–79. doi: 10.1046/j.1365-2958.1998.00773.x. [DOI] [PubMed] [Google Scholar]
- Wang Z., Wilson W. A., Fujino M. A., Roach P. J. The yeast cyclins Pcl6p and Pcl7p are involved in the control of glycogen storage by the cyclin-dependent protein kinase Pho85p. FEBS Lett. 2001;506:277–280. doi: 10.1016/s0014-5793(01)02914-3. [DOI] [PubMed] [Google Scholar]
- Waters N. C., Knight J. P., Creasy C. L., Bergman L. W. The yeast Pho80-Pho85 cyclin-CDK complex has multiple substrates. Curr. Genet. 2004;46:1–9. doi: 10.1007/s00294-004-0501-0. [DOI] [PubMed] [Google Scholar]
- Wilson W. A., Mahrenholz A. M., Roach P. J. Substrate targeting of the yeast cyclin–dependent kinase Pho85p by the cyclin Pcl10p. Mol. Cell. Biol. 1999;19:7020–7030. doi: 10.1128/mcb.19.10.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]