Abstract
The mechanisms regulating membrane recruitment of the p115 tethering factor in vivo are unknown. Here, we describe cycling of p115 between membranes and cytosol and document the effects of Golgi matrix proteins, Rab1, and soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein (SNAP) receptors (SNAREs) on this process. Rapid membrane/cytosol exchange is shown by swift (t1/2 ∼20 s) loss of Golgi-localized p115-green fluorescent protein (GFP) after repeated photobleaching of cell periphery and rapid (t1/2 ∼13 s) fluorescence recovery after photobleaching Golgi-localized p115-GFP. p115 mutant missing the GM130/giantin binding site exhibits analogous fluorescence recovery after photobleaching (FRAP) (t1/2 ∼13 s), suggesting that GM130 and giantin are not major determinants of p115 membrane dynamics. In contrast, p115-GFP exchanges more rapidly (t1/2 ∼8 s) in cells expressing the inactive Rab1/N121I mutant, indicating that p115 cycling is influenced by Rab1. p115-GFP dynamics is also influenced by the assembly status of SNAREs. In cells expressing an ATPase-deficient NSF/E329Q mutant that inhibits SNARE complex disassembly, the cycling kinetics of p115-GFP are significantly slower (t1/2 ∼21 s). In contrast, in cells incubated at reduced temperature (10°C) that inhibits vesicular traffic, the cycling kinetics of p115-GFP are faster (t1/2 ∼7 s). These data suggest that p115-binding sites on the membrane are provided by unassembled SNAREs. In agreement, biochemical studies show increased p115 recruitment to membranes in the presence of NSF and α-SNAP. Our data support a model in which recruitment of tethers is directly regulated by the assembly status of SNAREs.
INTRODUCTION
Membrane trafficking in eukaryotic cells requires the formation and delivery of transport intermediates to specific cellular locations. The fidelity of conveyance to a target membrane is ensured by the action of tethering factors and soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein (SNAP) receptors (SNAREs). Tethering factors seem to mediate initial, loose “tethering” of transport intermediates with target membranes. This loose interaction is followed by tighter, more stable “docking” interactions involving SNAREs (VanRheenen et al., 1999). It seems that tethering provides the initial level of recognition that is then amplified by SNARE pairings.
Mammalian and yeast tethering factors are either coiled-coil proteins or multisubunit complexes (reviewed in Whyte and Munro, 2002). Best studied of the coiled-coil proteins are p115/Uso1p, GM130, giantin, and golgin-84 that facilitate membrane tethering during endoplasmic reticulum (ER)–Golgi and intra-Golgi traffic; golgin-97 that acts during endosome–trans-Golgi network (TGN) traffic; and early endosome antigen (EEA) 1 that functions during endosomal traffic. Incomplete information is available on the structure-function relationships of the multisubunit TRAPPI/II and conserved oligomeric Golgi complexes that participate in membrane tethering during ER–Golgi and intra-Golgi traffic, the HOPS and GARP complexes that act during TGN–endosomal vacuolar traffic, and the sec6/8/exocyst complex that facilitates tethering to the plasma membrane.
The accepted model for tether function of coiled-coil proteins regards them as structural bridges spanning two membranes. In this model, a tether on the vesicle recognizes a cognate tether on the target membrane and forms a molecular link that holds the apposing membranes together to facilitate pairing of the vesicle- and target-SNAREs. The tethers are attached to the vesicle and target membranes by membrane-anchored receptors. The tethers may remain bound while the trans-SNARE complex forms and fusion occurs, or they may dissociate before fusion. In both cases, tethering is thought to proceed independently of subsequent SNARE events. Tethers show compartment-specific localization, and each tether could provide specificity to the bridging reaction and impose membrane selectivity before SNARE engagement.
Most tethering factors exist in cytosolic and membrane-associated pools. Association with membranes occurs by direct binding to specialized lipid domains, and/or interactions with proteins. In all examined cases, tethering proteins or components of tethering complexes seem to interact with small Ras-related GTPases and SNAREs. However, the exact roles these interactions play in tether recruitment and function remain under investigation.
p115 function in membrane traffic was first shown in an in vitro intra-Golgi transport assay established in the Rothman laboratory (Clary and Rothman, 1990; Waters et al., 1992; Wilson et al., 1992; Sapperstein et al., 1995; Aridor et al., 1998). The assay measures events subsequent to vesicle budding, predominantly membrane tethering and fusion. Removal of p115 from the assay inhibits cargo traffic, suggesting a critical role either in tethering or fusion. In vivo studies also document the requirement for p115 at an earlier pre-Golgi stage. Inactivating p115 by microinjecting anti-p115 antibodies, or depleting p115 from cells, prevents ER–Golgi traffic at ER exit sites (Alvarez et al., 1999; Puthenveedu and Linstedt, 2001, 2004). This finding suggests that p115 might be necessary for the fusion of ER-derived COPII vesicles to generate later transport intermediates. This conclusion is strongly supported by the presence of p115 on COPII vesicles generated in vitro and by direct evidence that p115 facilitates binding of such vesicles to Golgi membranes (Allan et al., 2000; Alvarez et al., 2001). Equally strong evidence for p115 function in membrane tethering comes from studies on its yeast homologue Uso1p. Uso1p was identified as an ER–Golgi transport factor because a temperature-sensitive mutant, uso1-1, blocks traffic to the Golgi (Nakajima et al., 1991). Subsequent in vitro studies provided insight into the secretory block by showing that Uso1p is required for tethering COPII vesicles to Golgi membranes (Barlowe, 1997).
p115 directly binds the Rab1 GTPase (Allan et al., 2000; Moyer et al., 2001; Beard et al., 2005) and the SNAREs syntaxin 5 (Allan et al., 2000; Shorter et al., 2002), GOS28 (Shorter et al., 2002), and membrin (Allan et al., 2000).
In cell fractionation assays, p115 is recovered in the cytosol (∼70%) and in stable association with membranes (∼30), suggesting that it may cycle between these compartments (Barroso et al., 1995; Nelson et al., 1998). Stable association of p115 with membranes seems to be regulated by Rab1. Specifically, recovery of p115 with COPII vesicles budded from semi-intact cells is abolished by Rab-GDP dissociation inhibitor (GDI) that extracts Rab proteins from membranes (Allan et al., 2000) or by dominant negative Rab1/N124I that interferes with function of endogenous Rab1 (Pind et al., 1994). Similarly, membrane association of the yeast p115 homologue Uso1p is reduced by the addition of GDI (Cao et al., 1998). Membrane association of Uso1p is also reduced in the ypt1-3 or the ypt1Δ strains that lack a functional Ypt1p, a yeast homologue of Rab1 (Ballew et al., 2005).
The role of SNAREs in p115 association with membranes has not been explored. In yeast, membrane association of Uso1p is normal in strains carrying thermosensitive mutations in Sed5p or Bos1p (yeast homologues of syntaxin-5 and membrin) (Cao et al., 1998). These results suggest that SNARE function is not required for Uso1p binding to membranes. However, whether interactions with SNAREs are required for Uso1p association with membranes has not been explored.
Here, we characterize a newly discovered rapid cycling of the tethering factor p115 between membranes and cytosol. Our results suggest that a major determinant of membrane association of a tether is its interaction with SNAREs. The p115 tether association with membranes seems linked to the SNARE complex disassembly cycle. This suggests a novel model in which tethering occurs through direct tether–SNARE interactions that may couple tethering and SNARE complex assembly.
MATERIALS AND METHODS
Antibodies and Plasmids
Rabbit polyclonal anti-p115 and anti-GBF1 antibodies have been described previously (Barroso et al., 1995, Garcia-Mata and Sztul, 2003, ref. 106). Anti-GM130 mouse monoclonal antibodies were from BD Biosciences (San Jose, CA). Anti-green fluorescent protein (GFP) antibodies were from Abcam (Cambridge, MA), and anti-Sec23 antibodies were from Affinity Bioreagents (Golden, CO). Monoclonal anti-giantin G1/133 antibody (Linstedt and Hauri, 1993) and monoclonal anti-ERGIC53 antibody G1/93 (Schindler et al., 1993) were a generous gift from Dr. Hans-Peter Hauri (University of Basel, Basel, Switzerland). Fluorescently labeled secondary antibodies were from Invitrogen (Carlsbad, CA).
Plasmid encoding myc-tagged Rab1b-N121I has been described previously (Alvarez et al., 2003). Plasmid encoding the glutathione S-transferase (GST)-GM130 N terminus has been described previously (Linstedt et al., 2000). GST-syntaxin 5 construct was generously provided by Dr. Wally Whiteheart (University of Kentucky College of Medicine, Lexington, KY). Plasmids encoding myc-tagged NSF and NSF-E329Q were a gift from Dr. Phyllis Hanson (Washington University, St. Louis, MO).
Generation of p115 Constructs
A 3.2-kb fragment encoding amino acids 1–959 of rat p115 was amplified by PCR to eliminate the stop codon at the C terminus. The PCR product was digested with Xho1/BamH1 and ligated into corresponding sites into pEGFP-N2 (Clontech, Mountain View, CA). This results in a chimera in which the GFP is added to the C terminus of p115. To generate myc-tagged full-length and truncated p115, the corresponding fragment of rat p115 was amplified by PCR, digested with Xho1/BamH1, and subcloned into pcDNA4 (Invitrogen). All plasmids were sequenced by University of Alabama at Birmingham Sequencing Core Facility on an Applied Biosystems Prism 377 (PerkinElmer Life and Analytical Sciences, Boston, MA) to ensure the fidelity of the PCR reaction.
Cell Culture, Transfection, and Preparation of Cell Lysates
HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 1.5 g/l sodium bicarbonate, and 1% penicillin/streptomycin at 37°C in 5% CO2. Between 0.5 and 1.0 μg of plasmid DNA was used in all transfections; plasmid was mixed with Mirus TransIT-LT1 transfection reagent (PanVera, Madison, WI) in RPMI-1640 serum-free medium according to the manufacturer’s instructions. Cotransfections of p115-GFP with Rab1/N121I were performed by mixing 0.5 μg of p115-GFP plasmid DNA with 0.5 μg of Rab1/N121I plasmid DNA, then following the manufacturer’s instructions for transfection. Expression of the NSF/E329Q construct is toxic; therefore, cells were first transfected with p115-GFP, and after 12 h they were transfected with the NSF/E329Q construct. Cells were cultured for another 10–12 h before imaging.
For preparing lysates, HeLa cells were transfected with full-length p115-GFP and cultured for 48 h at 37°C. Cells were lysed in NS buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2, and 1 mM dithiothreitol [DTT]) with 0.02% Triton X-100 by aspirating through a 26-gauge needle 10 times and rotating for 30 min at 4°C. Lysates were subjected to five freeze/thaw cycles. The lysate was centrifuged at 1000 × g at 4°C for 15 min, and the supernatant was centrifuged again at 100,000 × g at 4°C for 1 h. The resulting supernatant was used in GST pull-down assays.
Static Immunofluorescence Microscopy
HeLa cells grown on glass coverslips to 50–60% confluence in 60- or 35-mm dishes were transfected with plasmid and processed for immunofluorescence 24 h afterward, except for cells transfected with E329Q NSF. These cells were imaged 12 h after transfection. Cells were washed three times in phosphate-buffered saline (PBS) and fixed with 3% paraformaldehyde in PBS for 10 min at room temperature. Paraformaldehyde was quenched with 10 mM ammonium chloride, and cells were permeabilized with 0.1% Triton X-100 in PBS, for 7 min at room temperature. The coverslips were washed three times for 2 min each wash with PBS and then blocked with 0.4% fish skin gelatin in PBS, 0.2% Tween 20 for 5 min followed by a second block with 2.5% goat serum in PBS, 0.2% Tween 20 for 5 min. Coverslips were incubated with primary antibody diluted in PBS, 0.4% fish skin gelatin, 0.2% Tween 20 for 45 min at 37°C. Coverslips were washed five times for 5 min each wash with PBS, 0.2% Tween 20. Secondary antibodies coupled to fluorescein isothiocyanate or rhodamine were diluted in 2.5% goat serum in PBS, 0.2% Tween 20 and incubated on coverslips for 30 min at 37°C. Coverslips were washed as described above, rinsed briefly, and mounted on slides in 9:1 glycerol/phosphate-buffered saline with 0.1% q-phenylenediamine. Fluorescence patterns were visualized with an Orthoplan microscope (Leitz, Wetzlar, Germany) equipped for epifluorescence. Images were obtained using a high-resolution camera with a computer interface. Cell area, fluorescence intensity, and colocaliztion between specific markers were measured using IP Lab Spectrum software (Scanalytics, Fairfax, VA).
Live Imaging
HeLa cells grown on glass coverslips to 50–60% confluence in 35-mm dishes were transfected with plasmid and imaged 24 h later in a chamber sealed with a silicon gasket. During the imaging period, cell culture medium was buffered with 25 mM HEPES. Microscope stage was enclosed and heated so that temperatures ranged between 32 and 37°C. We used the 100× oil 1.4 numerical aperture (NA) objective of a Leica DMIRBE inverted epifluorescence microscope outfitted with Leica TCS NT Laser confocal optics for image acquisition.
Fluorescence Loss in Photobleaching (FLIP) and Fluorescence Recovery after Photobleaching (FRAP)
The same Leica DMIRBE microscope was used for FLIP and FRAP at 37°C. For FLIP, a 488-nm high-intensity argon laser beam was set up to photobleach a series of 2-μm-diameter spots in the cell periphery every 5 s. For FRAP, a single prebleach image was obtained, followed by an 8-s photobleach with the 488-nm argon laser. Postbleach images were obtained every 3 s. For both FLIP and FRAP, the laser was set on bidirectional scan at medium speed. Each round of scanning took 1.5 s in the 1024 × 1024 format. Every image was an average of two frames. Images were analyzed in ImageJ (http://rsb.info.nih.gov/ij/download.html), and fluorescence intensities were analyzed in Microsoft Excel (Microsoft, Redmond, WA). Graphs were made, and statistical analyses were performed in Microsoft Excel.
FRAP at 10°C was performed at the Keck Center for Cellular Imaging (University of Virginia, Charlottesville, VA). Nikon TE300 epifluorescent microscope with a 100-W Hg Arc lamp and a Plan Fluor 60× NA 1.2 water objective lens was used for image acquisition. TE300 was coupled to Bio-Rad Radiance2100 confocal/multiphoton system. A 10-W Verdi pumped, tunable modelocked ultrafast (78 MHz) pulsed (<150-fs) laser was coupled to the laser port of a Radiance2100. This laser is equipped with x-wave optics for easy tunable range of the entire wavelength (700–1000 nm). The system was equipped with laser spectrum analyzer (model E201) to monitor the excitation wavelength. LaserSharp2000 software was used to acquire the images using the internal detectors. The TC-202A bipolar temperature controller with open perfusion microincubators (PDMI-2) from Harvard Apparatus, Holliston, MA was used to cool the imaging plate. FRAP experiments were performed after 30 min of incubation at reduced temperature, when temperature inside plate reached 10 ± 1°C.
GST Pull-Down Assays
Bacteria harboring plasmids encoding GST fusion proteins were inoculated in 3-ml overnight cultures. Liquid cultures were expanded to 50 ml, and expression of proteins was induced at optical density (OD)0.5 with 1 mM isopropyl β-d-thiogalactoside for 3 h at 37°C (for Rab1b, N121I Rab1b, and GM130) or overnight at 22°C (for syntaxin 5). Cultures were centrifuged at 1000 × g, and the pellets were resuspended in 7.5 ml of PBS containing 5 mM MgCl2, 5 μg/ml DNAse and RNAse, 5 mM 2-β-mercaptoethanol, 0.2% NP-40, and protease inhibitors. For purification of Rab1b-GTP and Rab1b-GDP, 100 μM of either guanosine 5′-O-(3-thio)triphosphate (GTPγS) or GDP was added to all buffers. Cells were lysed by freezing in liquid N2 and thawing at 37°C six times. After the last thaw, bacterial lysates were incubated at 37°C for 10 min. and then centrifuged at 15,000 × g for 15 min at 4°C. For binding to glutathione-Sepharose, 3 ml of bacterial lysate was incubated with 80 μl of glutathione beads and rotated for 30 min. at 25°C. Beads were pelleted and washed 4X with PBS containing 5 mM MgCl2, 5 mM 2-β-mercaptoethanol, and protease inhibitors (and appropriate 100 μM nucleotide for Rab1b). Beads were resuspended in NS buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 1 mM GTPγS or GDP for Rab1b) and blocked in 0.1% bovine serum albumin (BSA) with 0.02% Triton X-100 for 30 min at 4°C. Beads were incubated at a 1:10 ratio of beads:HeLa lysate for 2 h at 4°C on a rotator. The beads were pelleted and washed four times with NS buffer containing 0.1% Triton X-100 and either 1 mM GTPγS or GDP for Rab1b. Bound proteins were eluted with Laemmli buffer and analyzed on 8% SDS-PAGE. In some experiments, proteins were transferred to nitrocellulose, and Western blotted, as described previously (Grabski et al., 2003).
p115-GBF1 Coimmunoprecipitation
HeLa cells were transfected with p115-GFP and cultured for 48 h at 37°C. Cells were washed twice with ice-cold PBS and then collected by scraping in 0.3 ml of immunoprecipitation (IP) buffer/plate (20 mM HEPES, pH 7.4, 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 1.0% Triton X-100, and 1 mM DTT). Cells were lysed by aspirating 30 times through a 26-gauge needle. Lysates were centrifuged at 1000 × g for 10 min at 4°C, and the resulting supernatant was used for the IPs. IPs were carried out in a total volume of 0.6 ml and contained 2 μg of antibody, 1% BSA, 200 μl of lysate, and 40 μl (50% buffer/50% beads) of washed protein A beads. Reactions were rotated 2 h at 4°C and then washed four times with IP buffer (same as above). Beads were resuspended in 1× Laemmli buffer, boiled 5 min, and loaded onto 8% SDS-PAGE. Ten percent of the starting material was also loaded on the gel. Proteins were transferred to nitrocellulose and were blotted with anti-GBF1 polyclonal antibodies.
Membrane Binding
Stacked Golgi (SG) membranes were isolated from rat liver as described previously (Salamero et al., 1990). In some experiments, SG membranes were incubated with 1 M KCl in HKM buffer (250 mM HEPES, pH 7.2, 50 mM magnesium acetate, and 75 mM potassium acetate) to remove peripheral membrane proteins. Radiolabeled recombinant p115 was prepared by using 1 μg of p115 in pCDNA3.1+ as a template in a coupled transcription/translation reaction containing [35S]methionine (0.02 mCi/reaction), Rabbit Reticulocyte In Vitro Translation kit (Promega), and T7 polymerase (Promega), according to the manufacturer’s protocol.
In experiments assaying p115 binding in the presence of endogenous NSF and SNAPs, 10 μl of SG membranes was supplemented with 5 μl of in vitro-translated p115, 2 μl of ATP-regenerating system (made by mixing 100 μl of 0.2 M creatine phosphate, 8 μl of 0.5 M Na-ATP, and 20 μl of 1000 U/ml creatine phosphokinase), and 4 μl of 10× HKM, in a total volume of 40 μl. In some reactions, the ATP-regenerating system was omitted. In some reactions, SG membranes were pretreated with 2.5 mM N-ethylmaleimide (NEM) (on ice for 15 min) and then quenched with 5 mM DTT (on ice for 15 min). Reactions were incubated at 34°C for 10 min and then centrifuged at 45,000 rpm for 1 h at 4°C in a TLX OPT ultracentrifuge (Beckman Coulter, Fullerton, CA), TLA100.2 rotor, to pellet membranes. Supernatants were removed, and the pellets were resuspended in 40 μl of RIPA buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 1% deoxycholate, and 0.1% SDS). Equivalent volumes of supernatant and pellet were analyzed on 8% SDS-PAGE.
In experiments assaying p115 binding in the presence of recombinant NSF and α-SNAP, 10 μl of SG membranes washed with 1 M KCl was supplemented with 5 μl of in vitro-translated p115, 2 μl of ATP-regenerating system (made by mixing 100 μl of 0.2 M creatine phosphate, 8 μl of 0.5 M Na-ATP, and 20 μl of 1000 U/ml creatine phosphokinase), 4 μl of 10× HKM, 4 μl of bacterially expressed and purified NSF (3 μM stock; kindly provided by Dr. Phyllis Hanson), and 0.5 μl of bacterially expressed and purified α-SNAP (80 μM stock; kindly provided by Dr. Phyllis Hanson), in a total volume of 40 μl. In some reactions, the ATP-regenerating system was omitted. In some reactions, NSF was omitted. Reactions were incubated at 34°C for 10 min and then centrifuged at 45,000 rpm for 1 h at 4°C in a TLX OPT ultracentrifuge, TLA100.2 rotor, to pellet membranes. Supernatants were removed and the pellets were resuspended in 40 μl of RIPA (see above). Equivalent volumes of supernatant and pellet were analyzed on 8% SDS-PAGE. The gels were transferred to nitrocellulose and Western blotted as described previously (Grabski et al., 2003). The same nitrocellulose filters were analyzed by autoradiography with a PhosphorImager screen (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) for 16 h. Densitometry was performed using NIH ImageQuant software, and graphs were created in Microsoft Excel.
Computational Evaluation and Modeling
Standard principles of chemical kinetics and differential equations were used to analyze the models. Details of computational procedures and the parameters used are described in Supplemental Materials.
RESULTS
Characterization of GFP-tagged p115
To generate a reagent for imaging in live cells, a chimera of p115 with GFP was constructed by fusing GFP to the C terminus of p115. The localization of p115-GFP was compared with that of endogenous p115 to ensure that it targeted to membranes correctly. Endogenous p115 is concentrated at the Golgi, the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), and ER exit sites (ERES), as shown by its colocalization with the Golgi matrix protein GM130, the ERGIC marker ERGIC53, and the Sec23/24 component of the COPII coat (Figure 1A; Nelson et al., 1998; Alvarez et al., 1999). Additional diffuse staining may represent cytosolic or ER-associated p115. Endogenous p115 and GM130 redistribute to dispersed punctate structures shown previously to represent ERES in cells treated with brefeldin A (BFA) (Figure 1A; Ward et al., 2001). Like endogenous p115, p115-GFP localizes to the Golgi and the ERGIC in control cells and to punctate ERES in BFA-treated cells (Figure 1B). In all cases, the p115-GFP pattern is analogous to that of endogenous p115, indicating that p115-GFP targets to membranes correctly.
Figure 1.
Characterization of p115-GFP. (A) Untreated HeLa cells or cells incubated in 5 μg/ml BFA for 30 min were processed for double-label immunofluorescence with antibodies against p115 and either GM130, ERGIC53, or the Sec23/24 component of the COPII coat. Endogenous p115 is detected in the Golgi (colocalization with the GM130 Golgi marker), the ERGIC (colocalization with the ERGIC53 ERGIC marker), and ER exit sites (colocalization with COPII components). GM130 and p115 relocate to ER exit sites in cells treated with BFA. (B) HeLa cells were transfected with p115-GFP. After 24 h, cells were left untreated or incubated in 5 μg/ml BFA for 30 min and then processed for double-label immunofluorescence with antibodies against GFP and either GM130 or ERGIC53. p115-GFP localizes to the Golgi and the ERGIC in untreated cells and to ER exit sites in BFA-treated cells. The localization of p115-GFP is analogous to that of endogenous p115 shown in A. (C) Fusions of GST with full-length syntaxin-5, the N-terminal p115-binding domain of GM130, the full-length Rab1b, or the full-length inactive Rab1/N121I mutant were expressed in bacteria, purified, and immobilized on SH beads. The beads were incubated with lysates from HeLa cells transfected with p115-GFP. The amount of p115-GFP bound to the various beads was quantitated by Western blotting with anti-p115 antibodies to detect p115-GFP and endogenous p115. p115-GFP as well as the endogenous p115 are recovered on GST-syntaxin5 beads (lane 4), GST-GM130 beads (lane 8), and GST-Rab1 beads loaded with GTP (lane 3). The same relative ratio of p115-GFP and p115 is detected in the starting material (lane 1) as in the bound material. Neither p115-GFP nor endogenous p115 is recovered on GST-Rab1b beads loaded with GDP (lane 5), GST-Rab1b/N121I beads (lane 6), SH beads alone (lane 7), or SH beads coated with GST (lane 2), attesting to the specificity of binding. The interactions of p115-GFP with its partner proteins are analogous to those of endogenous p115. (D) Lysates from HeLa cells expressing p115-GFP were immunoprecipitated with anti-GFP antibodies, nonimmune antibodies, or anti-GBF1 antibodies. The precipitates were analyzed by SDS-PAGE and Western blotting with anti-GBF1 antibodies. GBF1 is detected in the anti-GFP and the anti-GBF1 precipitates but not in the nonimmune precipitate. These results indicate that p115-GFP binds GBF1.
Some tethering proteins, exemplified by the endosomal EEA1, directly associate with membranes by binding phosphatidylinositol 3-phosphate via a FYVE domain (Mills et al., 2001; Lawe et al., 2002). Others, exemplified by GRASP55 and GRASP65 are fatty acylated (Barr et al., 1997; Kuo et al., 2000). p115 lacks a FYVE domain or modifications such as myristoylation or palmitylation known to mediate direct interactions with membrane lipids. In agreement, salt extraction experiments suggest that p115 binds membranes through protein–protein interactions (Waters et al., 1992; Barroso et al., 1995; Sapperstein et al., 1995; Nelson et al., 1998). p115 has been shown to bind directly to a number of membrane-associated proteins. In addition to Rab1 and SNAREs, p115 also binds the Golgi matrix proteins GM130 and giantin (Nakamura et al., 1997; Lesa et al., 2000; Linstedt et al., 2000), and the ARF guanine nucleotide exchange factor GBF1 (Garcia-Mata and Sztul, 2003). Interactions with any of these proteins may play a role in p115 association with membranes. To ensure that p115-GFP interacts with the relevant partner proteins, binding of the p115-GFP chimera to each partner protein was tested using GST pull-down assays or coimmunoprecipitation.
Fusion proteins of GST with full-length syntaxin-5, the N-terminal p115-binding domain of GM130 (Linstedt et al., 2000), full-length Rab1, or full-length inactive Rab1/N121I were expressed in bacteria, purified, and immobilized on glutathione (SH) beads. The beads were subsequently incubated with lysates from HeLa cells expressing p115-GFP. The amount of p115-GFP bound to the various beads was quantitated by Western blotting with anti-p115 antibodies. As shown in Figure 1C, p115-GFP binds to syntaxin-5 (lane 4), GM130 (lane 8), and GTP-loaded Rab1 (lane 3). The anti-p115 antibodies detect p115-GFP (migrating as a 138-kDa band) and the endogenous p115 (migrating as a 108 kDa band) that provides an internal positive control. p115-GFP does not bind to SH beads alone (lane 7), SH beads coated with GST (lane 2), SH beads coated with the inactive Rab1/N121I (lane 6), or SH beads coated with Rab1 loaded with GDP (lane 5). The same amount of each GST chimera was bound to beads (as determined by Coomassie blue staining), and the same amount of HeLa lysate was used in each pull-down assay.
The ability of p115-GFP to interact with GBF1 was confirmed by coimmunoprecipitation. Immunoprecipitation of a lysate from HeLa cells expressing p115-GFP with anti-GFP antibodies recovers p115-GFP and GBF1 (Figure 1D).
Together, the results suggest that the p115-GFP chimera interacts with its partner proteins in a manner indistinguishable from the endogenous p115. These results support the use of p115-GFP to analyze p115 membrane dynamics in live cells.
Cycling Kinetics of p115
The GFP-tagged p115 described above was used in FLIP and FRAP measurements to examine dynamics of p115-GFP in vivo. Repeated photobleaching of a 4-μm-diameter spot in a periphery of a cell expressing p115-GFP, depleted the cell of all but ∼10% of the fluorescence after 65 bleaches (Figure 2A). FLIP was rapid, with t1/2 of ∼20 s, and maximal fluorescence loss occurred in less than 2 min, suggesting that p115-GFP is in rapid equilibrium within the cell.
Figure 2.
Cycling kinetics of p115. (A) HeLa cells were transfected with p115-GFP. After 24 h, a cytosolic region of a transfected cell (arrows) was repetitively photobleached (bleach every 10 s), and fluorescence loss from the entire cell was measured over time. Bar, 5 μm. Graph shows FLIP as a function of time. (B) HeLa cells were transfected with p115-GFP. After 24 h, an area of the Golgi (arrows) was photobleached once, and fluorescence recovery into the bleached region was monitored by obtaining images every 3 s. Data from analogous experiments were analyzed and are presented in a graph in D. Bar, 10 μm. (C) HeLa cells were transfected with p115-GFP. After 24 h, cells were incubated on ice for 20 min and then warmed to 37°C in the presence of 1 μg/ml nocodazole to prevent microtubule assembly. This results in Golgi fragmentation. p115-GFP is detected on Golgi fragments dispersed throughout the cell. A single Golgi fragment was photobleached once (arrows), and fluorescence recovery into the bleached region was monitored by obtaining images every 3 s. Data from analogous experiments were analyzed and are presented in a graph in D. Bar, 10 μm. (D) Graph shows changes in fluorescence intensity over time. Data are presented as an average from at least eight individual cells.
FRAP measurements after photobleaching a 2-μm-diameter spot of Golgi-localized p115-GFP shows rapid recovery with a mean t1/2 of 12.6 ± 1.5 s (Figure 2, B and D, and Table 1). Parallel FRAP experiments on Golgi-localized Arf1-GFP show a mean recovery t1/2 of 20 ± 4.2 s (our unpublished data). This value is within the published range (∼15 s) for this protein (Vasudevan et al., 1998; Presley et al., 2002; Niu et al., 2004; Szul et al., 2005), suggesting that our FRAP measurements are comparable with those of other laboratories.
Table 1.
Half-times of p115 FRAP under different conditions
| Condition | FRAP t1/2 (s) |
|---|---|
| Control | 12.6 ± 1.5 |
| Expressing Rab1/N121I | 7.7 ± 1.1 |
| Expressing NSF/E329Q | 20.5 ± 1.8 |
| Incubated at 10°C | 7.2 ± 1.5 |
Golgi fluorescence may recover by p115-GFP diffusing from unbleached areas of the Golgi, by delivery via incoming transport intermediates, or by recruitment from the cytosol. To distinguish between these possibilities, FRAP of p115-GFP was measured in cells in which microtubules were depolymerized by nocodazole. Nocodazole treatment leads to the formation of Golgi mini-stacks adjacent to individual ERES (Storrie et al., 1998). FRAP after photobleaching p115-GFP in a peripheral punctate structure shows rapid recovery with a t1/2 of 16.9 ± 2.0 s (Figure 2, C and D). The data suggest that p115-GFP association with Golgi membranes is largely independent of incoming transport intermediates transported via microtubules. The slightly slower recovery after nocodazole is consistent with a small percentage of the overall FRAP being microtubule dependent. Together, the results suggest that p115 rapidly cycles on and off the membranes by repeated cycles of association and dissociation.
p115 dynamics seem similar to membrane cycling kinetics of peripherally associated transport factors involved in coat recruitment. The Arf1 and ε-COP components of the COPI coat and the GBF1 guanine nucleotide exchange factor for ARF rapidly exchange between membrane and cytosolic compartments (Vasudevan et al., 1998; Presley et al., 2002; Elsner et al., 2003; Niu et al., 2005; Szul et al., 2005). On-off cycling kinetics analogous to those of Arf1 also have been reported for GRASP65 (Ward et al., 2001). However, GRASP65 behavior remains controversial because another group reported that GRASP65 does not dissociate from membranes (Marra et al., 2001). Ours is the first report of cycling dynamics for a tethering factor shown to participate in SNARE complex assembly.
Effects of the Dominant Negative Inactive Rab1/N121I on Cycling of p115-GFP
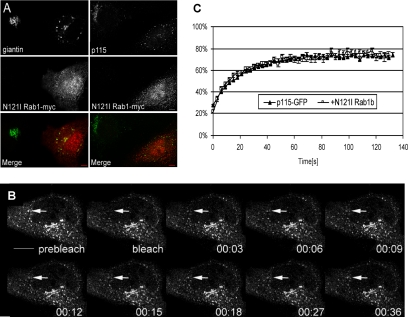
In vitro studies indicate that p115 recruitment to membranes of isolated COPII vesicles is inhibited by the Rab1/N121I mutant (Allan et al., 2000). The Rab1/N121I mutant has low affinity for both GTP and GDP and is thought to prevent the activation of endogenous Rab1 by binding to and sequestering the cognate guanine nucleotide exchange factor (Pind et al., 1994). In vivo studies show that expression of Rab1/N121I causes Golgi disruption. In cells expressing Rab1/N121I, mannosidase II and giantin redistribute to the ER, whereas GM130 and p115 localize to punctate structures dispersed throughout the cell (Alvarez et al., 2003 and Figure 3A). This suggests that p115 can associate with membranes when Rab1 function is compromised. These results support a model in which Rab1 may modulate membrane dynamics of p115, rather than p115 association. To test this hypothesis, FRAP of p115-GFP was measured in cells expressing Rab1/N121I. FRAP of p115-GFP in cells expressing Rab1/N121I shows a faster recovery, with t1/2 of 7.7 ± 1.1 s (Figure 3, B and C, and Table 1). A two-tailed Student’s t test for independent samples compared the mean t1/2 for recovery of p115-GFP in control and Rab1/N121I-expressing cells at α = 0.05 and showed a p value of 0.0256, indicating a significant difference in fluorescence recovery.
Figure 3.
Effect of Rab1 on cycling kinetics of p115. (A) HeLa cells were transfected with myc-tagged Rab1/N121I. After 24 h, cells were processed for double-label immunofluorescence with anti-myc and either anti-giantin or anti-p115 antibodies. Cells expressing Rab1/N121I have disrupted Golgi, and giantin and p115 are found in dispersed punctate structures. Bar, 10 μm. (B) HeLa cells were cotransfected with myc-tagged Rab1/N121I and p115-GFP. After 24 h, cells containing disrupted Golgi were selected for FRAP. A single punctate structure was photobleached once (arrow), and fluorescence recovery into the bleached region was monitored by obtaining images every 3 s. Data from analogous experiments were analyzed and are presented in a graph in C. Bar, 10 μm. (C) Graph shows changes in fluorescence intensity over time. Data are presented as an average from at least eight individual cells in at least two different experiments. p115-GFP shows faster recovery in cells expressing Rab1/N121I.
These results suggest that p115 associates with membranes in the absence of active Rab1, but the residency time of p115 on the membrane is compromised. Rab1 may directly link p115 to other Golgi proteins because Rab1 has been shown to interact with GM130 (Moyer et al., 2001), golgin-84 (Diao et al., 2003; Satoh et al., 2003), and the Sec34/35p components of the COG complex (Suvorova et al., 2002). However, it is currently unknown whether Rab1 can simultaneously bind two effector proteins. Alternatively, Rab1 binding may alter the conformation of p115 in such a way as to promote its interaction with the membrane (Beard et al., 2005). Rab1 may also alter the conformation of a membrane protein that facilitates p115 recruitment. It is also possible that the formation of a Rab1–p115 complex facilitates a process that results in a more stable association of p115 with the membrane.
Effects of GM130/Giantin Interactions on Cycling Kinetics of p115-GFP
Membrane-associated p115 is recovered in a stable complex with GM130 and giantin (Nakamura et al., 1997; Nelson et al., 1998; Lesa et al., 2000). GM130 and giantin are part of a detergent-resistant Golgi matrix (Linstedt and Hauri, 1993; Nakamura et al., 1995) and have been proposed to stabilize p115 within the Golgi. To explore the role of Golgi matrix proteins in p115 dynamics, a mutant p115 truncated at residue 934 (p115/1-934) that does not bind GM130 and giantin (Figure 4A; Linstedt et al., 2000) was tagged with myc or GFP. p115/1-934-myc and p115/1-934-GFP localize to the Golgi and pre-Golgi compartments in a manner analogous to that of full-length p115 (Figure 4B). FRAP of p115/1-934-GFP after photobleaching a 2-μm-diameter spot in the Golgi shows rapid recovery, with a t1/2 of 13.5 ± 1.9 s (Figure 4, C and E, and Table 1). This value is similar to that of full-length p115 (t1/2 of 12.6 ± 1.5 s), and a two-tailed Student’s t test for independent samples indicates that the difference is not significant. These results suggest that GM130/giantin do not play a major role in tethering p115 to membranes or in regulating p115 membrane dynamics in vivo. Interaction of p115 with GM130/giantin is not required for p115 function in ER–Golgi traffic (Puthenveedu and Linstedt, 2001, 2004). The cellular significance of p115 interactions with GM130 and giantin remains to be defined.
Figure 4.
Effect of GM130/giantin on cycling kinetics of p115. (A) Schematic of p115 constructs used to characterize membrane association. p115/1-934 lacks the acidic domain that contains the GM130/giantin-binding domain. p115/1-820 lacks the acidic domain and CC4. p115/1-766 lacks the acidic domain and CC3 and CC4. p115/1-655 lacks the acidic domain and CC1-CC4 that contains the SNARE-binding motif. AD, acidic domain. (B) HeLa cells were transfected with myc-tagged constructs diagrammed in A. After 24 h, cells were processed for double-label immunofluorescence with anti-myc and anti-ERGIC53 antibodies. All p115 constructs containing the SNARE-binding CC1 associate with membranes. p115/1-655 that lacks CC1 shows diffuse staining without discernible membrane association. Bar, 10 μm. (C) HeLa cells were transfected with p115/1-934-GFP. After 24 h, an area of the Golgi (arrows) was photobleached once, and fluorescence recovery into the bleached region was monitored by obtaining images every 3 s. Data from analogous experiments were analyzed and are presented in a graph in E. Bar, 10 μm. (D) HeLa cells were transfected with p115-YFP or p115/1-766-GFP. After 24 h, cells were processed for immunofluorescence with anti-GOS28 antibodies. Full-length p115 and p115/1-766 containing the SNARE-binding CC1 colocalize with GOS28. Bar, 10 μm. (E) Graph shows changes in fluorescence intensity over time. Data are presented as an average from at least 11 individual cells in at least two different experiments. Full-length p115 and p115/1-934 missing the GM130/giantin binding domain show analogous recovery.
Effects of SNARE Interactions on Cycling Kinetics of p115-GFP
A role for SNAREs in stabilizing tethering complexes on yeast vacuole membranes before fusion has been proposed previously (Price et al., 2000; Seals et al., 2000). p115 interacts with SNAREs in vitro (Allan et al., 2000; Shorter et al., 2002) through a SNARE-like motif within its coiled-coil 1 (CC1) region (Shorter et al., 2002). Full-length p115 and a p115/1-766 mutant containing the SNARE-binding CC1 (Figure 4A) colocalize with GOS28 SNARE within the Golgi and in pre-Golgi compartments (Figure 4D). To explore the role of SNAREs in membrane association of p115, p115 truncation mutants were generated (Figure 4A) and tagged with myc. The intracellular localization of each construct was explored after transfection into HeLa cells. Mutant p115/1-820 that lacks CC4 and p115/1-766 that lacks CC3 and CC4 associate with membranes (Figure 4B). In contrast, p115/1-655 that lacks CC1-CC4 shows diffuse staining throughout the cell (Figure 4B). The CC2 region has been shown not to bind SNAREs (Shorter et al., 2002). Together, the data suggest that the SNARE-binding CC1 is required for p115 association with membranes. Diffuse staining without discernible membrane association also was obtained with GFP-tagged p115/1-655. FRAP of cytosolic p115/1-655-GFP was intractable because fluorescence recovered completely within the 6 s necessary for image acquisition (our unpublished data). These results suggest that p115 association with membranes involves SNARE proteins.
The SNARE cycle of complex formation and disassembly can be disrupted in vivo by NEM, which inactivates the NSF (Wilson et al., 1989). NSF belongs to a family of chaperone-like ATPases (Junutula et al., 2004), and together with α-SNAP disassembles SNARE complexes (Whiteheart et al., 1994, 2001). FRAP of p115-GFP in cells treated with NEM is significantly delayed, with minimal recovery after 5 min (our unpublished data). NEM irreversibly alkylates sulfhydryl groups on cysteine residues and has been shown to inactivate the GTPases Arf1 (Yamaguchi et al., 1998) and tubulin (Mejillano et al., 1996; Phelps and Walker, 2000). Because NEM may act nonspecifically, its inhibition of p115-GFP dynamics is suggestive, but not indicative.
SNARE complex dissociation can be altered in vivo by expressing a dominant negative mutant of NSF. The ATPase activity of NSF is inactivated by the replacement of the second acidic residue in the Walker B DExx motif within the D1 domain (Whiteheart et al., 1994; Hung et al., 1998; Schmees et al., 1999). The NSF/E329Q mutant binds but does not hydrolyze ATP. NSF/E329Q also binds SNARE complexes. When expressed in cells, NSF/E329Q acts in a dominant negative manner by preventing binding of endogenous NSF to SNAREs. In NSF/E329Q-expressing cells, the Golgi is disrupted and Golgi proteins are detected in punctate structures clustered in the perinuclear region adjacent to the microtubule-organizing center (Dalal et al., 2004). In cells expressing NSF/E329Q, giantin and p115 are redistributed to perinuclear punctate structures (Figure 5A).
Figure 5.
Effect of SNARE cycle on cycling kinetics of p115. (A) HeLa cells were transfected with myc-tagged NSF/E329Q. After 24 h, cells were processed for double label immunofluorescence with anti-myc and either anti-giantin or anti-p115 antibodies. Expression of NSF/E329Q disrupts the Golgi, and giantin and p115 are found in dispersed punctate structures. Bar, 10 μm. (B) HeLa cells were cotransfected with myc-tagged NSF/E329Q and p115-GFP. After 24 h, cells containing disrupted Golgi were selected for FRAP. A single punctate structure was photobleached once (arrow), and fluorescence recovery into the bleached region was monitored by obtaining images every 3 s. Data from analogous experiments were analyzed and are presented in a graph in D. Bar, 10 μm. (C) HeLa cells were transfected with p115-GFP. After 24 h, cells were shifted to 10°C and maintained at 10°C during imaging. An area of the Golgi was photobleached once (arrow), and fluorescence recovery into the bleached region was monitored by obtaining images every 3 s. Data from analogous experiments were analyzed and are presented in a graph in E. Bar, 10 μm. (D) Graph shows changes in fluorescence intensity over time. Data are presented as an average from at least five individual cells in at least two different experiments. p115-GFP shows slower recovery in cells expressing NSF/E329Q. (E) Graph shows changes in fluorescence intensity over time. Data are presented as an average from at least eight individual cells in at least two different experiments. p115-GFP shows faster recovery in cells incubated at 10°C.
FRAP of p115-GFP in cells expressing NSF/E329Q shows delay in recovery (Figure 5, B and D, and Table 1), with a mean t1/2 of 20.5 ± 1.8 s; nearly twice that of control cells. A two-tailed Student’s t test for independent samples was performed to compare the mean t1/2 of p115-GFP recovery in control and in NSF/E329Q-expressing cells at α = 0.05. The obtained p value of 0.006 indicates the difference in FRAP is significant. The data suggest that the persistence of SNARE complexes in cells expressing NSF/E329Q markedly decreases p115 membrane association, linking SNARE complex disassembly and p115 binding to membranes.
Vesicular transport is known to be inhibited at temperatures below 15°C. Vesicle budding and fusion is slowed down, resulting in decreased formation of SNARE complexes. FRAP of p115-GFP in cells incubated at 10°C shows an increase in fluorescence recovery (Figure 5, C and E, and Table 1), with a mean t1/2 of 7.2 ± 1.5 s; nearly half that of control cells. A two-tailed Student’s t test for independent samples compared the mean t1/2 for recovery of p115-GFP in cells imaged at 30 and 10°C at α = 0.05 and showed a p value of 0.002, indicating a significant difference in fluorescence recovery.
Importantly, the p115 FRAP at 10°C is similar to that obtained in cells expressing the dominant negative Rab1/N121I (t1/2 of 7.7 ± 1.1 s; Table 1). Both treatments are expected to reduce SNARE complex formation and thereby increase levels of free SNAREs. The faster p115 FRAP under those conditions is consistent with an increase in p115-binding sites on the membrane and suggests that p115 preferentially binds to free SNAREs.
Effects of NSF on Membrane Binding of p115
The relationship between the status of SNAREs and p115 recruitment to membranes was further tested by in vitro binding experiments. Incubating Golgi membranes containing endogenous NSF and SNAP with in vitro transcribed/translated p115 results in ∼15% of the input p115 binding to membranes (Figure 6A, pellet). When analogous membranes containing endogenous NSF and SNAP, and in vitro-transcribed/translated p115, were supplemented with ATP-regenerating system, p115 was recovered with membranes (Figure 6B, lane 1). The amount of p115 bound under these conditions is taken as 100%. When analogous incubations were performed in the absence of ATP, or after NEM treatment of the membranes, significantly less p115 bound to the membranes (Figure 6B, lanes 2 and 3). In each incubation, the amount of bound p115 was normalized to the amount of recovered membranes (measured by the recovery of the transmembrane calreticulin). The reduced p115 binding in the absence of ATP or NEM treatment is consistent with NSF function in creating p115-binding sites on the membrane.
Figure 6.
Effect of NSF on membrane binding of p115. (A) Stacked Golgi membranes supplemented with an ATP-regenerating system were incubated with in vitro transcribed/translated p115. Membrane-bound (P, pellet fraction) and nonbound (S, supernatant fraction) p115 was quantitated and represents 100% input. Approximately 15% of the input p115 associates with membranes. (B) Binding reactions containing stacked Golgi membranes and in vitro-transcribed/translated p115 and either containing an ATP-regenerating system (lane 1), lacking an ATP-regenerating system (lane 2), or containing membranes pretreated with NEM (lane 3) were incubated at 34°C for 10 min. The membranes were collected by pelleting, and the amount of p115 associated in each sample was determined after SDS-PAGE, transfer to nitrocellulose, and autoradiography. The same nitrocellulose was subjected to Western blotting for calreticulin to provide a loading baseline for evaluating p115 association with membranes. Omitting ATP or treating membranes with NEM reduces p115 binding to membranes by ∼60%. (C) Binding reactions containing stripped SG membranes, in vitro-transcribed/translated p115 and recombinant α-SNAP, and supplemented with an ATP-regenerating system (lane 1), recombinant NSF (lane 2), or an ATP-regenerating system and NSF (lane 3) were incubated at 34°C for 10 min. The membranes were collected by pelleting, and the amount of p115 associated in each sample was determined after SDS-PAGE, transfer to nitrocellulose, and autoradiography. The same nitrocellulose was subjected to Western blotting for calreticulin to provide a loading baseline for evaluating p115 association with membranes. Omitting either ATP or NSF reduces p115 binding to membranes by ∼40 and 55%, respectively.
The participation of NSF in facilitating p115 binding to membranes was assessed directly by using membranes stripped of peripherally associated proteins and supplemented with recombinant NSF and α-SNAP. Radiolabeled p115 bound to membranes in the presence of ATP, NSF and α-SNAP (Figure 6C, lane 3). Significantly less p115 bound in the absence of ATP (lane 2) or in the absence of NSF (lane 1). The reduced p115 binding in the absence of ATP or NSF supports the participation of NSF in creating p115-binding sites on the membrane.
DISCUSSION
Based on the above-mentioned results and previous data, we formulate a simple model for p115 turnover at the membrane (Figure 7A). This model couples p115 and SNAREs and posits a direct feedback between them. The model includes the following processes: cytosolic p115 binds to free SNAREs (process 1). Binding can occur in the absence of active Rab1, but this results in rapid dissociation (process 7). Binding of the active form of Rab1 (process 2) stabilizes p115 association with membranes. Pairing and formation of SNARE complexes (process 3) results in release of p115 and Rab1 (process 4) into the cytosol. The NSF ATPase binds to the SNARE complex (process 5) and facilitates its disassembly (process 6). Processes 1 through 6 form a complete cycle of SNARE assembly and disassembly. Process 2 is interrupted by the dominant negative Rab1/N121I mutants. Process 3 is interrupted by cold 10°C temperature. Process 6 is interrupted by the dominant negative NSF/E329Q mutant.
Figure 7.
Model of p115 membrane dynamics. (A) Diagram showing the relationship between p115 binding to membranes and SNARE assembly/disassembly cycle. See text for discussion. (B) Mathematical model of p115 membrane dynamics based on mathematical formulations. See text and Supplemental Materials for description of the individual processes. (C) Fitting of data sets from FRAP experiments measuring p115 exchange on membranes in control cell or in cells expressing Rab1/N121I or NSF/E329Q or at 10°C to a mathematical formulation of the model shown in B. The circles represent data points. The solid line curves represent the model solution obtained with parameter values (see text) for the seven processes in the model.
This model was tested quantitatively on the basis of standard principles of chemical kinetics, with each process described by a system of differential equations (see Supplemental Methods). The model was tested against FRAP measurements while accounting for steady-state abundances of p115 and p115-binding SNAREs. The abundances of p115 (352,100 molecules/cell) and p115-binding SNAREs (320,950 molecules/cell) were assessed by immunoprecipitation and Western blotting (see Supplemental Methods). The model represented as a collection of rate constants (Figure 7B) fits all data sets (Figure 7C), with the theoretical values (plotted as smooth curves) correlating well with experimental FRAP data (plotted as circles). The model can account for all data sets using the same set of rate constants. The relevant rate constants are 0.11 s−1 for association of cytoplasmic p115 with Golgi membranes and 0.0384 s−1 for loss of p115 from Golgi membranes. The on/off rate constants are the same in control cells and in cells expressing mutant Rab1/N121I or NSF/E329Q or incubated at 10°C, suggesting that these treatments regulate only the availability of p115-binding sites on membranes.
Our in vivo and in vitro results and model evaluation suggest that p115 preferentially associates with free SNAREs. This is supported by FRAP data from two manipulations expected to prevent SNARE complex assembly (cells expressing Rab1/N121I or incubated at 10°C), and one manipulation expected to stabilize assembled SNARE complexes (cells expressing NSF/E329Q). Faster FRAP of p115 when SNARE complex assembly is slowed down suggests an increase in available binding sites. A decrease in FRAP when SNARE complexes are stabilized suggests a decrease in available binding sites. The data support the conclusion that p115 preferentially interacts with free SNAREs. Alternative modeling of p115 association with SNAREs does not fit the experimental data.
It is tempting to speculate that the p115–SNARE association has mutually positive feedback loops. Free SNAREs bind p115 that subsequently may facilitate their assembly into complexes. Disassembly of SNARE complexes after fusion regenerates p115-binding sites, priming the SNAREs for subsequent fusions.
Previous models for tether function postulate that tethers on apposing membranes link membranes in a process that is spatially and temporally independent of SNARE events. This type of tethering has been proposed for p115/Uso1p. p115/Uso1p has been shown to tether COPII vesicles to Golgi membranes by binding GM130 on cis-Golgi cisterna (Nakamura et al., 1995; Moyer et al., 2001). In addition, p115 has been proposed to form ”bridging“ tethers by linking giantin present in recycling COPI vesicles to GM130 present on cis-Golgi membrane (Sonnichsen et al., 1998). Both processes are assumed to be upstream of SNARE coupling. Uso1p-mediated tethering is assumed to be SNARE-independent because tethering of COPII vesicles to Golgi membranes is normal when SNARE function is inhibited by adding inhibitory anti-SNARE antibodies or by using temperature-sensitive inactive SNARE mutants (Barlowe, 1997) (VanRheenen et al., 1998). However, in both cases, SNARE proteins are still present and could theoretically bind Uso1p, despite their inability to catalyze fusion.
Our findings support a model in which tethers act within the context of SNAREs. Our results show that p115 associates with membranes in a SNARE-dependent manner because p115 lacking the SNARE-binding CC1 does not associate with membranes. Equally important, p115 association with membranes decreases significantly in cells expressing an ATPase-deficient NSF/E329Q mutant that prevents SNARE complex disassembly. That p115 associates with unassembled SNAREs is further supported by the finding that p115 recruitment to membranes is increased by NSF and α-SNAP known to promote SNARE complex disassembly. How can our findings be integrated with the current understanding of p115 function? p115 has been shown to directly bind SNAREs, and to facilitate SNARE-pin formation. A peptide mimetic of the CC1 of p115 stimulates the assembly of 3 Golgi SNARE-pins containing the t-SNARE syntaxin 5 and GS15, GOS28, and Ykt6, or syntaxin 5 and GOS28, Ykt6 and Bet1, or syntaxin 5 and membrin, Bet1 and Sec22 on isolated Golgi membranes (Shorter et al., 2002). The same peptide promotes the assembly of a complex between recombinant syntaxin 5 and GOS28 (Shorter et al., 2002). A direct role for p115 in SNARE assembly is in agreement with the finding that lack of functional Uso1p prevents SNARE complex formation in yeast in vivo (Sapperstein et al., 1996) and that overexpression of the SNAREs suppresses defects in tethering (Suvorova et al., 2002). For example, overexpression of the ER–Golgi SNAREs Bet1p and Sec22p suppresses the lethality of USO1 deletion (Sapperstein et al., 1996), indicating that Uso1p function is linked to SNAREs. Together, the findings suggest a model in which p115 binds to free SNAREs and facilitates SNARE complex formation. It is likely that p115 is displaced from the SNAREs as they pair and is released into the cytosol. This is supported by the inability to detect Uso1p in the SNARE complex in yeast (Sogaard et al., 1994; Sapperstein et al., 1996), p115 in a mammalian SNARE complex (Sollner et al., 1993), or after SNARE-pin formation (Shorter et al., 2002). Our data suggest that SNARE complexes must be disassembled by NSF to regenerate p115-binding sites on the membrane.
The model of SNARE-facilitated tethering is supported by recent findings. Specifically, the tethering function of p115 seems independent of binding GM130/giantin but requires its SNARE-interacting CC1 motif. In vivo experiments in mammalian cells show that p115 mutant unable to form a p115-GM130 or GM13-p115-giantin tether supports secretory traffic, whereas a mutant lacking the CC1 SNARE-binding motif does not support traffic (Puthenveedu and Linstedt, 2001, 2004). This suggests that p115 acts directly, rather than via other tethers, to facilitate vesicular traffic. In agreement, Drosophila cells depleted of the Drosophila homologue of the GM130 tether have morphologically normal secretory compartments and normal secretory traffic (Kondylis and Rabouille, 2003).
Mutations in tethers other than p115/Uso1p are also suppressed by overexpression of SNAREs active at that stage of traffic. For example, overexpression of the Ykt6p and Sec22p SNAREs suppresses defects in TRAPP function (Sacher et al., 1998). It is therefore likely that the model of tethers working in direct association with SNAREs is common to distinct stages of membrane trafficking.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Hans-Peter-Hauri, Phyllis Hanson, and Wally Whiteheart for kind gifts of antibodies, plasmids, and recombinant proteins. We are grateful to Drs. Jim Collawn, Anne Theibert, Brian Storrie, Jesse Hay, and Fred Hughson for constructive comments on this work. This work was supported by funding from National Institute of General Medical Sciences and American Heart Association. E. S. and C. A. are recipients of a Fogarty International Award from the National Institutes of Health.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0862) on April 19, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Allan B. B., Moyer B. D., Balch W. E. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Alvarez C., Fujita H., Hubbard A., Sztul E. ER to Golgi transport: requirement for p115 at a pre-Golgi VTC stage. J. Cell Biol. 1999;147:1205–1222. doi: 10.1083/jcb.147.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez C., Garcia-Mata R., Brandon E., Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol. Biol. Cell. 2003;14:2116–2127. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez C., Garcia-Mata R., Hauri H. P., Sztul E. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J. Biol. Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
- Aridor M., Weissman J., Bannykh S., Nuoffer C., Balch W. E. Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballew N., Liu Y., Barlowe C. A Rab requirement is not bypassed in SLY1-20 Suppression. Mol. Biol. Cell. 2005;16:1839–1849. doi: 10.1091/mbc.E04-08-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J. Cell Biol. 1997;139:1097–1108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F. A., Puype M., Vandekerckhove J., Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Barroso M., Nelson D. S., Sztul E. Transcytosis-associated protein (TAP)/p115 is a general fusion factor required for binding of vesicles to acceptor membranes. Proc. Natl. Acad. Sci. USA. 1995;92:527–531. doi: 10.1073/pnas.92.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard M., Satoh A., Shorter J., Warren G. A cryptic rab1-binding site in the p115 tethering protein. J. Biol. Chem. 2005 doi: 10.1074/jbc.M503925200. [DOI] [PubMed] [Google Scholar]
- Cao X., Ballew N., Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Rothman J. E. Purification of three related peripheral membrane proteins needed for vesicular transport. J. Biol. Chem. 1990;265:10109–10117. [PubMed] [Google Scholar]
- Dalal S., Rosser M. F., Cyr D. M., Hanson P. I. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao A., Rahman D., Pappin D. J., Lucocq J., Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner M., Hashimoto H., Simpson J. C., Cassel D., Nilsson T., Weiss M. Spatiotemporal dynamics of the COPI vesicle machinery. EMBO Rep. 2003;4:1000–1004. doi: 10.1038/sj.embor.embor942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R., Sztul E. The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucleotide-exchange factor. EMBO Rep. 2003;4:320–325. doi: 10.1038/sj.embor.embor762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabski R., Szul T., Sasaki T., Timpl R., Mayne R., Hicks B., Sztul E. Mutations in COCH that result in non-syndromic autosomal dominant deafness (DFNA9) affect matrix deposition of cochlin. Hum. Genet. 2003;113:406–416. doi: 10.1007/s00439-003-0992-7. [DOI] [PubMed] [Google Scholar]
- Hung L. W., Wang I. X., Nikaido K., Liu P. Q., Ames G. F., Kim S. H. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- Junutula J. R., Schonteich E., Wilson G. M., Peden A. A., Scheller R. H., Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 2004;279:33430–33437. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- Kondylis V., Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J. Cell Biol. 2003;162:185–198. doi: 10.1083/jcb.200301136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A., Zhong C., Lane W. S., Derynck R. Transmembrane transforming growth factor-alpha tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 2000;19:6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawe D. C., Chawla A., Merithew E., Dumas J., Carrington W., Fogarty K., Lifshitz L., Tuft R., Lambright D., Corvera S. Sequential roles for phosphatidylinositol 3-phosphate and Rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J. Biol. Chem. 2002;277:8611–8617. doi: 10.1074/jbc.M109239200. [DOI] [PubMed] [Google Scholar]
- Lesa G. M., Seemann J., Shorter J., Vandekerckhove J., Warren G. The amino-terminal domain of the Golgi protein giantin interacts directly with the vesicle-tethering protein p115. J. Biol. Chem. 2000;275:2831–2836. doi: 10.1074/jbc.275.4.2831. [DOI] [PubMed] [Google Scholar]
- Linstedt A. D., Hauri H. P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Jesch S. A., Mehta A., Lee T. H., Garcia-Mata R., Nelson D. S., Sztul E. Binding relationships of membrane tethering components. The giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J. Biol. Chem. 2000;275:10196–10201. doi: 10.1074/jbc.275.14.10196. [DOI] [PubMed] [Google Scholar]
- Marra P., Maffucci T., Daniele T., Tullio G. D., Ikehara Y., Chan E. K., Luini A., Beznoussenko G., Mironov A., De Matteis M. A. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat. Cell Biol. 2001;3:1101–1113. doi: 10.1038/ncb1201-1101. [DOI] [PubMed] [Google Scholar]
- Mejillano M. R., Shivanna B. D., Himes R. H. Studies on the nocodazole-induced GTPase activity of tubulin. Arch. Biochem. Biophys. 1996;336:130–138. doi: 10.1006/abbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- Mills I. G., Urbe S., Clague M. J. Relationships between EEA1 binding partners and their role in endosome fusion. J. Cell Sci. 2001;114:1959–1965. doi: 10.1242/jcs.114.10.1959. [DOI] [PubMed] [Google Scholar]
- Moyer B. D., Allan B. B., Balch W. E. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- Nakajima H., Hirata A., Ogawa Y., Yonehara T., Yoda K., Yamasaki M. A cytoskeleton-related gene, uso1, is required for intracellular protein transport in Saccharomyces cerevisiae. J. Cell Biol. 1991;113:245–260. doi: 10.1083/jcb.113.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Lowe M., Levine T. P., Rabouille C., Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. S., Alvarez C., Gao Y. S., Garcia-Mata R., Fialkowski E., Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J. Cell Biol. 1998;143:319–331. doi: 10.1083/jcb.143.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T. K., Pfeifer A. C., Lippincott-Schwartz J., Jackson C. L. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 Exchange Factor at the Golgi. Mol. Biol. Cell. 2004 doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T. K., Pfeifer A. C., Lippincott-Schwartz J., Jackson C. L. Dynamics of GBF1, a brefeldin a-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps K. K., Walker R. A. NEM tubulin inhibits microtubule minus end assembly by a reversible capping mechanism. Biochemistry. 2000;39:3877–3885. doi: 10.1021/bi992200x. [DOI] [PubMed] [Google Scholar]
- Pind S. N., Nuoffer C., McCaffery J. M., Plutner H., Davidson H. W., Farquhar M. G., Balch W. E. Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J. Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley J. F., Ward T. H., Pfeifer A. C., Siggia E. D., Phair R. D., Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- Price A., Wickner W., Ungermann C. Proteins needed for vesicle budding from the Golgi complex are also required for the docking step of homotypic vacuole fusion. J. Cell Biol. 2000;148:1223–1229. doi: 10.1083/jcb.148.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 2001;155:227–238. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Linstedt A. D. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:1253–1256. doi: 10.1073/pnas.0306373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Jiang Y., Barrowman J., Scarpa A., Burston J., Zhang L., Schieltz D., Yates J. R., 3rd, Abeliovich H., Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamero J., Sztul E. S., Howell K. E. Exocytic transport vesicles generated in vitro from the trans-Golgi network carry secretory and plasma membrane proteins. Proc. Natl. Acad. Sci. USA. 1990;87:7717–7721. doi: 10.1073/pnas.87.19.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein S. K., Lupashin V. V., Schmitt H. D., Waters M. G. Assembly of the ER to Golgi SNARE complex requires Uso1p. J. Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein S. K., Walter D. M., Grosvenor A. R., Heuser J. E., Waters M. G. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc. Natl. Acad. Sci. USA. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Wang Y., Malsam J., Beard M. B., Warren G. Golgin-84 is a rab1 binding partner involved in Golgi structure. Traffic. 2003;4:153–161. doi: 10.1034/j.1600-0854.2003.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler R., Itin C., Zerial M., Lottspeich F., Hauri H. P. ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif. Eur. J. Cell Biol. 1993;61:1–9. [PubMed] [Google Scholar]
- Schmees G., Stein A., Hunke S., Landmesser H., Schneider E. Functional consequences of mutations in the conserved ‘signature sequence’ of the ATP-binding-cassette protein MalK. Eur. J. Biochem. 1999;266:420–430. doi: 10.1046/j.1432-1327.1999.00871.x. [DOI] [PubMed] [Google Scholar]
- Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Beard M. B., Seemann J., Dirac-Svejstrup A. B., Warren G. Sequential tethering of golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J. Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard M., Tani K., Ye R. R., Geromanos S., Tempst P., Kirchhausen T., Rothman J. E., Sollner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Sollner T., Whiteheart S. W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., Rothman J. E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B., Lowe M., Levine T., Jamsa E., Dirac-Svejstrup B., Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J. Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B., White J., Rottger S., Stelzer E. H., Suganuma T., Nilsson T. Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova E. S., Duden R., Lupashin V. V. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szul T., Garcia-Mata R., Brandon E., Shestopal S., Alvarez C., Sztul E. Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic. 2005;6:374–385. doi: 10.1111/j.1600-0854.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- VanRheenen S. M., Cao X., Lupashin V. V., Barlowe C., Waters M. G. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J. Cell Biol. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen S. M., Cao X., Sapperstein S. K., Chiang E. C., Lupashin V. V., Barlowe C., Waters M. G. Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J. Cell Biol. 1999;147:729–742. doi: 10.1083/jcb.147.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan C., Han W., Tan Y., Nie Y., Li D., Shome K., Watkins S. C., Levitan E. S., Romero G. The distribution and translocation of the G protein ADP-ribosylation factor 1 in live cells is determined by its GTPase activity. J. Cell Sci. 1998;111:1277–1285. doi: 10.1242/jcs.111.9.1277. [DOI] [PubMed] [Google Scholar]
- Ward T. H., Polishchuk R. S., Caplan S., Hirschberg K., Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. G., Clary D. O., Rothman J. E. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J. Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart S. W., Rossnagel K., Buhrow S. A., Brunner M., Jaenicke R., Rothman J. E. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart S. W., Schraw T., Matveeva E. A. N-ethylmaleimide sensitive factor (NSF) structure and function. Int. Rev. Cytol. 2001;207:71–112. doi: 10.1016/s0074-7696(01)07003-6. [DOI] [PubMed] [Google Scholar]
- Whyte J. R., Munro S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Wilson D. W., Whiteheart S. W., Wiedmann M., Brunner M., Rothman J. E. A multisubunit particle implicated in membrane fusion. J. Cell Biol. 1992;117:531–538. doi: 10.1083/jcb.117.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. W., Wilcox C. A., Flynn G. C., Chen E., Kuang W. J., Henzel W. J., Block M. R., Ullrich A., Rothman J. E. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Nakayama K., Hatsuzawa K., Tani K., Himeno M., Tagaya M. ADP-ribosylation factor-1 is sensitive to N-ethylmaleimide. J. Biochem. 1998;124:1229–1236. doi: 10.1093/oxfordjournals.jbchem.a022242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.