Abstract
The endoplasmic reticulum (ER) contains both cisternal and reticular elements in one contiguous structure. We identified rtn1Δ in a systematic screen for yeast mutants with altered ER morphology. The ER in rtn1Δ cells is predominantly cisternal rather than reticular, yet the net surface area of ER is not significantly changed. Rtn1-green fluorescent protein (GFP) associates with the reticular ER at the cell cortex and with the tubules that connect the cortical ER to the nuclear envelope, but not with the nuclear envelope itself. Rtn1p overexpression also results in an altered ER structure. Rtn proteins are found on the ER in a wide range of eukaryotes and are defined by two membrane-spanning domains flanking a conserved hydrophilic loop. Our results suggest that Rtn proteins may direct the formation of reticulated ER. We independently identified Rtn1p in a proteomic screen for proteins associated with the exocyst vesicle tethering complex. The conserved hydophilic loop of Rtn1p binds to the exocyst subunit Sec6p. Overexpression of this loop results in a modest accumulation of secretory vesicles, suggesting impaired exocyst function. The interaction of Rtn1p with the exocyst at the bud tip may trigger the formation of a cortical ER network in yeast buds.
INTRODUCTION
The endoplasmic reticulum (ER) is a large, elaborate structure that is typically spread throughout the cytoplasm of eukaryotic cells (Terasaki and Jaffe, 1991; Greenfield and High, 1999). In higher eukaryotic cells, the ER has classically been divided into three distinct regions: the nuclear envelope; the rough ER, characterized by the presence of ribosomes and a cisternal morphology; and the smooth ER, a polygonal network of membrane tubules that seems to emanate from the cisternae of the rough ER (Palade, 1956). These morphologically distinct membrane structures define a continuous lumen, indicating that they are all part of one organelle (Terasaki et al., 1996). In the yeast Saccharomyces cerevisiae, the ER is composed of the nuclear envelope and a meshwork of membrane tubules closely apposed to the cell cortex with only a few cytoplasmic tubules connecting the cortical ER to the nuclear envelope (Preuss et al., 1991; Prinz et al., 2000).
In all cells, the ER structure is very dynamic. The smooth ER can increase in volume, as in hepatocytes (Remmer and Merker, 1963), or undergo structural rearrangements of three types: a new tubule can stem from an existing tubule and fuse with another one to form new alveolus, an existing alveolus can shrink in a process akin to ring closures, and a tubule can slide along another tubule (Lee and Chen, 1988; Prinz et al., 2000). The reorganization and maintenance of the ER in higher eukaryotes depends on microtubules (Terasaki et al., 1986; Terasaki, 1990), whereas in yeast, ER reorganization depends on actin (Prinz et al., 2000).
In yeast, actin also plays a defining role in the inheritance of the ER between mother and daughter cell, a process that occurs before nuclear inheritance and that is independent of microtubules (Fehrenbacher et al., 2002; Estrada et al., 2003). Indeed, early in the cell cycle, an ER tubule emanating from the mother cell is pulled along actin cables into the nascent bud by the type V myosin Myo4 and its adaptor protein She3 (Du et al., 2001; Fehrenbacher et al., 2002; Estrada et al., 2003). Once the tubule is in the bud, it anchors at the bud tip (Fehrenbacher et al., 2002), a process that requires at least three subunits of the exocyst complex, Sec3, Sec5, and Sec8 (Wiederkehr et al., 2003, 2004; Reinke et al., 2004). The exocyst is an octameric complex found at sites of secretion, such as the bud tip, where it acts to tether secretory vesicles to the plasma membrane in preparation for exocytic fusion. After anchoring at the bud tip, the ER spreads along the cortex of the bud by an unknown mechanism.
A key question concerns the mechanism by which the ER forms and maintains its meshwork structure. In this study, we provide compelling evidence for a role of Rtn1, an integral ER membrane protein, in structuring the ER. This protein belongs to the Rtn family of proteins that is ubiquitously found in eukaryotes and is characterized by a reticulon domain consisting of two transmembrane domains flanking a hydrophilic loop (Oertle et al., 2003). In mammalian cells, four RTN genes are present and several splice variants for each have been identified. In yeast, only one paralog has been found and named Rtn2 with no known function (Oertle et al., 2003). RTN genes are differentially expressed in different cell types (Roebroek et al., 1998; Moreira et al., 1999; GrandPre et al., 2000). All homologues of Rtn1 are localized on the ER, yet they have no identified function there (Van de Velde et al., 1994; Chen et al., 2000; Hamada et al., 2002). A small fraction of Rtn4, also called Nogo, is also localized at the plasma membrane where it is involved in the negative regulation of neuronal growth (Chen et al., 2000; GrandPre et al., 2000; Prinjha et al., 2000). We also show that Rtn1, through its conserved hydrophilic loop, interacts with Sec6p, a subunit of the exocyst.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Growth Conditions
S. cerevisiae strains used in this study are listed in Table 1. Cells were grown in either YP (rich medium) or SC (complete medium) with the appropriate amino acids omitted to ensure plasmid maintenance and the mentioned carbon source to a final concentration of 2%. Standard techniques were used for sporulation and tetrad analysis of yeast strains (Sherman, 1991). Yeast cells were transformed using the modified lithium acetate method (Gietz et al., 1992).
Table 1.
Yeast strains used in this work
| Strain | Genotype | Reference |
|---|---|---|
| NY2647 | MATα SEC61:(GFP LEU2) ura3 | This study |
| NY2648 | MATα rtn1::KanMX6 SEC61:(GFP LEU2) ura3 | This study |
| NY2649 | MATa SSH1:(GFP HIS3) his3 leu2 ura3 | This study |
| NY2650 | MATa rtn1::KanMX6 SSH1:(GFP HIS3) leu2 ura3 | This study |
| NY1210 | MATa his3 leu2 ura3 | Lab strain |
| NY2651 | MATa rtn1::KanMX6 leu2 ura3 | This study |
| NY2652 | MATα SEC7:(GFP URA3) leu2 | This study |
| NY2653 | MATa rtn1::KanMX6 SEC7:(GFP URA3) leu2 | This study |
| NY2654 | MATα rtn1::KanMX6 VPH1:(GFP HIS3) leu2 ura3 | This study |
| NY2655 | MATα VPH1:(GFP HIS3) leu2 ura3 | This study |
| NY2656 | MATα rtn1::KanMX6 ura3::(ATP9-DsRed URA3) his3 leu2 | This study |
| NY2657 | MATα ura3::(ATP9-DsRed URA3) his3 leu2 | This study |
| NY2658 | MATα RTN1:(GFP KanMX6) leu2::(HDEL-DsRed LEU2) ura3 | This study |
| NY2659 | MATα RTN1:(3HA KanMX6) leu2 ura3 | This study |
| NY2660 | MATα Sec61:(GFP LEU2) ura3::(GAL1pGST URA3) | This study |
| NY2661 | MATa Sec61:(GFP LEU2) ura3::(GAL1pGST-RTN1 URA3) his3 | This study |
| NY2662 | MATa Sec61:(GFP LEU2) ura3::(GAL1p GST-RTN1 loop URA3) his3 | This study |
| NY2463 | MATa SEC10:(TAP LEU2) his3 ura3 | This study |
| NY2664 | MATa ura3::(GAL1p GST URA3) his3 leu2 | This study |
| NY2665 | MATa ura3::(GAL1p GST-RTN1 URA3) his3 leu2 | This study |
| NY2666 | MATa ura3::(GAL1p GST URA3) his3 leu2/MATα his3 leu2 ura3 | This study |
| NY2667 | MATa ura3::(GAL1p GST-RTN1 URA3) his3 leu2/MATα his3 leu2 ura3 | This study |
| NY2668 | MATa ura3::(GAL1p GST URA3) his3 leu2/MATα ura3::(GAL1p SEC6 URA3) leu2 | This study |
| NY2669 | MATa ura3::(GAL1p GST-RTN1 URA3) his3 leu2/MATα ura3::(GAL1p SEC6 URA3) leu2 | This study |
| NY2670 | MATa leu2::(GAL1p GST-SNC1 LEU2) his3 ura3/MATα ura3::(GAL1p SEC6 URA3) leu2 | This study |
| NY2671 | MATa rtn1::KanMX6 ura3::(RTN1-HA URA3) leu2 | This study |
| NY2672 | MATa rtn1::KanMX6 ura3::(RTN1A94S-HA URA3) leu2 | This study |
| NY2673 | MATa rtn1::KanMX6 ura3::(RTN1R116N-HA URA3) leu2 | This study |
| NY2674 | MATa rtn1::KanMX6 ura3::(RTN1K125N-HA URA3) leu2 | This study |
| NY2675 | MATa rtn1::KanMX6 ura3::(RTN1V215S-HA URA3) leu2 | This study |
| NY2676 | MATα Sec61:(GFP LEU2) ura3(pNB1282) | This study |
| NY2677 | MATα Sec61:(GFP LEU2) ura3 (pNB1284) | This study |
| NY2678 | MATa his3 leu2 ura3 (pNB1285) | This study |
| NY2679 | MATa his3 leu2 ura3 (pNB1286) | This study |
| NY2680 | MATα RTN2:(GFP kanMX6) leu2 ura3 | This study |
| NY2681 | MATα RTN2:(3HA kanMX6) his3 leu2 ura3 | This study |
| NY2682 | MATα rtn1::(kanMX6) rtn2::(kanMX6) leu2::(SEC61-GFP LEU2) ura3 | This study |
Genomic insertions were done by amplifying kanMX6 cassettes flanked by 5′ and 3′ recombination regions. The PCR template used for the deletion of the RTN1 gene was genomic DNA prepared from the deletion library corresponding mutant. The PCR template for C-terminal tagging of Rtn1 was pFA6a3HA-kanMX6 or pFA6aGFP-kanMX6 (Longtine et al., 1998). The PCR template for C-terminal tagging of Vph1 was pFA6aGFP-HIS5MX6.
To tag Sec61 with GFP, we used plasmid SFNB1018 (LEU2 marker) linearized by PmlI and Ssh1 by SFNB1068 (LEU2 marker) linearized by EcoRI (Wiederkehr et al., 2003). To tag Sec7p with GFP, plasmid SFNB798 (pUSE-URA3) was linearized by SpeI (Seron et al., 1998).
Plasmid pNRB1264 was constructed by digesting plasmid HDEL-DsRed (a gift of the Glick laboratory, University of Chicago, Chicago, IL) with HindIII and NdeI, and this last site was blunted by Klenow. The fragment was inserted in pRS305 (LEU2 marker) between sites SmaI and HindIII and linearized by EcoRV. Plasmid pNRB1265 was constructed by digesting plasmid Atp9-DsRed (Mozdy et al., 2000) with ApaI and SacI. The fragment was inserted in pRS303 (HIS3 marker) between sites KpnI and SacI and linearized by HindIII.
Isolation of rtn1Δ Mutant
The yeast MATa haploid deletion library from Research Genetics (ResGen/Invitrogen, Carlsbad, CA) was screened to identify genes displaying a defect in the inheritance of the cortical ER. The transformation of this library was performed as described in Estrada et al. (2003) in 96-well blocks. The cells were transformed with plasmid SFNB1000 (HMG1-GFP CEN LEU2; Estrada et al., 2003). A total of 4080 mutants were screened.
Fluorescence Microscopy
To visualize the ER, cells expressing GFP or DsRed fusion proteins were grown overnight in SC medium at 25°C to optical density (OD)600 of 0.3. 1 ml of culture was pelleted and resuspended in 50 μl of growth medium. Then, 3 μl of cells was mixed with an equal volume of growth medium containing 0.6% of low melting agarose.
Epifluorescence microscopy was performed using a Zeiss Axioplan2 microscope with a 63× oil immersion objective. Images were collected with an Orca ER digital camera (Hamamatsu, Bridgewater, NJ) and the OpenLab 4.02 imaging software (Improvision, Lexington, MA).
Confocal microscopy was performed using a laser scanning microscope (LSM 510; Carl Zeiss MicroImaging, Thornwood, NY) with a 100× oil immersion objective. All images were processed for display using Photoshop and Illustrator from Adobe Systems (Mountain View, CA). Three dimensional (3D) rendering was performed using Imaris from BitPlane (St. Paul, MN).
Electron Microscopy
For ER analysis, yeast cells were harvested, resuspended, and incubated for 20 min at room temperature (RT) in 1.5% KMnO4. After five washes with distilled water, cells were stained with 2% uranyl acetate for 4 h at RT in the dark.
For vesicle density, cells were fixed in 3% glutaraldehyde with 0.1 M sodium cacodylate buffer, pH 6.8, overnight. After two washes in 50 mM phosphate buffer, pH 7, cells were resuspended in 2 ml of 0.25 mg/ml Zymolyase 100T (Seigaku Corporation, Tokyo, Japan) for 25 min. After two washes in 0.1 M sodium cacodylate, cells were resuspended in 0.5 ml of cold 2% OsO4 in 0.1 M sodium cacodylate for 1 h on ice. After three washes with water, cells were resuspended in 2% uranyl acetate for 1 h at RT and washed five times with water.
All samples were dehydrated by incubation with 50, 70, 95 (2 times for 5 min each), and 100% ethanol (2 times for 15 min each). The cells were then washed briefly in acetone before embedding in Spurr resin (Electron Microscopy Science, Fort Washington, PA). For polymerization, the cell pellets in Spurr were cured for 48 h at 80°C. Ultrathin (60-nm) sections were cut on a Reichert ultramicrotome and collected on Formvar- and carbon-coated grids. The samples were poststained with 2% uranyl acetate and lead citrate and examined on a Philips Tecnai 12 electron microscope.
Subcellular Fractionation
The ER and plasma membrane were fractionated on a linear sucrose density gradient in the presence of EDTA as described previously (Roberg et al., 1997). In brief, cells were grown to an OD600 of 1.0 in YP glucose, harvested and resuspended in 0.5 ml of STE buffer [10% (wt/wt) sucrose, 10 mM EDTA, and 10 mM Tris-HCl, pH 7.5] containing a protease inhibitor cocktail (10 μM antipain, 30 μM leupeptine, 30 μM chymostatin, 1 μM pepstatin A, 1 μM phenylmethylsulfonyl fluoride [PMSF], and 1 μg/ml aprotinin). Cells were then lysed by vortexing with glass beads. After lysis, 1 ml of STE buffer was added, and the lysate was cleared by centrifugation at 500 × g for 10 min. Then, 15 OD600 units (∼300 μl of the lysate) were layered on top of a 20–60% linear sucrose gradient (5 ml) prepared in TE (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA). Samples were then centrifuged at 100,000 × g for 18 h at 4°C using a SW50.1 rotor (Beckman Coulter, Fullerton, CA). Fractions (∼440 μl each) were collected from the top of the gradient, and proteins were precipitated with 500 μl of 50% trichloroacetic acid (TCA), and pelleted by centrifugation at 14,000 rpm for 5 min at 4°C. Protein pellets were resuspended in 66 μl of Tris base and 99 μl of 2× SDS sample buffer. Then, 30 μl of each fraction was resolved by SDS-PAGE and subjected to Western blot analysis. The antibodies were subsequently detected by enhanced chemiluminescence (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and the bands were quantified using BioImage software (Yale University, New Haven, CT). The data are plotted as a percentage of the maximum of antigenic material.
Membrane Extraction Experiment
Cells expressing Rtn1-3xHA were used for preparation of microsomes as described in Gunthrie and Fink (1991). Cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, and 150 mM NaCl) and complete protease inhibitors (Roche Diagnostics, Indianapolis, IN). Equal volumes of lysate were treated either with 0.5 M NaCl, 0.1 M Na2CO3, 1% TritonX-100, or 0.1% SDS to a finale volume of 100 μl and incubated for 30 min on ice. The treated lysates were centrifuged for 1 h at 100,000 × g. The supernatant fractions were mixed to equal volume of 2× loading buffer, and pellets were resuspended in 200 μl of loading buffer. Ten microliters of each sample was loaded on a 10% SDS-PAGE and after Western blotting was probed with anti-hemagglutinin (HA) (BabCO, Richmond, CA).
Tandem Affinity Purification (TAP) Tag Purification of Exocyst Proteins
Exocyst proteins were C-terminally fused to a TAP tag (NY2463; Sec10p-TAP LEU2). Fusion proteins were isolated using the method described in Puig et al. (2001) with some modifications. Instead of using a French press, 2 liters of cells (grown at 25°C to OD600 1.5) were lysed in a buffer of 20 mM PIPES, pH 6.8, 150 mM NaCl, 1 mM EDTA, 0.2 mM PMSF, 10 μM antipain, 20 μM aprotinin, 20 μM chymostatin, 20 μM leupeptin, 20 μM pepstatin A, and 10 mM β-mercaptoethanol (β-ME) using a Bead Beater (Biospec Products, Bartlesville, OK). The 30-ml chamber was filled halfway with 0.5-mm glass beads (Biospec Products) and run four times for 1 min on ice. NP-40 (IGEPAL CA-630; Sigma-Aldrich, St. Louis, MO) was added [0.5% (vol/vol)], and the lysate was incubated at 4°C for 15 min and then centrifuged at 30,000 × g for 15 min. The method for protein isolation in Puig et al. (2001) was followed except 20 mM PIPES, pH 6.8, was substituted for Tris-HCl in all buffers, the TEV protease incubation was allowed to proceed overnight at 4°C, and the concentration of EGTA in the elution buffer was increased to 10 mM.
Mass Spectrometry
Fifty percent of the exocyst TAP tag purification was precipitated using 20% (vol/vol) TCA, washed five times with 100% acetone (−20°C), and sent to the Proteomic Mass Spectrometry Laboratory at the Scripps Research Institute (La Jolla, CA). Samples were suspended in 8 M urea, 100 mM Tris, pH 8.5, reduced with 100 mM Tris(2-carboxyethyl)phosphine (Pierce, Rockford, IL), and cysteines were alkylated with 55 mM iodoacetamide. Then, Lys-C was used to digest the proteins for 4 h at 37°C at a concentration of 1 μg/100 μl. CaCl2 was added to ensure tryptic specificity at 1 mM, and trypsin was used to digest the samples further at 1 μg/100 μl. The digests were then analyzed by micro-liquid chromatography (μLC)/μLC-tandem mass spectrometry (MS)/MS using an LCQ Deca ion trap mass spectrometer (Thermo Electron, Waltham, MA). Multidimensional chromatography was performed online using the following salt steps of 500 mM ammonium acetate: 10, 25, 35, 50, 65%, 80, and 100% (MacCoss et al., 2002). Tandem mass spectra were collected in data-dependent manner by collecting one full MS scan (m/z range = 400–1600) followed by MS/MS spectra of the three most abundant precursor ions. The collection of resulting spectra was then searched against a database of yeast open reading frames obtained from Saccharomyces Genome Database (release date August 27, 2004) using the SEQUEST algorithm (Eng et al., 1994). Peptide identifications were organized and filtered using the DTASelect program (Tabb et al., 2002).
Expression of Exocyst Proteins in Escherichia coli
Exocyst genes were PCR-amplified and cloned using the pET-46 Ek/LIC cloning kit (strains NRB1266–1273; Novagen, Madison, WI). Constructs were transformed into Rosetta 2 cells (Novagen) and grown in Terrific Broth (Sambrook et al., 1997) at 25°C to an OD600 of 1. Cultures were chilled on ice for 30 min and then induced using 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h at 15°C. The cultures were pelleted, resuspended in a buffer of 1× phosphate-buffered saline (PBS), 160 mM NaCl, 15 mM imidazole, and 1 mM PMSF, and lysed on ice using a Branson Sonifier 450 sonicator (4 times for 1 min each). Triton X-100 was added [1% (vol/vol)], and the lysates were incubated at 4°C for 30 min and then centrifuged at 30,000 × g for 15 min. Fusion proteins were isolated using Ni-NTA agarose (QIAGEN, Valencia, CA) for 1.5 h at 4°C. The resin was washed with 1× PBS, 160 mM NaCl, 1% Triton X-100, and 25 mM imidazole, and fusion proteins were eluted with the same buffer, except the imidazole concentration was raised to 400 mM.
In Vitro Binding Assays for Rtn1p
RTN1 was PCR amplified from the yeast strain NY1210 using primers that created a BamHI and SmaI site at the N and C terminus, respectively, and cloned into pGEX4T-1 (NRB1274; Promega, Madison, WI). A deletion series of Rtn1p was constructed using PCR on the full-length clone with primers that allowed for cloning into the BamHI and XhoI sites of pGEX4T-1. The first construct contains the small N terminus and the first predicted transmembrane domain (amino acids 1–58; NRB1275). The second construct includes the N terminus and the entire reticulon domain (aa 1–168; NRB1276). The third construct contains only the reticulon domain (aa 27–168; NRB1277). The fourth construct includes the hydrophilic loop and the second predicted transmembrane domain (aa 59–168; NRB1278). The fifth construct contains only the hydrophilic loop (aa 59–142; NRB1279). The sixth construct contains the C terminus of the protein (aa 169–295; NRB1280). As a control, the Snc1p coding sequence was amplified from pADH-cSNC1 (Gerst et al., 1992) and cloned into pGEX4T-1 using the same restriction enzymes (NRB1281).
Glutathione S-transferase (GST) fusion proteins were induced at OD600 1.0 with 1 mM IPTG for 1.5 h at 37°C. Cultures were pelleted and resuspended in a buffer of 1× PBS, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 0.5 mM PMSF and lysed on ice using a sonicator (4 times for 1 min each). Triton X-100 was added [1% (vol/vol)], and the lysates were incubated at 4°C for 30 min and then centrifuged at 30,000 × g for 15 min. Glutathione beads (GE Healthcare) were incubated with soluble proteins for 1.5 h at 4°C followed by five washes.
Approximately equal amounts (100 nM) of soluble exocyst proteins were mixed with either GST or GST-Rtn1 protein immobilized on beads in a buffer of 20 mM PIPES, pH 6.8, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM PMSF, and 10 mM β-ME. Reactions were incubated at RT for 2 h and then washed five times. Proteins were boiled off beads and run on a 7% gel, transferred to membrane, and detected using a His monoclonal antibody (mAb) (Novagen). The same protocol was used for binding Sec6p to the deletion series of Rtn1p.
Overproduction of Rtn1p and Sec6p
To overproduce Rtn1p, the gene was PCR amplified from NRB1274 and inserted into a vector (pNB530) containing the GAL1 promoter and an ADH1 terminator using BamHI and SmaI. The gene encoding GST was cloned upstream in frame as a BamHI fragment (NRB1283 URA3). The same methodology was used for GST (NRB1282 URA3) and GST-Snc1p (NRB1292 LEU2), which both act as negative controls in this experiment, except SNC1 was cloned into pNB529 using the restriction sites BamHI and PstI. The plasmids were integrated into the yeast genome, and the strains were crossed to wild type or a strain overexpressing Sec6p to create diploids.
Strains overproducing either GST alone, GST-Rtn1p alone, GST and Sec6p, GST-Rtn1p and Sec6p, or GST-Snc1p and Sec6p were grown 16 h at 25°C to an OD600 of 1.0 in 50-ml cultures of YP containing 2% galactose. Proteins were lysed in the same buffers used for TAP purification, and glutathione beads were incubated with soluble proteins for 2 h at 4°C followed by five washes. Proteins were boiled off the beads and loaded on a 7% polyacrylamide gel, transferred to a membrane, and a Sec6p antibody (Potenza et al., 1992) was used for detection.
To overproduce the loop of Rtn1p, the sequence expressed in NRB1279 was PCR amplified and cloned into a vector containing the GAL1 promoter (pNB530) and a 2μ vector (pRS426 URA3; Mumberg et al., 1995) containing the GPD1 promoter using BamHI and SmaI. The sequence encoding GST was cloned upstream of the loop, in frame as a BamHI fragment creating plasmids NRB1284 URA3 and NRB1286 URA3, respectively. As a control, GST was cloned after the GPD1 promoter by itself (NRB1285 URA3).
Generation of Glycosylation Sites in Rtn1p
Glycosylation sites were created in the hydrophilic loop and C terminus of Rtn1p to address the topology of the protein. RTN1, along with 516 base pairs of its own promoter and a single C-terminal HA tag, was amplified and cloned into a vector (pRS306; Sikorski and Hieter, 1989) using the KpnI and SacII sites. The ADH1 terminator was added using SacI and SacII, and PstI was used to integrate the plasmid (NRB1287 URA3) into the genome of an rtn1Δ strain (NY 2651) at the URA3 gene (NY2671). PCR mutagenesis was used to create glycosylation sites by changing the Rtn1p amino acids 94, 116, 125, and 215 to a serine, asparagine, asparagine, and serine, respectively (NRB1288–1291). These plasmids were also integrated at URA3 in an rtn1Δ strain using PstI (NY2672–2675, respectively). Soluble protein from a 50-ml culture was isolated in a buffer of 20 mM PIPES, pH 6.8, 150 mM NaCl, 1 mM CaCl2, 1 mM MnCl2, 1% Triton X-100, 0.2 mM PMSF, 10 μM antipain, 20 μM aprotinin, 20 μM chymostatin, 20 μM leupeptin, 20 μM pepstatin A, and 10 mM β-ME, and concanavalin A (ConA)-Sepharose 4B (Sigma-Aldrich) was added for 1 h at 4°C. The beads were washed in the same buffer, and proteins were eluted by boiling, loaded on a 10% polyacrylamide gel, transferred to membrane, and detected using either an antibody to the HA tag or carboxypeptidase Y.
RESULTS
rtn1Δ Affects the Structure of the Cortical ER
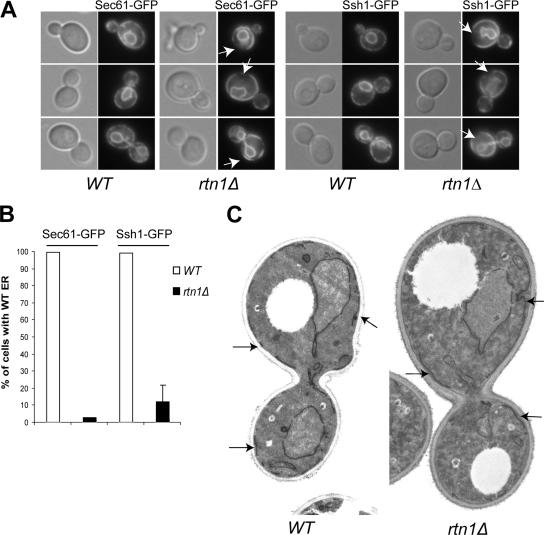
To identify new mutants affected in cortical ER structure and inheritance, we screened the yeast deletion library as described in Estrada et al. (2003) using the ER marker Hmg1-GFP. We selected the rtn1Δ mutant for further analysis because the ER was clearly aberrant in this strain. Although it did not have an ER inheritance defect as shown by characterization using two other ER markers, Sec61-GFP and Ssh1-GFP, both subunits of the translocon in yeast, the structure of the ER was dramatically altered in this mutant. In almost 100% of the wild-type cells, both markers show a very even punctate distribution along the cell cortex as well as continuous labeling of the perinuclear ER (Figure 1, A and B). The punctate pattern reflects a cross section view of the cortical network of ER tubules. In contrast, the cortical ER staining in the rtn1Δ mutant seemed to be more cisternal than reticular, as indicated by the continuous stretches of fluorescence, and separated by large cortical regions with little or no fluorescence (Figure 1A). Indeed <5% of the cells examined showed a wild-type structure of the cortical ER (Figure 1B). Although the cortical ER is contiguous with the nuclear envelope, there was no change evident in the distribution of the fluorescence around the nucleus.
Figure 1.
Rtn1p is required for the structure of the ER. (A) Wild type (NY2647, NY2649) and rtn1Δ mutant (NY2648, NY2650) cells expressing the ER marker Sec61-GFP or Ssh1-GFP were grown at 25°C in SC medium. The cells were examined using an epifluorescence microscope. Arrows point to cortical regions devoid of fluorescence. (B) Quantification of the altered cortical ER phenotype was done by scoring at least 300 cells with wild-type or altered morphology based on the presence of large gaps in fluorescence at the cell cortex and a more cisternal appearance of the cortical ER in an rtn1Δ mutant. The graph shows the percentage of cells having wild-type ER in each strain. (C) Thin section electron microscopy micrographs of wild-type (NY1210) and rtn1Δ (NY2651) cells grown in YPD and fixed with KMnO4 and embedded in Spurr. Arrows point to ER membrane stretches. Magnification, 4700×.
To better ascertain exactly how the cortical ER was altered, rtn1Δ cells were examined by thin section electron microscopy (EM) after permanganate treatment to highlight the ER. Using a double lattice grid, we measured the average length of cortical ER membrane segments in wild-type and in rtn1Δ cells (Figure 1C). In wild-type cells, the average length of an ER membrane segment is 0.6 ± 0.2 μm, whereas in the rtn1Δ cells the average length is 1.7 ± 1.3 μm. The ER segments in the mutant are thus threefold longer than in the wild-type, but also more variable, confirming that in an rtn1Δ mutant, the structure of the cortical ER is altered. This modification of the ER does not reflect a change in the overall surface area of the cortical ER. Indeed, the average surface of the ER in rtn1Δ cells is 1.6 ± 0.7 μm2/μm3, which is not significantly different from the 1.5 ± 0.6 μm2/μm3 measured in wild type. Other membrane-bound organelles were normal in appearance.
We also performed a 3D optical reconstruction of the cortical ER in wild-type and rtn1Δ cells using Sec61-GFP as an ER marker. As expected, in wild-type cells, Sec61-GFP localizes to a reticulated structure at the cell cortex that corresponds to the cortical ER as well as to a spherical structure in the cell that corresponds to the nucleus (Supplemental Figure S1). In contrast, in the rtn1Δ mutant, the nuclear structure is not affected, but the cortical ER seems more cisternal with large areas of the cortex devoid of ER (Supplemental Figure S2).
Previous work by Prinz et al. (2000) has shown that mutants affecting ER-to-Golgi transport in yeast result in a more cisternal ER morphology. More recently, Wakana et al. (2005) have shown that overexpression of RTN3 results in a block of the retrograde transport of ERGIC-53 in HeLa cells. Furthermore, Geng et al. (2005) have shown that Rtn1 interacts with Yip3, a membrane protein that interacts with the ER-to-Golgi Rab GTPase Ypt1 in S. cerevisiae (Schmitt et al., 1986; Bacon et al., 1989). Nonetheless, in that study, they did not observe alterations in trafficking of several biosynthetic markers in an rtn1Δ mutant. We have confirmed this by comparing the secretion of invertase in an rtn1Δ mutant to a wild type strain and found no difference (our unpublished data). This is consistent with the normal generation time of an rtn1Δ mutant. In total, these results suggest a role for Rtn1p in the maintenance of the cortical ER structure but not in ER membrane proliferation.
Effect of rtn1Δ on Other Organelles
Because the rtn1Δ mutant has a strong effect on the structure of the cortical ER, we looked at the structure of other organelles in the cell. We used Sec7-GFP as a late Golgi marker, which in wild type occurs as small punctae distributed throughout the cell (Seron et al., 1998). In the rtn1Δ mutant, we could not see any alteration in size, number, or distribution of the Sec7-GFP punctae compared with the wild type (Supplemental Figure S3A).
We next assessed the role of Rtn1p in the morphology of the vacuole using the vacuolar ATPase subunit Vph1-GFP (Urbanowski and Piper, 1999). In wild-type cells, the vacuole often occurs as a multilobed structure. In the rtn1Δ mutant, we could not see any alteration in size or distribution of the vacuole compared with the wild type (Supplemental Figure S3B).
Finally, we assessed the role of Rtn1p in the morphology of mitochondria using the mitochondrial ATPase subunit Atp9-DsRed as a marker. In wild-type cells, the mitochondria form a tubular structure that is apposed to the plasma membrane (Mozdy et al., 2000). In the rtn1Δ mutant, we could not see any alteration in shape or distribution of mitochondria compared with wild type (Supplemental Figure S3C). These results indicate that Rtn1p is specifically involved in the structure of the ER.
Rtn1p Is Enriched at the Cell Cortex
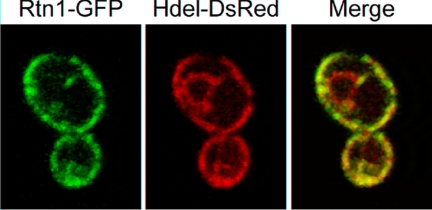
Previous studies have shown that homologues of Rtn1p in mammalian cells or in Caenorhabditis elegans cells are localized to the ER with only a small fraction at the plasma membrane in the case of the mammalian Rtn4 or Nogo (reviewed in Oertle et al., 2003). To localize Rtn1p in yeast, we fused GFP to its carboxy terminus and expressed this construct as the sole copy of RTN1. As shown in Figure 2, Rtn1-GFP was prominently localized at the cell cortex and in cytoplasmic tubules, but it was only marginally detected in the perinuclear region. This localization is consistent with an association of Rtn1-GFP with the cortical ER. However, it was important to establish that the chimeric protein was functional. Having no direct assay for function, we have relied on the fact that if Rtn1-GFP were mislocalized or otherwise nonfunctional, the ER would show a similar structure to that observed in an rtn1Δ mutant using Sec61-GFP as an ER marker. Because we have not observed any such alteration of the ER structure in strains expressing Rtn1-GFP, we conclude that the GFP tag does not significantly affect Rtn1p function. Geng et al. (2005) have reported a similar localization of Rtn1p. To quantify the enrichment of Rtn1-GFP at the cortex of the cell versus the perinuclear region, we performed a ratiometric analysis as described in Wang et al. (2002). We determined that there is a twofold enrichment of Rtn1-GFP at the cell cortex compared with the perinuclear region.
Figure 2.
Rtn1-GFP is enriched at the cortical ER. Wild-type cells (NY2658) expressing ER markers Rtn1-GFP and HDEL-DsRed were grown at 25°C in SC medium. The cells were examined using a confocal microscope.
Further proof of Rtn1-GFP functionality was obtained when we performed a 3D reconstruction of Rtn1-GFP localization (Supplemental Figure S4). This showed that Rtn1p is localized in a reticulated structure very similar to that observed with Sec61-GFP in wild-type cells (Supplemental Figure S1), supporting our hypothesis that Rtn1p is on the cortical ER. We also observe tubules emanating from this cortical structure oriented toward the center of the cell where the unlabeled nucleus resides.
Rtn1p Is an Integral ER Membrane Protein
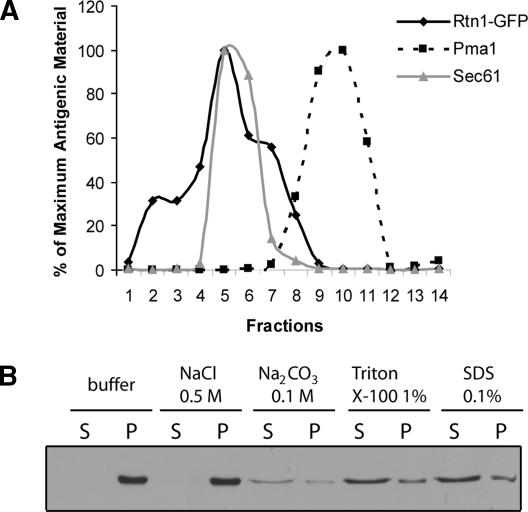
Our results so far strongly suggest that Rtn1p is a cortical ER protein. But to definitively rule out a plasma membrane localization of Rtn1-GFP, we fractionated a wild-type lysate on a sucrose density gradient and compared the distribution of Rtn1-GFP with that of markers for the ER and plasma membrane (Figure 3A). We found that Rtn1p fractionates as a single peak that overlaps with the Sec61p peak, a marker for the ER, and is resolved from the Pma1p peak, a plasma membrane marker. This confirms that Rtn1p is an ER-associated protein.
Figure 3.
Rtn1p is an ER integral membrane protein. (A) Wild-type cells (NY2658) expressing Rtn1-GFP were grown at 25°C in YPD. The lysate was loaded on top of a sucrose density gradient containing EDTA. Rtn1-GFP is represented by the solid black line, the ER marker Sec61p by the solid gray line, and the plasma membrane marker Pma1p by the dashed black line. (B) Wild-type cells (NY2659) expressing Rtn1-3HA were grown at 25°C in SC medium with. Cell lysates were incubated with reagents as indicated and separated into a pellet fraction P and a supernatant fraction S. The fractions were immunoblotted with anti-HA antibodies.
We then set out to determine whether Rtn1p is loosely associated with or tightly bound to the ER membrane, as are its mammalian homologues (Senden et al., 1994; Van de Velde et al., 1994). The region that identifies a protein as a member of the Rtn family is called the reticulon, a domain defined by two large (>30 aa) transmembrane domains separated by a conserved, hydrophilic loop of ∼60 amino acids (Oertle et al., 2003). In Rtn1p, the two transmembrane domains are poorly defined because they are predicted to be only weakly hydrophobic and the loop is ∼80 amino acids. To assess whether Rtn1p is indeed an integral membrane protein, we performed an extraction study on a total yeast lysate using high salt, high pH, nonanionic detergent and anionic detergent (Figure 3B). Rtn1p remained predominantly in the pellet in the presence of 0.5 M NaCl or 0.1 M Na2CO3 but shifted to the supernatant in the presence of either detergent, indicating that although it has nonconventional transmembrane domains, it is an integral membrane protein.
Overproduction of Rtn1p Alters the Distribution of Sec61-GFP
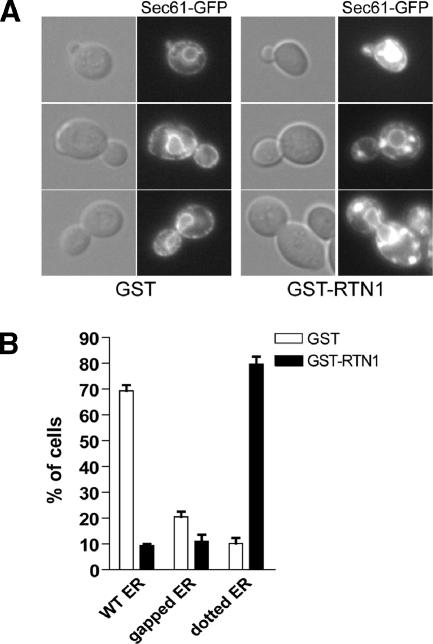
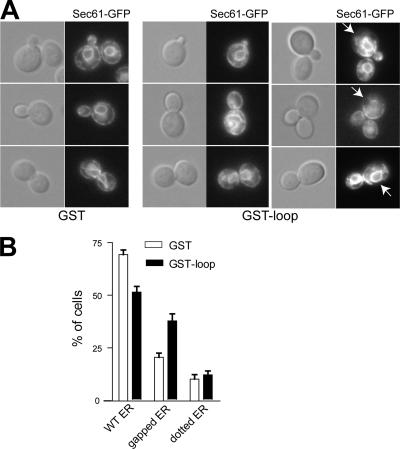
We have so far addressed the consequence of the loss of Rtn1p function, but it would also be interesting to determine whether the overproduction of Rtn1p has a demonstrable phenotype. To this effect, we transformed a wild-type strain expressing the ER marker Sec61-GFP with an intergrating plasmid containing the GAL1 promoter upstream of either GST or GST-Rtn1p. As shown in Figure 4, A and B, when the strain overproducing GST was grown on galactose-containing medium, 70% of the cells showed a wild-type structure for the ER. But when GST-Rtn1p was overproduced in the same strain, we observed round and bright Sec61-GFP–containing structures in the cytoplasm. These structures differ in appearance from karmellae, the multilayered ER membrane surrounding the nucleus that occurs in response to overproduction of Hmg1p (Wright et al., 1988).
Figure 4.
Rtn1 overproduction alters ER structure. (A) Wild-type strain expressing the ER marker Sec61-GFP and either GST (NY2660), GST-Rtn1 (NY2661) under the control of the GAL1 were grown at 25°C in SC galactose medium. The cells were examined using an epifluorescence microscope. Arrows point to fluorescence patches. (B) Quantification of the altered cortical ER phenotype was done by scoring at least 300 cells with wild-type or altered morphology based on the presence of large puncta or the presence of large gaps in fluorescence at the cell cortex. The graph shows the percentage of cells having either ER structure.
TAP Tag Purification of the Exocyst and Mass Spectrometry
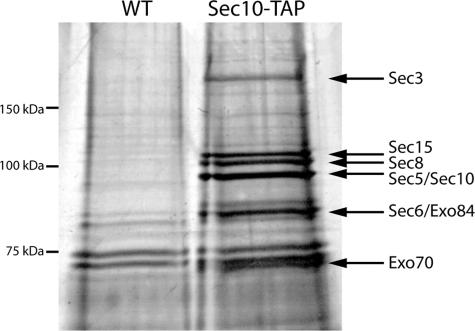
In an independent study we sought to identify novel proteins that interact with the exocyst complex. The Sec10p subunit was tagged with a C-terminal TAP tag, and this construct was expressed as the sole copy of SEC10. Sec10-TAP was purified from a 2-liter culture of using a variation of the published methods (Puig et al., 2001). In preliminary trials, we performed the isolation over a pH range and found pH 6.8 to be optimal for maintaining the octameric complex intact. This is consistent with our earlier observations regarding immunoisolation of the exocyst (Terbush et al., 1996). Twenty percent of the purification was loaded on an SDS gel and after silver staining, eight bands were identified above the background bands that were present in a mock isolation from an untagged strain (Figure 5). Four of the bands were confirmed as exocyst subunits by Western blot using Sec6p, Sec8p, Sec10p, and Sec15p polyclonal antibodies (Salminen and Novick, 1989; Bowser et al., 1992; Potenza et al., 1992; our unpublished data). Antibodies to the other four subunits were not available.
Figure 5.
TAP tag isolation of the intact exocyst complex. The exocyst complex was purified from 2 liters of culture using a C-terminal TAP tag on Sec10p. Twenty percent of the sample was run on a 7% polyacrylamide gel and silver stained. The eight exocyst subunits are identified with arrows. Proteins isolated from an untagged strain are shown for comparison.
The yield from 2 liters of culture was ∼2–4 μg of intact exocyst complex based on a comparison with known amounts of bovine serum albumin protein. Therefore, each subunit comprises 0.06% of the 600 mg of soluble protein used in the purification. A similar abundance was calculated when the exocyst was purified using a 3xmyc tag (Terbush et al., 1996). Based on band intensity of the silver stain gel (Figure 5), all of the subunits except Sec3p seem to be approximately stoichiometric. The reduced amount of Sec3p may be a result of degradation, as was observed by Terbush et al. (1996), or it may partially dissociate during purification.
Tandem mass spectrometry was performed on the purified exocyst preparation and on the control sample from the untagged strain to identify any novel proteins that were coisolated with the complex (Eng et al., 1994). One-half of the above-mentioned purification was precipitated and sent to the Proteomic Mass Spectrometry Laboratory at the Scripps Research Institute. Peptides present in the mixture were identified through a search of the protein database using the algorithm SEQUEST. An array of 135 proteins was identified in the tagged sample as opposed to 105 in the untagged control. All eight exocyst subunits were identified with the percent of coverage being higher (31–36%) for the smaller proteins (Exo70 and Exo84), and in the range of 17–26% for the other six proteins (Table 2). Most of the background proteins (44%) were found to be ribosomal in origin. Sixteen additional proteins specific to the exocyst isolation were noted and are available on the Yeast Resource Center’s public data repository Web page (http://www.yeastrc.org/pdr/). Among the 16 proteins specific to the exocyst TAP tag purification was Rtn1p. Mass spectrometry identified 13.2% coverage of the 34-kDa protein, approximately one-half the coverage identified for most exocyst subunits.
Table 2.
Results of mass spectrometry on the exocyst complex purification
| Isolated protein | % Coverage |
|---|---|
| Sec3 | 22.5 |
| Sec5 | 25.8 |
| Sec6 | 17.3 |
| Sec8 | 21.5 |
| Sec10 | 21.5 |
| Sec15 | 18.8 |
| Exo70 | 35.8 |
| Exo84 | 30.9 |
| Rtn1 | 13.2 |
Tandem mass spectrometry was performed on 50% of the exocyst purification in Figure 5. All eight exocyst subunits were identified as well as the novel protein Rtn1. The percentage of coverage of the proteins isolated is listed.
In Vitro Binding Assays for Rtn1
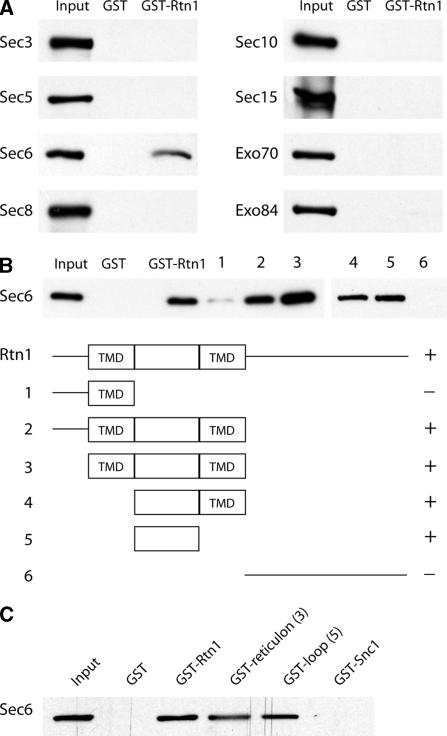
To determine whether there was a direct interaction between the exocyst complex and Rtn1 and to define the interacting subunit, all eight exocyst subunits were separately expressed in bacteria, and purified using a 6xHis tag. Expression levels varied, with Sec6p, Exo70p, and Exo84p being purified in higher quantities than the other five subunits from the same volume of culture (our unpublished data). Rtn1p was N-terminally tagged with GST, expressed in bacteria and purified with glutathione beads. As a control, the GST protein was also purified.
Approximately equal amounts of soluble exocyst proteins and either GST or GST-Rtn1 immobilized on glutathione beads was incubated together for 2 h. The beads were washed, and bound proteins were eluted, loaded on gels, transferred to membranes, and probed with anti-His antibody. Figure 6A shows that Sec6p is the only exocyst subunit that detectably binds Rtn1p. This result also implies that there is a direct interaction between the exocyst complex and Rtn1p. By comparison to a lane loaded with 30% of the amount of Sec6 input, we conclude that ∼15% of Sec6 binds to GST-Rtn1p under the conditions used. None of the exocyst proteins were found to bind to GST alone.
Figure 6.
(A) Rtn1p binds to the Sec6p subunit of the exocyst. Soluble recombinant exocyst proteins tagged with 6xHis were incubated with recombinant GST or GST-Rtn1p immobilized on glutathione beads. Approximately 15% of the available Sec6p binds to Rtn1p based in comparison with the 30% input loaded on Western blots. (B) Sec6p binds to the hydrophilic loop of Rtn1p. A deletion series of the Rtn1 protein was constructed and expressed in E. coli. The proteins, immobilized on glutathione beads, were incubated with recombinant 6xHis-Sec6. The hydrophilic loop between the two transmembrane domains (TMD) of Rtn1 was capable of binding 30% of the available Sec6 protein. (C) As a control, GST-Snc1p was included in the binding assay. Again, the Rtn1 loop was sufficient to bind 30% of the available Sec6p, whereas no binding was observed between Sec6p and GST-Snc1p or GST alone.
To determine which domain of Rtn1p binds to Sec6p, six different sections of the protein were used in an in vitro binding assay. Two transmembrane domains in Rtn1p are predicted according to the Saccharomyces Genome Database (http://www.yeastgenome.org/). These two domains, surrounding an 84-amino acid hydrophilic loop, constitute the reticulon domain. Recombinant proteins containing the reticulon domain were found to bind Sec6p, whereas proteins with only the N or C terminus did not (Figure 6B). When the reticulon domain was broken down even further, the hydrophilic loop between the transmembrane domains was found to be sufficient to bind Sec6p. Snc1p, a small transmembrane domain protein involved in fusion of secretory vesicles, was expressed as a GST fusion protein and used as a control. No binding between Sec6p and Snc1p was observed under the conditions used (Figure 6C).
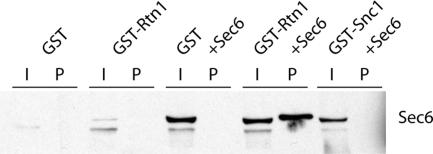
Overexpression of Sec6p and Rtn1p
We next tested whether Rtn1p binds to Sec6p in yeast. Diploid strains were constructed using the GAL1 promoter to overexpress either GST alone, GST-Rtn1p alone, GST and Sec6p, GST-Rtn1p and Sec6p, or GST-Snc1p and Sec6p as a control. If a direct interaction between Sec6p and Rtn1p occurs in vivo, it should be exaggerated when the levels of both these proteins are increased. Glutathione beads were used to isolate GST, GST-Snc1p, or GST-Rtn1p from a culture grown overnight in 2% galactose. Figure 7 shows a Western blot detecting Sec6p in the five strains. The amount of Sec6p is much higher when driven by the GAL1 promoter, and therefore not as readily detected in the strains expressing endogenous levels. When Rtn1p and Sec6p were cooverexpressed, ∼0.5% of the Sec6p copurified with Rtn1p. The band seems to run slightly higher in the last lane due to the coincidence that Sec6p is similar in size to the large amount of Rtn1p isolated, causing the Sec6p band to slightly shift. No copurification of Sec6p with Snc1p was identified under these conditions. This finding, along with the mass spectrometry result, suggests that the exocyst and Rtn1p can interact in vivo.
Figure 7.
Coprecipitation of Rtn1p and Sec6p when both proteins are overexpressed. The GAL1 promoter was used to overexpress GST, GST-Snc1p, GST-Rtn1p, Sec6p, or combinations of these proteins. GST, GST-Snc1p, or GST-Rtn1p was isolated on glutathione bead, and Sec6p was detected on a Western blot using a Sec6p antibody. The amount of Sec6p is much lower when not driven by the GAL1 promoter, and therefore not as readily detected in samples with endogenous levels. When both Rtn1p and Sec6p are overexpressed in the same strain, ∼0.5% of the available Sec6p (I) is purified (P) with Rtn1p. This band is slightly higher because Sec6p is similar in size to the large amount of Rtn1p isolated, thus causing a slight shift in mobility. No copurification between GST or GST-Snc1 and Sec6 was observed.
Generation of Glycosylation Sites in Rtn1
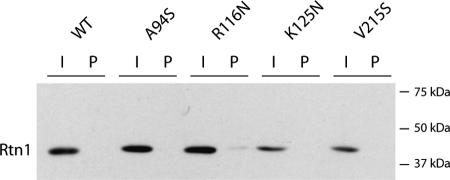
If the hydrophilic loop within the reticulon domain of Rtn1p interacts with the exocyst, as our binding experiments imply, then we would predict that loop to be oriented toward the cytosol rather than toward the lumen of the ER, because the exocyst is a cytosolic complex. To determine the orientation of the yeast reticulon loop, three different asparagine-linked glycosylation sites were added to the Rtn1p loop at amino acids 92, 116, and 125. A fourth glycosylation site was added near the C terminus of the protein at amino acid 213. If any of these regions were exposed to the lumen of the ER, we would expect them to be glycosylated. Such glycosylation would be revealed by a shift in mobility on a Western blot using a C-terminal HA tag on Rtn1p and by precipitation using ConA-Sepharose beads, which bind specifically to mannosyl and glucosyl residues of polysaccharides and glycoproteins in the presence of calcium and manganese ions.
All four mutagenized constructs, as well as wild-type RTN1, were introduced by transformation into an rtn1Δ strain. Soluble protein was isolated and bound to ConA-Sepharose. Unbound protein (0.25% of total volume) was loaded beside protein (2.5%) that bound to ConA beads on a polyacrylamide gel and a C-terminal HA tag was used to detect Rtn1p. Not only did none of the engineered glycosylation sites cause a shift in mobility of Rtn1p, none of them caused Rtn1p to bind to the ConA beads (Figure 8). As a control, a parallel filter was immunoblotted using an antibody directed against carboxypeptidase Y, a protein that is known to be glycosylated. Carboxypeptidase Y was efficiently bound by the ConA beads. Although this evidence does not conclusively prove that these domains of Rtn1p are exposed to the cytosol, they do suggest that they are not exposed to the lumen of the ER and therefore are likely to be appropriately oriented to interact with the exocyst.
Figure 8.
Orientation of the Rtn1 loop. Three different asparagine-linked glycosylation sites were created in the loop by mutating amino acids 94, 116, and 125 to a serine, asparagine, and asparagines, respectively. A fourth glycosylation site was added near the C terminus of the protein by mutating amino acid 215 to a serine. Soluble protein was isolated and unbound protein (I; 0.25% of total volume) was loaded beside protein (P; 2.5%) that bound to ConA beads. A C-terminal HA tag was used to detect Rtn1p. None of the engineered glycosylation sites caused a shift in mobility, and likewise none of them caused Rtn1p to bind to ConA beads. This negative result suggests these domains are not exposed to the lumen and therefore likely oriented to the cytosol.
The Rtn1p Loop Is Involved in Organizing the Structure of the ER
So far, we have shown that the loop of Rtn1p is cytoplasmic and interacts with the exocyst subunit Sec6p. To assess the function of this loop in ER organization, we transformed a wild-type strain expressing Sec61-GFP with an integrative plasmid carrying the GAL1 promoter to overproduce either GST or GST-loop. As shown in Figure 9A, overproduction of GST alone did not strongly affect the perinuclear and cortical localization of Sec61-GFP (Figure 9B). We did note an increase in the percentage of cells showing a gap in the cortical ER that correlated with growth in galactose; however, in general, the ER structure was very similar to that observed when cells are grown on glucose. Alternatively, the overproduction of the loop resulted in a significant increase in the number of cells having an altered ER structure, similar to that observed in rtn1Δ cells (Figure 9A), yet not as dramatic. Indeed, 50% of the cells had a wild-type ER structure and 40% had a structure reminiscent of the rtn1Δ mutant (Figure 9B). This supports the possibility that this loop is involved in organizing the structure of the ER.
Figure 9.
Rtn1p loop overproduction affects ER structure. (A) Wild-type strain expressing the ER marker Sec61-GFP and either GST (NY2660) or GST-loop (NY2662) under the control of the GAL1 was grown at 25°C in SC galactose medium. The cells were examined using an epifluorescence microscope. Arrows point to fluorescence patches. (B) Quantification of the altered cortical ER phenotype was done by scoring at least 300 cells with wild-type or altered morphology based on the presence of large puncta or the presence of large gaps in fluorescence at the cell cortex. The graph shows the percentage of cells having either ER structure.
Rtn1p Loop Overproduction Leads to an Accumulation of Secretory Vesicles
Last, we addressed the question as to whether this interaction between the loop of Rtn1p and Sec6p has any effect on the role of Sec6p in secretion. We compared the percentage of invertase secreted by wild-type cells transformed with a 2μ plasmid bearing GST under the strong GPD1 promoter to wild type transformed with a plasmid bearing the GST-loop in a parallel construct. We measured no significant difference in the secretion of invertase between the wild type overproducing GST and the wild type overproducing GST-loop (our unpublished data). This indicated that no major effect on secretion resulted from the overproduction of GST-loop. We did notice that overproduction of the GST-loop resulted in slower growth compared with GST overproduction.
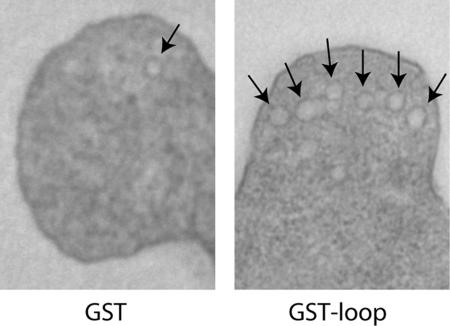
As a more sensitive test of secretory function, we examined thin sections of the same strains by electron microscopy. By quantitative analysis of the number of vesicles in buds of both strains, we determined that in the strain overproducing GST, the vesicle density was 0.82 vesicles/μm2, whereas overproduction of GST-loop led to a density of 2.84 vesicles/μm2 (Figure 10). This suggests a very mild but significant increase.
Figure 10.
Rtn1p loop overproduction interferes with secretion. Thin section EM micrographs of wild type transformed either with a 2μ plasmid bearing GST or GST-loop under the control of the GPD1 promoter (NY2678 and NY2679). Cells were grown on SC medium and fixed with glutaraldehyde and stained with OsO4 and embedded in Spurr. Arrows point to vesicles. Magnification, 6500×.
Finally, we assessed the effect of the overproduction of GST or GST-loop on the cut-off temperature of all late acting sec mutants as well as one mutant affecting each of the earlier steps of the secretory pathway. As shown in Table 3, mutants sec2-41, sec6-4, sec8-9, sec5-24, and sec15-1 are the most strongly affected by the overproduction of the loop. Alternatively, sec1-1 and more surprisingly sec3-2 are only weakly affected, if at all. We could not assess the effect on sec9-4 because its cut-off temperature is 30°C. In all, we did observe a genetic interaction between the late-acting sec mutants and Rtn1 loop. These results support our hypothesis that the hydrophilic loop of Rtn1p interacts with Sec6p.
Table 3.
Rtn1 loop overproduction decreases the cut-off temperature of most sec mutants
| Mutant | GST (°C) | GST-loop (°C) |
|---|---|---|
| sec1-1 | 34 | 34 |
| sec2-41 | 34 | 30 |
| sec3-2 | 34 | 34 |
| sec5-24 | 34 | 30 |
| sec6-4 | 34 | 30 |
| sec8-9 | 34 | 30 |
| sec9-4 | 30 | 30 |
| sec10-2 | 30 | 34 |
| sec15-1 | 34 | 30 |
DISCUSSION
We have addressed the role of Rtn1p, a member of the Rtn family of proteins, in the structure and organization of the ER. In yeast, under normal circumstances, the cortical ER is exclusively reticulated and closely apposed to the plasma membrane (reviewed in Baumann and Walz, 2001; Prinz et al., 2000). We have shown that deletion of the RTN1 gene results in the reorganization of the cortical ER from a highly reticulated structure to a more cisternal one, with no significant change in the total membrane surface area of the ER. A similar observation was recently reported by Voeltz et al. (2006), although with some notable differences. To see a similar phenotype, they had to delete both RTN1 and its paralog RTN2 and either add 1 M NaCl to the growth medium or delete YOP1. As shown in Supplemental Figure S5A and S5B), in our strain background, RTN2 is not expressed in normal media and is induced by 1 M NaCl, as reported previously (Posas et al., 2000; Causton et al., 2001). We did not see any significant reversal of the rtn1Δ phenotype upon induction of RTN2 with 1 M NaCl (Supplemental Figure S6A and S6B). In constrast, we did note a weakening of the rtn1Δ phenotype during growth in the presence of salt (Supplemental Figure S6A and S6B). It is likely that additional components contribute to the reticular structure of the ER, and there seem to be some differences in strain backgrounds with respect to the relative importance of these additional components. Nonetheless, both studies point to a central role for Rtn1p in the structure of the ER.
Prinz et al. (2000) have observed that mutations that inhibit ER-to-Golgi transport result in the formation of a largely cisternal ER. Such mutations also result in dramatic expansion of the surface area of the ER as membrane destined for export accumulates in the ER (Novick et al., 1980). In contrast, rtn1Δ cells exhibit no significant increase in the surface area of the ER, despite the dramatic change in ER organization. This is consistent with the observation that there is no intracellular accumulation of the secreted protein invertase (our unpublished data) or any slowing in the growth rate of rtn1Δ cells, as would be expected of a mutation affecting ER export. Therefore, it is likely that the change in ER organization observed in rtn1Δ cells reflects a direct role for Rtn1p, rather than an indirect effect of a transport block.
Rtn proteins are defined by the presence of a reticulon domain, consisting of two transmembrane domains linked by a conserved hydrophilic loop (Oertle et al., 2003). All members analyzed to date localize exclusively to the ER with the sole exception of the Rtn4 protein, which is also partially localized to the plasma membrane. The localization of Rtn1-GFP that we report here and that Geng et al. (2005) have recently reported is consistent with this pattern. Interestingly, Rtn1-GFP is enriched in the cortical ER relative to the nuclear envelope. This enrichment could either reflect the partial exclusion of Rtn1-GFP from the nuclear envelope or the partial retention of Rtn1-GFP at the cell cortex. Additional studies will be needed to resolve these two possibilities.
We have also shown that Rtn1p, like its mammalian homologues, behaves as an integral membrane protein in extraction studies. Furthermore, we show that the hydrophilic loop connecting the two transmembrane domains is most likely cytoplasmic in its orientation, because it does not seem to be accessible to the glycosylation machinery in the lumen of the ER. This orientation is opposite to that proposed for its mammalian homologues (GrandPre et al., 2000). However, that proposal was based on the orientation of the small pool of Rtn4 that reaches the plasma membrane, which may differ in orientation from the bulk of Rtn4p that remains in the ER. More recently, Voeltz et al. (2006) have shown in COS cells that the NogoC loop as well as its hydrophilic N terminus faces the cytoplasm, in agreement with our topology for Rtn1p in yeast.
The identification of Rtn1p as a binding partner of the exocyst is very interesting in light of the documented role of the exocyst in ER inheritance. Previous work has shown that at least three subunits of the exocyst, Sec3p, Sec5p, and Sec8p, are involved in ER inheritance (Wiederkehr et al., 2003, 2004; Reinke et al., 2004). In sec3Δ cells, an ER tubule often migrates into the bud, but it fails to be retained at the bud tip, as it is in wild-type cells (Wiederkehr et al., 2003). This phenotype has suggested a model in which the exocyst serves as an anchor for the ER at the bud tip early in the cell cycle; however, the mechanism by which the ER becomes attached to this anchor has been unclear. One suggestion has been that the translocon serves as the link to the exocyst, because it was also shown that the translocon subunit Sec61p interacts with the exocyst (Toikkanen et al., 2003; Lipschutz et al., 2003). Our binding data suggest that Rtn1p might also serve as a receptor for the exocyst on the ER. The cytoplasmic orientation of the hydrophilic loop of Rtn1p is consistent with our identification of this loop as a binding partner for the Sec6p subunit of the exocyst. In addition, the apposition of the ER to the plasma membrane in yeast is also consistent with an interaction of the exocyst with Rtn1p. The elongated shape of the exocyst (Hsu et al., 1998; Dong et al., 2005) would allow it to bridge the small gap between the two membranes. Nonetheless, we did not observe a clear ER inheritance defect in rtn1Δ cells, suggesting either that Rtn1p is redundant with the translocon in this function or that the Rtn1–exocyst interaction serves a function other than anchoring the ER at the bud tip. The exocyst might signal to Rtn1p to direct the formation of reticular ER along the cortex of the bud from the ER tubules that have attached to the bud tip. Thus, in an rtn1Δ mutant, the ER would still migrate into the bud and spread along the cortex, but it would not be able to form a reticulum.
It is noteworthy that in mammalian tissues, cell function dictates the structure of the ER. Cell types that specialize in protein secretion have a greater amount of rough ER, which is typically cisternal in structure. In contrast, cells types that are involved in hormone synthesis, fat metabolism, calcium storage, and detoxification have more smooth ER, which is typically reticulated in structure. Thus, the morphology of this organelle is regulated by cell function (reviewed in Baumann and Walz, 2001). In some cell types, this regulation is dynamic. In hepatocytes for instance, the ER is more cisternal in unchallenged cells but upon drug challenge, the reticulated portion increases (Remmer and Merker, 1963). It will be important to determine whether any or all of the Rtn proteins in mammalian cells act to regulate the structure of the ER and whether the differential expression of the four Rtn proteins and their many isoforms (Roebroek et al., 1998; GrandPre et al., 2000) determines the structure of the ER in different cell types.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Chalouni for help with confocal microscopy and A. Hartley for the 3D reconstructions. We also like thank Prof. A. Merz for help with ratiometrics. This work was supported by National Institutes of Health Grants GM-73892 and GM35370 (to P. N.) and CA-46128 (to S.F.-N.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0080) on April 19, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bacon R. A., Salminen A., Ruohola H., Novick P., Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J. Cell Biol. 1989;109:1015–1022. doi: 10.1083/jcb.109.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O., Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- Bowser R., Muller H., Govindan B., Novick P. Sec8p and Sec15p are components of a plasma membrane-associated 19.5S particle that may function downstream of Sec4p to control exocytosis. J. Cell Biol. 1992;118:1041–1056. doi: 10.1083/jcb.118.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., Jennings E. G., Lee T. I., True H. L., Lander E. S., Young R. A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. S., Huber A. B., van der Haar M. E., Frank M., Schnell L., Spillmann A. A., Christ F., Schwab M. E. Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for mAb IN-1. Nature. 2000;403:434–439. doi: 10.1038/35000219. [DOI] [PubMed] [Google Scholar]
- Dong G., Hutagalung A. H., Fu C., Novick P., Reinisch K. M. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat. Struct. Mol. Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- Du Y., Pypaert M., Novick P., Ferro-Novick S. Aux1p/Swa2p is required for cortical endoplasmic reticulum inheritance in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2614–2628. doi: 10.1091/mbc.12.9.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J., McCormack A., Yates J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spetrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Estrada P., Kim J., Coleman J., Walker L., Dunn B., Takizawa P., Novick P., Ferro-Novick S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher K. L., Davis D., Wu M., Boldogh I., Pon L. A. Endoplasmic reticulum dynamics, inheritance, and cytoskeletal interactions in budding yeast. Mol. Biol. Cell. 2002;13:854–865. doi: 10.1091/mbc.01-04-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Shin M. E., Gilbert P. M., Collins R. N., Burd C. G. Saccharomyces cerevisiae Rab-GDI displacement factor ortholog Yip3p forms distinct complexes with the Ypt1 Rab GTPase and the reticulon Rtn1p. Eukaryot. Cell. 2005;4:1166–1174. doi: 10.1128/EC.4.7.1166-1174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst J. E., Rodgers L., Riggs M., Wigler M. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc. Natl. Acad. Sci. USA. 1992;89:4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St. Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GrandPre T., Nakamura F., Vartanian T., Strittmatter S. M. Identification of the Nogo inhibitor of axon regeneration as a reticulon protein. Nature. 2000;403:439–444. doi: 10.1038/35000226. [DOI] [PubMed] [Google Scholar]
- Greenfield J. J., High S. The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J. Cell Sci. 1999;112:1477–1486. doi: 10.1242/jcs.112.10.1477. [DOI] [PubMed] [Google Scholar]
- Gunthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. London: Academic Press; 1991. [Google Scholar]
- Hamada N., Iwahashi J., Suzuki K., Ogi H., Kashiwagi T., Hara K., Toyoda M., Yamada T., Toyoda T. Molecular cloning and characterization of the mouse reticulon 3 cDNA. Cell. Mol. Biol. (Noisy-le-grand) 2002;48:163–172. [PubMed] [Google Scholar]
- Hsu S. C., Hazuka C. D., Roth R., Foletti D. L., Heuser J., Scheller R. H. Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron. 1998;20:1111–1122. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Lee C., Chen L. B. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54:37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- Lipschutz J. H., Lingappa V. R., Mostov K. E. The exocyst affects protein synthesis by acting on the translocation machinery of the endoplasmic reticulum. J. Biol. Chem. 2003;278:20954–20960. doi: 10.1074/jbc.M213210200. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- MacCoss M. J., et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira E. F., Jaworski C. J., Rodriguez I. R. Cloning of a novel member of the reticulon gene family (RTN3): gene structure and chromosomal localization to 11q13. Genomics. 1999;58:73–81. doi: 10.1006/geno.1999.5807. [DOI] [PubMed] [Google Scholar]
- Mozdy A. D., McCaffery J. M., Shaw J. M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Oertle T., Klinger M., Stuermer C. A., Schwab M. E. A reticular rhapsody: phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 2003;17:1238–1247. doi: 10.1096/fj.02-1166hyp. [DOI] [PubMed] [Google Scholar]
- Palade G. E. The endoplasmic reticulum. J. Biophys. Biochem. Cytol. 1956;2:85–98. doi: 10.1083/jcb.2.4.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Chambers J. R., Heyman J. A., Hoeffler J. P., De N. E., Arino J. The transcriptional response of yeast to saline stress. J. Biol. Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Potenza M., Bowser R., Muller H., Novick P. SEC6 encodes an 85 kDa soluble protein required for exocytosis in yeast. Yeast. 1992;8:549–558. doi: 10.1002/yea.320080706. [DOI] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Kaiser C. A., Orlean P., Albright C., Rose M. D., Robbins P. W., Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Prinjha R., Moore S. E., Vinson M., Blake S., Morrow R., Christie G., Michalovich D., Simmons D. L., Walsh F. S. Inhibitor of neurite outgrowth in humans. Nature. 2000;403:383–384. doi: 10.1038/35000287. [DOI] [PubMed] [Google Scholar]
- Prinz W. A., Grzyb L., Veenhuis M., Kahana J. A., Silver P. A., Rapoport T. A. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Reinke C. A., Kozik P., Glick B. S. Golgi inheritance in small buds of Saccharomyces cerevisiae is linked to endoplasmic reticulum inheritance. Proc. Natl. Acad. Sci. USA. 2004;101:18018–18023. doi: 10.1073/pnas.0408256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmer H., Merker H. J. Drug-induced changes in the liver endoplasmic reticulum: association with drug-metabolizing enzymes. Science. 1963;142:1657–1658. doi: 10.1126/science.142.3600.1657. [DOI] [PubMed] [Google Scholar]
- Roberg K. J., Rowley N., Kaiser C. A. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebroek A. J., Contreras B., Pauli I. G., Van de Ven W.J.M. cDNA cloning, genomic organization, and expression of the human RTN2 gene, a member of a gene family encoding reticulons. Genomics. 1998;51:98–106. doi: 10.1006/geno.1997.5175. [DOI] [PubMed] [Google Scholar]
- Salminen A., Novick P. J. The Sec15 protein responds to the function of the GTP binding protein, Sec4, to control vesicular traffic in yeast. J. Cell Biol. 1989;109:1023–1036. doi: 10.1083/jcb.109.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manuel. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Schmitt H. D., Wagner P., Pfaff E., Gallwitz D. The ras-related YPT1 gene product in yeast: a GTP-binding protein that might be involved in microtubule organization. Cell. 1986;47:401–412. doi: 10.1016/0092-8674(86)90597-0. [DOI] [PubMed] [Google Scholar]
- Senden N. H., Van de Velde H., Broers J. L., Timmer E. D., Kuijpers H. J., Roebroek A. J., Van de Ven W.J.M., Ramaekers F. C. Subcellular localization and supramolecular organization of neuroendocrine-specific protein B (NSP-B) in small cell lung cancer. Eur. J. Cell Biol. 1994;65:341–353. [PubMed] [Google Scholar]
- Seron K., et al. A yeast t-SNARE involved in endocytosis. Mol. Biol. Cell. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D. L., McDonald W. H., Yates J. R., III DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M. Recent progress on structural interactions of the endoplasmic reticulum. Cell Motil. Cytoskeleton. 1990;15:71–75. doi: 10.1002/cm.970150203. [DOI] [PubMed] [Google Scholar]
- Terasaki M., Chen L. B., Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J. Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M., Jaffe L. A. Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J. Cell Biol. 1991;114:929–940. doi: 10.1083/jcb.114.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M., Jaffe L. A., Hunnicutt G. R., Hammer J. A., III Structural change of the endoplasmic reticulum during fertilization: evidence for loss of membrane continuity using the green fluorescent protein. Dev. Biol. 1996;179:320–328. doi: 10.1006/dbio.1996.0263. [DOI] [PubMed] [Google Scholar]
- Terbush D. R., Maurice T., Roth D., Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Toikkanen J. H., Miller K. J., Soderlund H., Jantti J., Keranen S. The beta subunit of the Sec61p endoplasmic reticulum translocon interacts with the exocyst complex in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:20946–20953. doi: 10.1074/jbc.M213111200. [DOI] [PubMed] [Google Scholar]
- Urbanowski J. L., Piper R. C. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 1999;274:38061–38070. doi: 10.1074/jbc.274.53.38061. [DOI] [PubMed] [Google Scholar]
- Van de Velde H., Roebroek A. J., Senden N. H., Ramaekers F. C., Van de Ven W.J.M. NSP-encoded reticulons, neuroendocrine proteins of a novel gene family associated with membranes of the endoplasmic reticulum. J. Cell Sci. 1994;107:2403–2416. doi: 10.1242/jcs.107.9.2403. [DOI] [PubMed] [Google Scholar]
- Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Wakana Y., Koyama S., Nakajima K., Hatsuzawa K., Nagahama M., Tani K., Hauri H. P., Melancon P., Tagaya M. Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem. Biophys. Res. Commun. 2005;334:1198–1205. doi: 10.1016/j.bbrc.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Wang L., Seeley E. S., Wickner W., Merz A. J. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- Wiederkehr A., De Craene J. O., Ferro-Novick S., Novick P. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J. Cell Biol. 2004;167:875–887. doi: 10.1083/jcb.200408001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., Du Y., Pypaert M., Ferro-Novick S., Novick P. Sec3p is needed for the spatial regulation of secretion and for the inheritance of the cortical endoplasmic reticulum. Mol. Biol. Cell. 2003;14:4770–4782. doi: 10.1091/mbc.E03-04-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R., Basson M., D’Ari L., Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.