Abstract
The unfolded protein response (UPR) regulates the protein-folding capacity of the endoplasmic reticulum (ER) according to cellular demand. In mammalian cells, three ER transmembrane components, IRE1, PERK, and ATF6, initiate distinct UPR signaling branches. We show that these UPR components display distinct sensitivities toward different forms of ER stress. ER stress induced by ER Ca2+ release in particular revealed fundamental differences in the properties of UPR signaling branches. Compared with the rapid response of both IRE1 and PERK to ER stress induced by thapsigargin, an ER Ca2+ ATPase inhibitor, the response of ATF6 was markedly delayed. These studies are the first side-by-side comparisons of UPR signaling branch activation and reveal intrinsic features of UPR stress sensor activation in response to alternate forms of ER stress. As such, they provide initial groundwork toward understanding how ER stress sensors can confer different responses and how optimal UPR responses are achieved in physiological settings.
INTRODUCTION
The endoplasmic reticulum (ER) is an initial checkpoint for the folding and modification of proteins that reside within the secretory pathway, because only properly folded and modified proteins can exit the ER (Gilmore, 1993; Walter and Johnson, 1994; Voeltz et al., 2002). Depending upon environmental changes or developmental stages, the capacity of the ER to perform protein folding must adjust according to cellular needs. The unfolded protein response (UPR) signaling pathway plays a major role in this regulation by sensing the ER lumen environment and transmitting this information to the nucleus to up-regulate transcription of genes that increase both ER protein folding and secretory capacities (Kaufman, 1999; Mori, 2000; Patil and Walter, 2001; Harding et al., 2002).
The yeast S. cerevisiae senses unfolded proteins with a single sensor, IRE1 (Cox, 1993; Mori, 1993). In mammalian cells, the UPR response is induced by three initiator/sensor molecules on the ER membrane, IRE1, PERK, and ATF6. Each sensor undergoes activation in response to elevated levels of unfolded proteins. IRE1 is an ER transmembrane receptor with kinase and endoribonuclease domains in its cytoplasmic C-terminal portion. IRE1 senses the protein folding needs of the ER through its N-terminal lumenal domain (Cox et al., 1993; Mori et al., 1993; Tirasophon et al., 1998; Wang et al., 1998). Once sensed, this signal is transmitted across the ER membrane to the IRE1 cytosolic domain, causing IRE1 oligomerization and autophosphorylation (Shamu and Walter, 1996; Papa et al., 2003). A unique feature of IRE1 is its function as an endoribonuclease (RNase) which, after activation by unfolded proteins, mediates the cleavage step in the nonconventional splicing of XBP1 mRNA encoding a bZiP transcription factor (Sidrauski and Walter, 1997; Shen et al., 2001; Yoshida et al., 2001; Calfon et al., 2002). Removal of the UPR intron in XBP1 causes a frame-shift, producing an XBP1 protein that is a more potent transcription factor and is an obligate step in the UPR pathway (Yoshida et al., 2001).
A second initiator, PERK, is a type-I ER-transmembrane kinase that phosphorylates the α subunit of eukaryotic translation initiation factor 2 (eIF2α; Shi et al., 1998; Harding et al., 1999). Phosphorylated eIF2α interferes with the formation of the 43S translation initiation complex, leading to overall translational repression in UPR-induced cells; presumably to alleviate ER stress by reducing the influx of the newly synthesized proteins into the ER (Harding et al., 2000a). Moreover, when eIF2α is phosphorylated, a second transcription factor, ATF4, is produced preferentially (Harding et al., 2000b; Scheuner et al., 2001a). In addition to XBP1, ATF4 also participates in transcription of some UPR target genes.
A third component transmitting UPR activating signals from the ER in mammalian cells is ATF6, a 90-kDa type-II ER-membrane transcription factor also required for activating many UPR target genes (Li et al., 2000; Ye et al., 2000; Yoshida et al., 2000). On UPR induction, ATF6 is released from the membrane by sequential cleavage by Site 1 (S1P) and Site 2 Proteases (S2P; Ye et al., 2000), producing a 50-kDa cytosolic fragment that moves to the nucleus to activate genes that ameliorate ER stress. S1P and S2P also mediate cleavage of the ER membrane transcription factor sterol response element binding protein (SREBP), involved in regulating cholesterol biosynthesis (Wang et al., 1994; Sakai et al., 1996).
In most studies of UPR function, elevated levels of ER unfolded proteins are typically induced by treating cells with pharmacological agents including dithiothreitol (DTT), which disrupts or prevents protein disulfide bonding; thapsigargin (Tg), an inhibitor of the ER Ca2+-dependent ATPase; or tunicamycin (Tm), an inhibitor of protein glycosylation. Although UPR induction by pharmacological agents has proven extremely valuable for characterizing UPR pathway signaling components, functional roles for the UPR in physiological settings are not well defined. A well-studied example among the few known settings where the UPR is active is during the terminal differentiation of mature B lymphocytes (B-cells) into antibody secreting plasma cells (Calfon et al., 2002; Gass et al., 2002; Gunn and Brewer, 2003; Iwakoshi et al., 2003; van Anken et al., 2003; Zhang et al., 2005). It is reported that during this process IRE1 and ATF6, but not PERK, are activated, and further that activation of ATF6 is preceded by activation of IRE1 (Gass et al., 2002; Rutkowski and Kaufman, 2004). These observations indicate that activation of the tripartite UPR signaling branches is not a concerted process in vivo and suggest differences in the abilities of individual UPR initiators to recognize or respond to various forms of ER stress. In turn, individual branches may be specialized to respond to particular conditions.
As the ER performs a variety of protein maturation functions, the actual demand for specific protein folding functions may differ depending on prevailing conditions. An ability to selectively modulate individual UPR signaling branches might allow UPR responses to be customized to provide the most appropriate response. The contribution of each signaling branch to a custom response could be determined by the sensitivity of a UPR initiator to a given stress type alone or with the help of additional “discriminator” components.
Previous studies have focused on the individual signaling events mediated by each of the UPR components to understand their roles in the UPR. Very little is known about the relationships between the signaling branches initiated by IRE1, PERK, and ATF6. The activation behavior of UPR sensors in B-cell differentiation, however, highlights the importance of such studies in order to provide better understanding of UPR functions in physiological settings. Direct comparisons of the kinetic responses of UPR signaling branch activation, particularly during the experimentally well-defined ER stress caused by pharmacological agents, will provide a first step toward uncovering intrinsic differences in the behavior of, and relationships between signaling branches mediated by IRE1, PERK, and ATF6.
Here, we have compared the activation kinetics of IRE1, PERK, and ATF6 signaling branches in response to altered forms of ER stress induced by three pharmacological agents. ER stress induced by ER calcium release in particular revealed fundamental differences between UPR sensors; during Tg-induced UPR, both IRE1 and PERK were activated quickly, whereas ATF6 responded with significantly delayed kinetics. Furthermore, we observed that phosphorylation of the PERK kinase substrate, eIF2α, was modulated independent of PERK activation in Chinese hamster ovary (CHO) cells, suggesting the presence of an additional regulatory mechanism mediating events downstream of PERK kinase. Curiously, PERK-independent eIF2α phosphorylation may be specific to certain cell types because it was observed only in CHO cell. Further experiments also suggested that the observed kinetics were unlikely to be caused by disruption of the UPR sensor structure. Thus, our studies directly compare UPR signaling branch activation and reveal similarities, but importantly, fundamental differences between UPR sensors in their recognition of alternate forms of ER stress.
MATERIALS AND METHODS
Cell Culture and Induction of ER Stress
CHO cells were maintained in DMEM/F12 media (Cellgro, Herndon, VA) supplemented with 5% fetal calf serum (FCS; Invitrogen, Carlsbad, CA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Cellgro). Cells were maintained at 37°C in a 5% CO2 incubator. To induce ER stress, cells were seeded onto 10-cm plates at a density of 2–2.5 × 106. Twenty-four hours after seeding, cells were incubated for 2–3 h in fresh media without antibiotics before treating with 2 mM DTT (Invitrogen), or 200 nM Tg (Calbiochem, La Jolla, CA), or 10 μg/ml tunicamycin (Calbiochem). Although cells used in experiment shown in Figures 1B and 2 were treated with 20 μM MG-132 (Calbiochem) for 1 h before addition of 2 mM dithiothreitol, cells for all the rest of time-course experiments were not treated with this proteasome inhibitor. NIH3T3, mouse embryonic fibroblasts (MEFs), and HeLa cells were cultured in DMEM (Cellgro) supplemented with 10% FCS (Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin (Cellgro).
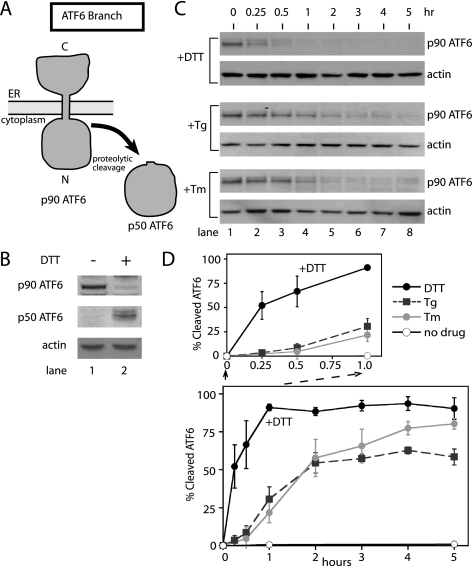
Figure 1.
The ATF6 branch of the UPR is most responsive to the accumulation of unfolded proteins disrupted by disulfide bond formation in the ER. (A) ATF6 processing during the UPR induction. ATF6 (p90ATF6) is proteolytically cleaved during UPR induction. The transcriptional transactivation domain (p50ATF6) is released in the cytoplasm and subsequently migrates into the nucleus. (B) Conversion of p90ATF6 to p50ATF6 upon DTT treatment of CHO cells. Total cell lysate prepared from CHO cells pretreated with proteasome inhibitor was incubated with (lane 2; 2 mM for 1 h) or without DTT (lane 1) and analyzed by immunoblotting using anti-ATF6 antiserum raised against the N-terminal portion of ATF6 (Imgenex) and β-actin as a control. (C) Immunoblots of ATF6 from lysates of CHO cells treated with 2 mM DTT, 200 nM thapsigargin (Tg), and 10 μg/ml tunicamycin (Tm) for the indicated amount of time. At each time point, treated cells were collected and divided into two samples (for protein and RNA analysis) before quick freezing in liquid nitrogen. β-Actin immunoblotting of the same lysate, from each time course was carried out by reprobing blots with anti-β-actin antibody. (D) Quantitation of p90ATF6 cleavage over the time courses shown in C. p90ATF6 cleavage was quantitated with a Typhoon 9400 phosphorimager (Amersham Biosciences) and normalized with levels of β-actin from corresponding treatments. Percent cleaved ATF6 was calculated by subtracting the p90ATF6 at each time point from the level of p90ATF6 at time zero. The graph shown represents three independent time-course experiments carried out with each drug. Differences in the initial phases of the responses were shown with the close-up of the first hour of treatment; DTT (black solid line), Tg (black dashed line), and Tm (gray solid line). Untreated is represented as a solid black line with open circles.
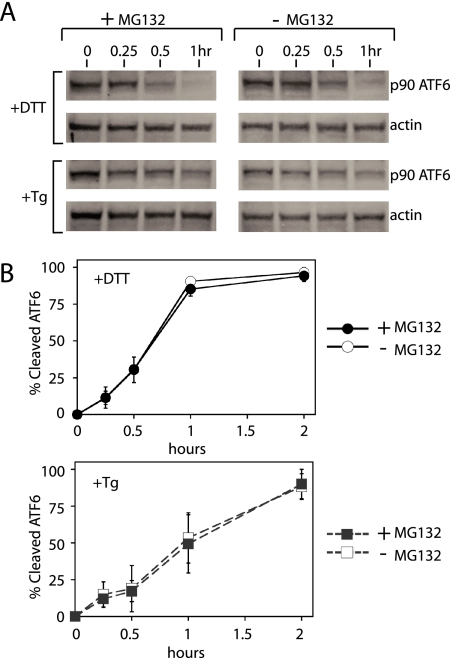
Figure 2.
Rate of p90 ATF6 disappearance during the UPR is similar with or without proteasome inhibitor, MG132. (A) Immunoblots of ATF6 from HeLa cell lysates treated with 2 mM DTT, or 200 nM thapsigargin (Tg) in the presence of proteasome inhibitor, MG132, (lanes 1–4) or without MG132 (lane 5–8). β-Actin immunoblotting of the same lysates was used to normalize p90ATF6 quantitation. (B) Quantitation of p90ATF6 shown in A. Levels of p90ATF6 were quantitated using a Typhoon phosphorimager and normalized with levels of β-actin. Percent cleaved ATF6 was calculated as described in Figure 1 and were shown during DTT treatment with (●) or without (○) MG132 or during Tg treatment with (■) or without (□) MG132.
XBP1 Splicing Assay
Total RNA was prepared using RNeasy mini kit (Qiagen, Chatsworth, CA) and treated twice with RQ1 DNase (Promega, Madison, WI). DNase was removed by phenol chloroform extraction before ethanol precipitation. RNA was reverse-transcribed with ThermoScript reverse transcriptase (Invitrogen), and PCR was performed with iTaq polymerase (Bio-Rad, Hercules, CA). XBP1 mRNA was amplified using (CACCTGAGCCCCGAGGAG) and (TTAGTTCATTAATGGCTTCCAGC). PCR products were run on 1.5% agarose gels.
Western Blotting
For Western blotting anti-ATF6 (Imgenex, San Diego, CA), anti-β-actin (Sigma, St. Louis, MO), anti-BiP (StressGen, Victoria, British Columbia, Canada), anti-phospho-eIF2α (Cell Signaling, Beverly, MA), anti-total eIF2α and anti-ATF4 (both from Santa Cruz Biotechnology, Santa Cruz, CA) were purchased commercially. Antibodies against mouse PERK and mouse IRE1α were directed against the C-terminal residues of PERK (residues 1079–1100) and a protein fragment containing both kinase and endoribonuclease domains, respectively. Whole cell extracts were resolved by SDS-PAGE and transferred to nitrocellulose. Membranes were developed with ECL Plus Western blotting detection reagent (Amersham Biosciences, Piscataway, NJ).
Immunoprecipitation
Immunoprecipitation of PERK and IRE1α were carried out based on the protocols described in Bertolotti et al. (2000). Briefly, treated CHO cells (see above) were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in 200 μl 1% Triton buffer on ice. All following steps were performed at 4°C unless otherwise stated. Extracts were rotated end-over-end for 15 min, clarified by centrifugation at 16,000 × g for 10 min, and preincubated for 1 h with 10 μl protein G-agarose (Upstate Biotechnology, Lake Placid, NY). PERK and IRE1 were incubated overnight with anti-PERK (Santa Cruz Biotechnology) or anti-IRE1α and then incubated for 3 h with 10 μl protein G-agarose. Beads were washed three times in 1% Triton buffer and once in ice-cold PBS with 100 mM NaF. Proteins were boiled 10 min in 2× Laemmli buffer with 100 mM DTT and resolved by SDS-PAGE. Proteins were transferred to nitrocellulose and probed with antibodies.
Detection and Quantification
Chemifluorescence of Western blots and ethidium staining of agarose gels were visualized using a Typhoon 9400 imager (Amersham Biosciences). Bands were quantified using ImageQuant 5.2 software (Amersham Biosciences).
RESULTS
To uncover potential differences or similarities in their responses to UPR-activating agents, we first monitored the activation kinetics of UPR initiators (ATF6, IRE1, PERK) in CHO cells after ER-stress induction with pharmacological agents that induce unfolded proteins by different mechanisms. We used DTT to disrupt disulfide bond formation, Tg to inhibit ER Ca2+-dependent ATPase, and Tm to inhibit glycosylation of newly synthesized proteins in the ER. We then compared the activation status IRE1, PERK, and ATF6 over time.
ATF6 Responds Most Quickly to DTT-induced UPR
ATF6 activation was measured by following its proteolytic cleavage over time by Western blotting with a monoclonal antibody raised against an N-terminal region of ATF6. This antibody was previously used and characterized by others and allowed observation of both p90 full-length and p50 cleaved ATF6 (Thuerauf et al., 2002, 2004; Hong et al., 2004). Thus, using this antibody, we detected a 90-kDa protein (p90 ATF6) in untreated CHO cells (Figure 1B, lane 1). Using the same antibody, UPR activation causes the disappearance of p90 ATF6 and the concomitant appearance of a 50-kDa fragment (p50 ATF6; Figure 1B, lane 2). Robust detection of p50 ATF6 required the presence of a proteasome inhibitor, indicating that the cleaved fragment is relatively unstable as previously reported (Haze et al., 1999). Thus, ATF6 activation was quantitated by calculating the disappearance of full-length ATF6 over time. We found ∼50% of p90 was cleaved at 15 min after DTT treatment (Figure 1C, lane 2, +DTT). Beyond 1 h essentially all full-length ATF6 had disappeared, indicating ATF6 activation is sustained throughout the time course (Figure 1, C and D).
During UPR induction by either Tm or Tg, ATF6 activation was comparatively slow. Almost no ATF6 was cleaved 15 min after Tm or Tg exposure and only ∼50% was cleaved after 2 h (Figure 1, C and D, +Tg and +Tm). Beyond 2 h a more gradual increase in ATF6 cleavage was observed. Taken together, these results demonstrated that ATF6 responded to DTT- induced ER stress more quickly and more efficiently than to stress induced by Tg or Tm.
Activation of the UPR also induces ER-associated degradation (ERAD) to clear permanently misfolded proteins from the ER (Kaufman, 1999; Mori, 2000; Patil and Walter, 2001; Harding et al., 2002; Meusser et al., 2005; Sayeed and Ng, 2005). Thus, if a portion of p90ATF6 was degraded by ERAD instead of conversion to p50ATF6, quantitation of p90ATF6 might have overestimated the ATF6 activation rate. This could become a problem particularly if ERAD degradation of p90ATF6 differed depending on the agents used to induce ER stress. Thus, we measured p90ATF6 disappearance during DTT and Tg treatment in the presence of a well-characterized proteasome inhibitor, MG132, to inhibit ERAD (Figure 2). During both DTT- and Tg-induced UPR, slightly higher levels of p90ATF6 were detected in the presence of MG132. Furthermore, with DTT or Tg, quantitation of p90ATF6 showed similar rate of activation (p90ATF6 disappearance) between cells treated with or without MG132. Thus, ERAD mediated degradation of ATF6 is unlikely to account for differences in the rate of ATF6 activation during DTT- and Tg-induced UPR.
We also measured p90ATF6 disappearance in mouse embryonic fibroblasts derived from Perk knockout embryo (Perk−/− MEFs) in order to test if UPR-induced translation repression contributed to the disappearance of p90ATF6 (Supplementary Figure 1). During UPR induction by either DTT or Tg in Perk−/− MEFs, phosphorylation of the eIF2α translational initiator did not take place to repress translation (see below and Supplementary Figure 2). In Perk−/−MEFs, we found that the differences in the rates of P90ATF6 activation between DTT- and Tg-induced ER stress were similar to those of wild-type mouse 3T3 fibroblast (Supplementary Figure 1, and unpublished data). Together, these experiments provided further support for the use of p90ATF6 disappearance as a measure of p90ATF6 activation, and thus, that ATF6 responds more efficiently to DTT-induced ER stress than to Tg.
The PERK Signaling Branch Responds Efficiently to ER Stress Induced by Tg and by DTT
To allow direct comparisons, we analyzed the activation kinetics of both IRE1 and PERK signaling branches in the same lysates used for the ATF6 time-course experiments. UPR induction by DTT, Tm and Tg has been shown to induce PERK autophosphorylation that can be monitored by the appearance of a slower migrating band on SDS-PAGE (Shi et al., 1998; Harding et al., 1999; Bertolotti et al., 2000). Thus, we examined PERK autophosphorylation to probe PERK activation after UPR induction. By immunoprecipitating with an antibody against the PERK N-terminal region followed by immunoblotting with an antibody raised against the C-terminal kinase domain, we could detect PERK efficiently in untreated or UPR-activated CHO cells (Figure 3). A slower migrating form of PERK was detected within 15 min of incubation (Figure 3B, indicated as p-PERK) with DTT and Tg treatment of CHO cells. Furthermore, this form continued to increase over a 1-h incubation. Phosphatase treatment of immunoprecipitated PERK changed its mobility on SDS-PAGE, consistent with the previous reports that phosphorylation was responsible for the slow mobility form of PERK (unpublished data). Together, kinetic appearance of autophosphorylated PERK (p-PERK) during DTT-induced UPR was almost identical to that during Tg-induced UPR (Figure 3, B and C).
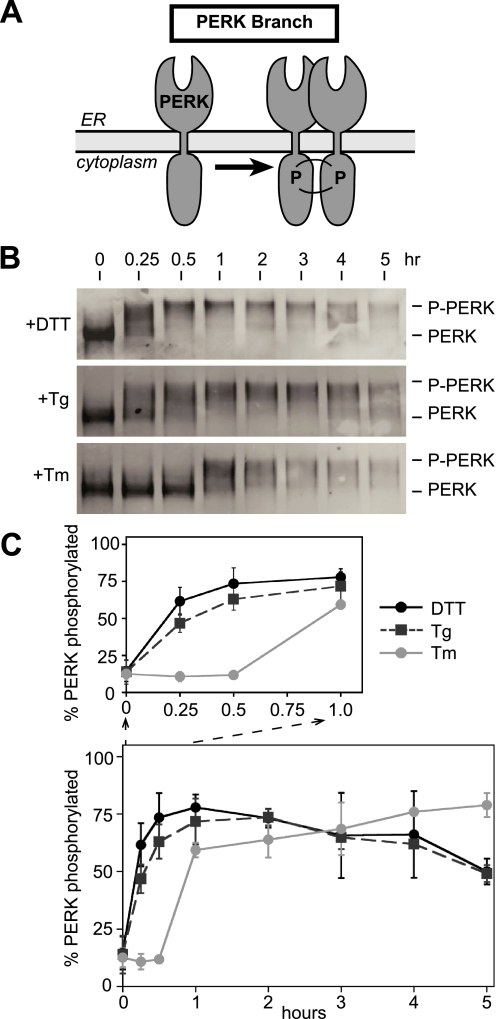
Figure 3.
PERK activation measured by autophosphorylation is responsive to ER stress caused by both ER calcium release and disruption of disulfide bonds. (A) PERK autophosphorylates during the UPR response. (B) Immunoblots, after immunoprecipitation of PERK from CHO cells treated with thapsigargin (Tg; 200 nM), tunicamycin (Tm; 10 μg/ml), and DTT (2 mM) for indicated amounts of time. Immunoprecipitated PERK using anti-PERK antibody from total cell lysate of time points from each treatment were Western blotted with anti-PERK antiserum. The slower mobility PERK formed during ER stress caused by DTT, Tg, and Tm treatment is indicated as p-PERK. (C) Quantitation of p-PERK appearance shown in B. DTT (black solid line), Tg (black dashed line), and Tm (gray solid line).
Treatment of CHO cells with Tm also produced autophosphorylated PERK, but with significantly slower kinetics; autophosphorylated PERK was not detected until 1 h after Tm treatment. Together, these results suggested that PERK could sense UPR induced by either DTT or Tg equally well, whereas ATF6 response to Tg was much slower than that to DTT (compare Figures 1 and 3). Thus, ER stress caused by Tg revealed differences in intrinsic abilities of ATF6 and PERK to sense lumenal conditions.
Phosphorylation of the PERK Kinase Substrate, eIF2α, Occurs with Kinetics Different from Activation of PERK Itself
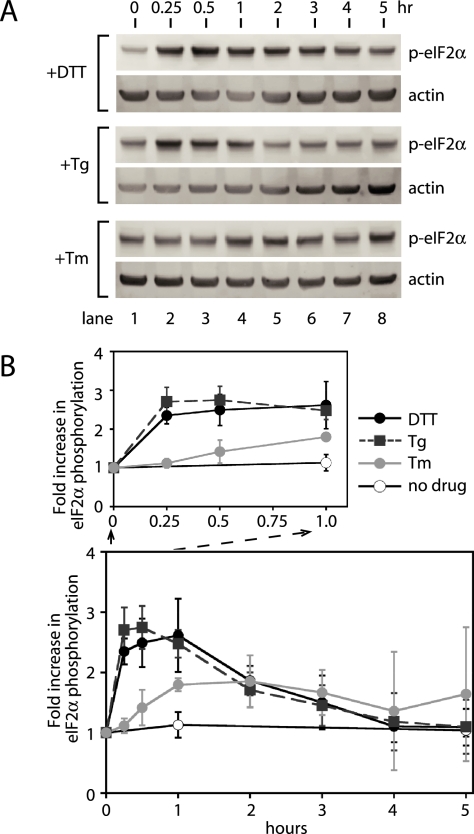
To further confirm the above observation, we followed the kinetics of PERK activation by quantitating its ability to phosphorylate eIF2α (p-eIF2α) by Western blotting using an antibody capable of specifically recognizing p-eIF2α in total cell lysates (Figure 4). A basal level of p-eIF2α was observed in uninduced CHO cells (Figure 4B, lane 1). We also observed differences in the amount of basal p-eIF2α in other uninduced cell lines (see Figure 4 and Supplementary Figure 2). Tg treatment produced the most rapid and extensive response, within 15 min of incubation p-eIF2α was detected, increasing to fivefold within 30 min and reaching maximum induction by 1 h (Figure 4, B and C, +Tg). The rapid phosphorylation of eIF2α was consistent with the rapid autophosphorylation of PERK induced by Tg (Figure 3). Furthermore, this form continued to increase over a 1-h incubation, again correlating well with the time course of PERK autophosphorylation (compare Figure 3 with Figure 4).
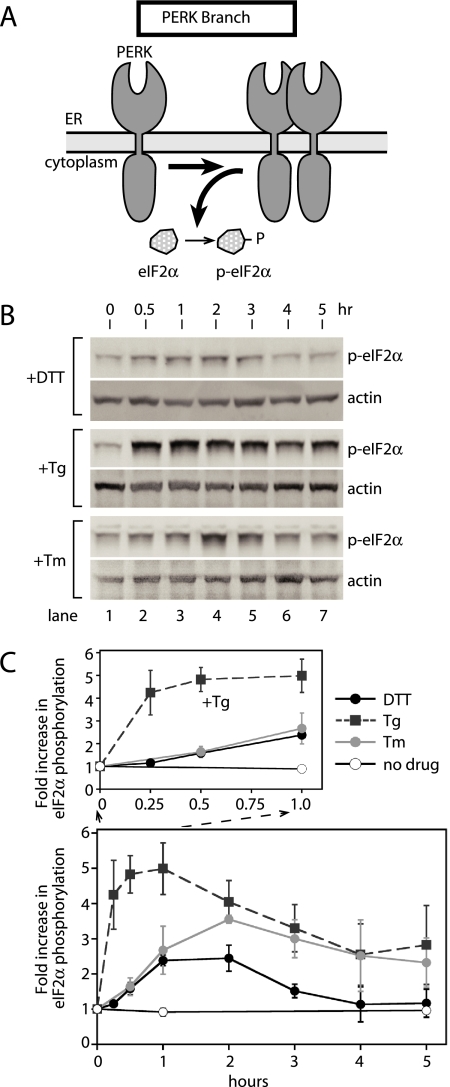
Figure 4.
Phosphorylation of eIF2α, a PERK kinase substrate, is most responsive to ER stress caused by ER calcium release. (A) PERK phosphorylates eIF2α during UPR induction. (B) Immunoblots of phosphorylated eIF2α (p-eIF2α) and β-actin from lysates of CHO cells treated with DTT, thapsigargin (Tg), and tunicamycin (Tm) for the indicated amounts of time. For direct comparisons with ATF6- and IRE1-signaling branch activation, the same cell lysates used for Figure 1 were examined for phosphorylation of eIF2α. (C) Quantitation of the increase in phosphorylated eIF2α (p-eIF2α) levels over the time course shown in B. The levels of p-eIF2α were quantitated with a Typhoon 9400 phosphorimager and normalized to levels of β-actin. Fold induction was calculated by taking the ratio between the levels of normalized p-eIF2α at time zero and each time point. A close-up of the first hour is shown for comparison in the initial phases of the responses. The graph represents three independent time course experiments carried out with DTT (black solid line), Tg (black dashed line), and Tm (gray solid line). Untreated is represented as a solid black line with open circles.
The appearance of p-eIF2α after DTT and Tm treatment was both slower and less extensive (Figure 4, B and C). The increase in eIF2α phosphorylation produced by Tm was significant, but smaller and delayed compared with Tg. To our surprise, DTT treatment induced only a minimal increase in eIF2α phosphorylation throughout the time course (Figure 4, B and C). This was unexpected because autophosphorylation of PERK was much more efficient during DTT-induced UPR (Figure 3). With each inducing agent, eIF2α phosphorylation reached a maximum and then declined slowly. This decline is presumably caused by the accumulation of GADD34, a p-eIF2α specific phosphatase, as reported previously (Novoa et al., 2001). In all three cases, using antibody that detects total eIF2α protein, we found that eIF2α protein levels remains unchanged after UPR induction (Supplementary Figure 3), consistent with a previous report (Harding et al., 2000a). From these results, we conclude that activation of the PERK-induced UPR signaling branch or phosphorylation of a PERK kinase target at least is most responsive to ER stress caused by Tg, less responsive to that by Tm, and least responsive to DTT. Thus, PERK appeared to respond equally well to ER stress induced by DTT and Tg, based on the kinetic appearance of p-PERK, whereas PERK activation was significantly delayed with DTT when phosphorylation of eIF2α was used to determine the order of PERK activation.
BiP Dissociation from PERK Lumenal Domain Correlates with Autophosphorylation of PERK
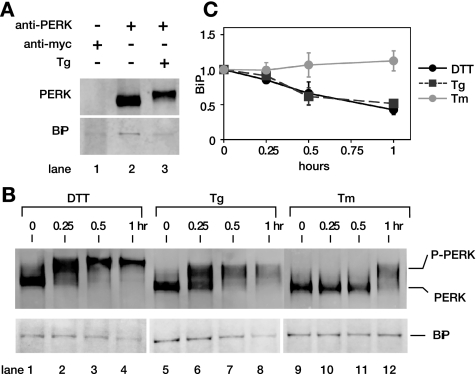
To further investigate the discrepancy between the kinetics of PERK autophosphorylation and PERK phosphorylation of eIF2α, we examined the kinetics of BiP dissociation from PERK lumenal domain. BiP, is a major ER chaperone, that has been identified as PERK-interacting protein coimmunoprecipitating with PERK (Bertolotti et al., 2000; Ma et al., 2002). In addition, BiP dissociation from PERK lumenal domains has been shown to induce PERK autophosphorylation and oligomerization (Bertolotti et al., 2000; Ma et al., 2002). Thus, we tested if the kinetics of BiP dissociation from PERK reflected the appearance of either phosphorylated PERK or phosphorylated eIF2α in response to ER stress agents. We examined BiP association with PERK by blotting PERK immunoprecipitates with anti-BiP antibody (Figure 5, B and C), as previously established (Bertolotti et al., 2000). We found that levels of coimmunoprecipitated BiP in uninduced cells (Figure 5A, lane 2; and 5B, lanes 1, 5, and 9) were comparable with these previous reports and was reduced upon UPR induction with treatment of cells with Tg (Figure 5A, lanes 2 vs. 3, or 5B, lanes 5 vs. 8). Furthermore, BiP coimmunoprecipitation was specific to anti-PERK antibody, because we detected no significant levels of BiP coimmunoprecipitated with anti-myc antibody (Figure 5A, lanes 1 vs. 2). Normalizing the levels of coimmunoprecipitated BiP to immunoprecipitated PERK revealed a significant reduction in PERK-associated BiP at 30 min after exposure of CHO cells to Tg (Figure 5C). We detected a similar reduction in BiP-associated PERK during the DTT-induced ER stress. In contrast, Tm treatment did not cause changes in the levels of BiP associated with PERK. Thus, kinetic changes in BiP levels were in general agreement with the appearance of phosphorylated PERK.
Figure 5.
PERK activation measured by dissociation of BiP correlates with PERK activation measured by PERK autophosphorylation. (A) Immunoprecipitation of BiP was specific to uninduced PERK. BiP was specifically immunoprecipitated with anti-PERK antibody from uninduced lysate (lane 2), but not with anti-myc antibody (lane 1). Furthermore, UPR induction upon Tg treatment diminished levels of BiP-associated PERK (lane 3). (B) BiP associated with PERK during treatment with DTT (lanes 1–4), Tg (lanes 5–8), and Tm (lanes 9–12) is shown. Levels of BiP associated with PERK in each immunoprecipitated fraction were detected by blotting the same membranes using anti-BiP antibody. (C) Quantitation of BiP associated with PERK. Levels of BiP associated with PERK were quantitated with a phosphorimager and normalized against immunoprecipitated PERK from CHO cells treated with DTT (black solid line), Tg (black dashed line), and Tm (gray solid line). Error bars and values of each time point were averaged over three independent experiments.
In addition to BiP dissociation, we also examined the kinetics of ATF4 appearance. Translation of ATF4 is regulated by small open reading frames (ORFs) present within the 5′ untranslated region (UTR) of ATF4 mRNA. When eIF2α is not phosphorylated, ATF4 translation is prevented by stop codons within the small ORFs causing translation initiation complexes to fall off the template (Vattem and Wek, 2004). In response to ER stress, phosphorylation of eIF2α suppresses initiation complex release from ATF4 mRNA, allowing ATF4 translation (Harding et al., 2000b). Thus, we examined the appearance of ATF4 by Western blot in the same extract used above to analyze eIF2α phosphorylation (Supplementary Figure 4). Appreciable levels of ATF4 were detected after 3 h of incubation with DTT, whereas ATF4 started to accumulate after 1–2 h during the Tg treatment of cells. Because phosphorylation of eIF2α occurred more rapidly and efficiently with Tg than with DTT, the production of ATF4 further correlates with kinetics of eIF2α phosphorylation. Together, PERK appeared to respond equally well to ER stress induced by DTT and Tg, and thus, an additional regulatory step might exist to account for the altered kinetics of events downstream of PERK activation, including eIF2α phosphorylation and ATF4 production.
PERK Activation Kinetics in Mouse Fibroblast NIH3T3 Cells
Because differences in the kinetics of PERK activation measured by autophosphorylation and by eIF2α phosphorylation have not been reported previously, we examined eIF2α phosphorylation kinetics during UPR activation in cell lines other than CHO cells. On treatment of mouse NIH3T3 cells with DTT, Tg, or Tm, lysates were probed for phosphorylated forms of eIF2α with the same antibody used to detect p-eIF2α in CHO cells. Similar to CHO cells, we observed a robust increase in p-eIF2α levels that reached a maximum within 1 h of Tg treatment (Figure 6, A and B). NIH3T3 cells also produced a robust increase in p-eIF2α during DTT-induced ER stress. Thus, in 3T3 cells, the kinetics and the extent of increase in p-eIF2α was similar for both DTT- and Tg-induced ER stress. Therefore, a potential regulatory step conferring differences between the kinetics PERK kinase activation and of eIF2α phosphorylation might be specific to CHO cells.
Figure 6.
eIF2α phosphorylation during UPR induction in NIH3T3 cells. (A) Immunoblots of phosphorylated eIF2α (p-eIF2α) and β-actin from lysates prepared from NIH3T3 cells treated with DTT, thapsigargin (Tg), or tunicamycin (Tm) for the indicated amounts of time. (B) Quantitation of the fold increase in p-eIF2α levels over the time course shown in A. The levels of p-eIF2α were quantitated as described in Figure 2 and fold induction was calculated by taking the ratio between the levels of normalized p-eIF2α at time zero and each time point. A close-up of the first hour is shown for comparisons in the initial phase of the responses. The graph represents three independent time course experiments carried out with DTT (black solid line), Tg (black dashed line), and Tm (gray solid line). Untreated is represented as a solid black line with open circles.
IRE1 Discriminates between ER Stress Types
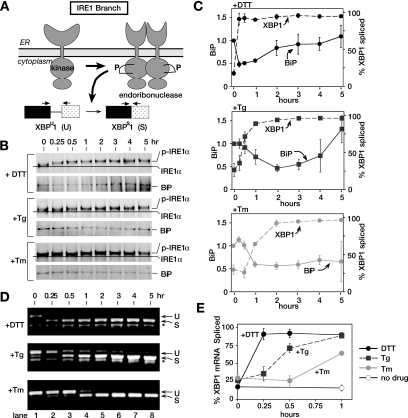
To monitor activation of IRE1, we examined IRE1 autophosphorylation after UPR induction by DTT, Tg, and Tm. IRE1 autophosphorylation can be monitored by mobility shift on SDS-polyacrylamide gels (SDS-PAGE; Tirasophon et al., 1998; Bertolotti et al., 2000; Lee et al., 2002; Harding et al., 2003) and has been used previously to analyze IRE1 activation. Endogenous IRE1 was detected by immunoprecipitation with a polyclonal antibody against the C-terminal portion of IRE1 followed by immunoblotting with same antibody (Figure 7), allowing efficient detection of IRE1 in lysates from untreated and UPR-induced CHO cells. In response to DTT, we observed a decrease in IRE1 mobility starting after 15 min, consistent with the time frame we observed for XBP1 mRNA splicing (Figure 7, B and C, and see below). The small mobility shifts upon IRE1 autophosphorylation has been reported previously, and we have found it to be consistent over multiple experiments (Tirasophon et al., 1998; Bertolotti et al., 2000; Lee et al., 2002; Harding et al., 2003). Phosphorylated IRE1 appeared at 30 min in Tg-treated CHO cells and at 1 h in Tm-treated cells.
Figure 7.
The IRE1 signaling branch of the UPR can respond efficiently to all types of ER stress, but is most sensitive to the accumulation of unfolded proteins due to disrupted disulfide bonds in the ER. (A) IRE1 autophosphorylates upon UPR induction. On activation, IRE1 becomes oligomerized and is an active endoribonuclease in the splicing of XBP1 mRNA. Schematic representation of both the unspliced (U) and spliced (S) forms of XBP1 mRNA and the PCR primers used. (B) Immunoblots, after immunoprecipitation, of IRE1α from CHO cells treated with DTT (2 mM), thapsigargin (Tg; 200 nM), and tunicamycin (Tm; 10 μg/ml) for indicated amounts of time using anti-IRE1α antiserum raised against the C-terminal portion of IRE1α. The slower mobility IRE1α formed during ER stress caused by DTT, Tg, and Tm treatment is indicated as p-IRE1α. BiP associated with IRE1α was detected by blotting the same membrane using anti-BiP antibody. During the analyses, we consistently observed that the extent of IRE1α mobility shift was much more pronounced during DTT-induced ER stress than during other treatments. Currently, the molecular bases for these differences are not clear, although they may represent differential phosphorylation in response to the altered forms of ER stress. (C) Quantitation of immunoprecipitated BiP. Levels of BiP associated with IRE1α were quantitated with a phosphorimager and normalized against immunoprecipitated IRE1α (shown as a solid line). Error bars and values of each time point were averaged over three independent experiments. IRE1α RNase activity indicated as % spliced XBP1 is shown as dashed line. (see E for detail). (D) RT-PCR of RNA isolated from CHO cells treated with 2 mM DTT, 200 nM thapsigargin (Tg), and 10 μg/ml tunicamycin (Tm). Total RNA was isolated from samples collected for Figures 1 and 3. cDNA was prepared using oligodT primers. PCR with primers flanking the 26-nt UPR intron of hamster XBP1 RNA. PCR products were analyzed on 1.5% agarose gels and stained with ethidium bromide. DNA fragments derived from unspliced (U) and spliced (S) are indicated. Bands marked as (*) are nonspecific PCR fragments. (E) Quantitation of the spliced XBP1 shown in D. Both unspliced and spliced forms of XBP1 were quantitated with a phosphorimager. Percent spliced at each time point was calculated by S/(S + U) × 100. The graph represents three independent time-course experiments carried out with DTT (black solid line), thapsigargin (black dashed line), and tunicamycin (gray solid line). Untreated is represented as a solid black line with open circles. Quantitation of the spliced XBP1 for the entire 5-h time course was shown in C as dashed lines, whereas that for the first hour is shown in E.
BiP Dissociation from IRE1 Confers Specificity for Different ER Stress
In addition to autophosphorylation of IRE1, we examined the kinetics of BiP dissociation from IRE1 lumenal domains. BiP is a major ER chaperone, identified as an IRE1-interacting protein coimmunoprecipitating with IRE1 (Bertolotti et al., 2000; Okamura et al., 2000; Kimata et al., 2004). In addition, BiP dissociation from IRE1α lumenal domains was shown to induce IRE1 autophosphorylation and oligomerization (Bertolotti et al., 2000; Okamura et al., 2000; Kimata et al., 2004). Thus, we expected the kinetics of BiP dissociation to reflect the appearance of phosphorylated IRE1α in response to ER stress agents. We examined BiP association with IRE1α by blotting IRE1α immunoprecipitates with anti-BiP antibody (Figure 7B), as previously established (Bertolotti et al., 2000). We found that levels of coimmunoprecipitated BiP were compatible with previous reports. Normalization of coimmunoprecipitated BiP levels to immunoprecipitated IRE1 revealed a significant reduction in IRE1-associated BiP 15 min after exposure of CHO cells to DTT (Figure 7, B and C), in agreement with the appearance of both phosphorylated IRE1α and spliced XBP1 mRNA (see below). Curiously however, levels of IRE1-asscociated BiP returned to unstimulated levels by 2 h after DTT treatment, contrasting with the sustained presence of phosphorylated IRE1α and spliced XBP1 over the 5-h time course. These results suggest that BiP reassociation precedes inactivation of IRE1α. Similarly, in direct agreement with the delayed appearance of phosphorylated IRE1α and spliced XBP1 mRNA after Tg treatment compared with DTT, we observed a significant decrease in IRE1-associated BiP at 1 h after treatment with Tg. This was followed by BiP reassociation starting at the 3-h time point (Figure 7, B and C). Thus, taken together, these results suggest that affinity of BiP for IRE1α differ depending upon the types of ER stress imposed.
The IRE1 Signaling Branch Responds Rapidly to Different Forms of Stress
To further monitor IRE1 activation, we assayed splicing of the 26-nucleotide UPR-specific intron (UPR intron) from XBP1 mRNA; an event dependent on activation of the IRE1 endoribonuclease domain. Splicing was quantitated by reverse transcription followed by PCR (RT-PCR) of cellular RNA isolated from cells at each time point (Figure 7, C–E). In untreated CHO cells, PCR amplification of cDNA using primers flanking the UPR intron produced the 599-base pair fragment predicted for unspliced XBP1 mRNA (U; Figure 7, A and D) and the 573-base pair fragment corresponding to spliced XBP1 RNA after UPR induction (S; Figure 7, A and D). Quantitation of the PCR fragments produced in untreated cells shows that ∼20% of cellular XBP1 mRNA is present in the spliced form (Figure 7D, lane 1). This finding suggested that IRE1 was constitutively activated at a low level in normal cells, consistent with the low level of p-eIF2α also observed in these cells before UPR induction (Figures 4B and 7D).
IRE1 was maximally and nearly completely activated (∼90% spliced) by DTT within 15 min of DTT treatment (Figure 7, C–E) and remained fully activated throughout the time course. As reported previously (Yoshida et al., 2001), we also noted an increase in XBP1 mRNA at later time points (Figure 7D), consistent with the idea that XBP1 is a UPR target gene induced by ATF6. Because the splicing efficiency of XBP1 mRNA remained high throughout the time course, these results suggest that IRE1 endoribonuclease activity remains active and capable of splicing all newly synthesized XPB1 mRNA.
In contrast to DTT, Tg treatment produced only a minimal increase in XBP1 splicing at 15 min after treatment (30% spliced), taking about 1 h to reach maximal levels, which were sustained throughout the time course (Figure 7, C–E). Tm-induced splicing did not occur until 1 h after incubation but did reach maximal levels by 2 h, which were also sustained through the time course (Figure 7, C–E). Thus, the timing of IRE1 autophosphorylation in response to UPR induction by DTT, Tg, and Tm correlated with the activation of IRE1-dependent splicing of XBP1 mRNA (compare Figure 7, B and D). Together these results showed that in contrast to the ATF6 and PERK signaling branches, which responded maximally to distinct ER stressors, the IRE1 signaling branch responded to each stressor in comparatively short time periods.
Differential Activation of UPR Signaling Branches Reflects Intrinsic Properties of UPR Sensors
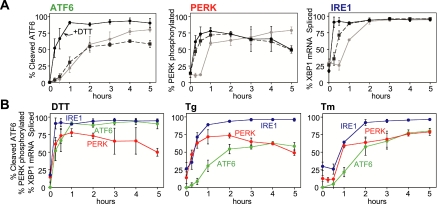
A direct comparison of our findings, which were essentially all obtained from a single cell extract for each inducing agent, is shown in Figure 8A. In summary, the ATF6 branch responds rapidly to disulfide bond disruption and slowly to other forms of ER stress. The PERK signaling branch responds most rapidly to changes in ER Ca2+ release, whereas the IRE1 branch responds efficiently and relatively rapidly to each ER stress agent, although some discrimination was seen at early time points. When data from Figures 1, 3, and 7 were replotted by individual UPR-inducing agent (Figure 8B), it was clearly seen that activation of IRE1 and ATF6 by DTT was rapid and robust. Furthermore, although eIF2α phosphorylation was significantly delayed and inefficient in CHO cells, activation of PERK itself judged by its autophosphorylation occurred rapidly. Presumably, disruption of disulfide bonds by DTT results in accumulation of unfolded proteins quickly. On the other hand, ER stress due to Tg treatment revealed different properties of the initiators; a quick response to PERK and IRE1, but a significantly slower response to ATF6.
Figure 8.
Preferential activation of UPR signaling branches by alternate types of ER stress. (A) ATF6 is activated quickly to disruption of disulfide bonds of ER proteins caused by DTT (black solid line), whereas both IRE1 and PERK are activated rapidly and extensively in response to ER stress caused by thapsigargin (black dashed line), an ER Ca2+ release agent. For all ER stress tested, IRE1 reacts relatively fast and efficiently, resulting in splicing of XBP1 mRNA. (B) Because all activation profiles for ATF6, PERK, and IRE1α described in Figures 1, 2, and 6 were performed with the same extract samples, the kinetic induction of the three UPR initiators was reanalyzed by the type of ER stress imposed. ER stress caused by DTT activated both IRE1α (blue) and ATF6 (green) rapidly and efficiently. UPR induced by thapsigargin was detected by both PERK (red) and IRE1 (blue), whereas all three UPR sensors respond to ER stress activated by tunicamycin with relatively similar kinetics.
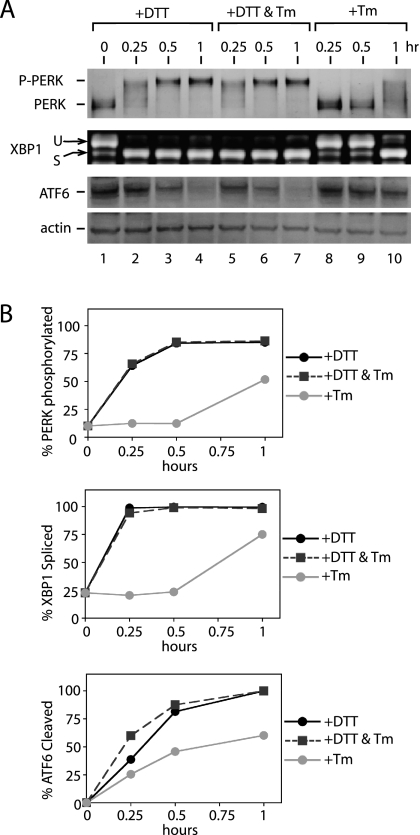
A trivial explanation for the differential responses we observed is structural damage to UPR sensors caused by the UPR-inducing agent. The lumenal domains of IRE1, PERK, and ATF6 each contain conserved cysteines as well as glycosylation sites. Thus, for example, the delayed activation of PERK by Tm could be due to structural disruption of its sensor domain. To test this possibility, we asked if Tm affected the rapid activation of PERK by DTT. Accordingly, we treated CHO cells simultaneously with DTT (2 mM) and Tm (10 μg/ml) and followed the activation of PERK, IRE1, and ATF6 activation by measuring PERK autophosphorylation, XBP1 mRNA splicing, and the disappearance of p90ATF6, respectively. If slow activation of PERK were caused by structural damage to PERK during Tm treatment, we would expect that its presence during DTT-induced ER stress should delay the rapid activation of PERK observed during DTT alone. As shown in Figure 9, we found that the addition of Tm did not change PERK, IRE1, or ATF6 activation kinetics compared with DTT treatment alone (compare DTT only, lanes 2–4, with DTT+Tm, lanes 5–7). Similar results were obtained when CHO cells were pretreated with Tm for 1 h before DTT addition (unpublished data). Thus, together, these results suggest that differences we observed in UPR sensor activation are likely to reflect differences intrinsic to the manner in which each sensor recognizes different stress types.
Figure 9.
Differential activation of three UPR sensors is likely to reflect their intrinsic properties. (A) Activation kinetics of PERK, IRE1, and ATF6 signaling branches during DTT (2 mM) or tunicamycin (Tm; 10 μg/ml) treatment, or during treatment with both DTT (2 mM) and tunicamycin (Tm; 10 μg/ml) together for indicated amounts of time. Immunoblots, after immunoprecipitation, of PERK using anti-PERK antiserum, and of ATF6 using anti-ATF6 antibody and RT-PCR of XBP1 mRNA from total RNA isolated, are shown. (B) Quantitation of PERK autophosphorylation, ATF6 p90 fragment disappearance, and XBP1 mRNA splicing, shown in A during ER stress caused by DTT only (black solid line), both DTT and tunicamycin (black dashed line), and tunicamycin only (gray solid line).
DISCUSSION
The ER performs a variety of protein maturation steps including chaperone-assisted folding, modification, and complex assembly, for secreted proteins and proteins residing in the secretory pathway. Cellular demand for such functions fluctuates according to environmental or other specific growth conditions. In mammalian cells, at least three ER-proximal molecules (IRE1, PERK, and ATF6) sense ER protein-folding needs and initiate cellular responses to meet increased demand, collectively referred to as the UPR. Activation of each sensor results in the activation of specific transcription factors: XBP1 for IRE1, ATF4 for PERK, and activated ATF6 for ATF6—and thus, a major consequence of UPR activation is transcriptional induction of UPR target genes (Harding et al., 2000a, 2003; Scheuner et al., 2001b; Okada et al., 2002; Lee et al., 2003; Shen et al., 2005). XPB1, ATF4, and ATF6 share some targets, but also activate unique sets of genes. Thus, selective activation and/or kinetic differences in the activation of specific transcription factors may allow the ER to mount a “best-fit” response to prevailing ER conditions. Future experiments will be required to delineate how the observed differences in activation of the UPR sensors dictate overall transcription outputs, depending on types of ER stress.
Alternate transcriptional profiles of UPR target genes in response to different forms of ER stress could be achieved by regulation at several levels. Here we show that such regulation begins at the ER lumen. We show that although IRE1 and PERK display similar sensitivities to alternate stress types, ATF6 behaves differently. IRE1 and PERK were activated with nearly equal efficiency during UPR induction by DTT (disulfide bond inhibition) and also similarly to Tg (ER calcium efflux). However ATF6 was significantly less sensitive than IRE1 or PERK to Tg-induced ER stress. Because IRE1, PERK, and ATF6 each respond with approximately equal efficiencies to DTT, the inefficient response of ATF6 to Tg may reflect intrinsic differences in how ATF6 senses ER stress. In physiological settings, UPR activation could be caused by multiple forms of ER stress, possibly obscuring an objective analysis of UPR initiator properties. Thus, our use of pharmacological agents to induce defined types of stress in tissue culture cells was key to observing differences in the properties of UPR initiators. Because these similarities and differences between the properties of UPR initiators have never been examined side-by-side, our findings have paved a road for further studies to determine detail molecular mechanisms underlying these differences.
How can the kinetic responses we observed for each ER sensor be explained? A trivial explanation could postulate that pharmacological agents used to induce the UPR produce direct structural “damage” to UPR sensors. For example, the PERK lumenal (sensor) domain contains at least one conserved glycosylation site. If Tm disrupted proper PERK folding, PERK function could be diminished and cause delayed activation kinetics. However, based on our two drug experiments, this would seem unlikely because Tm did not prevent the rapid appearance of phosphorylated PERK caused by DTT (Figure 9). Thus, kinetic differences between sensor activation seem more likely to reflect intrinsic differences in stress-type recognition by the sensors themselves.
Topologically, IRE1 and PERK are similar; both are transmembrane receptor kinases with N-terminal domains that sense conditions within the ER lumen (Liu et al., 2000, 2002). The recently published crystal structure of the yeast IRE1 (yIre1p) lumenal domain provides exciting insights into how UPR sensors may recognize ER lumenal conditions (Credle et al., 2005). yIre1p contains a structural element similar to the peptide-binding groove of major histocompatibility complexes (MHC). Modeling experiments followed by mutagenesis predicted that a 10-residue poly-valine peptide can fit in the yIre1p MHC-like peptide-binding groove without steric-hindrance. Interestingly, a conserved stretch of hydrophobic and hydrophilic residues facing the groove floor appear important for IRE1 function because mutations within this conserved stretch diminish IRE1 function (Credle et al., 2005). Thus, the model allows that unfolded polypeptides or partially folded protein could bind directly to the IRE1 groove to affect Ire1 activation. Critical determinants for peptide binding to the IRE1 groove might therefore reside with specific amino acids or with steric features of “unfolded” or “partially unfolded” polypeptides. Amino acid sequence conservation between IRE1 and PERK infers a similarly sized peptide-binding groove in the PERK lumenal domain. Thus, regardless of specific requirements for binding, the mechanism of polypeptide recognition by PERK seems likely to be similar to IRE1. The similarity we observed between IRE1 and PERK activation kinetics is therefore consistent with this structure-based model. It would be interesting to determine if “unfolded” polypeptides produced by different forms of ER stress have similar binding properties for the IRE1 (or PERK) polypeptide-binding groove.
Based on the amino acid alignments, however, two specific loops (between β10 and β11 and between β17 and β18, based on the yIre1 lumenal structure) differ significantly in length when compared between IRE1 and PERK (Credle et al., 2005). For yIre1, these loops appear to be exposed to the surface of the peptide-binding groove. Thus, differences in sizes of these loops could provide subtle alterations in peptide binding. On close inspection, our data suggest that IRE1 responds slightly faster to DTT than Tg, whereas PERK responds almost identically to each drug. In addition to differences in loop size, one of the PERK loops contains two cysteines that are conserved between species. Although mutational studies have not assigned significant structural importance to these residues, they may be involved in discriminating between different types of ER stress (Ma et al., 2002; Narasimhan et al., 2004).
Finally, structural differences between MHC class I (MHC I) and class II (MHC II) molecules may also provide interesting mechanistic insights into the specificity of polypeptide binding between IRE1 and PERK. Although the overall structures of the MHC I and MHC II peptide-binding grooves are similar, amino acids at the ends of the two α-helices comprising the MHC I groove are in closer proximity to each other and are involved in peptide binding (Bjorkman, 1997). This structural feature seems to place constraints on the size and sequence of polypeptides available for binding. In addition to size, most peptides bound by MHC I contain hydrophobic or basic residues at their C-termini. In contrast, the MHC II groove is more open, allowing longer peptides to bind. Furthermore, residues within the MHC I groove are directly involved in peptide binding. These structural variations contribute the differences in polypeptide bindings between MHC I and II such that MHC I binds smaller peptides, whereas MHC II can accommodate larger polypeptides (Bjorkman, 1997). Thus, structural differences between IRE1 and PERK determined by residues at the end of the groove, or differences in the types of amino acid residues surrounding the groove surface could play a role in their differential activation kinetics with specific types of ER stress.
In contrast to IRE1 and PERK, less structure-function information is available for the lumenal portion of ATF6. On activation, ATF6 might create polypeptide-binding pockets different from IRE1 or PERK. ATF6 is a type II transmembrane protein, with its C-terminus in the ER lumen and a cytoplasmic N-terminus. Although an MHC-like peptide binding pocket is not predicted by the ATF6 amino acid sequence, two stretches of homologous sequence approximately 40 residues each in the ATF6 lumenal domain appear to be conserved across species (Shen et al., 2002, 2005) and may thus be involved in binding unfolded polypeptides when dimerized/oligomerized. Alternatively, they might provide a binding site for potential UPR-associated regulatory factors. In addition, these ATF6 also contains conserved cysteines and conserved sites of potential glycosylation. In contrast to IRE1, mutations in conserved ATF6 glycosylation sites caused either constitutive activation or increased rate of ATF6 responses (Hong et al., 2004). Thus, these conserved glycosylation sites, as well as the conserved cysteines might be located on the surface of a polypeptide-binding pocket and play a role in regulating the kinetic response of ATF6 to different stress types. Further structural analysis of the ATF6 lumenal domain will help to answer such mechanistic questions.
Before the IRE1 crystal structure, the most widely discussed model of UPR sensor activation proposed a single mechanism for activating all three sensors. This model proposes that under nonstressed conditions, sensor activation is repressed by binding of the major ER chaperone BiP to sensor lumenal domains, whereas under stressed conditions, high levels of unfolded proteins cause BiP dissociation from UPR sensors, allowing activation to proceed (Bertolotti et al., 2000; Ma et al., 2002; Shen et al., 2002).
Based on the BiP dissociation model then, the kinetic behaviors of UPR sensor activation could be governed by the affinity of BiP for unfolded proteins or the dissociation constants of BiP from each UPR sensor. Our observations, however, suggest a more complicated scenario. On one hand, rapid dissociation of BiP from both IRE1 and PERK during Tg-induced UPR might suggest a high-affinity of BiP for unfolded protein induced by Tg (Figures 5 and 7). On the other hand, the slower activation of ATF6 by Tg suggests the opposite. In contrast, if the affinity of BiP for UPR sensors plays a major role, the kinetic order of IRE1, PERK, and ATF6 should be the same for any type of ER stress. However, this was not what we observed because ATF6 activation was rapid in response to DTT but slow in response to Tg (Figures 1 and 8). In addition, although the antibody we used to detect ATF6 by Western blot was unable to immunoprecipitate ATF6 (and thus subsequent examination of BiP association was not possible), the behavior of BiP with both IRE1 and PERK provide further support for the presence of an additional control(s). We observed that robust PERK autophosphorylation within 15 min of DTT or Tg treatment, but did not observe significant BiP dissociation until at least 30 min (Figure 5). We have also observed BiP reassociation at time points when IRE1 was still active (Figure 7). Thus, although BiP dissociation clearly occurs during UPR activation, the kinetic behavior of all three UPR sensors suggests the presence of additional regulatory mechanisms or components. Presumably, such additional control mechanism(s) would allow UPR sensors to fine tune both the speed and extent of activation, depending on specific types and magnitude of ER stress. Together, our observation is consistent with both recent x-ray crystal structural study and deletion analysis where the putative BiP-binding domain within yIre1 lumenal domain can be deleted without significant changes in Ire1 activity (Kimata et al., 2004).
Events downstream of sensor activation may also be regulated by additional components. During DTT-induced ER stress in CHO cells, PERK phosphorylation of eIF2α was inefficient although PERK activation, measured by either autophosphorylation or by BiP dissociation, occurred efficiently (Figures 3–5). Although several kinases are reported to phosphorylate eIF2α, we and others have shown that eIF2α phosphorylation does not occur in PERK knockout MEFs treated with UPR-inducing agents, indicating that PERK is the only kinase that phosphorylates eIF2α during the UPR (Supplementary Figure 2; Harding et al., 2000a, 2002b). Thus, additional components or regulatory mechanisms must also exist to explain the kinetics of eIF2α phosphorylation we observed. Curiously, because this alteration occurred only in CHO cells but not in mouse 3T3 cells, this regulatory machinery may be specific to certain cell types or species. Perhaps, the kinetics of UPR sensor activation can be further modulated this machinery, again to meet specific demands, as seen by kinetic production of ATF4 transcription factor followed the eIF2α phosphorylation kinetics (Supplementary Figure 4). Such localized modulation could also influence final outcomes of the UPR without drastically changing the signaling pathway.
Time-dependent expression of mammalian genes has been described previously (Yoshida et al., 2003). Transcription of certain genes regulated specifically by XBP1, including EDEM (involved in degrading glycosylated proteins), are up-regulated at much later times than some ATF6-regulated genes, including ER chaperones. This is due to the time required to produce UPR-specific transcription factors. The time required to produce active ATF6 transcription factor is considerably shorter than that to produce XBP1 because ATF6 requires only proteolytic cleavage of a pre-existing ATF6 protein, whereas XBP1 production requires mRNA splicing (by activated IRE1) and translation. The differential kinetics of UPR signaling branch activation could therefore alter the time and/or the magnitude of the downstream events they control while maintaining the overall order of ATF6 and XBP1 activation. For example, the time to produce functional XBP1 protein after ATF6 activation varies with the UPR-inducing agent. Depending on type of ER stress, this could be caused by modulating either the absolute amount of activated transcription factor produced or by controlling the extent of translational repression, with either one resulting in a best-fit UPR response.
Finally, our work also provides important insights to further understand the behavior of UPR sensors in physiological settings. During terminal differentiation of mature B-cells to plasma cells, IRE1 and ATF6, but not PERK, were found to be activated (Gass et al., 2002; Rutkowski et al., 2004). Our results here show that IRE1 and PERK respond similarly to different type of ER stress (disruption of glycosylation, disruption of disulfide bonds, ER calcium release). Thus, the lack of PERK activation during B-cell differentiation suggests an additional level of regulatory control in physiological settings. For example, a specific inhibitor might prevent PERK activation during B-cell differentiation. Future studies will be needed to uncover such additional regulatory mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Doug Cavener for providing Perk−/− and +/+ MEFs and Dr. Douglass Forbes for discussion and critical comments on the manuscript. We also thank Drs. Douglass Forbes, Randy Hampton, and Vivek Malhotra for critical reading of the manuscript and Alicia Bicknell, Aditi Chawla, and Ryan Brunsing for valuable discussion and their helpful comments throughout this work. This work was supported by a National Institutes of Health predoctoral training grant to J.B.D. and by the Searle Scholar Program to M.N. via the University of California, Cancer Research Committee and American Cancer Society Grant RSG-05-059-01-GMC to M.N.
Abbreviations used:
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- Tg
thapsigargin
- Tm
tunicamycin
- UPR
unfolded protein response.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0055) on May 3, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J. MHC restriction in three dimensions: a view of T cell receptor/ligand interactions. Cell. 1997;89:167–170. doi: 10.1016/s0092-8674(00)80195-6. [DOI] [PubMed] [Google Scholar]
- Calfon M., Zeng H. Q., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic-reticulum resident proteins requires a transmembrane protein- kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M., Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J. N., Gifford N. M., Brewer J. W. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- Gilmore R. Protein translocation across the endoplasmic-reticulum—a tunnel with toll booths at entry and exit. Cell. 1993;75:589–592. doi: 10.1016/0092-8674(93)90476-7. [DOI] [PubMed] [Google Scholar]
- Gunn K. E., Brewer J. W. The unfolded protein response in marginal zone and follicular zone B cells. FASEB J. 2003;17:C209–C209. [Google Scholar]
- Harding H. P., Calfon M., Urano F., Novoa I., Ron D. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Novoa I., Zhang Y. H., Zeng H. Q., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000a;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y. H., Bertolotti A., Zeng H. Q., Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 2000b;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding H. P., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M., Luo S. Z., Baumeister P., Huang J. M., Gogia R. K., Li M. Q., Lee A. S. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J. Biol. Chem. 2004;279:11354–11363. doi: 10.1074/jbc.M309804200. [DOI] [PubMed] [Google Scholar]
- Iwakoshi N. N., Lee A. H., Vallabhajosyula P., Otipoby K. L., Rajewsky K., Glimcher L. H. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Kaufman R. J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kimata Y., Oikawa D., Shimizu Y., Ishiwata-Kimata Y., Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire 1. J. Cell Biol. 2004;167:445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. H., Iwakoshi N. N., Glimcher L. H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen X. H., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R. J. IREI-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Q., Baumeister P., Roy B., Phan T., Foti D., Luo S. Z., Lee A. S. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 2000;20:5096–5106. doi: 10.1128/mcb.20.14.5096-5106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. Y., Schroder M., Kaufman R. J. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- Liu C. Y., Wong H. N., Schauerte J. A., Kaufman R. J. The protein kinase/endoribonuclease IRE1alpha that signals the unfolded protein response has a luminal N-terminal ligand-independent dimerization domain. J. Biol. Chem. 2002;277:18346–18356. doi: 10.1074/jbc.M112454200. [DOI] [PubMed] [Google Scholar]
- Ma K., Vattem K. M., Wek R. C. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J. Biol. Chem. 2002;277:18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- Mori K., Ma W. Z., Gething M. J., Sambrook J. A transmembrane protein with a Cdc2+/Cdc28-related kinase-activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Narasimhan J., Staschke K. A., Wek R. C. Dimerization is required for activation of eIF2 kinase Gcn2 in response to diverse environmental stress conditions. J. Biol. Chem. 2004;279:22820–22832. doi: 10.1074/jbc.M402228200. [DOI] [PubMed] [Google Scholar]
- Novoa I., Zeng H. Q., Harding H. P., Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2 alpha. J. Cell Biol. 2001;153:1011–1021. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Kimata Y., Higashio H., Tsuru A., Kohno K. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000;279:445–450. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- Papa F. R., Zhang C., Shokat K., Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- Patil C., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Rutkowski D. T., Kaufman R. J. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Sakai J., Duncan E. A., Rawson R. B., Hua X. X., Brown R. S., Goldstein J. L. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Sayeed A., Ng D.T.W. Search and destroy: ER quality control and ER-associated protein degradation. Crit. Rev. Biochem. Mol. Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001a;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Scheuner D., Song B. B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 2001b;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Shamu C. E., Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Shen J. S., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of golgi localization signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shen X., et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shen X. H., Ellis R. E., Sakaki K., Kaufman R. J. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C-elegans. PLoS Genet. 2005;1:355–368. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. G., Vattem K. M., Sood R., An J., Liang J. D., Stramm L., Wek R. C. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Thuerauf D. J., Morrison L., Glembotski C. C. Opposing roles for ATF6 alpha and ATF6 beta in endoplasmic reticulum stress response gene induction. J. Biol. Chem. 2004;279:21078–21084. doi: 10.1074/jbc.M400713200. [DOI] [PubMed] [Google Scholar]
- Thuerauf D. J., Morrison L. E., Hoover H., Glembotski C. C. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J. Biol. Chem. 2002;277:20734–20739. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- Tirasophon W., Welihinda A. A., Kaufman R. J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Anken E., Romijn E. P., Maggioni C., Mezghrani A., Sitia R., Braakman I., Heck A.J.R. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- Vattem K. M., Wek R. C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G. K., Rolls M. M., Rapoport T. A. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Johnson A. E. Signal sequence recognition and protein targeting to the endoplasmic-reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wang X. D., Sato R., Brown M. S., Hua X. X., Goldstein J. L. Srebp-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Wang X. Z., Harding H. P., Zhang Y., Jolicoeur E. M., Kuroda M., Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Rawson R. B., Komuro R., Chen X., Dave U. P., Prywes R., Brown M. S., Goldstein J. L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Hosokawa N., Kaufman R. J., Nagata K., Mori K. A time-dependent phase shift in the Mammalian unfolded protein response. Dev. Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Wong H. N., Song B., Miller C. N., Scheuner D., Kaufman R. J. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J. Clin. Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.