Abstract
Using molecular dynamics simulations, we examine the behavior of lipids whose preferred curvature can be systematically varied. This curvature is imposed by controlling the headgroup size of a coarse-grained lipid model recently developed by us. To validate this approach, we examine self-assembly of each individual lipid type and observe the complete range of expected bilayer and micelle phases. We then examine binary systems consisting of lipids with positive and negative preferred curvature and find a definite sorting effect. Lipids with positive preferred curvature are found in greater proportions in outer monolayers with the opposite observed for lipids with negative preferred curvature. We also observe a similar, but slightly stronger effect for lipids in a developing spherical bud formed by adhesion to a colloid (e.g., a viral capsid). Importantly, the magnitude of this effect in both cases was large only for regions with strong mean curvature (radii of curvature <10 nm). Our results suggest that lipid shape must act in concert with other physico-chemical effects such as phase transitions or interactions with proteins to produce strong sorting in cellular pathways.
INTRODUCTION
There are hundreds of different naturally occurring lipids that make up the internal and enclosing membranes of the cell. They are not just homogeneously distributed but are found in different concentrations in different organelles. Remarkably, this specificity occurs in a highly dynamic environment in which lipids are constantly recycled throughout the cellular machinery, yet lipids themselves lack lock-and-key type binding sites found on proteins and other function specific biomolecules. Examples of lipid sorting are numerous and include the differentiation between apical and basal regions of Golgi and endoplasmic reticulum (1), the function of sorting endosomes (2), and lipid selection during viral budding (3–5). Just what drives this sorting is currently one of the key mysteries of cell biology (6).
Consider the example of sorting that occurs between the endoplasmic reticulum and the plasma membrane. These regions differ markedly in composition although they are connected via vesicular endocytic and exocytic pathways. In fact, it is these pathways that are thought to be responsible for the sorting, and an indicator toward the mechanism by which they differentiate between lipids is the presence of highly curved tubular or vesicular regions that form and bud off from the parent membrane (1,2,6). The basic principle is that highly curved regions may selectively include or exclude certain lipids on the basis of their shape or stiffness. Neither the influence of shape or membrane stiffness is yet adequately understood, but recent key experiments involving simple model membranes have demonstrated that lipids with unsaturated tails (that typically form less stiff bilayers) become concentrated in tubular regions pulled from a vesicle (7), and in vivo experiments have demonstrated that lipids with greater tail unsaturation are sorted into pathways involving highly curved tubular intermediates (8). While these experiments have begun to address the issue of lipid fluidity/stiffness in partitioning different lipids, the influence of lipid shape has so far received little attention (2,8) and to verify theoretical predictions (9) it is essential that its coupling with membrane curvature be quantified.
It is important to remember that lipids can be sorted in different ways; either between leaflets of the bilayer (interleaflet) or between highly curved regions and flat regions of the membrane (interregion). Interregion and interleaflet sorting involve processes on different timescales (diffusion versus flip-flop) and depend differently on the makeup of the lipid mixture that comprises the membrane. To see why this is the case, consider the fact that there are always two leaflets to a bilayer and that wherever one leaflet has a curvature K, the other will have curvature ≈−K. This means that in a binary lipid system the curvature coupling will be approximately equal but opposite in each bilayer sheet. Such a situation is conducive to strong interleaflet sorting when there are two lipid species with opposing preferred curvature. On the other hand, interregion sorting is likely to require a third species with intermediate curvature that will partition into flat regions of the membrane. Other possible mechanisms for interregion sorting can be envisaged in biological systems where flip-flop is very slow, but our purpose here is not to examine such mechanisms. Instead we will focus on the fundamental effect that is common to both interleaflet and interregion sorting, that is, the coupling between lipid shape and membrane curvature.
As a final point of interest it is worthwhile considering the role of lipid shape when combined with nonequilibrium processes such as active concentration of lipids via flippases (10). This is essentially the reverse situation to passive sorting and represents a mechanism by which the shape of cellular components may be altered. Such active sorting is now thought to be commonplace and represents an important component in vesicle budding and movement processes (11).
Regardless of whether we are concerned with active or passive lipid sorting, one of the key physical processes underlying biological function is the coupling between lipid shape and membrane curvature. The aim of this study is to systematically examine the physics of this coupling. Firstly we would like to determine whether lipids with different shapes will preferentially distribute into monolayers with a different curvature. More importantly, we aim to quantify this effect, both as a function of the curvature and of the deviation of the lipid from a cylindrical shape.
MODEL SURFACTANTS AND SIMULATION METHODOLOGY
The goals of this study represent a particularly difficult simulation task. On the one hand, very large systems (10,000 lipids) and very long timescales are required. Yet at the same time, our goal of observing the effects of molecular shape necessitates the use of a particle-based model to represent individual lipids. This requirement clearly excludes non-particle-based techniques used for large-scale systems such as dynamic triangulation, and alternatively, our need for large systems rules out the use of fully atomistic simulations. This leaves us with coarse-grained particle-based lipid models, which cover a wide range of complexity from single beads per lipid (12) up to models that capture some chemical detail (13,14), and including many that lie in between (15–17). Most of these coarse-grained models employ explicit solvent particles and for certain two-dimensional geometries such as a flat bilayer this does not pose a serious performance drawback. For three-dimensional geometries such as vesicles and budding bilayers, however, the solvent typically accounts for >95% of computation time and limits the extent to which large length and timescales may be studied. Of course there remain situations where the effects of the solvent itself are of interest, or where hydrodynamics are important. In such cases the solvent particles are treated explicitly (15,18). An alternative to the explicit approach is to model the solvent implicitly via attractive interactions between tails, thereby achieving more than an order-of-magnitude gain in efficiency. This aim has been pursued for more than a decade (12) but only recent advances have led to models with well understood and tunable properties suitable for a wide variety of applications (see the review by Brannigan et al. (17) and references therein). In this study, we shall employ a model developed by us that is based on the use of broad tail attractions (19). Since this has previously been described in detail (20) we provide only a brief description here.
Lipids
Throughout this study we employ lipids consisting of a single head bead (spherical particle) and two tail beads (see Fig. 1). The size of these beads is fixed by using a purely repulsive (Weeks-Chandler-Andersen) potential
 |
(1) |
with rc = 21/6b and where ε is our unit of energy. The effective size of individual lipids is determined by the parameter b, which may be set to different values for head beads (bh) or tail beads (bt) to achieve different types of conical shapes. For simplicity, we kept the size of tail beads fixed at bt = σ, and varied the size of heads over the range bh = 0.7…1.4σ where σ is our unit of length. We shall see later that this encompasses a range of lipid spontaneous curvatures from strongly negative to strongly positive.
FIGURE 1.
Representation of the lipids used in this study showing the conical and inverted conical shapes obtained by varying the headgroup size bh.
The three beads are linked by two almost inextensible bonds and the lipid is made stiff through the use of a harmonic spring with rest length 4σ between head bead and second tail bead. Further details may be found in the reference (20).
Since no explicit solvent molecules are used, we introduce an attractive interaction between the tail beads to account for their hydrophobicity. Although the overall width of this attraction is the key to obtaining a realistic fluid bilayer phase we have previously shown that the precise details of this potential are of minor importance. We chose a cosine-based interaction whose width can be easily varied:
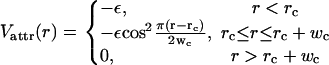 |
(2) |
This describes an attractive potential with depth ε that for r > rc smoothly tapers to zero. By tuning the range wc it is possible to alter the stiffness and fluidity of the resulting bilayer, which effectively mimics real lipid characteristics such as tail length and saturation.
Comparison with real membranes
Given the strongly coarse-grained nature of our lipids it is remarkable just how well the physical properties of the resulting simulated membranes correspond with those of real systems. Parameters such as stiffness, area stretching modulus, and rupture tension all lie within their experimentally observable ranges (20). Nevertheless, there is one important parameter that is different between simulated and real lipid systems. That parameter is the flip-flop rate, which is much faster in the simulation (20), and would be of concern if our study were to focus on the dynamics of exchange between leaflets. Since this study is not concerned with dynamics, a fast flip-flop rate is actually very useful since it allows us to reach equilibrium quickly. Interestingly, in biological (as opposed to synthetic experimental) systems, flippases catalyze the flip-flop process (21). Although some types of flippases are unidirectional—that is, they catalyze flip into one leaflet only—our simulations are also relevant for the passive case where catalysis is bidirectional.
Colloidal particles
In the section “Lipid Shape Coupling During Budding”, we simulate the partial wrapping of a colloidal particle by a section of bilayer membrane. This situation rather closely models the acquisition of membrane coats via budding as seen in many common animal viruses (22). In such cases the viral particles are wrapped due to their adhesion with the parent membrane. Nevertheless, our purpose was much more general. We simply wanted to obtain a membrane geometry close to that which would be found generally for all types of budding events, i.e., a spherical region, a neck, and a flat parent membrane.
For various technical reasons it was not convenient to model the colloid as a single large particle with an offset repulsive potential. Instead, it was constructed by creating a spherical mesh of small hard spheres enclosing a collection of soft filler particles. The mesh itself was generated by placing a single bead of size σ at each point in an optimal spherical covering (23). FENE bonds were then imposed between each bead and its near neighbors. To make the colloid rigid it was inflated with soft spheres with radius 2.5σ. These spheres interacted with each other via a simple harmonic repulsive potential U(r) = (kf/2)(rcut – r)2 where r is the interparticle distance, kf = 20ε is the strength of interaction, and rcut = 2.5σ is the cutoff distance. In each of the wrapping simulations in this work a single colloid of radius Rcolloid = 10σ consisting of 1002 cage atoms and 1000 filler spheres was used. Using a particle-based colloid model such as this entails several advantages. The first is that there are no particularly large particles in the system, which allows optimizations such as cell and particle lists in the integration to retain their efficiency gains. The second is that we now have additional control over the particle shape and interaction. In particular, we have constructed colloids in which one part of the cage surface adheres to the membrane and the other part does not. Such attractions, when used, were always imposed via a simple Lennard-Jones attractive potential with well-depth ε and equilibrium-distance σ.
Simulation details and units
We simulated systems consisting of the above-mentioned model lipids and possibly colloids in the absence of any explicit solvent particles. The attractive tail-width, wc, was set to 1.6σ, which corresponds to bilayers with a bending stiffness on the order of 12 kBT. Molecular dynamics simulations with a Langevin thermostat (fixing the temperature at kBT = 1.1ε) were used throughout, with a time step of δt = 0.01 τ and a friction constant Γ = τ−1 (in Lennard-Jones units). Constant volume simulations were performed using a box with sides Lx = Ly, Lz subject to periodic boundary conditions. If needed, constant tension conditions were also implemented via a modified Andersen barostat (24) allowing box resizing in x and y dimensions only (with a box friction Γbox = 2 × 10−4 τ−1 and box mass Q = 10−5).
All simulations were performed using the ESPResSo program (25) with additional bilayer-specific analysis and setup being performed by the MBtools add-on package.
The basic unit of length in our simulations is σ. As detailed by us in a previous article (20) it is possible to map this onto real lengths with the convenient result that σ ≈ 1 nm. Throughout this article, we shall use “nm” when referring to biologically relevant quantities such as vesicle size, but when referring to simulation details we shall continue to refer to σ.
SYSTEMS WITH HOMOGENEOUS LIPID COMPOSITION
The role of shape in determining the behavior of pure amphiphillic systems such as lipids or surfactants is quite complex, but the most basic effects may be summarized rather beautifully by calculating the packing parameter introduced by (26)
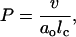 |
(3) |
where v and lc are the tail volume and length, respectively, and ao is the preferred head area per lipid. For spherical micelles we have the prediction that P ≲ 1/3, whereas for cylindrical micelles one expects 1/3 ≲ P ≲ 1/2, and for bilayers, P ≳ 1/2 (26). In the spirit of this geometrical approach recent work has analyzed the more complicated case of branched micelles (27) and several simulations (28–30) and experiments (31–34) have varied headgroup size and chain length to obtain a range of P sufficient to induce changes to micelle structure. Such variations can be obtained through a change in salt concentration, a change in pH, or by using specific lipid types such as diacylglycerol or lysophospholipids, which have strong intrinsic curvature that depends on acyl-chain length and head chemistry (35–37).
Since our study focuses on lipid shape and its interaction with membrane curvature, it is essential that our model reproduce the major known surfactant phases and their relationship with P. One of the objectives of this section will therefore be to check that this is the case by examining the self-assembly of pure systems of our lipids with varying headgroup size. This will also serve as a reference point for matching different lipids to form mixed systems.
By making the headgroup larger than the tail beads one can achieve a conical shape, and by using small headgroups, a cone of the opposite orientation can be obtained. To encompass a wide variety of surfactant behaviors we varied the headgroup size bh over the range 0.7…1.4σ. Head size is evidently related to the packing parameter P; however, the relation is not always easy to calculate accurately since ao and v/lc must be obtained from the model. In our case, we know that the transition from wormlike to spherical micelles occurs for bh = 1.4σ and therefore, in that case, P ≈ 1/3. We also know that for a flat membrane with bh = 1.0σ we have ao = 1.24σ2. Assuming that for all our phases 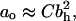 where C is a constant, we can now obtain the relation
where C is a constant, we can now obtain the relation 
In Fig. 2, we present a snapshot of each of our self-assembled systems, taken at the end of a long simulation run where the energy of the system indicated that it had reached equilibrium (after ∼15,000τ). Remarkably, our simple model was not only able to capture the most basic phases such as bilayers, worms, and spherical micelles but also encompasses the more subtle phenomena of branched wormlike micelles. The values of P at which our micellar phases occur sit within their expected ranges (see Fig. 2 legend), which is pleasing, given the rather approximate calculation of ao used. What is more important, however, is that each phase appears in its correct relative position along a gradient of P. Starting with values of P > 1 we first observe an unstructured globular phase. Given the importance of the headgroup in determining lipid ordering, the existence of such a globule for lipids with very small heads is unsurprising. As P is decreased, one finds a range of values over which bilayer phases are observed, 0.6 ≲ P ≲1.0 , and then for P ≈ 0.55, one finds a cross-linked wormlike micelle phase followed by non-cross-linked worms, 0.33 ≲ P ≲ 0.5, and eventually spheres P < 0.33.
FIGURE 2.
Various single component self-assembled phases corresponding to lipids with headgroup sizes bh in units of σ: (a) 0.7; (b) 0.9; (c) 1.1; (d) 1.2; and (e) 1.4. Corresponding values of the shape parameter P are (a) 1.4; (b) 0.83; (c) 0.55; (d) 0.46; and (e) 0.33. A schematic representation of the constituent lipids is shown next to each of the phases. All simulations correspond to 4000 lipids in a cubic box of side length 50σ. In the case of snapshot e, a simulation with only 800 lipids was used for visual clarity (shown) but identical spherical micelles were obtained with 4000 lipids. Note that although it is not completely clear from the snapshot, none of the wormlike micelles in simulation d have any branches.
One particular point of interest in Fig. 2 is the presence of both branched and unbranched cylindrical micelles. Using fluorescence imaging and electron microscopy experiments (33,34,38), it is possible to directly view wormlike micelles, and both branched and unbranched structures have been observed. It is also well known that the addition of salt to certain surfactant solutions results in the formation of branched or cross-linked micelle structures rather than entangled worms (32,38,39). This is thought to occur because the addition of salt acts to screen repulsions between ionic headgroups leading to a decrease in effective headgroup area (27). Previous simulation results on dimeric surfactants would tend to agree with this geometrical explanation (30) but also permitted more complicated theories based on the presence of two surfactant tails (30). Our observation of branched micelles in a simple, single-chained surfactant model confirms that two tails are not required for the formation of branched wormlike micelles and that pure packing considerations are the most likely cause. The ability of our very simple model to reproduce such subtle changes in morphology is remarkable, and gives us further confidence in our implementation of preferred lipid curvature.
Another notable feature of Fig. 2 is the sheer range of phases that can be observed. This is particularly useful because it has been achieved within a single consistent model framework and depends (as predicted) simply on headgroup size. We therefore anticipate that a good deal remains to be learned by applying the present simulation model to micellar systems.
The final point to note about Fig. 2 is that a bilayer will only form for relatively small deviations from the ideal head-size, and only where such deviations are toward smaller heads. Since our goal was to study mixed binary systems forming stable bilayers, we designed pairs of lipid types such that the average of their head-bead sizes would be the ideal value 1.0σ. We tested three such mixtures with head-bead ratios (1.1:0.9), (1.2:0.8), and (1.3:0.7) for their ability to self-assemble to bilayer structures (vesicles or lamellae). Only the two least asymmetric mixtures actually self-assembled to bilayers on the timescales of our simulations. In the case of the (1.3:0.7) mixture, spherical micelles rapidly formed but coalesced extremely slowly and we could not be certain that a bilayer would ever form.
MIXED LIPID SYSTEMS: CURVATURE COUPLING ON SPHERICAL VESICLES
The possibility that lipid shape could couple with membrane curvature to produce a sorting mechanism presents us with the following questions: How strongly curved must a membrane be to produce a sizeable sorting effect? How does this effect vary with the degree of asymmetry of the lipid? The purpose of this section is to examine these two questions systematically by using the simplest possible curved systems, namely spherical vesicles.
We simulated vesicles ranging in size from 1000 up to 16,000 constituent lipids, corresponding to a range of radii from ≈5 nm up to ≈30 nm. All vesicles were composed of equal parts large-headed lipids and small-headed lipids, where the two head sizes were matched to give an overall neutral curvature effect. In the previous section we mentioned two cases where such binary mixtures self-assemble to flat bilayer sheets. These corresponded to the head-size ratios (1.1:0.9) and (1.2:0.8). We restricted our efforts to these two cases.
After preformed vesicles had reached equilibrium we were able to examine the profile of bead densities as a function of radius. Fig. 3 shows a typical plot of such a profile in which the outer and inner leaflets as well as the densities of heads and tails of each lipid type can be seen. Due to shape fluctuations, the peaks in this plot are broadened; however, one can nevertheless see clear evidence of a coupling between curvature and shape. The density of large-headed lipids is appreciably larger in the outer leaflet than the inner one, and the opposite is true for small-headed lipids. This simple result indicates that lipid shape can act as a sorting criterion. What remains to be seen is precisely how strong an effect it is, particularly for vesicles on the order of the sizes seen in the endosomal pathway, 50 nm. To quantify the enhancement or sorting effect between bilayer leaflets, we calculated the ratios
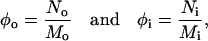 |
(4) |
where No and Ni indicate the number of large-headed lipids in the outer and inner leaflets, respectively, and where Mo and Mi are the total number of lipids in these leaflets. If a curvature-shape coupling exists, then these ratios should be related to the local curvature of the bilayer K and to the curvature Kℓ that would be optimal for the lipids. We constructed a simple theory to model this, based on the following two assumptions. First, K and Kℓ are harmonically coupled; i.e., the energy of a lipid with its own curvature Kℓ embedded into a monolayer of curvature K is given by
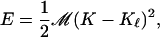 |
(5) |
where the modulus ℳ measures the strength of this coupling. Second, the curved lipids behave like an ideal lattice gas in the membrane plane; i.e., the entropy of mixing is given by
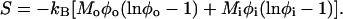 |
(6) |
FIGURE 3.
Radial density profile across the two monolayers of a vesicle constructed from 2000 lipids with head size 1.1σ and 2000 lipids with head size 0.9σ. Lipids with large heads are represented by bold lines and lipids with small heads are represented by light lines. In both cases, head beads correspond to solid lines and tail beads to dashed lines.
Minimizing the resulting free energy subject to the constraint of a given total number of lipids results in the pleasingly simple relation
 |
(7) |
where β = 1/kBT is the inverse thermal energy and ℳ is the coupling constant between lipid and bilayer curvatures. In the limit of zero curvature, i.e., a flat bilayer, the term on the left goes to 0. Based on the present analysis it is impossible to determine either ℳ or Kℓ independently, so we shall refer instead to their product, 𝒥 = ℳKℓ, which summarizes the strength of curvature coupling for a particular lipid type.
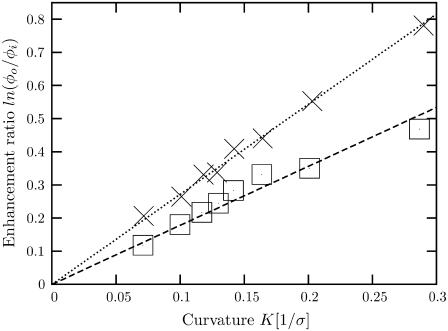
To test the validity of Eq. 7 and also to determine the strength of coupling effects for our binary systems, we calculated time averages of φi and φo for equilibrated vesicles ranging in radius from ≈5 nm up to ≈30 nm. In each case, the curvature was calculated from K = 2/R, where R is the radius at the bilayer midplane. The results are plotted in Fig. 4 for the two different lipid mixtures used. We find a remarkable agreement with theory even for very tightly curved vesicles and observe that the range of vesicle sizes used (5–30 nm) gives rise to a good spread of the possible values of the enhancement ratio (left-hand side of Eq. 7). Naturally we also found that the more asymmetric the lipid curvatures in our mixture, the stronger the coupling. We obtained 𝒥 = 5.58 pN nm2 for the lipid mixture with head ratio (1.2:0.8) and 𝒥 = 3.65 pN nm2 for the lipid mixture with head ratio (1.1:0.9). The large difference in coupling constants between these two mixtures suggests that very strong effects should be observed for lipids with more extreme curvature. In that case, however, one would require a lipid mixture with a small volume fraction of the extremely curved species in order to maintain bilayer stability.
FIGURE 4.
Degree of interleaflet lipid sorting as a function of curvature for two different lipid asymmetries. The logarithm of the ratio of outer and inner lipid fractions φo and φi (see text) is plotted against curvature K, which is computed from the mean vesicle radius at the bilayer midplane. Open squares correspond to lipids with head size 1.1σ and crosses correspond to those with head size 1.2σ. The lines are fits to Eq. 7.
Let us now take note of the lipid sorting effect in light of biological function. Our results convincingly show that lipids can be partitioned on the basis of curvature. This could potentially be important in any instance where strong membrane bending is involved, such as, for example, in endosomal tubules or vesicles, pearling of tubules and fission, or budding processes. Our results indicate, however, that the magnitude of any real effect will depend strongly on the curvatures of membrane and lipids involved. In particular, if we consider tubules and vesicles in the endosomal pathway whose radii of curvature are on the order of 50 nm, the strength of shape-induced partitioning is likely to be quite weak (≲5%) compared with the very efficient sorting (≈95%) seen in experiments (8). It therefore seems doubtful whether curvature alone could give rise to the sorting seen in endosomal tubules and vesicles. Taking lipids with very strong asymmetry such as lysophospholipids (35) or diacylglycerol (36) would clearly increase the effect, but in order to maintain a stable bilayer membrane, such lipids would need to be present in relatively small fractions. Such cases may well be biologically important but are beyond the scope of our study.
At the interface between a tubule or partially formed vesicle and the parent membrane, curvatures can become very strong indeed. In such cases, the mean curvature is typically zero, although its two principle components can be very large. Just how shaped lipids would interact with such an environment remains an important question and one which we shall attempt to address in the following section.
LIPID SHAPE COUPLING DURING BUDDING
Having noted in the previous section that the coupling between lipid shape and curvature is weak when curvature radii exceed values of ∼20 nm, we now turn to the question of whether effects can be seen in the peculiar membrane geometries that occur during budding. To generate such a state we shall examine the wrapping of a membrane around an adhesive colloid. In so doing, we shall also observe any influence of the adhesive potential itself on the shape-curvature coupling.
As detailed in the section “Colloidal Particles”, we constructed a model spherical colloid from many small beads. Since budding is a dynamical process and eventually proceeds to the complete pinching off of a bud, we needed to take steps to prevent our system from proceeding to completion. (Note that this process occurs in the absence of fission proteins. The most likely reason is that our colloid adhesion potential is comparatively strong. The strength of colloid adhesion has little bearing on this study.) To do this and to obtain a partially wrapped state at equilibrium, we coated 85% of the area of our colloid with beads that adhere to the surface of our membrane (i.e., an equally strong adhesion to all head beads). The remaining 15% was coated with simple hard spheres that did not interact with the membrane. This small 15% region was located as a patch at the top of the colloid (see inset to Fig. 5).
FIGURE 5.
Snapshot of lipid membrane consisting of 4000 lipids with head size 1.1σ (light beads) and 4000 lipids with head size 0.9σ (dark beads). All tail beads are shown in white. The colloid itself is shown as an inset to clearly display the adhesive (light) and nonadhesive (dark) regions. Black circles indicate the two principle curvatures in the neck region.
Although we would ultimately like to have simulated wrapping for systems with a radius on the order of 50 nm, this was simply not practical, due to the very long equilibration times required for shape deformations, bilayer fluctuations, and lipid diffusion. As a compromise, we simulated the wrapping of a colloid with radius 10 nm by a patch of membrane with side ≈75 nm (8000 lipids) at constant vanishing lateral tension. The system was set up with the colloid just touching the surface of a flat bilayer consisting of a random distribution of the two lipid types. The adhesion between bilayer and colloid quickly lead to the desired partially wrapped state (after ∼10,000τ; see Fig. 5), and we then simulated for a further 20,000τ to allow the system to reach equilibrium. Time-averaged quantities were then taken over the remaining 40,000τ of simulation time.
We performed wrapping simulations on both of the lipid mixtures used in the previous section, but the more asymmetric of these underwent rupture at the interface between colloid and parent membrane. That such rupture should occur is not surprising, given the strong curvatures of the constituent lipids; and it is also consistent with the behavior of biological membranes. It was shown long ago that the structural integrity of plasma membrane in Acholeplasma laidlawii depends heavily on the shape of its constituent lipids (40). We present results here only for the mixture with lipid/head ratio (1.1:0.9).
For the section of membrane that adheres to our colloid, it is easy to obtain a profile of lipid densities across the bilayer. Applying the same analysis as we did previously for vesicles, we obtained a value of ∼0.2 for the natural log of the ratio φo/φi. This is actually considerably greater than the value (0.15) expected for a vesicle with the same radius as the bud that wraps our colloid (12.5σ; as obtained from Fig. 4). This additional enhancement is most likely due to the influence of the adhesion potential between colloid and membrane: Small-headed lipids are able to pack more tightly at the surface of the colloid and are therefore favored over those with large heads, simply due to the head-colloid attraction. This acts in concert with the effect of curvature, and an additional sorting is produced.
Since the geometry of the nonattached portion of the bilayer is considerably more complex than that of a simple sphere, we could not perform our trans-bilayer density analysis for all regions of the system. In particular, we wanted to examine the distribution of big and small-headed lipids at the neck region. To do this we created a density map utilizing the cylindrical symmetry of the system to reduce it to two dimensions. In Fig. 6, the density distribution of tail beads for large-headed lipids is shown. For clarity, we also show the ratio of densities between tail beads for large-headed and small-headed lipids. One can clearly identify the colloid and the relative preference of large-headed lipids for the outer monolayer in the part of the bilayer that wraps around it. What is more interesting, however, is that this segregation of large- and small-headed lipids is restricted to the colloidally wrapped region. Looking at our two-dimensional representation, this might at first seem surprising since the neck region at the interface between colloid and membrane appears to be very strongly curved and should therefore sort our lipids on the basis of their shape. In fact, however, our two-dimensional projection shows only one component of the curvature. There is another component whose radius is the radius of the neck itself and which has the opposite sign. Indeed, according to theoretical predictions that model the membrane as a simple elastic sheet (41), its shape at equilibrium is such that both opposing curvatures in the neck will essentially average to zero. Given that this theory ignores any possibilities arising due to mixtures of differently shaped lipids, it is interesting to see that its predictions appear to be borne out despite the additional complexities in our system.
FIGURE 6.
Radially projected density maps. The left plot shows the logarithm of the density of tail beads tb for large-headed lipids. The right plot shows the differential density ln(ts/tb) between large- and small-headed lipid tails, where ts is the density of tail beads for small lipids. In both maps a perfectly circular region can be seen that corresponds to the location of the wrapped colloid. The bar shown indicates correspondence between shading and relative values but its meaning is different for left and right plots. In the left plot, lighter hues indicate lower densities and darker hues, higher densities. In the right plot, the extremes of the spectrum indicate a strong difference in densities whereas hues in the center indicate identical values. The key point to note in the left plot is a dark band along the outer monolayer of the spherical region, which indicates a high density of large-headed lipids. In the right plot, one should note the extreme contrast between layers in the spherical region compared with the completely identical layers outside this region.
CONCLUSIONS
We have investigated the coupling between local membrane curvature and the distribution of lipids with noncylindrical shapes. Our study has focused on a simple two-component lipid system and analyzed the partitioning between individual monolayers of the membrane. We have demonstrated that appreciable sorting of this type can occur where the curvature is very high such as in vesicles or tubules with radii on the order of 10 nm. For radii of curvature more likely to be encountered in the endosomal pathways of living cells, 50 nm, this sorting effect was relatively minor. By examining their self-assembly in the pure phase, we were able to determine a reference point against which the behavior of the model lipids used to make these measurements could be checked. By varying lipid headgroup size we were able to obtain all major known lipid micelle and bilayer phases including branched wormlike micelles. The lipids used to measure the strength of sorting effects were those that formed branched and unbranched wormlike micelles in the pure phase. This suggests that the model lipids examined by us correspond to real lipids of moderate to strong curvature but not the strongest possible in a biological system.
Since regions of high curvature may form at the junction of vesicles or cylinders with a parent membrane we examined the distribution of differently shaped lipids in the neck of a partially formed bud. Although the individual components of the curvature in this region were undoubtedly high, no difference in lipid composition was observed between monolayers. We attribute this to the fact that the mean curvature is close to zero in such regions. This result suggests that it is the mean curvature and not its individual components that determine the degree of curvature-shape coupling.
The main conclusion of this work is that although coupling between curvature and shape may occur, it is likely to be a relatively weak effect in biologically sized systems. This does not mean, however, that the curvature of lipids is unimportant. It may be the case that lipid shape acts in concert with other physico-chemical properties to produce a sorting mechanism. For example, a system of differently shaped lipids that is poised close to a phase boundary could be pushed over it by an increase in the local curvature. The occurrence of a relatively weak cause that triggers a subsequent strong effect is indeed a motif frequently encountered in biology.
Acknowledgments
We thank Eva Sinner, Niko van der Vegt, and Kurt Kremer for useful comments on the manuscript.
We gratefully acknowledge financial support by the German Science Foundation under grants No. DE775/1-2 and No. DE775/1-3.
References
- 1.van Meer, G. 2005. Cellular lipidomics. EMBO J. 24:3159–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukherjee, S., and F. R. Maxfield. 2000. Role of membrane organization and membrane domains in endocytic lipid trafficking. Traffic. 1:203–211. [DOI] [PubMed] [Google Scholar]
- 3.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038–2044. [DOI] [PubMed] [Google Scholar]
- 4.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen, D. H., and J. E. K. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holthuis, J. C. M., and T. P. Levine. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6:209–220. [DOI] [PubMed] [Google Scholar]
- 7.Roux, A., D. Cuvelier, P. Nassoy, J. Prost, P. Bassereau, and B. Goud. 2005. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 24:1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee, S., T. T. Soe, and F. R. Maxfield. 1999. Endocytic sorting of lipid analogues differing solely in the chemistry of their hydrophobic tails. J. Cell Biol. 6:1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shemesh, T., A. Luini, V. Malhotra, K. N. K. Burger, and M. M. Kozlov. 2003. Prefission constriction of Golgi tubular carriers driven by local lipid metabolism: a theoretical model. Biophys. J. 85:3813–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farge, E., D. M. Ojcius, A. Subtil, and A. Dautry-Varsat. 1999. Enhancement of endocytosis due to aminophospholipid transport across the plasma membrane of living cells. Am. J. Physiol. 276:C725–C733. [DOI] [PubMed] [Google Scholar]
- 11.McMahon, H. T., and J. L. Gallop. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438:590–596. [DOI] [PubMed] [Google Scholar]
- 12.Drouffe, J.-M., A. C. Maggs, and S. Leibler. 1991. Computer simulations of self-assembled membranes. Science. 254:1353–1356. [DOI] [PubMed] [Google Scholar]
- 13.Marrink, S.-J., A. H. de Vries, and A. E. Mark. 2004. Coarse-grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 108:750–760. [Google Scholar]
- 14.Shelley, J. C., M. Y. Shelley, R. C. Reeder, S. Bandyopadhyay, and M. L. Klein. 2001. A coarse-grain model for phospholipid simulations. J. Phys. Chem. B. 105:4464–4470. [Google Scholar]
- 15.Shillcock, J. C., and R. Lipowsky. 2005. Tension-induced fusion of bilayer membranes and vesicles. Nat. Mater. 4:225–228. [DOI] [PubMed] [Google Scholar]
- 16.Groot, R. D., and K. L. Rabone. 2001. Mesoscopic simulation of cell membrane damage, morphology change and rupture by nonionic surfactants. Biophys. J. 81:725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brannigan, G., L. C. Lin, and F. L. H. Brown. 2005. Implicit solvent simulation models for biomembranes. Eur. Biophys. J. 35:104–124. [DOI] [PubMed] [Google Scholar]
- 18.Smit, B., P. A. J. Hilbers, K. Esselink, L. A. M. Rupert, N. M. van Os, and A. G. Schlijper. 1990. Computer simulations of a water/oil interface in the presence of micelles. Nature. 348:624–625. [Google Scholar]
- 19.Cooke, I. R., K. Kremer, and M. Deserno. 2005. Tuneable, generic model for self-assembling fluid bilayer membranes. Phys. Rev. E. 72:011506. [DOI] [PubMed] [Google Scholar]
- 20.Cooke, I. R., and M. Deserno. 2005. Simulation of fluid bilayers without solvent: an approach based on broad attractive potentials. J. Chem. Phys. 123:224710. [DOI] [PubMed] [Google Scholar]
- 21.Pomorski, P., J. C. M. Holthuis, A. Herrmann, and G. van Meer. 2004. Tracking down lipid flippases and their biological functions. J. Cell Sci. 117:805–813. [DOI] [PubMed] [Google Scholar]
- 22.Garoff, H., R. Hewson, and D.-J. E. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardin, R. H., N. J. A. Sloane, and W. D. Smith. Tables of spherical codes with icosahedral symmetry. Published electronically at http://www.research.att.com/∼njas/icosahedral.codes/.
- 24.Kolb, A., and B. Dünweg. 1999. Optimized constant pressure stochastic dynamics. J. Chem. Phys. 111:4453–4459. [Google Scholar]
- 25.Limbach H. J., A. Arnold, B. A. Mann, and C. Holm. 2006. ESPResSo—an extensible simulation package for research on soft matter systems. Comp. Phys. Comm. 174:704–727. [Google Scholar]
- 26.Israelachvili, J. N., D. J. Mitchell, and B. W. Ninham. 1976. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc., Faraday Trans. 2. 72:1525–1566. [Google Scholar]
- 27.May, S., Y. Bohobot, and A. Ben-Shaul. 1997. Molecular theory of bending elasticity and branching of cylindrical micelles. J. Phys. Chem. B. 101:8648–8657. [Google Scholar]
- 28.Nelson, P. H., G. C. Rutledge, and T. A. Hatton. 1997. On the size and shape of self-assembled micelles. J. Chem. Phys. 107:10777–10781. [Google Scholar]
- 29.Maillet, J. B., V. Lachet, and P. V. Coveney. 1999. Large-scale molecular dynamics simulation of self-assembly processes in short and long chain cationic surfactants. Phys. Chem. Chem. Phys. 1:5277–5290. [Google Scholar]
- 30.Karaborni, S., K. Esselink, P. A. J. Hilbers, B. Smit, J. Karthäuser, N. M. van Os, and R. Zana. Simulating the self-assembly of Gemini (dimeric) surfactants. Science. 266:254–256. [DOI] [PubMed]
- 31.Porte, G., R. Gomati, O. El Haitamy, J. Appell, and J. Marignan. 1986. Morphological transformations of the primary surfactant structures in brine-rich mixtures of ternary systems (surfactant/alcohol/brine). J. Phys. Chem. 90:5746–5751. [Google Scholar]
- 32.Khatory, A., F. Kern, F. Lequeux, J. Appell, G. Porte, N. Morie, A. Ott, and W. Urbach. 1993. Entangled versus multiconnected network of wormlike micelles. Langmuir. 9:933–939. [Google Scholar]
- 33.Gustafsson, J., G. Orädd, G. Lindblom, U. Olsson, and M. Almgren. 1997. A defective swelling lamellar phase. Langmuir. 13:852–860. [Google Scholar]
- 34.Dalhaimer, P., H. Bermudez, and D. E. Discher. 2003. Biopolymer mimicry with polymeric wormlike micelles: molecular-weight scaled flexibility, locked-in curvature, and coexisting microphases. J. Polym. Sci. [B]. 42:168–176. [Google Scholar]
- 35.Rand, R. P., and N. Fuller. 2001. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophys. J. 81:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szule, J. A., N. L. Fuller, and R. P. Rand. 2002. The effects of acyl chain length and saturation of diacylglycerols and phosphatidylcholines on membrane monolayer curvature. Biophys. J. 83:977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuller, N., C. R. Benatti, and R. P. Rand. 2003. Curvature and bending constants for phosphatidylserine-containing membranes. Biophys. J. 85:1667–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danino, D., Y. Talmon, H. Levy, G. Beinert, and R. Zara. 1995. Branched thread-like micelles in an aqueous-solution of a trimeric surfactant. Science. 269:1420–1421. [DOI] [PubMed] [Google Scholar]
- 39.Kadoma, I. A., and W. van Egmond. 1998. Flow-induced nematic string phase in semidilute wormlike micelle solutions. Phys. Rev. Lett. 90:5679–5682. [Google Scholar]
- 40.Wieslander, A., A. Christiansson, L. Rilfors, and G. Lindblom. 1980. Lipid bilayer stability in membranes. regulation of lipid composition in Acholeplasma laidlawii as governed by molecular shape. Biochemistry. 19:3650–3655. [DOI] [PubMed] [Google Scholar]
- 41.Fourcade, B., L. Miao, M. Rao, and M. Wortis. 1994. Scaling analysis of narrow necks in curvature models of fluid lipid-bilayer vesicles. Phys. Rev. E. 49:5276–5286. [DOI] [PubMed] [Google Scholar]