Abstract
The Hsp70 chaperone system is the major molecular chaperone system that assists protein-folding processes in all cells. To understand these processes, we analyzed the kinetic characteristics of the Escherichia coli homologs of this chaperone system during folding of a denatured protein using computer simulations and compared the results with in vitro refolding experiments. Rate constants used for the model were derived from recent literature or were determined and scrutinized for their applicability to the refolding reaction. Our simulation results are consistent with reported laboratory experiments, not only simulating the refolding reaction of wild-type proteins but also the behavior of mutant variants. Variation of kinetic parameters and concentrations of components of the Hsp70 system demonstrate the robustness of the chaperone system in assisting protein folding. Furthermore, the importance of the synergistic stimulation of the ATPase activity of Hsp70 is demonstrated. The limitations of our kinetic model indicate sore spots in our understanding of this chaperone system. Our model provides a platform for further research on chaperone action and the mechanism of chaperone-assisted refolding of denatured proteins.
INTRODUCTION
Although the entire information for the precise three-dimensional structure of a protein is encoded in its amino acid sequence, in vivo, in the crowded environment of a cell, many proteins depend on the assistance by molecular chaperones such as Hsp70 and Hsp60 heat-shock proteins for folding from nascent state into their correct structure (1,2). Once in the native, active state, a protein is not safe but constantly endangered to loose its active conformation by denaturation and misfolding due to collisions with other cellular components as result of Brownian motion. This is particularly prevalent under stress conditions such as elevated temperature, which is counteracted by living organisms through the induction of a small set of proteins, called heat-shock proteins, that protect denatured proteins from aggregation and assist their refolding into the native state.
The Escherichia coli Hsp70 chaperone machinery, consisting of the Hsp70 homolog DnaK and the cochaperones DnaJ and GrpE, was shown to be the most efficient chaperone system in E. coli preventing aggregation of many proteins of different sizes in vivo at elevated temperatures and refolding them to the native state after return to optimal growth temperature (3,4).The DnaK system can efficiently repair denatured model proteins such as Photinus pyralis luciferase both in vivo and in vitro but cannot protect it from heat induced activity loss (5). Experimental findings suggest that this refolding process is achieved by ATP-dependent transient interaction between the DnaK chaperone with a short peptide stretch within the substrate polypeptide (S) (6). DnaJ and GrpE function as regulators in this system by stimulating DnaK's ATP hydrolysis activity and subsequent nucleotide exchange (7–10).
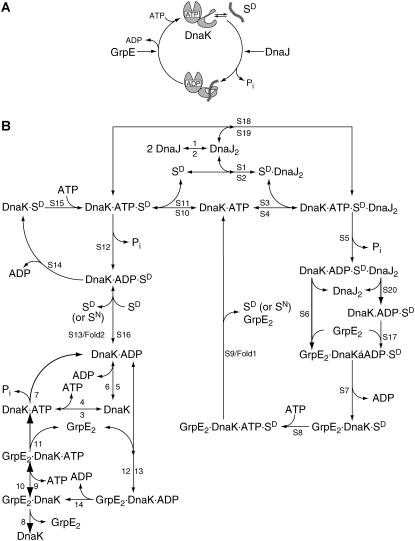
The kinetics of the DnaK chaperone system has been studied extensively in vitro. These studies elucidated that the nucleotide-regulated transition between two conformational states is crucial to the function of DnaK. These states are the ATP-bound state with high association and dissociation rates but low affinity for substrates and the ADP-bound state with two and three orders of magnitude lower association and dissociation rates but high affinity for substrates (Fig. 1 A) (11–13). The spontaneous transition between the two states is extremely slow but stimulated by substrates and DnaJ synergistically. Under physiological conditions of high ATP concentrations, nucleotide exchange is rate limiting for substrate release. However, nucleotide exchange of DnaK is slow but stimulated 5000-fold by the nucleotide exchange factor GrpE. Based on the results of a number of laboratories, Schröder et al. (5) suggested a chaperone folding cycle, which was further elaborated by McCarty et al. (8), Karzai and McMacken (14), and Laufen et al. (10) and is meanwhile widely accepted. In this proposed mechanism, an unfolded protein substrate (e.g., P. pyralis luciferase) first associates with DnaJ, which will present it to DnaK·ATP and induce the formation of a trimeric DnaK·ATP·DnaJ·substrate complex. DnaJ and substrate synergistically stimulate ATP hydrolysis by DnaK and thereby trigger the transition of DnaK from the ATP state with low affinities for substrates to the high-affinity ADP state. GrpE binds to the latter complex and catalyzes the release of ADP. Subsequent ATP binding induces conformational changes in the ATPase domain and substrate-binding domain leading to a rapid dissociation of GrpE and substrate from the complex. These steps form the cycle of DnaK-assisted protein folding (Fig. 1 A). With enough ATP and all the chaperone molecules, after many cycles, the substrate can be refolded back to its active state (Fig. 2 A; see also (5)).
FIGURE 1.
DnaK chaperone cycle. (A) Schematic ATPase cycle of the DnaK system. (B) Elemental chemical equations for the refolding of a denatured protein substrate SD by the DnaK system. With a certain probability the substrate SD can be refolded to the native state SN within any given cycle of binding, ATP hydrolysis, and release simulated as alternative reactions at the indicated positions. Numbers at the arrows indicate the reaction number in Table 1, where by, at horizontal arrows, the top number indicates the reaction from left to right, and the bottom number indicates the reaction from right to left; at vertical arrows, the left number indicates the reaction from top to bottom, and the right number indicates the reverse reaction.
FIGURE 2.
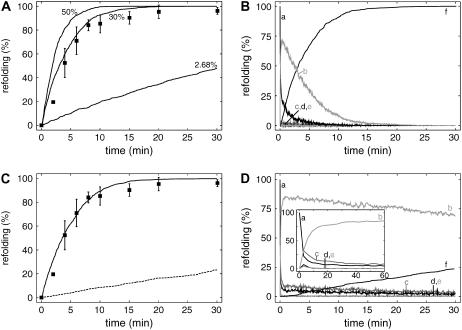
Refolding probability and model validation. (A) Refolding reaction in dependence of refolding probability as indicated. Experimental data of in vitro refolding chemically denatured luciferase (▪; data from (13)). Error bars represent standard deviation of three independent experiments. (B) Distribution of the substrate between the different pools: (a) SD; (b) DnaK·ATP·SD; (c) DnaK·ADP·SD·DnaJ2; (d), DnaK·ATP·SD·DnaJ2; (e) GrpE2·DnaK·ATP·SD; and (f) SN. (C) Dependence of ATP consumption on refolding probability as indicated. Basal ATP consumption of the system without substrate (dashed line).
Here, we describe a kinetic model for the chaperone action of E. coli DnaK in protein refolding based on the different interactions of the five components of the systems DnaK, DnaJ, GrpE, substrate, and nucleotide reported in the literature (Fig. 1 B). Due to a lack of experimental data, this model is currently restricted to the simulation of the in vitro situation excluding effects of crowding and diffusion inside the cell. The rate constants were derived from the literature or completed by our experiments. Our model correctly simulates the behavior of DnaK's chaperone action including the behavior of DnaK mutant proteins with altered kinetic parameters. The sensitivity of refolding productivity to alterations in activity and concentration of the chaperones and ATP consumption are discussed.
MATERIALS AND METHODS
Reaction and parameters
All of the reactions and parameters used in this computer model were either based on published literature or measured in the laboratory of M.P.M. The list of reaction equations, including parameters and references, are given in Table 1, together with the buffer conditions used in the measurements. Parameters that were determined in different laboratories using buffers that were different from the buffer used for firefly luciferase were checked exemplarily in our laboratory in the appropriate buffer and found to be similar. For reactions that start with the same reactants but have alternative products depending on a probability factor, rate constants are assigned to accommodate the desired probability. The reaction volume is set to a typical volume of an E. coli cytoplasm.
TABLE 1.
Reaction list for the kinetic model of the DnaK chaperone system
| No.* | Reactions | Rates | References/conditions |
|---|---|---|---|
| 1 | 2 DnaJ → DnaJ2 | 105 M−1 s−1 | Estimated† |
| 2 | DnaJ2 → 2 DnaJ | 10−4 s−1 | Estimated† |
| 3 | DnaK + ATP → DnaK·ATP | 1.3 × 105 M−1 s−1 | (47) A |
| 4 | DnaK·ATP → DnaK + ATP | 1.33 × 10−4 s−1 | (47) A |
| 5 | DnaK + ADP → DnaK·ADP | 2.67 × 105 M−1 s−1 | (47) A |
| 6 | DnaK·ADP → DnaK + ADP | 0.022 s−1 | (47,48) A, B |
| 7 | DnaK·ATP → DnaK·ADP + Pi | 6 × 10−4 s−1 | (8) C |
| 8 | GrpE2·DnaK → DnaK + GrpE2 | 0.033 s−1 | (49) C |
| 9 | GrpE2·DnaK + ATP → GrpE2·DnaK·ATP | 6 × 106 M−1 s−1 | (9) B |
| 10 | GrpE2·DnaK·ATP → GrpE2·DnaK + ATP | 127 s−1 | Estimated to be similar to No. 14 |
| 11 | GrpE2·DnaK·ATP → DnaK.ATP + GrpE2 | 0.1 s−1 | Estimated based on Fig. 4 A in (49) |
| 12 | DnaK·ADP + GrpE2 → GrpE2·DnaK·ADP | 3.4 × 105 M−1 s−1 | (49) C |
| 13 | GrpE2·DnaK·ADP → DnaK·ADP + GrpE2 | 0.033 s−1 | Estimated to be similar to No. 8 |
| 14 | GrpE2·DnaK·ADP → GrpE2·DnaK + ADP | 127 s−1 | (9) B |
| S1 | DnaJ2 + SD → SD·DnaJ2 | 3.3 × 105 M−1 s−1 | (50) D |
| S2 | SD·DnaJ2 → DnaJ2 + SD | 0.0062 s−1 | (50) D |
| S3 | DnaK·ATP + SD·DnaJ2 → DnaK·ATP·SD·DnaJ2 | 4.5 × 105 M−1 s−1 | Estimated to be similar to No. S10 |
| S4 | DnaK·ATP·SD·DnaJ2 → SD·DnaJ2 + DnaK·ATP | 2 s−1 | Estimated to be similar to No. S11 |
| S5 | DnaK·ATP·SD·DnaJ2 → DnaK·ADP·SD·DnaJ2 + Pi | 1.8 s−1 | (10) C |
| S6 | DnaK·ADP·SD·DnaJ2 + GrpE2 → GrpE2·DnaK·ADP·SD + DnaJ2 | 3.4 × 105 M−1 s−1 | Estimated to be similar to No. 12 |
| S7 | GrpE2·DnaK·ADP·SD → GrpE2·DnaK·SD + ADP | 127 s−1 | Estimated to be similar to No. 14 |
| S8 | GrpE2·DnaK·SD + ATP → GrpE2·DnaK·ATP·SD | 6 × 106 M−1 s−1 | Estimated to be similar to No. 9 |
| S9 | GrpE2·DnaK·ATP·SD → SD + GrpE2 + DnaK·ATP | 2 s−1 | Estimated to be similar to No. S11 |
| S10 | DnaK·ATP + SD → DnaK·ATP·SD | 4.5 × 105 M−1 s−1 | (11,12) E |
| S11 | DnaK·ATP·SD → DnaK·ATP + SD | 2 s−1 | (11,12) E |
| S12 | DnaK·ATP·SD → DnaK·ADP·SD + Pi | 0.001 s−1 | (10,13) C |
| S13 | DnaK·ADP·SD → DnaK·ADP + SD | 4.7 × 10−4 s−1 | (13) C |
| S14 | DnaK·ADP·SD → DnaK·SD + ADP | 0.022 s−1 | Estimated to be similar to No. 6 |
| S15 | DnaK·SD + ATP → DnaK·ATP·SD | 1.3 × 105 M−1 s−1 | Estimated to be similar to No. 3 |
| S16 | DnaK·ADP + SD → DnaK·ADP·SD | 1000 M−1 s−1 | estimated based on (13)‡ C |
| S17 | DnaK·ADP·SD + GrpE2 → GrpE2·DnaK·ADP·SD | 3.4 × 105 M−1 s−1 | Estimated to be similar to No. 12 |
| S18 | DnaK·ATP·SD + DnaJ2 → DnaK·ATP·SD·DnaJ2 | 3.3 × 105 M−1 s−1 | Estimated to be similar to No. S1 |
| S19 | DnaK·ATP·SD·DnaJ2 → DnaK·ATP·SD + DnaJ2 | 0.0062 s−1 | Estimated to be similar to No. S2 |
| S20 | DnaK·ADP·SD·DnaJ2 → DnaK·ADP·SD + DnaJ2 | 0.0062 s−1 | Estimated to be similar to No. S2 |
| Fold 1 | GrpE2·DnaK·ATP·SD → SD + GrpE2 + DnaK·ATP | Depend on probability | |
| Fold 2 | DnaK·ADP·SD → DnaK·ADP + SD | Depend on probability |
All reactions designated with S contain substrates; fold 1/2, reactions to introduce the refolding probability per chaperone cycle.
Association and dissociation rates for DnaJ dimerization were estimated to fit the concentration dependence of the luciferase refolding reaction at low DnaJ concentrations.
Association rate of a denatured protein to DnaK·ADP was estimated to be in between the association rate of a peptide and the rate for a native protein. Buffer conditions: A, 40 mM Hepes/KOH, pH 7.6, 11 mM Mg-acetate, 200 mM K-glutamate; B, 50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 100 mM KCl, 2 mM EDTA, 2 mM dithioerythritol; C, 25 mM Hepes-KOH (pH 7.6), 5 mM MgCl2, 50 mM KCl; D, 25 mM Hepes/KOH pH 7.0, 5 mM MgCl2, 50 mM KCl, 5 mM 2-mercaptoethanol, and 0.005% Tween 20; and E, 25 mM Hepes/NaOH pH 7.0, 5 mM MgCl2, 100 mM KCl.
Simulation
The simulations are based on an improved version of Gillespie's exact stochastic simulation algorithm (15) by Gibson and Bruck (16), as implemented in E-CELL version 3, an open source computer software package for simulation of large-scale cellular events developed at Keio University (http://www.e-cell.org) (17,18).
RESULTS
Model construction and validation
To construct a kinetic model for the chaperone-assisted refolding of a denatured protein, the refolding process was dissected into its elemental chemical equations (Fig. 1 B) with a set of kinetic parameters for the individual steps. For most of the simple binary reactions, kinetic constants can be extracted from the literature (Table 1). For many of the more complex reactions involving three or more components of the system, no parameters were determined experimentally, but they could be approximated by the values for similar simpler reactions. However, most available kinetic constants were determined using model compounds like peptides and native protein substrates of DnaK instead of denatured proteins. The use of denatured proteins for the determination of the kinetic constants is unfeasible for a number of obvious reasons. Therefore, a first goal of our simulation study was to determine whether the kinetic constants known so far can be fitted into a refolding reaction resulting in refolding rates that are comparable with the rates found in in vitro-refolding experiments of a model substrate. In addition, for some kinetic constants, a wide range of possible values have been reported (e.g., peptide association to DnaK·ATP is sequence dependent with bimolecular rates from 44 M−1 s−1 to 106 M−1 s−1), and it is not clear which range of values is suitable to support refolding (for most simulations, 4.5 × 105 M−1 s−1 was used according to (11), which is the association rate for a peptide rich in hydrophobic residues as they are found in the hydrophobic core of a protein and presumably exposed upon denaturation). Therefore, a second goal of our simulation study was to evaluate the different parameters to elucidate the most critical steps in the refolding reaction.
To simulate refolding of denatured proteins using our kinetic model, we introduced a given number of denatured protein molecules SD into our system and calculated the number of molecules bound to the chaperones and refolded to the native state SN over time. We made no assumptions about the nature of the denatured state SD other than that it is inactive and that it is recognized by the chaperones DnaK and DnaJ as a substrate. DnaK and DnaJ do not bind to the substrate in its native state, which is consistent with in vitro observations and explained by the binding preference of DnaK and DnaJ for hydrophobic peptide stretches. These stretches are buried in the hydrophobic core of native proteins but exposed during de novo folding or in the denatured state (19,20). The interaction of the misfolded protein with the chaperones eventually leads to refolding to the native, active state. To allow for scaling of the refolding kinetics, we assigned a probability value expressing the likelihood with which a denatured molecule is refolded in a single cycle of binding and release by the chaperones. Thereby, ATP hydrolysis was considered crucial, and only a substrate that was bound to DnaK in the ATP state and released subsequent to ATP hydrolysis had a chance to refold to the native state. Because most experimental data of the chaperone action of DnaK are available for the in vitro-refolding of guanidinium-Cl denatured firefly luciferase, we tried to model these reactions and compared our simulation results with data published in Mayer et al. (13) to validate our model. Except for this probability value, all other parameters have been reported in literature (see references in Table 1). As shown in Fig. 2 A, with a refolding probability of 2.68% in a single cycle, results from our model are consistent with in vitro-refolding data for chemically denatured firefly luciferase.
Partition of the misfolded substrate
To analyze the refolding reaction in more detail, we determined the distribution of the substrate protein between the free pool and the different chaperone complexes (Fig. 2 B). After ∼3 s, almost 80% of the substrate molecules disappeared from the free pool and was found in different complexes with chaperones (Fig. 2 B, inset). Unexpectedly, the most prevalent of these complexes was the DnaK·ATP·SD complex and not the DnaJ2·SD or the DnaK·ADP·SD·DnaJ2 complex as generally assumed in the Hsp70 field. After the initial extremely rapid establishing of the chaperone-system equilibrium, the concentration of the free denatured protein as well as the concentrations of all chaperone-substrate complexes decreased slowly at a pace at which the protein is refolded to the native state.
ATP consumption of the system
To our knowledge, one parameter for refolding was never determined in the past: the number of ATP molecules consumed by the Hsp70 system for protein refolding. This is a very important parameter because, under stress conditions, ATP production by catabolic process may cease and ATP concentrations may drop considerably as was reported for heat-stress conditions (21). Therefore, we determined the ATP consumption of the Hsp70-refolding system per 1000 molecules of denatured proteins using our kinetic model. Even in the absence of any chaperone substrate, the Hsp70 system consumed a basal amount of ATP as given by the black dashed line in Fig. 2 C. Depending on the refolding probability, ATP consumption increased significantly during the refolding reaction, indicating that low refolding probabilities by constant rates for the chaperone cycle could be detrimental for survival under stress conditions.
Critical parameters for the refolding reaction
As mentioned above, many of the kinetic constants published for the Hsp70 system and used in our simulations were determined for model reactions, which did not contain all the components of the refolding machinery, and for peptide substrate instead of denatured protein substrates. Therefore, we wanted to determine which are the critical parameters of the refolding reaction and which values for the kinetic constants would support refolding. Where possible, we compared the simulation results with in vitro data for DnaK mutant proteins defect in the investigated parameter.
Association of substrate with DnaK
One characteristic of the refolding reaction is the fast drop in the concentration of free denatured protein and the rapid increase in the concentration of the DnaK·ATP·SD complex. To analyze the influence of the association rate constant of substrate binding to DnaK·ATP, we varied this parameter and compared the simulation curves with data for DnaK variants, which have amino acid replacements in the substrate-binding pocket and were earlier shown to be defective in their interaction with substrates to different degrees (13). In our simulation, we assumed that the association of the DnaJ2·SD complex to DnaK·ATP is determined by the association rate of the substrate because there is evidence that the functional interaction of DnaJ with DnaK is very weak (22,23). Therefore, we varied the values for the association rate of the two reactions DnaK·ATP + SD and DnaK·ATP + DnaJ2·SD, simultaneously keeping the refolding probability factor constant at 2.68% (Fig. 3 A). Interestingly, decreasing the association rate to 1/10th slows the refolding reaction only slightly, and a decrease by a factor of 100 or 1000 is necessary to affect the refolding reaction significantly. Overall, the experimental data for the mutant DnaK proteins could be retraced by the simulation curves fairly well. However, the association rate values, which had to be used to fit the data, are not identical to the values calculated for peptide association to these DnaK mutant proteins. There are two possible reasons for this observation. First, the association rates were calculated from the dissociation equilibrium constant and the dissociation rate constant for peptide interaction with the DnaK wild-type and mutant proteins. Denatured protein substrates may interact differently with DnaK. Second, the mutant proteins may have additional defects affecting other reactions of the refolding cycle and in particular the refolding probability, which were considered to be constant in our simulations.
FIGURE 3.
Critical parameters for the refolding reaction. (A) Dependence of refolding efficacy on the bimolecular rate constants for the reactions DnaK·ATP + SD·DnaJ2 → DnaK·ATP·SD·DnaJ2 and SD + DnaK·ATP → DnaK·ATP·SD as indicated (M−1 s−1). Symbols represent experimental data for DnaK wild-type (▪, same data as in Fig. 2 A) and DnaK-M404A (•) and DnaK-V436F (▴) mutant proteins, which are defective in their interaction with substrates to different degrees (data from (13)). Error bars represent standard deviation of three independent experiments. Error bars for the DnaK-V436F mutants are too small to be visible. (B) Dependence of refolding efficacy on the bimolecular rate for association of DnaJ2 to SD and to DnaK·ATP·SD, which were varied simultaneously as indicated (M−1 s−1). Experimental data (▪; same as in Fig. 2 A). (C) Importance of the individual reaction of DnaJ2 association to SD (b) and DnaK·ATP·SD (c) with an association rate of 30 M−1 s−1 as compared to the wild-type situation of a rate of 3.3 × 105 M−1 s−1 (a). (D) Importance of synergistic stimulation of the ATP hydrolysis rate of DnaJ and substrate. Dashed and dashed-dotted lines represent simulations without stimulation of the ATPase activity of DnaK. The solid line corresponds to the simulation result for wild-type DnaK. The ATPase rates were set to the basal ATPase rate (0.0006 s−1, dashed line) or fully stimulated rate (1.8 s−1, dashed-dotted line). Symbols represent experimental data for DnaK wild-type (▪) and DnaK-P143G (▴) mutant proteins, for which the stimulated ATPase activity is close to the basal activity for the wild-type protein (data from (25)). (E) ATP consumption for the wild-type (khyd stimulated; same as in Fig. 2 C) and the always fully active (khyd = 1.8 s−1) DnaK proteins. Basal ATP consumption in the wild-type situation (dashed curve; same as in Fig. 2 C). (F) dependence of refolding efficacy on the bimolecular rate for the reactions DnaK·ADP + GrpE2 → GrpE2·DnaK·ADP, DnaK·ADP·SD·DnaJ2 + GrpE2 → GrpE2·DnaK·ADP·SD + DnaJ2, and DnaK·ADP·SD + GrpE2 → GrpE2·DnaK·ADP·SD as indicated (M−1 s−1). Experimental data for the wild-type situation (▪; same data as in Fig. 2 A). The refolding probability was kept constant at 2.68% for all simulations.
Association of substrates with DnaJ
The interaction of DnaJ with the denatured protein substrate is generally considered to be the first step in the refolding reaction (6). Therefore, it was surprising that neither the DnaJ2·SD complex nor one of the subsequent DnaJ2·SD complexes with DnaK·ATP or DnaK·ADP were the most abundant complex during the refolding reaction in our simulation experiments. To determine the importance of the fast association of DnaJ with the denatured substrate, we varied the corresponding association rate constant and simulated the refolding kinetics (Fig. 3 B). As above, we varied the rate constants for the two reactions DnaJ2 + SD and DnaJ2 + DnaK·ATP·SD simultaneously because both reactions are considered to be dominated by the association of DnaJ with the substrate and therefore to occur with the same rates. The simulation results indicate that decreasing the association rate of DnaJ to substrate affects the refolding reaction more seriously than the association of DnaK with substrate. This becomes apparent when comparing the respective curves in Fig. 3, A and B, which simulates a 10- and 100-fold decrease in association rate for substrates to DnaK·ATP and DnaJ2, respectively.
The investigation of the functional role of the Zn2+-binding sites of DnaJ revealed that one site is important for the interaction with substrates because its mutational removal rendered DnaJ incapable to prevent aggregation of a denatured substrate, whereas the other site seemed to be more important for the interaction with DnaK (24). Interestingly, the mutational removal of the site important for substrate binding had only a minor effect on the refolding of chemically denatured luciferase, in contrast to the removal of the other site, which affected refolding more seriously. To model these observations, we changed the association rates for the two reactions DnaJ2 + SD and DnaJ2 + DnaK·ATP·SD separately (Fig. 3 C). Even the reduction of the association rates by four orders of magnitude resulted in only a minor reduction of the refolding efficacy whereby the second reaction, in which DnaJ2 interacts with the DnaK·ATP·SD complex, was clearly affected more (curve c in Fig. 3 C).
Synergistic stimulation of the ATPase activity of DnaK
The simultaneous interaction of DnaJ and substrate with DnaK·ATP stimulates the ATP hydrolysis rate of DnaK greatly (10,14). This is considered to be a key feature of the Hsp70 chaperone cycle. Our kinetic model of the refolding cycle fully supports this intuitive concept. When the substrate and DnaJ-mediated stimulation of DnaK's ATPase activity is abolished by reducing all ATP hydrolysis rates to the rate of the intrinsic ATPase activity of DnaK no refolding of the denatured substrate is observed (Fig. 3 D). Our simulation results fit in vitro data with a DnaK variant (DnaK-P143G), the ATPase activity of which in the presence of DnaJ and substrate (9 × 10−4 s−1) is similar to the unstimulated rate of DnaK wild-type (6 × 10−4 s−1) (25).
These data clearly show that the low basal rate is not sufficient for effective refolding of chemically denatured luciferase. The question remains whether a DnaK protein, which always has an ATPase activity that corresponds to the high, stimulated rate, would be as efficient as the natural DnaK. Therefore, we set the rates for all reactions where ATP is hydrolyzed to 1.8 s−1. We kept the refolding probability at 2.68% because it is not expected that such a parameter would be dependent on the ATP hydrolysis rate in the absence of substrates. We simulated the refolding reaction (Fig. 3 D) and determined the ATP consumption during the process (Fig. 3 E). Our results demonstrate that refolding efficacy goes down significantly, yielding only 50% refolding after 30 min, whereas ATP consumption rises dramatically being linear with time independent of the refolding success.
Together, our data demonstrate the importance of the stimulation of DnaK's ATPase activity by DnaJ and substrate.
Importance of nucleotide exchange for the refolding reaction
Under physiological conditions of high ATP concentrations, nucleotide exchange is rate limiting for substrate release in the DnaK chaperone cycle, and the importance of the nucleotide exchange factor GrpE for substrate release and the refolding reaction is well established (9,26,27). To analyze the importance of the GrpE-DnaK interaction for the refolding of a denatured protein, we varied the association rate constant for this reaction (Fig. 3 F). Our simulation results demonstrate that lower association rates for GrpE dramatically affect the refolding reaction underlining the importance of the nucleotide exchange factor for the refolding activity of the DnaK system.
Robustness of the chaperone system
One property of many natural regulatory systems is their robustness, i.e., their insensitivity to changes in the concentration of some of their components. In vitro experiments with fluorescently labeled peptides demonstrated that substrate binding by DnaK was affected by substoichiometric concentrations of DnaJ and GrpE within a relatively narrow range (28). In contrast, refolding of chemically denatured luciferase was supported optimally within a broad range of DnaJ and GrpE concentrations (9,10). Interestingly, high concentrations of either DnaJ or GrpE reduced refolding efficacy significantly. The reason for this behavior is unclear, and we wanted to analyze whether such a behavior is inherent to this system. Therefore, we varied the concentration of DnaJ and GrpE in a range identical to the published data and simulated the refolding of the denatured protein (Fig. 4, A and C). The refolding yield after 30 min was plotted versus the DnaJ and GrpE concentrations and compared to the published data (Fig. 4, B and D). Originally, we had simulated DnaJ as a monomeric protein without the reaction 2 DnaJ ⇔ DnaJ2 but could not simulate the dramatic concentration dependence of the refolding reaction at low concentrations of DnaJ. After introduction of the dimerization reaction of DnaJ, a dimerization rate constant could be found to allow the simulation results to follow closely the published data. However, at high concentrations of DnaJ, our simulations did not result in decreased refolding yields as found in in vitro-experiments.
FIGURE 4.
Robustness of the DnaK system. (A) Refolding reactions at different DnaJ concentrations as indicated (nM). (B) Dependence of refolding yields after 30 min on DnaJ concentrations. Simulations (▪); Experimental data (•; from (10)). (C) Refolding reactions at different GrpE concentrations as indicated (nM). (D) Dependence of refolding yields after 30 min on GrpE concentrations. Simulations (gray bars); experimental data (black bars; from (9)). (E) Refolding reactions at different DnaK concentrations as indicated (nM). (F) Dependence of refolding yields after 10 min on DnaK concentrations. Simulations (▴); experimental data (▪; from (13)). Error bars represent standard deviation of three independent experiments.
The variation of the GrpE concentration also followed reasonably well the published refolding data at low concentrations of GrpE, though not at high concentrations, similar to the results with DnaJ.
We also varied the concentration of DnaK and simulated the refolding reaction. As shown in Fig. 4, E and F, in our kinetic model, the DnaK system is very robust with respect to DnaK concentrations supporting the refolding reaction even at low concentrations of DnaK. However, this behavior is not consistent with in vitro data (Fig. 4 F) where at least a fivefold excess of DnaK over luciferase is necessary for efficient refolding.
Taken together, although our kinetic model simulates the refolding of chemically denatured luciferase by the DnaK chaperone system fairly well and, in particular, reproduces the effects of changes in several kinetic parameters on the refolding efficacy, concentration effects are not simulated with the same accuracy, clearly indicating the limitations of our model and our current understanding of the system.
DISCUSSION
In this study, we established a kinetic model for the refolding action of Hsp70 chaperones. Experimental data of in vitro refolding chemically denatured luciferase could be simulated fairly well by introducing a probability factor of 2.68% for the denatured protein to be refolded successfully in any given chaperone cycle. This factor means that, on average, any denatured protein needs 38 cycles for successful refolding. This value is much smaller than earlier estimations based on the maximal rates for each individual step of the cycle yielding a cycle time of ∼1 s and the refolding rate determined experimentally resulting in a half-life for refolding of 300 s (29). The refolding probability is most likely a characteristic parameter depending on the nature of the substrate, its sequence, fold, and energy landscape for folding. Experimental evidence for this hypothesis is found by comparing different substrates, for example luciferase, malat dehydrogenase, lactate dehydrogenase, and glucose-6-phosphate dehydrogenase, which are refolded by the DnaK chaperone system at largely different refolding kinetics (30). The method of denaturation, for example chemical denaturation versus heat denaturation, also seems to influence the refolding probability because heat-denatured luciferase is refolded by the DnaK system much slower than guanidinium-denatured luciferase (compare (5) with (13)) The reason for the effect of different denaturation methods may be the formation of different conformers or oligomeric states as substrates for the refolding reaction.
The distribution of the denatured protein in the different chaperone complexes was surprising. It was generally assumed that DnaJ interacts with the substrate first and subsequently transfers it to DnaK·ATP. The substrate and DnaJ-mediated synergistic stimulation of ATP hydrolysis would then lead to the DnaK·ADP·SD·DnaJ2 complex (6). In such a scenario, the DnaK·ATP·SD complex would not be expected to be a major species. However, our simulation results show that, with the available association rate constants, the DnaK·ATP·SD complex is by far the most abundant chaperone-substrate complex (Fig. 2 B). The substrate association rates used in our kinetic model have been determined for substrate peptides. If the association of substrate proteins would not be limited by the direct binding of the binding site into the substrate binding pocket of DnaK but by conformational fluctuations of the substrate that expose the binding sites within the substrate polypeptide for binding by DnaK, these association rates could also be significantly lower. Such lower association can also be fitted in our model; however, the refolding probability would then have to be much higher to fit the in vitro data. In such a scenario, the SD·DnaJ2 complex becomes the dominant species indicating that it would form first and start the refolding reaction (Fig. 5, A and B).
FIGURE 5.
Variations of the refolding model. (A and B) Simulating the refolding reaction with low association rates for the interaction of DnaK·ATP with substrates. (A) The refolding reaction is shown in dependence of refolding probability as indicated with the bimolecular rate for the association of SD and SD·DnaJ2 to DnaK·ATP (reactions S3 and S10) set to 4000 M−1 s−1 (compared to 4.5 × 105 M−1 s−1 in the original model). Experimental data of in vitro refolding chemically denatured luciferase (▪; data from (13); same as in Fig. 2 A). (B) Distribution of the substrate between the different pools in the refolding reaction shown in A at a refolding probability of 30%: a, SD; b, SD·DnaJ2; c, DnaK·ADP·SD·DnaJ2; d, GrpE2·DnaK·ATP·SD; e, DnaK·ATP·SD; and f, SN. (C and D) Dependence of the refolding reaction on the exit point of DnaJ. (C) Elimination of reaction S6 reduces refolding efficacy significantly. Kinetic model including reaction S6 (solid line); Kinetic model without reaction S6 (dashed line). In vitro refolding of chemically denatured luciferase (▪; data from (13); same as in Fig. 2 A). (D) distribution of the substrates between different pools in the refolding process without reaction S6 as shown in C (dashed line): a, SD; b, DnaK·ADP·SD·DnaJ2; c, DnaK·ATP·SD; d, DnaK·ATP·SD·DnaJ2; e, SD·DnaJ2; and f, SN.
Based on the currently available data for substrate association to DnaK·ATP, our model suggests that the chaperone cycle in the majority of cases starts with DnaK·ATP interacting with the substrate; DnaJ2 enters this complex subsequently. Nevertheless, the interaction of DnaJ2 with substrate is very critical as already a 10-fold decrease of the association rate for the respective reactions reduces the refolding rate to 50%. This change in refolding efficacy is more dramatic than changes observed by decreasing the association rates for the interaction of DnaK·ATP with substrates or the association of GrpE2 to the DnaK·ADP·SD complex. However, the interaction of DnaJ with SD becomes unimportant when interaction of DnaJ with DnaK·ATP·SD is fast enough and not governed by DnaJ-substrate interaction. To test this finding, experimentally mutants of DnaJ with amino acid replacements in the substrate-binding site could be used. Alternatively, CbpA, a homolog of DnaJ in E. coli, which is not able to prevent luciferase aggregation and, therefore, is not considered to interact with luciferase, could be used. However, in both cases (the DnaJ variants or CbpA), it is essential to demonstrate by a method different from aggregation prevention that they indeed do not interact with substrates.
One of the key features of the DnaK chaperone systems is the ability of substrate and DnaJ to synergistically stimulate DnaK's ATPase activity (10,14). Not surprisingly, the basal ATPase activity of DnaK of one ATP molecule hydrolyzed per 30 min is much too slow to support growth of an organism, which divides every 20 min under optimal conditions. Mutational replacements of residues in DnaK or DnaJ that prevents the synergistic stimulation therefore result in chaperone proteins that are inactive in refolding in vitro and cannot complement the ΔdnaK phenotypes in vivo (10,25,31). A DnaK protein that always hydrolyses ATP at top rates would, as expected, increase the ATP consumption tremendously and render it independent of cochaperone needs in the cell. Interestingly, such a system would also be less effective. One could have assumed that speeding up the left (Fig. 1 B) DnaJ-independent side of our model would increase the refolding efficiency. However, increasing the basal ATPase activity would shift the relative distribution of DnaK between the ATP- and the ADP-bound states, the former having high substrate association and dissociation rates, whereas the latter having low substrate association rates. This distribution is obviously critical for the refolding reaction. Therefore, it is not surprising that similar synergistic stimulation of the ATPase activity was also found in other Hsp70 proteins (32–34).
Our simulations also demonstrate that the DnaK chaperone system is robustly designed. Relatively large (10-fold) changes in individual rate constants affect the overall refolding rate only to a minor extent (by a factor of 2 at most). This is also true for changes in the concentrations of the chaperone and cochaperones. Albeit, our simulations failed to reproduce the effects of cochaperone imbalance at high concentrations. The reason for this can only be in side reactions that we have not yet included in our model because variations of many rate constants did not lead to a better fit of the data.
One side reaction for which good experimental evidence exists is the competition of DnaJ at high concentrations with substrates for binding to itself and to DnaK (23,35,36). DnaJ was shown by gelfiltration and analytical ultracentrifugation to form a wide range of oligomers at high concentrations (37). DnaJ presumably interacts with its own exposed hydrophobic amino acid side chains, which are part of its substrate binding site to interact with hydrophobic peptide stretches exposed by denatured proteins. One DnaJ could therefore present a second DnaJ molecule as substrate to DnaK·ATP. In addition, one DnaJ could also present itself to DnaK·ATP as a substrate because it was shown that the J-domain, the short N-terminal domain of DnaJ that is essential to stimulate DnaK's ATPase activity, fused to a peptide substrate is able to fully stimulate DnaK's ATPase activity similar to the combination of DnaJ with a substrate protein and to bind to DnaK like a substrate peptide (38).
In principal, there are three possibilities of how DnaJ could inhibit the refolding reaction: 1), depletion of refolding active DnaJ species by oligomerization; 2), depletion of refolding active DnaK species by sequestration; and 3), sequestration of the substrate. Depletion of refolding active DnaJ species does not seem possible for reasons of the law of mass action. Any oligomerization should be concentration dependent, and forward and back reactions increase with increasing concentrations. Consequently, all forms, monomeric, dimeric, and oligomeric, will increase to different degrees with increasing protomer concentrations. Therefore, it is not possible to create an optimum curve as observed in Fig. 4 B from such a system. How could increasing DnaJ lead to a depletion of available refolding-active DnaK? Because DnaJ inhibits already at equimolar concentrations, dimers or higher order oligomers of DnaJ cannot deplete DnaK sufficiently by mere competition for binding because DnaJ can only occupy as many DnaK proteins as DnaJ complexes are around. Only DnaJ as a monomer could theoretically occupy all of the DnaK molecules at equimolar concentrations. Already as a dimer, it would only occupy half of the DnaK molecules, which would not lead to a decrease of the refolding reaction as shown in the simulations with decreasing DnaK concentrations (Fig. 4, E and F). For DnaJ oligomers to be effective, they would have to act catalytically on DnaK depletion. DnaJ proteins were shown to catalyze Hsp70 oligomerization in the presence of ATP (39). Such a scenario would be able to deplete active DnaK sufficiently to explain the observed reduction of refolding yields. The third alternative is that DnaJ could sequester the substrate at high concentrations, maybe in oligomeric form. DnaJ oligomers could compete with DnaK for substrate. Indication that this might be the mechanism can be found in an earlier study in which DnaJ variants in the Zn2+-binding domain were investigated. One of the DnaJ variants, DnaJΔZnI, did not bind to substrates very well, as demonstrated by its inability to prevent aggregation of substrates. Its optimal concentration for refolding was much higher than for DnaJ wild-type, and an inhibition of the refolding reactions was not observed until ∼10-fold more of this mutant protein as compared to wild-type DnaJ was used in the reaction. The affinity of this mutant toward luciferase seems to be decreased by a factor of 10, and a 10-fold higher concentration is needed to inhibit the refolding reaction. Therefore, several alternative schemes need to be explored individually or in combination to solve the behavior of the refolding reaction at high concentrations of DnaJ.
The functional oligomeric state of DnaJ was also under debate (20). Our simulations support that DnaJ acts as a dimer, which is consistent with recent crystallographic work on related proteins in yeast (40,41).
The reason for declining refolding activity at high GrpE concentrations may be due to GrpE interacting with DnaK·ATP, leading to the premature dissociation of ATP. Preliminary simulations of such a reaction showed that premature dissociation of ATP in DnaK·ATP caused by GrpE would decrease refolding efficacy at very high concentrations of GrpE but makes the simulations extremely slow (data not shown). Alternatively, high GrpE concentrations could also affect the refolding probability. Because nucleotide exchange is rate limiting for substrate release at physiological concentrations of ATP, GrpE regulates the lifetime of the DnaK-substrate complex. A sufficiently long lifetime allowing domain movements and other structural rearrangements could be a prerequisite for successful refolding. Decreasing the lifetime of the DnaK-substrate complex by increasing GrpE concentrations would then decrease the refolding probability and, hence, the overall refolding efficacy.
In contrast to the effects at high concentrations, the influence of low concentrations of DnaJ and GrpE were simulated fairly well by our kinetic model. However, this was not the case for lower DnaK concentrations, which had no influence in our simulations but significantly affects in vitro refolding of luciferase (13). The reason for this inconsistency may be that we have not considered the possibility of more than one molecule of DnaK binding simultaneously to a single substrate, because, to our knowledge, there is no conclusive experimental evidence published on this issue so far; albeit, this has been suggested (42). However, we hypothesize that such a mechanism could enhance the refolding process and would be more sensitive to a reduction of DnaK concentration and activity. Further in vitro experiments are needed to validate this hypothesis.
One question of debate concerning the DnaK chaperone cycle was the dissociation of DnaJ. Based on the fact that DnaJ is only 1/10th to 1/30th as abundant as DnaK in vivo (43) and can act substoichiometrically in vitro (10,28), it was assumed that DnaJ leaves the cycle just after the transfer of the substrate onto DnaK and ATP hydrolysis (reaction S20) and before GrpE binds to the complex (reaction S17) or, alternatively, is displaced by GrpE (reaction S6) (6,28). Both scenarios were used in our simulations; however, reaction S6 contributed more prominently to the overall refolding reaction than reactions S20 and S17. If reaction S6 is eliminated, i.e., DnaJ has to leave the DnaK·ADP·SD·DnaJ2 complex before GrpE can bind, refolding becomes very slow, and a much higher refolding probability would be necessary to simulate the in vitro data (Fig. 5 C). Furthermore, the DnaK·ADP·SD·DnaJ2 complex accumulates because DnaJ dissociation becomes rate limiting for the refolding reaction (Fig. 5 D). To distinguish the two scenarios, in vitro experiments are necessary that clarify whether GrpE association to DnaK is inhibited by DnaJ.
The elucidation of the binding sites for GrpE in the cocrystal structure with DnaK (44) and for DnaJ through genetic and biochemical analyses (45,46) indicated that both DnaJ and GrpE could eventually bind at the same time to DnaK and possibly to the DnaK-substrate complex. Therefore, we asked whether the refolding efficacy for the substrate would change when DnaJ leaves the cycle together with GrpE upon binding of ATP (reaction S9) instead of before or upon binding of GrpE (reaction S6/S20). However, the results did not change when only the exit point for DnaJ was varied (data not shown). Therefore, despite the substoichiometric concentration of DnaJ, the actual exit point of DnaJ is not critical for the refolding efficacy as long as a quaternary complex of DnaK with DnaJ, substrate, and GrpE does not change the dissociation kinetics significantly or has any other additional effect on the refolding probability of the substrate.
One major difference between our simulations and in vitro data is the endpoint of the reaction. In our kinetic model, the refolding reaction always reaches 100% refolding after a sufficiently long time. However slow the refolding reaction may be, the denatured protein will finally be refolded because the native state is modeled as a sink, and the denatured protein has no other way to go. This is not the case in vitro. Refolding reactions generally level off between 60% and 80% of the total protein substrate. There are two alternative and mutually not exclusive explanations for such observations. First, during the initial dilution of the substrate out of the chemical denaturant, a certain portion of the substrate converts into an inactive state, which is not substrate of DnaK or cannot be refolded by DnaK. Second, the denatured substrate converts continuously by a slow and largely irreversible reaction into an inactive state, which cannot be refolded by the DnaK system. By introducing the reaction SD → SD* with a rate constant of 7 × 10−4 s−1, and SD* not being a substrate for DnaK and DnaJ, we were able to simulate such a reaction and, thereby, to fit the in vitro data much better (Fig. 6). Irreversible aggregation could be the biological cause for these in vitro observations. We have not formally excluded aggregation in our kinetic model as long as aggregation does not interfere with the interaction with chaperones and refolding by them. However, this may not be the case, and the likelihood of interaction with the chaperones and of being refolded may vary largely between aggregated and nonaggregated species and within aggregates. Further experiments in vitro and in silico are needed to elucidate these reactions.
FIGURE 6.
Simulating a reduced refolding yield. Introduction of the reaction SD → SD*, with SD* not being a substrate for DnaK, and DnaJ simulates the refolding yields lower than 100% observed in experimental data in vitro. Refolding reactions with bimolecular rate constants for the reactions DnaK·ATP + SD·DnaJ2 → DnaK·ATP·SD·DnaJ2 and SD + DnaK·ATP → DnaK·ATP·SD as indicated with (solid lines) and without (dashed lines) the reaction SD → SD* at a rate of 7 × 10−4 s−1. Symbols represent experimental data for DnaK wild-type (▪) and DnaK-M404A (•) (data from (13) same as in Fig. 3 A).
Acknowledgments
The authors thank all the members of E-CELL project at Keio University for their support.
This work was supported in part by grants from The 21st Century COE Program “Understanding and Control of Life's Function via Systems Biology” and Leading Project for Biosimulation, Keio University, by The Ministry of Education, Culture, Sports, Science and Technology of Japan and CREST, JST “Development of modeling/simulation environment for systems biology” by Japan Science and Technology Agency (JST). This work was also supported by the Deutsche Forschungsgemeinschaft (SFB638 to M.P.M).
Bin Hu and Matthias P. Mayer contributed equally to this work.
References
- 1.Gething, M. J. 1991. Molecular chaperones: individualists or groupies? Curr. Opin. Cell Biol. 3:610–614. [DOI] [PubMed] [Google Scholar]
- 2.Langer, T., G. Pfeifer, J. Martin, W. Baumeister, and F.-U. Hartl. 1992. Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J. 11:4757–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mogk, A., T. Tomoyasu, P. Goloubinoff, S. Rüdiger, D. Röder, H. Langen, and B. Bukau. 1999. Identification of thermolabile E. coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18:6934–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomoyasu, T., A. Mogk, H. Langen, P. Goloubinoff, and B. Bukau. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia cytosol. Mol. Microbiol. 40:397–413. [DOI] [PubMed] [Google Scholar]
- 5.Schröder, H., T. Langer, F. U. Hartl, and B. Bukau. 1993. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12:4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell. 92:351–366. [DOI] [PubMed] [Google Scholar]
- 7.Liberek, K., J. Marszalek, D. Ang, C. Georgopoulos, and M. Zylicz. 1991. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA. 88:2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarty, J. S., A. Buchberger, J. Reinstein, and B. Bukau. 1995. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249:126–137. [DOI] [PubMed] [Google Scholar]
- 9.Packschies, L., H. Theyssen, A. Buchberger, B. Bukau, R. S. Goody, and J. Reinstein. 1997. GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry. 36:3417–3422. [DOI] [PubMed] [Google Scholar]
- 10.Laufen, T., M. P. Mayer, C. Beisel, D. Klostermeier, J. Reinstein, and B. Bukau. 1999. Mechanism of regulation of Hsp70 chaperones by DnaJ co-chaperones. Proc. Natl. Acad. Sci. USA. 96:5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid, D., A. Baici, H. Gehring, and P. Christen. 1994. Kinetics of molecular chaperone action. Science. 263:971–973. [DOI] [PubMed] [Google Scholar]
- 12.Gisler, S. M., E. V. Pierpaoli, and P. Christen. 1998. Catapult mechanism renders the chaperone action of Hsp70 unidirectional. J. Mol. Biol. 279:833–840. [DOI] [PubMed] [Google Scholar]
- 13.Mayer, M. P., H. Schröder, S. Rüdiger, K. Paal, T. Laufen, and B. Bukau. 2000. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7:586–593. [DOI] [PubMed] [Google Scholar]
- 14.Karzai, A. W., and R. McMacken. 1996. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271:11236–11246. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie, D. T. 1976. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22:403–434. [Google Scholar]
- 16.Gibson, M. A., and J. Bruck. 2000. Efficient exact stochastic simulation of chemical systems with many species and many channels. J. Phys. Chem. A. 104:1876–1889. [Google Scholar]
- 17.Takahashi, K., K. Kaizu, B. Hu, and M. Tomita. 2004. A multi-algorithm, multi-timescale method for cell simulation. Bioinformatics. 20:538–546. [DOI] [PubMed] [Google Scholar]
- 18.Tomita, M., K. Hashimoto, K. Takahashi, T. S. Shimizu, Y. Matsuzaki, F. Miyoshi, K. Saito, S. Tanida, K. Yugi, J. C. Venter, and others. 1999. E-CELL: software environment for whole-cell simulation. Bioinformatics. 15:72–84. [DOI] [PubMed] [Google Scholar]
- 19.Rüdiger, S., L. Germeroth, J. Schneider-Mergener, and B. Bukau. 1997. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16:1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rüdiger, S., J. Schneider-Mergener, and B. Bukau. 2001. Its substrate specificity characterizes the DnaJ chaperone as scanning factor for the DnaK chaperone. EMBO J. 20:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirmer, E. C., C. Queitsch, A. S. Kowal, D. A. Parsell, and S. Lindquist. 1998. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J. Biol. Chem. 273:15546–15552. [DOI] [PubMed] [Google Scholar]
- 22.Greene, M. K., K. Maskos, and S. J. Landry. 1998. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. USA. 95:6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer, M. P., T. Laufen, K. Paal, J. S. McCarty, and B. Bukau. 1999. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J. Mol. Biol. 289:1131–1144. [DOI] [PubMed] [Google Scholar]
- 24.Linke, K., T. Wolfram, J. Bussemer, and U. Jakob. 2003. The roles of the two zinc binding sites in DnaJ. J. Biol. Chem. 278:44457–44466. [DOI] [PubMed] [Google Scholar]
- 25.Vogel, M., B. Bukau, and M. P. Mayer. 2006. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol. Cell. 21:359–367. [DOI] [PubMed] [Google Scholar]
- 26.Brehmer, D., S. Rüdiger, C. S. Gässler, D. Klostermeier, L. Packschies, J. Reinstein, M. P. Mayer, and B. Bukau. 2001. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 8:427–432. [DOI] [PubMed] [Google Scholar]
- 27.Brehmer, D., C. Gassler, W. Rist, M. P. Mayer, and B. Bukau. 2004. Influence of GrpE on DnaK-substrate interactions. J. Biol. Chem. 279:27957–27964. [DOI] [PubMed] [Google Scholar]
- 28.Pierpaoli, E. V., E. Sandmeier, H.-J. Schönfeld, and P. Christen. 1998. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J. Biol. Chem. 273:6643–6649. [DOI] [PubMed] [Google Scholar]
- 29.Mayer, M. P., D. Brehmer, C. S. Gassler, and B. Bukau. 2001. Hsp70 chaperone machines. Adv. Protein Chem. 59:1–44. [DOI] [PubMed] [Google Scholar]
- 30.Diamant, S., and P. Goloubinoff. 1998. Temperature-controlled activity of DnaK-DnaJ-GrpE chaperones: protein folding arrest and recovery during and after heat shock depends on the substrate protein and the GrpE concentration. Biochemistry. 37:9688–9694. [DOI] [PubMed] [Google Scholar]
- 31.Wall, D., M. Zylicz, and C. Georgopoulos. 1994. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for λ replication. J. Biol. Chem. 269:5446–5451. [PubMed] [Google Scholar]
- 32.Barouch, W., K. Prasad, L. Greene, and E. Eisenberg. 1997. Auxilin-induced interaction of the molecular chaperone Hsc70 with clathrin baskets. Biochemistry. 36:4303–4308. [DOI] [PubMed] [Google Scholar]
- 33.Silberg, J. J., T. L. Tapley, K. G. Hoff, and L. E. Vickery. 2004. Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron-sulfur cluster assembly protein IscU. J. Biol. Chem. 279:53924–53931. [DOI] [PubMed] [Google Scholar]
- 34.Misselwitz, B., O. Staeck, and T. A. Rapoport. 1998. J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell. 2:593–603. [DOI] [PubMed] [Google Scholar]
- 35.Wawrzynów, A., and M. Zylicz. 1995. Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J. Biol. Chem. 270:19300–19306. [DOI] [PubMed] [Google Scholar]
- 36.Suh, W.-C., C. Z. Lu, and C. A. Gross. 1999. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem. 274:30534–30539. [DOI] [PubMed] [Google Scholar]
- 37.Schönfeld, H.-J., D. Schmidt, and M. Zulauf. 1995. Investigation of the molecular chaperone DnaJ by analytical ultracentrifugation. Prog. Colloid Polym. Sci. 99:7–10. [Google Scholar]
- 38.Wittung-Stafshede, P., J. Guidry, B. E. Horne, and S. J. Landry. 2003. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry. 42:4937–4944. [DOI] [PubMed] [Google Scholar]
- 39.King, C., E. Eisenberg, and L. Green. 1995. Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP. J. Biol. Chem. 270:22535–22540. [DOI] [PubMed] [Google Scholar]
- 40.Sha, B., S. Lee, and D. M. Cyr. 2000. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Struct. Fold. Des. 8:799–807. [DOI] [PubMed] [Google Scholar]
- 41.Li, J., X. Qian, and B. Sha. 2003. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 11:1475–1483. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Zvi, A., P. De Los Rios, G. Dietler, and P. Goloubinoff. 2004. Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual Hsp70 chaperones. J. Biol. Chem. 279:37298–37303. [DOI] [PubMed] [Google Scholar]
- 43.Tomoyasu, T., T. Ogura, T. Tatsuta, and B. Bukau. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in E. coli. Mol. Microbiol. 30:567–581. [DOI] [PubMed] [Google Scholar]
- 44.Harrison, C. J., M. Hayer-Hartl, M. Di Liberto, F.-U. Hartl, and J. Kuriyan. 1997. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 276:431–435. [DOI] [PubMed] [Google Scholar]
- 45.Gässler, C. S., A. Buchberger, T. Laufen, M. P. Mayer, H. Schröder, A. Valencia, and B. Bukau. 1998. Mutations in the DnaK chaperone affecting interaction with the DnaJ co-chaperone. Proc. Natl. Acad. Sci. USA. 95:15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suh, W.-C., W. F. Burkholder, C. Z. Lu, X. Zhao, M. E. Gottesman, and C. A. Gross. 1998. Interactions of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. USA. 95:15223–15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, R., R. Jordan, and R. McMacken. 1998. Kinetic Characterization of the ATPase Cycle of the DnaK Molecular Chaperone. Biochemistry. 37:596–607. [DOI] [PubMed] [Google Scholar]
- 48.Theyssen, H., H.-P. Schuster, B. Bukau, and J. Reinstein. 1996. The second step of ATP binding to DnaK induces peptide release. J. Mol. Biol. 263:657–670. [DOI] [PubMed] [Google Scholar]
- 49.Chesnokova, L. S., S. V. Slepenkov, I. I. Protasevich, M. G. Sehorn, C. G. Brouillette, and S. N. Witt. 2003. Deletion of DnaK's lid strengthens binding to the nucleotide exchange factor, GrpE: a kinetic and thermodynamic analysis. Biochemistry. 42:9028–9040. [DOI] [PubMed] [Google Scholar]
- 50.Gamer, J., G. Multhaup, T. Tomoyasu, J. S. McCarty, S. Rudiger, H. J. Schonfeld, C. Schirra, H. Bujard, and B. Bukau. 1996. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J. 15:607–617. [PMC free article] [PubMed] [Google Scholar]