Abstract
The hepatitis delta virus (HDV) ribozyme is an RNA enzyme from the human pathogenic HDV. Cations play a crucial role in self-cleavage of the HDV ribozyme, by promoting both folding and chemistry. Experimental studies have revealed limited but intriguing details on the location and structural and catalytic functions of metal ions. Here, we analyze a total of ∼200 ns of explicit-solvent molecular dynamics simulations to provide a complementary atomistic view of the binding of monovalent and divalent cations as well as water molecules to reaction precursor and product forms of the HDV ribozyme. Our simulations find that an Mg2+ cation binds stably, by both inner- and outer-sphere contacts, to the electronegative catalytic pocket of the reaction precursor, in a position to potentially support chemistry. In contrast, protonation of the catalytically involved C75 in the precursor or artificial placement of this Mg2+ into the product structure result in its swift expulsion from the active site. These findings are consistent with a concerted reaction mechanism in which C75 and hydrated Mg2+ act as general base and acid, respectively. Monovalent cations bind to the active site and elsewhere assisted by structurally bridging long-residency water molecules, but are generally delocalized.
INTRODUCTION
The hepatitis delta virus (HDV) ribozyme is an RNA enzyme, found in the RNA genome of a human pathogen, the hepatitis delta virus. The ribozyme catalyzes a site-specific transesterification reaction generating 5′-hydroxyl and 2′,3′-cyclic phosphate termini, and plays an essential role in the formation of antigenomic and genomic strands during the viral replication of multimeric intermediates (1). The HDV ribozyme was the first RNA enzyme for which direct involvement of its own nucleobase C75 in catalysis was suggested (2,3).
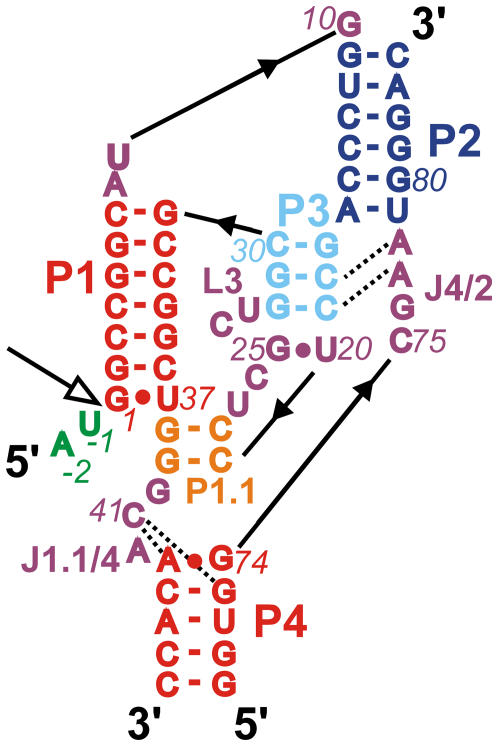
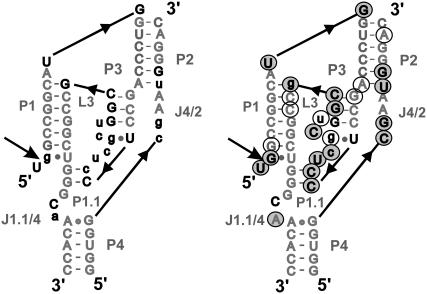
Several crystal structures of the ribozyme are currently available, including wild-type and C75U mutant precursor and product forms (4–6). The global structure of the HDV ribozyme is characterized as a double-nested pseudoknot, stabilized by stacking of five helical stems and an extensive network of hydrogen bonds among helices, loops, and joiners (Fig. 1) (4–6). Cytosine C75 is located close to the cleavage site. The most significant conformational difference in the otherwise very similar tertiary structures of the precursor and product forms (root mean-square deviation (RMSD) = 2 Å) has been identified as a collapse of stem P1 and loop L3 toward the center of the ribozyme (6), resulting in a deeper placement of cytosine C75 into the catalytic pocket, close to L3 in the product form. The flexible L3 is bridged by a noncanonical G25/U20 basepair that shows an anti-to-syn flip of G25 between the precursor and product structures.
FIGURE 1.
Secondary structure of the simulated genomic HDV ribozyme with structural elements color-coded. The product form lacks A-2 and U-1. Open arrow, cleavage site; broken lines, quadruple and A-minor motifs of A77 and A78.
The crystallographic data suggest that C75 is well poised to act as the general base to deprotonate the nucleophilic 2′ OH at the cleavage site (6). The mechanistic data are contradictory, with the latest study suggesting that C75 may act as the general acid to protonate the leaving group, which requires C75 to be N3-protonated just before cleavage (7). However, since a chemical modification was introduced at the scissile phosphate group in the latter study to accelerate leaving group departure, the reaction pathway may be affected. Alternatively, it cannot be ruled out that the transition state structure of the catalytic pocket differs from the ground state structure observed in x-ray diffraction studies, which may lead to the contradictory picture obtained by structural and mechanistic studies. Finally, the HDV ribozyme may utilize distinct C75-based catalytic strategies depending on the exact RNA construct and metal ion conditions (8,9).
Metal ions critically contribute to RNA folding and function (10). Active tertiary conformation and catalytic function are remarkably sensitive to the concentration and type of cation(s) present (11,12). HDV ribozyme activity has long been known as exclusively dependent on the presence of divalent cations (12–14). The majority of available crystal structures of the HDV ribozyme in its precursor form (6) reveals a consistent picture of cation binding with two typical binding sites, formed by L3 and the major groove of P4. In contrast, there are nine RNA-bound Mg2+ cations suggested by the crystal structure of the wild-type product form (Fig. 2 A) (15). For comparison, the number of folding-specific Mg2+ cations indicated by solution experiments for various medium-sized RNAs is only in the range of 0–4 (16,17). As demonstrated recently, assessment of cations in x-ray structures is complex. Some observed cations are related to the crystallization process (packing, etc.) and their occurrence in the crystal is highly sensitive to experimental details (18,19). Occasionally, water molecules or even sulfate anions were suggested to be mislabeled as magnesium dications (19). Variability of Mg2+ binding patterns in otherwise similar structures could also reflect competition of divalent and monovalent cations for the same binding sites. Specifically, bound divalent cation clearly seen in one x-ray structure may be substituted by fluctuating divalent or monovalent cations that would evade detection by x-ray crystallography (20).
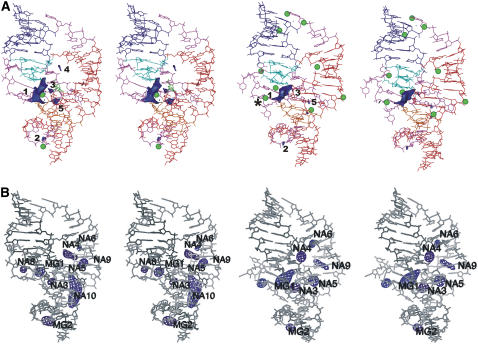
FIGURE 2.
(A) ESP contour maps of precursor (left) and product (right) ribozyme crystal structures, contoured at −25 kT/e. Nucleotides are color-coded as in Fig. 1; crystal positions of Mg2+ are shown as green balls. Position of Mg2+ closest to the active site in product crystal is marked by an asterisk. (B) Na+ binding maps contoured at 10 σ in simulations PreC41+A-2 (left) and ProC41+A-2 (right). Na+ binding sites located at the major NESP sites have the same numbering as their associated NESP sites. Sites NA7 and NA11 (Table S2) are not seen at this contour level due to low occupancy.
A critically located hydrated magnesium ion is thought to be an important chemical participant in the reaction mechanism (2,3,9,21). The change in divalent metal-ion preference upon alteration of the linkage at the scissile bond provided the first biochemical evidence for coordination of a divalent metal ion in the active-site region of the cis-(self-)cleaving genomic ribozyme (22). Two equivalent reaction mechanisms have been proposed wherein a hydrated magnesium ion could either donate a proton to the 5′-oxygen leaving group acting as the general acid or deprotonate the 2′ hydroxyl group acting as the general base, playing a complementary role to C75 so that both general acid and base catalysis are utilized in a concerted fashion (2,3,7). Consistent with such models, anticooperative interactions between a protonated C75 and a magnesium cation have been demonstrated by pH and magnesium titration studies (3). Although cleavage activity of the genomic HDV ribozyme can also be observed in the absence of magnesium ions, it is greatly reduced even at molar concentrations of sodium cations in comparison to millimolar magnesium concentrations (2,3,8,23). Nakano et al. have detected structural and catalytic ions with 125-fold and 25-fold contributions, respectively, to cleavage rate enhancement (8). Additional studies have tentatively suggested that the structural site shows an inner sphere interaction with a preference for Mg2+ and that the catalytic site has outer sphere binding with little preference for a particular divalent ion (9).
In addition to cations, hydrating water is thought to be an integral part of nucleic acid structure (24–34). Despite limitations imposed by force field approximations and limited simulation timescales (sampling), molecular dynamics (MD) techniques are well suited to analyze hydration, especially regarding predictions of highly specific long-residency hydration sites (20,27–33,35). MD simulations are capable of providing qualitative insights into cation binding to nucleic acids, including detection of complex cation binding pockets that contain delocalized cations (20,29,32,36).
Recently, we analyzed ∼120 ns of explicit solvent MD simulations of the HDV ribozyme (37). Precursor simulations with unprotonated C75 revealed weak dynamic binding of C75 in the catalytic pocket with spontaneous transient formation of a hydrogen (H-) bond between U-1(O2′) and C75(N3). This H-bond would be required for C75 to act as the general base. Protonated C75H+ moved deeper toward loop L3 (resembling its product location) and established a firm H-bonding network. However, a C75H+(N3)-G1(O5′) H-bond, which would be expected if C75 acted as a general acid catalyst without substantial structural rearrangement, was not observed. The simulations confirmed that loop L3 is dynamical and may serve as a flexible structural element, possibly gated by the closing U20/G25 basepair, to facilitate a conformational switch induced by a protonated C75H+. In this study, we i), substantially extend our simulations to more than 200 ns, and ii), provide a detailed analysis of hydration, cation binding, electrostatic potential, and backbone dynamics of the HDV ribozyme.
METHODS
Initial structures
Starting geometries were based on crystal structures of the C75U mutant precursor HDV ribozyme (Protein Data Bank (PDB) codes 1SJ3 and 1VC7, resolution 2.30 Å and 2.45 Å) and the wild-type product (PDB code 1CX0, 2.30 Å) (Table 1) (4,6,15). The remaining crystal structures were inspected to reveal the most typical crystal cation and water binding sites, if available. The structure of the catalytically inactive C75U mutant precursor was modified using InsightII (38) to carry C75 of the wild-type or a protonated C75H+. Note that the lower-resolution crystal structure of wild-type precursor in the absence of divalents (PDB 1VC5, 3.4 Å resolution) has essentially the same structure as the C75U mutant (RMSD of 0.30 Å) (6).
TABLE 1.
Overview of simulations discussed in this study (bold, simulations of unmodified crystal structures)
| Simulation | Initial structure PDB | 5′-sequence | 75 nucleotide | Duration (ns) | Ions |
|---|---|---|---|---|---|
| PreC41+ | 1SJ3* | U-1 | C75 | 13 | 60 Na+ |
| PreC41+Mg | 1SJ3* | U-1 | C75 | 15 | 2 Mg2+, 56 Na+ |
| PreC41+A-2† | 1VC7‡ | U-1/A-2 | C75 | 15 | 61 Na+ |
| PreC41+A-2Mg§ | 1VC7‡ | U-1/A-2 | C75 | 15 | 2 Mg2+, 57 Na+ |
| PreC41+C75+ | 1SJ3* | U-1 | C75H+ | 13 | 59 Na+ |
| PreC41+C75+Mg§ | 1SJ3* | U-1 | C75H+ | 15 | 2 Mg2+, 55 Na+ |
| PreC41+U75 | 1SJ3* | U-1 | U75 | 13 | 60 Na+ |
| PreC41+U75A-2† | 1VC7‡ | U-1/A-2 | U75 | 15 | 61 Na+ |
| PreC41+U75A-2Mg§ | 1VC7‡ | U-1/A-2 | U75 | 15 | 2 Mg2+, 57 Na+ |
| ProC41+ | 1CX0‖ | – | C75 | 15 | 59 Na+ |
| ProC41+Mg§ | 1CX0‖ | – | C75 | 10 | 2 Mg2+, 55 Na+ |
| ProC41+C75+ | 1CX0‖ | – | C75H+ | 15 | 58 Na+ |
| ProMg** | 1CX0‖ | – | C75 | 15 | 9 Mg2+, 42 Na+ |
| ProC41+9Mg | 1CX0‖ | – | C75 | 15 | 9 Mg2+, 41 Na+ |
| PreC41+Trc†† | 1SJ3* | – | C75 | 10 | 59 Na+ |
C75U mutated precursor HDV ribozyme crystallized in the presence of Mg2+ (resolution 2.20 Å).
Initial positions of Na+ were shifted away from the electronegative pockets.
C75U mutated precursor HDV ribozyme crystallized in presence of Sr2+ with A-2 resolved (resolution 2.45 Å).
Two Mg2+ cations were initially placed as in the corresponding precursor crystal structure (1SJ3 or 1VC7).
Wild-type product HDV ribozyme crystallized in the presence of Mg2+ (resolution 2.30 Å).
C41 was not protonated; wild-type product x-ray and random distribution of 9 Mg2+ were used in ProMg and ProC41+9Mg simulations, respectively.
U-1 was removed.
Inclusion of Mg2+ cations
We tested different variants for initial positioning of Mg2+ ions (Fig. 2 A, Table 1). Most simulations were carried out with two Mg2+ ions initially placed as in the precursor x-ray structure. Inclusion of nine Mg2+ cations (as suggested by the wild-type product crystal) results in a Mg2+ concentration of 0.1 M. Such simulations provide an upper limit for possible Mg2+-related structural effects seen in nanosecond-scale simulations.
Divalent cations are poorly described by pair additive potentials and also sample insufficiently in simulations (29,39). Since Na+ are better parameterized and demonstrate rather satisfactory sampling (20,29,40), we have also performed MD simulations in the presence of just sodium ions to identify and compare cation binding sites between the precursor and product forms of the ribozyme. Na+-only simulations are justified since the C75U mutant ribozyme that was prepared and crystallized in the absence of divalent metal ions (2.7 Å, PDB 1VBX) has the same structure as it does in the presence of divalents (RMSD = 0.31 Å). Our unpublished terbium(III) footprinting data on the genomic cis-acting ribozyme also support the notion that divalent ions are not required for HDV ribozyme folding. Finally, the same HDV ribozyme shows residual cleavage activity in molar concentration of Na+ in the absence of divalents (3,8,41). Anyway, the simulations are too short to reveal an unfolding caused by the lack of divalent cations (34). In the Supplementary Material, we provide justification for using minimal neutralizing Na+ concentrations.
Molecular dynamics
All MD simulations (Table 1) were carried out using the AMBER 7.0 program package (42) with the parm99 Cornell et al. force field (43–45). The RNA was solvated in a rectangular box of TIP3P waters (46) extended to a distance of ≥10 Å from any solute atom. The simulated system was neutralized by a minimal number of sodium cations (47) initially placed by the LeaP module at points of favorable electrostatic potential close to the RNA. This corresponds to an ion concentration of ∼0.2 M. In several simulations, sodium cations were initially moved away from the solute (after the initial electrostatic placement) to prevent trapped cations. The ions then spontaneously locate to the binding sites starting from bulk. The Sander module of AMBER 7.0 was used for the equilibration and production simulations using standard protocols (see, e.g., Reblova et al. (29) and Razga et al. The particle mesh Ewald method (48) was applied with a heuristic pair list update, using a 2.0-Å nonbonded pair list buffer and a 9.0 Å cutoff. The particle mesh Ewald charge grid dimensions are products of powers of 2, 3, and 5, resulting in a grid spacing of ∼1.0 Å. The direct sum tolerance of 10−5 and the nonbonded cutoff of 9.0 Å lead to an Ewald coefficient of 0.30 Å−1 (see Supplementary Material for further details).The production runs were carried out at 300 K with constant-pressure boundary conditions using the Berendsen temperature coupling algorithm (49) with a time constant of 1.0 ps.
Analysis of MD trajectories
The trajectories were analyzed using the Ptraj module of the AMBER 7.0 package and our own scripts and visualized by the programs PyMOL (50) and VMD (51). Long residency cation-binding and hydration sites were identified by means of calculation of cation and water density maps by a Fourier-averaging method (52). Individual solvent particles were traced up to the distance cutoff of 3.4 Å for water molecules and 2.5 Å for Na+ from ribozyme electronegative atoms. The positions of cations were taken in regular time intervals and then Fourier-transformed into pseudoelectron densities. The solvent density contour maps were visualized using the program Xfit (53). The electrostatic potentials of crystal structures and a series of simulated averaged structures were calculated using the program Delphi (54) by solving the nonlinear Poisson-Boltzmann equation for ionic strength 0.2 M, and were visualized using InsightII (38). Each atom was placed in a medium with a dielectric constant of 2.0 in the solvent inaccessible surface-enclosed volume, which was obtained using a probe radius of 1.4 Å, whereas solvent was treated as a continuum with a dielectric constant of 80. Our backbone analysis was based on systematic monitoring of the backbone torsion angles followed by comparison with known backbone conformational families (55).
RESULTS AND DISCUSSION
Crystal structures and MD simulations: Mg2+ is consistently accommodated in the catalytic pocket of the wild-type precursor
Locations of the deepest negative electrostatic surface potentials (located within the −25 kT/e contour and further referred to as NESP sites) are similar in crystal structures of the precursor and product forms of the ribozyme (Fig. 2 A) and do not change significantly in our MD simulations. The widest NESP site 1 (Fig. 2 A) is found at the pocket formed by the compact fold of loop L3, which encompasses the cleavage site and catalytic residue C75. The strong bend of the L3 backbone at the C21-U23 segment results in clustering of phosphates. NESP site 1 is associated with the global ESP minimum in the precursor (∼−63 kT/e), whereas it is reduced to ∼−45 kT/e in the product, becoming a local minimum. The NESP weakening is a consequence of the departure of the scissile phosphate (G1) and rearrangement of the active site after the cleavage reaction. At the same time, the collapse of P1 and L3 toward the center of the product and the shift of C75 deeper into the active site result in a shallower catalytic pocket. The presence of the −1 phosphate strengthens the NESP at the active site by only 2 kT/e (test calculations not shown). NESP site 5 (at the major groove of the P1/P1.1 helices with peak near G1/U37 pair in the immediate vicinity of the active site) is also significantly amplified in precursor.
Two divalent cation-binding sites (marked as MG1 and MG2 in this article) observed in precursor crystal structures are located within NESP sites 1 and 2. On the contrary, only one of the nine Mg2+ cations seen in crystal structure of wild-type product locates within NESP site 5. Further, in this product structure, the Mg2+ closest to the catalytic pocket is 7 Å away from the NESP site 1 (Fig. 2 A). The calculated ESP is thus consistent with cation binding in precursor crystal structures, but inconsistent with the distribution of Mg2+ cations in the wild-type product crystal. Binding of cations in simulations is substantially determined by ESP (see below).
Mg2+ possesses an octahedral coordination sphere (56,57). Hexahydrated Mg2+ interacts with RNA via nonspecific electrostatic interactions, whereas interactions including direct contact (inner shell) between RNA and Mg2+ require partial dehydration of the cation (58,59). The crystal structures, however, do not reveal positions of water molecules. Six functional groups (four phosphoryl oxygen and two uracil keto oxygen atoms) are located within the outer sphere coordination distance from the MG1 metal ion in crystals of C75U precursor (6). Further, a short contact between U75(O4) and Mg2+ suggests likely inner sphere binding.
In the precursor simulation PreC41+U75A-2Mg (cf. Table 1), the whole coordination sphere was in basic agreement with the crystal structure, including the inner shell Mg…U75(O4) contact (1.98 Å). However, additional Mg…G1(O2P) (1.87 Å) inner shell contact was formed (Fig. 3) in contrast to outer shell (water-mediated) coordination suggested by the crystal structure, accompanied by shift of the cation by ∼2.0 Å from the initial position. The octahedral ligand shell is completed by four water molecules, bridging the cation with phosphates of U-1, C22, and U23. The first hydration shell waters do not exchange with the bulk solvent in agreement with the experimentally measured microsecond residency time (60) and computational studies (20,29,61).
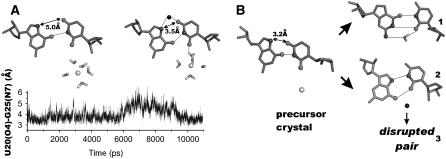
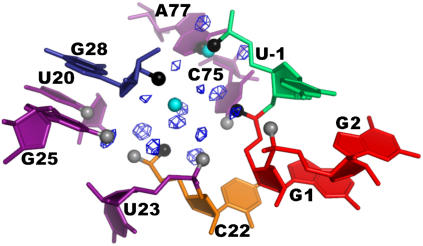
FIGURE 3.
First hydration shell of Mg2+ bound at the catalytic pocket in simulations PreC41+U75A-2Mg (left) and PreC41+A-2Mg before the G1(O5′)…Mg2+ water bridge was broken (middle). (Right) First and second hydration shells of the catalytic Mg2+ in the simulation PreC41+A-2Mg (after the G1(O5′)…Mg2+ water bridge was lost). The residues are color-coded as shown in Fig. 1. Mg2+ is represented by a green ball; dark blue and cyan mesh represent first and second hydration shells of Mg2+, respectively. Functional groups of the catalytic pocket directly bound to Mg2+ are black balls, those bound to the first hydration shell waters are gray balls, and those bound to the second hydration shell waters are white balls. See also Fig. S1.
A metal ion is also identified in the active site of the low-resolution x-ray structure of the wild-type (C75) precursor ribozyme (6), ∼3.0 Å away from the Mg2+ position in the C75U mutant precursor. This difference in the metal ion location has been attributed to the loss of a favorable contact between the keto oxygen of U75 and the metal ion (6). In all wild-type precursor simulations, C75 is shifted by 2 Å within the active site compared to U75. This shift creates enough space for Mg2+ binding in the catalytic pocket while avoiding a direct contact with the N4-amino group of C75 (Fig. 3). Indeed, both wild-type simulations PreC41+Mg and PreC41+A-2Mg reveal stable binding of Mg2+ in the pocket. The average displacement of the Mg2+ from its initial position is 1.8 Å and 1.3 Å in the two simulations, respectively, and Mg2+ forms a direct 1.85 Å contact with U23(O1P). The first hydration shell now consists of five water molecules. Four of these waters are equatorial and the fifth is positioned axially with respect to the U23(O1P) atom while being bound to U20(O2). Only the U20 nucleotide interacts with the first Mg2+ hydration shell through a nucleobase atom (O2), consistent with an experimentally suggested role of the U20 base in Mg2+ binding by the active site of the HDV ribozyme (62). The equatorial water molecules are involved in H-bonding with phosphates of G1, C22, and U23, and only one water molecule does not have contact with the ribozyme (Fig. 3). It should be noted that competitive inhibition of the wild-type HDV ribozyme by cobalt hexammine has been used to suggest that the catalytic metal ion binds into a region of low charge density and/or by outer-sphere contacts, in contrast to our simulation results (9).
The G1(O5′)…H-O…Mg bridge, critical for ribozyme cleavage in mechanistic proposals involving general acid catalysis by a hydrated Mg2+, was formed only at the beginning of the C75 wild-type simulations (Fig. 3, Fig. S1). This water bridge disappeared in simulations due to rearrangement of U-1/G1 backbone, described in Fig. 2 A of Krasovska (37). After this rearrangement, G1(O5′) becomes occasionally hydrated by water molecules from the Mg2+ second hydration shell. Currently, we cannot decide whether this local backbone rearrangement is correct or reflects a force field imbalance. The MD backbone geometry basically corresponds to the RNA backbone family 29 (55), whereas the x-ray geometry does not match any established RNA backbone geometry class.
Twelve water molecules reside in the second shell of a bulk Mg2+ ion with average distances of 4.25 Å between water oxygen and the magnesium (56,57). The second hydration shell of the Mg2+ bound at the catalytic pocket includes on average only nine water molecules, whereas the outer coordination sphere is completed by contacts with the ribozyme, filling the whole catalytic pocket with a dense network of H-bonds (Fig. 3). Binding of Mg2+ in the catalytic pocket results in a significant increase in the residence time of water molecules in the second hydration shell (by almost two orders of magnitude to up to >10 ns, Table 2). The average water residency time in the second shell of the bulk Mg2+ is ∼15 ps with the present force field, in line with literature data using a specialized force field (56), see also Auffinger et al. (61).
TABLE 2.
Averaged (tave) and maximal (tmax) binding times of water molecules in the second hydration shell of Mg2+ (simulation PreC41+A-2Mg) and the first hydration shell of Na+ (simulation PreC41+A-2) permanently bound at the catalytic pocket, as compared to bulk cations
| Mg2+bound | Mg2+bulk | Na+bound | Na+bulk | |
|---|---|---|---|---|
| tmax (ns) | 13.0 | 0.180 | 4.30* | 0.200 |
| tave (ns) | 0.23 | 0.015 | 0.18 | 0.017 |
Binding times up to 7 ns are observed in simulations PreC41+U75 and PreC41+U75A-2 with position of Na+ additionally stabilized due to binding to U75(O4).
In summary, our MD simulations are in basic agreement with the crystal structure of C75U precursor. The simulations reveal smooth accommodation of the divalent ion in the catalytic pocket of the wild-type (C75) precursor, and a dense network of long-residency water molecules bridging the magnesium and the RNA.
Mg2+ is expelled from the active site in simulations of the C75H+ protonated precursor and the C75 product form
Protonation of C75 would be required for it to play a role as the general acid during catalysis. Protonated C75H+ moves toward its product-like location in all precursor simulations and establishes a stable H-bonding network (37). When placing the Mg2+ into the active site, the cation is expelled within 1 ns, consistent with the known competition between C75 protonation and Mg2+ binding at the active site (3). No magnesium cation is observed at the cleavage site in wild-type and C75U product crystal structures. In a product simulation with an Mg2+ cation initially placed at the pocket, the cation is expelled during the equilibration.
The very swift expulsions of the ions indicate that the ion binding is substantially destabilized. Further details can be found in the Supplementary Material.
Loop L3, bridged by the noncanonical U20/G25 basepair, is rearranged in the absence of divalents
The dynamic loop L3 regulates the negative electrostatic potential of the catalytic pocket (37). In addition, the residues U-1, G1, U20, C22, and U23 contribute specific coordination sites for catalytic cation binding. The U20/G25 basepair forms a rigidifying bridge across L3 and supports a compact fold of the otherwise flexible loop. The bifurcated cis-W.C./W.C. (Watson-Crick) configuration (63) of the U20/G25 basepair observed in the precursor crystal structures is preserved in all simulations with the catalytic Mg2+ bound in the pocket. It has two substates, one of them additionally mediated by Na+ cation (Fig. 4 A) for ∼60% of the basepair lifetime. In the absence of Mg2+, the U20/G25 basepair is immediately rearranged into a trans-W.C./H. (Hoogsteen) or cis-W.C./W.C. basepair (37) (Fig. 4 B). The shape of L3 is significantly affected by the type of U20/G25 basepair (Fig. S2).
FIGURE 4.
Interactions of the U20/G25 basepair with cations in precursor simulations. Basepair edge oriented inside the catalytic pocket is directed downward. Gray and black balls represent Mg2+ and Na+ cations, respectively. (A) Two substates seen in precursor simulations with Mg2+. The U20(O4)-G25(N7) distance trajectory in simulation PreC41+A-2 is shown below. (B) Rearrangement of the U20/G25 basepair in the absence of Mg2+; arrows show transitions observed in simulations. Substates 1–3 were observed in the following simulations: 1), PreC41+ and PreC41+A-2; 2), PreC41+C75+, PreC41+U75, PreC41+A-2 and PreC41+U75A-2; and 3), PreC41+Trc and PreC41+U75.
The U20/G25 basepair is different in the product crystal structure due to the anti-to-syn flip of G25. The anti-conformation of G25 observed in the precursor leads to a larger size of the catalytic pocket, required to accommodate the Mg2+ and U-1/A-2 residues. An actual anti-to-syn transition of G25 between the precursor and product configurations upon cleavage would require disruption of the U20/G25 basepair and at least a partial unfolding of L3. Indeed, disruption of the basepair accompanied by unfolding of L3 was observed in simulation PreC41+Trc, initiated using the precursor crystal structure but deleting U-1 (Table 1) as well as in the last 2 ns of simulation PreC41+U75, both in the absence of Mg2+. Similarly, simulations of product show formation of two different types of U20/G25 basepairs and the effect of its occasional disruption on L3 unfolding (37). However, G25 always remains in the syn conformation in product simulations and anti in precursor simulations. The timescale of the simulations is unlikely to allow a spontaneous flipping, even in the simulation PreC41+Trc, attempting to mimic the precursor to product switch.
We suggest that the U20/G25 configuration observed in the precursor crystal structure requires binding of Mg2+. Furthermore, after cleavage and expulsion of Mg2+ from the cleavage site, L3 becomes more dynamic and its unfolding could allow for the anti-to-syn transition of G25 on a longer timescale.
A major Na+ binding site is located at the catalytic pocket
Cleavage rates above the background level have been measured for the genomic HDV ribozyme in molar concentrations of Na+ ions only (3,8,41). In all simulations lacking the Mg2+ ion at the active site, an exceptionally strong Na+ binding is seen at the catalytic pocket, which accommodates up to two Na+ cations simultaneously. A higher occupancy and slower exchange of cations between the pocket and bulk solvent were clearly observed in precursor simulations (Table 3).
TABLE 3.
Na+ binding at the catalytic pocket (MG1 site): number of cations simultaneously bound at the pocket averaged over the whole simulation (Tot) and number of distinct cations visiting the pocket during simulation (N).
| Simulation | Tot | N |
|---|---|---|
| PreC41+ | 1.3 | 3 |
| PreC41+A-2 | 1.9 | 3 |
| PreC41+C75+ | 1.0 | 1 |
| PreC41+U75* | 2.0 | 2 |
| PreC41+U75A-2 | 2.0 | 2 |
| ProC41+ | 1.4 | 4 |
| ProC41+C75+ | 0.8 | 6 |
Last 2 ns of simulation PreC41+U75 when L3 was unfolded were not considered.
The occupancy of the pocket by Na+ is sensitive to arrangement of the electronegative atoms pointing into the pocket. Thus, unfolding of L3 in simulation ProC41+C75+ results in reduced occupancy (Table 3). Furthermore, occupancy of monovalent ions is affected by orientation of the U20/G25 basepair. For example, a U20/G25 trans-W.C./H. basepair shows long residency (6–13 ns) inner-shell Na+ binding in the pocket, whereas a cis-W.C./W.C. basepair seen in simulation PreC41+ is only involved in outer shell cation binding (Fig. 4 B). This fact explains a relatively low occupancy of the pocket in simulation PreC41+. Protonation of C75 is another factor that disfavors simultaneous binding of two Na+ ions (Table 3).
Although in most simulations there was essentially no exchange of ions between catalytic pocket and bulk solvent (Table 3), this observation does not reflect incidental trapping of ions at the beginning of the simulations (64). The outcome of the simulations does not depend on the initial location of the ions, as some simulations were initiated with all ions shifted away from the solute (Table 1). A detailed analysis of the ion binding in one representative precursor simulation is given in Table 4.
TABLE 4.
Balance between inner-shell Na+ binding and long-residency water bridges involving the Na+ first-shell waters to individual atoms in the catalytic pocket (MG1 site); simulation PreC41+A-2
| Direct Na+ binding
|
Water binding
|
||
|---|---|---|---|
| Atom | Occup.* | tmax of Na† | tmax of water† |
| U-1(O2P) | 62 | 9.0 | 4.8 |
| G1(O2P) | 38 | 2.5 | 4.5 |
| G2(O2P) | – | – | 5.0 |
| U20(O2) | – | – | 2.3 |
| C22(O1P) | – | – | 2.4 |
| C22(O2P) | 23 | 2.5 | 5.1 |
| C23(O1P) | 14 | 1.6 | 3.3 |
| C25(N2) | – | – | 2.0 |
| G28(O2′) | 19 | 1.7 | 1.5 |
| C75(N4) | – | – | 4.7 |
| A77(N1) | 55 | 6.5 | 2.0 |
Total inner-shell occupancies (%) higher than 10% are shown.
Longest binding time (ns).
The first hydration shell of Na+ shows residency times comparable to the second hydration shell of Mg2+ (Table 2). Although maximal binding times of water molecules in the ligand shell of bulk Na+ cations do not exceed 200 ps, they significantly increase when the waters bridge the ribozyme backbone with bound Na+ ions (Fig. 5).
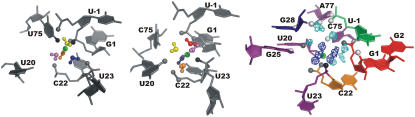
FIGURE 5.
Two Na+ cations (cyan balls) and their hydration shell (blue mesh) bound at the catalytic pocket in simulation PreC41+A-2. Positions of cations in the figure are determined by points with highest pseudo-electron density of Na+. The residues are color-coded as in Fig. 1; functional groups of the catalytic pocket directly bound to the Na+ are shown as black balls and those interacting with the first hydration shell waters as gray balls.
In contrast to divalents, two Na+ freely migrate along the wide catalytic pocket, making multiple direct and water-mediated long residency contacts with the ribozyme (Table 4). The hydration shells of two Na+ cations fill the whole pocket; however, no long residency hydration of G1(O5′) is observed. It thus does not seem that a specific Na+-stabilized water molecule is poised to facilitate protonation of G1(O5′), at least within the approximations of the present force field.
In summary, there is a strong Mg2+ or Na+ binding at the active site in the wild-type precursor, whereas only Na+ cations show stable binding at the active site in the product and C75H+ precursor simulations. Despite 100% occupancy of the precursor pocket by 1–2 Na+ cations, the binding is variable and the cations are delocalized (Table 4), which would make them difficult to detect in crystal structures.
Monovalent cations compete with Mg2+ at the MG2 binding site
Divalent cation binding to J1.1/4 and P4 was suggested by Pb2+ induced scission at the MG2 site (65) and was also revealed in crystal structures of precursor and C75U mutant product forms. Location of this cation close to C41/C41H+ indicates that it is a plausible candidate for the pH-sensitive structural cation associated with C41 (8). Mg2+ is located within NESP site 2 in the crystals (Fig. 2 A) and makes no inner-sphere contact to the ribozyme. In our simulations, the cation remains in the pocket and typically interacts via inner shell binding with A42(O2P) and five water molecules, which do not exchange with the second shell. The axial water has a fixed position and makes no contact to the ribozyme, whereas the equatorial water molecules easily rotate, alternately bridging the cation with C41(O2′) and A43(O2P) (Fig. S3). The second hydration shell of this Mg2+ is considerably more dynamical than the second hydration shell of the Mg2+ at the active site pocket (Fig. S3). The binding times of water molecules in the second shell are up to 5.5 ns. All water molecules with binding time above 1 ns mediate the C41H+/A43 basepair. Although this basepair is stable and water mediated in Na+-only simulations, the water binding times are much shorter in the absence of bound Mg2+ (see below).
In the absence of Mg2+, a prominent Na+ binding site is located at the NESP site 2 (Fig. 2 B). Three to five Na+ ions exchange in the MG2 site in our simulations, leading to a total occupancy close to 100%. The cations bind alternately via inner and outer shell binding to C41H+(O2′), A42(O2P), A43(O2P), and G71(N7) (Table 5). The MG2 site may act as a structural site and together with the MG1 site could be the site of the strong competition between magnesium and sodium, affecting HDV ribozyme catalytic activity (9).
TABLE 5.
Inner-shell Na+-binding occupancies (%) of individual atoms higher than 10% and number of exchanged cations (in parentheses) in binding sites MG2, NA3, NA4, and NA5.
| Simulation
|
|||
|---|---|---|---|
| Site | Atom | PreC41+A-2 | ProC41+ |
| MG2 | C41(O2′) | 34(4) | 57(5) |
| A42(O2P) | 12(3) | 15(3) | |
| G43(O2P) | 22(2) | 15 (3) | |
| G71(N7) | 8(3) | 28(4) | |
| NA3 | U20(O1P) | 36(5) | 24(4) |
| C21(O1,2P) | 17(1) | 34(1) | |
| C75(O2P) | 22(2) | 84(3) | |
| NA4 | C32(O2P) | 23(9) | 23(9) |
| G31(O1P) | 13(5) | 28(3) | |
| A78(O1P) | – | 22(8) | |
| A77(O2P) | 11(3) | – | |
| A77(N7) | 34(3) | – | |
| A78(N7) | 28(4) | 80(3) | |
| NA5 | G1(N7) | 44(4) | 25(8) |
| G1(O1P) | 30(3) | – | |
| G2(N7) | 15(2) | 10(4) | |
| C75(N3) | 16(4) | – | |
Other major cation binding sites
There are several other remarkable Na+ cation-binding sites in the HDV ribozyme (Table 5). Cation binding regions are similar in all precursor and product simulations and are primarily determined by ESP (Fig. 2, Fig. S4). The single-stranded junction J4/2 bearing the catalytic C75 is associated with two Na+ binding sites to phosphate. A rather compact inner-shell binding site NA3 bridges the trefoil turn with the C19/U20 backbone (Table 5, Fig. 6). It fits NESP site 3 and is in agreement with metal ion-induced cleavage experiments that have observed scission sites at positions G74, C75, and G76 induced by Mg2+ (66), Pb2+ (65), and Tb3+ (67,68) cations.
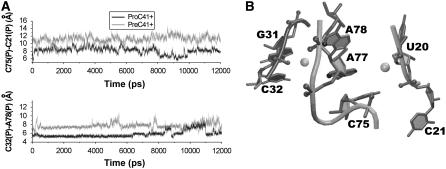
FIGURE 6.
Na+ binding sites associated with the J1.1/4 junction. (A) Interphosphate distances C32(P)-A78(P) (top) and C75(P)-C21(P) (bottom) in product and precursor simulations. (B) Na+ (gray balls) binding in sites NA3 and NA4 represented by cation density maxima in simulation PreC41+.
The cation binding site NA4 is located at the arching junction of P1 and P3 and fits NESP site 4. This inner-shell phosphate Na+ binding site is further extended by cation binding in the major groove of consecutive A77 and A78 (Table 5, Fig. 6). Occupancies of sites NA3 and NA4 are affected by the C32/A78 and C75/C21 interphosphate distances, which in turn are sensitive to arrangement of joiner J4/2. These sites have higher Na+ occupancy (Table 5, Table S1) in product simulations with deeply buried and firmly bound C75 and J4/2 placed closer to P1 and L3 (Fig. 6).
Cation binding in double-helical segments is described in the Supplementary Material.
Product simulations with nine Mg2+ cations
We have carried out several simulations with nine Mg2+ cations, which are described in more detail in the Supplementary Material. Assessing all the data including Na+ simulations that provide sufficient sampling, we suggest that our simulations are inconsistent with the overall positioning of nine Mg2+ cations in the wild-type product crystal structure. In contrast, dynamics of cations seen in simulations is entirely consistent with the two Mg2+ binding sites in the precursor and C75U product crystal structures.
Water-mediated interactions stabilize noncanonical basepairs and the A-minor motif
The available crystal structures of the HDV ribozyme provide essentially no information about hydration. The simulations reveal a variety of hydration sites, including common phosphate hydration sites (25) and specific hydration sites related to noncanonical basepairs and complex elements of tertiary structure (Fig. 7).
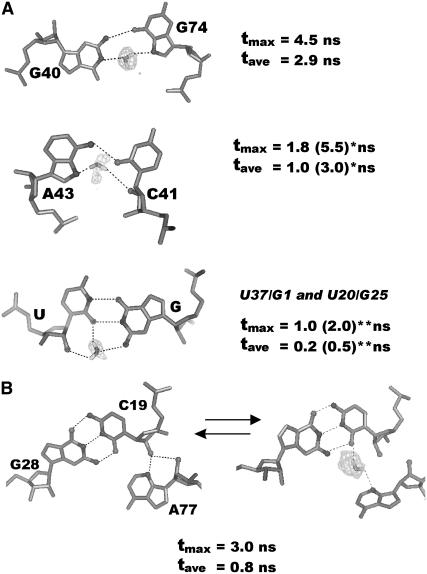
FIGURE 7.
(A) Long-residency water bridges in noncanonical basepairs. (*) Binding times of water molecules are significantly longer when they belong to the second hydration shell of Mg2+ (in parentheses, see text). (**) Two cis-W.C./W.C. basepairs show different water binding times, which are significantly longer in the U20/G25 basepair exposed into the catalytic pocket (in parentheses). (B) Closed (left) and open (right) substates of A-minor type II. Water density at contour level >10 σ (gray mesh); typical orientation of the water molecules, maximal (tmax) and averaged (tave) water binding times, are shown.
G1/U37, trans-W.C./H., or cis-W.C./W.C. U20/G25, G40/G74, and C41/A43 (Fig. 7 A) represent known types of water-mediated basepairs commonly observed in crystal structures (63,69,70). Water-mediated basepairs show water binding times 0.5–5.5 ns (27,29,32). Water binding times of a given basepair, however, can significantly vary depending on its environment. Thus, vicinity of hydrated mono- and especially divalent cation results in a significant increase of water binding times as observed for C41/A43 and cis-W.C./W.C. U20/G25 basepairs (Fig. 7 A). Interestingly, the trans-W.C./H. U20/G25 basepair was observed in our simulations as either water- or Na+-mediated, with the cation or water molecule bridging U20(O2) and G25(O6) (Fig. 4 B). The cation mediated form is more stable (37).
The A-minor motif is the most important recurring RNA tertiary interaction formed by adenines interacting with the minor groove edges of G=C basepairs (71). Dynamical water insertion into A-minor motifs was recently reported for ribosomal kink-turns (34). A-minor motif in the HDV ribozyme involves the consecutive stacked A78 and A77, interacting with two W.C. G=C pairs in helix P3 (Fig. 1). A78 optimally fits into the minor groove of the G29=C18 basepair as an A-minor type I motif (71). This interaction is entirely stable in the simulations. A77 stacks on C75 and forms an A-minor type II interaction. Its position is therefore related to the positioning of C75 in the active site. As a result, this A-minor type II interaction occurs in two substates in simulations (Fig. 7 B). The x-ray-like geometry is stable in simulations PreC41+U75, PreC41+U75A-2, and ProC41+C75+, whereas in all other simulations it is alternating with an open A-minor interaction mediated by a long-residency (up to 3 ns) C19(O2)/C19(O2′)/A77(N3) water bridge. Thus, insertion of a water molecule stabilizes the open conformation of the A-minor motif and is likely related to subtle regulation of base 75 positioning in the active site. A77 is the only base that makes vertical (stacking) contact with the 75 base. The closed A-minor interaction observed in simulation PreC41+U75 leads to maximal mutual overlap of the aromatic rings of U75 and A77 (Supplementary Fig. S5a). The closed A-minor interaction was also observed in simulation PreC41+C75+, but in this case A77 and protonated C75H+ bases were shifted against each other. The base-base stacking was supplemented by sugar-base stacking between the A77 five-membered ring and the C75+ ribose, which also is a quite efficient dispersion-controlled interaction similar to base stacking (72). Presence of the open A-minor interaction and canonical C75 (e.g., in simulations PreC41+Mg, PreC41+, and ProC41+) is characterized by reduced base-base overlap and improved sugar-base stacking (Supplementary Fig. S5b).
The backbone dynamics
When analyzing results of MD simulations, it is important to assess their quality. The simulations appear to provide good agreement with the available x-ray structures of the HDV ribozyme, as judged, for example, by RMSD, positions of bases, etc. (37). The backbone is inherently more difficult to describe by force fields compared to interactions involving the rigid nucleobases, since i), the backbone is anionic (and thus polarizable), and ii), the constant point atomic charges might not work equally well for distinct combinations of backbone torsion angles. Thus, the quality of the backbone description is becoming one of the main issues for nucleic acids simulations. Recent B-DNA simulations reported unexpected α-γ flips of the B-DNA backbone. Such switches lead to long-lived backbone substates with concomitant changes in B-DNA geometry (40,73,74). Major problems with DNA backbone topology were reported for G-DNA loops (75).
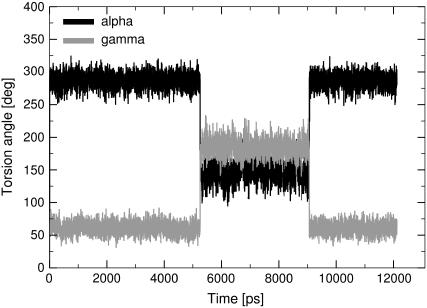
In the helical segments of our simulated structures, the backbone is rather rigid, with backbone torsion angles close to those of canonical A-RNA. There are, however, backbone flips mainly in nonterminal residues of the double-helical segments. These flips entail simultaneous transitions of the α (from 295 to 155°) and γ (from 55 to 180°) torsion angles, which are compensatory and do not affect arrangement of the helix. Switching occurs stochastically in all analyzed simulations and is reversible. The resulting geometry matches a modified A-type backbone conformation commonly observed in RNA crystal structures and described as family number 24 (55). Fig. S6 in the Supplementary Material represents the population of phosphate switches observed in double-helical segments in two representative simulations (PreC41+ and ProC41+). Lifetimes of the individual flipped substates range from 2 to 10 ns, and the number of flipped nucleotides happens to be smaller at the end of the simulations than in the middle of them. Fig. 8 shows a typical reversible backbone switch. Similar behavior was noticed also for multiple 25 ns simulations of the 23S rRNA sarcin-ricin loop motif (76). Thus, for RNA, we do not observe any cumulation of the backbone flips, clearly contrasting behavior of B-DNA simulations (40,74).
FIGURE 8.
Characteristic values of backbone torsion angles α (black line) and γ (gray line) connected with the phosphate switch in the canonical helix P1 (residuum C33) in the simulation PreC41+.
There is a wide range of other backbone geometries in the nonhelical segments of the HDV ribozyme (Fig. 9). Many of them fall into typical backbone families observed in crystal structures of 23S and 5S ribosomal RNAs (55). By contrast, the residues U23-U27 of loop L3 do not match any established backbone family, which may reflect their dynamic disorder, their uniqueness, or it may be due to a limited crystallographic resolution. In fact, many nucleotide conformations as refined in x-ray crystal structures cannot be classified as one of the typical conformational classes (55). Our simulations reveal multiple transient and permanent backbone switches in the nonhelical regions (Fig. 9) which, however, do not distort the ribozyme. Most such switches represent transitions between some known RNA backbone families. In many other cases, the switch shifts the backbone from crystallographically observed conformation that does not belong to an established backbone family to a conformation that can be assigned to a specific RNA backbone family. Thus, our simulations reduce the number of nucleotides with unidentified backbone geometry compared to crystal structures. Such shifts were observed, for instance, in the L3 region (Fig. 9) and were especially apparent in simulation ProC41+C75+ with unfolded L3, where the unfolded L3 backbone became mostly canonical (not shown). The simulations preserve key noncanonical backbone arrangements of several residues. For instance, C41H+ retained its backbone topology that is specific for low-rise (A-platform like) dinucleotide steps (55).
FIGURE 9.
Secondary structure of the HDV ribozyme, with nucleotide backbone families (55) shown in the starting C75U precursor crystal (left) and PreC41+ simulated structure (right). Gray and black upper cases represent canonical and established noncanonical backbone families, respectively; black lower case letters represent an unidentified backbone angle combination. Temporary backbone flips seen in the simulation are marked by open circles, whereas permanent backbone switches are indicated by gray circles.
CONCLUSIONS
We have carried out a set of explicit solvent molecular dynamics simulations to investigate the structural dynamics of the HDV ribozyme, with special emphasis on its interactions with ions and water. Careful analysis of the backbone behavior in the simulations suggests that the force field is performing reasonably well. Specifically, there are no irreversible backbone flips in the canonical regions, whereas key backbone segments that are critical for the complex 3D fold of the molecule are fairly stable. These observations obviously do not rule out the occurrence of force-field related problems on a longer timescale; in fact, it is unlikely that simple classical molecular mechanics would fully describe an anionic sugar-phosphate with its intricate electronic structure.
The compact tertiary structure of the HDV ribozyme is associated with multiple regions of deep negative electrostatic surface potential, which lead to several prominent and often uniquely structured cation-binding sites. When integrating all the available experimental and computational data, we suggest that the two most significant cation binding sites correspond to Mg2+ cations bound at the active site (MG1) and the major groove of P4 (MG2) as seen in the precursor crystal structures. The simulations reveal that the ability of the active site to accommodate the catalytic Mg2+ is determined by the ESP and the size of the pocket. Very low (negative) ESP and a larger catalytic pocket in the precursor structure are favorable for Mg2+ binding. In accord with the crystal structures, the divalent cation remains stably bound at the catalytic pocket in simulations of the C75U mutant and C75 wild-type precursors. It makes direct and water mediated contacts with the scissile phosphate, including a temporary G1(O5′)…Mg2+ water bridge as necessary for the general acid catalysis by the hydrated Mg2+ cation. Protonation of C75 in the precursor results in product-like deep binding of C75H+ in the active pocket and appears to be incompatible with binding of the catalytic Mg2+ cation at the active site. These findings provide additional support for a role of C75 as the general base during catalysis complemented by concerted action of a hydrated Mg2+ ion as general acid. The Mg2+ cation is not stable at the active site of the product form of ribozyme, in agreement with the crystallographic data. This observation is likely due to the significantly higher (less negative) ESP and shallower catalytic pocket of the product relative to the precursor.
A bound MG1 Mg2+ cation also stabilizes the structure of loop L3 (particularly the U20/G25 basepair), which shows rearrangements in the absence of Mg2+. We thus propose that the collapse of P3 and L3 toward the center of the ribozyme and the related flip of the noncanonical U20/G25 basepair as observed in crystal of product ribozyme might be a consequence of Mg2+ expulsion from the active site after the cleavage. We believe that the picture of the Mg2+ binding emerging from our simulations is qualitatively correct, though it is fair to admit that description of divalent cations as simple van der Waals probes with central point charge of +2 is a crude approximation. Thus, details of Mg2+ binding should be assumed to be imperfect. In addition, the simulation timescale is very limited for divalent cations.
Simulations carried out entirely in the absence of Mg2+ reveal that the MG1 and MG2 sites are occupied by monovalent cations with occupancy close to 100%. The catalytic pocket (MG1 site) typically accommodates two monovalent cations simultaneously. In contrast to Mg2+, bound monovalents are fluctuating in the pocket and thus are not likely to be detected by x-ray crystallography. The exchange of Na+ between the pocket and the bulk is slow in the simulations, so that no statistics could be computed. However, simulations utilizing different initial ion distributions show that the ions in the pocket are not incidentally trapped. When comparing with literature data, the catalytic pocket in the precursor HDV ribozyme is the most prominent Na+ binding site in RNA characterized by MD simulations so far (20,29,36). The simulations also suggest the presence of two significant cation-binding sites located near the J4/2 junction, as indicated by Pb2+ induced scission experiments (65). The description of monovalent ions is considerably more accurate compared with divalents and we suggest that the simulations are fairly sufficient to deal with highly occupied monovalent binding sites in RNA molecules.
Structural dynamics of the HDV ribozyme is associated with long residency hydration sites, including several water-mediated basepairs. A water-mediated interaction is also suggested as a substate for the fluctuating A-minor type II interaction involving A77, which is coupled with positioning of the catalytically critical nucleotide 75 in the catalytic pocket. The long-residency water molecules usually make contacts with electronegative RNA atoms and regions, and thus compete with cations for the same binding sites, as exemplified in the trans-Watson-Crick/Hoogsteen U20/G25 basepair. We suggest that selectivity of the binding site with respect to water molecules, monovalent, or divalent cations is affected by the geometry of the binding site. Monovalent cations may readily substitute for divalents. This is to be considered when analyzing experimental structural data, since a bound divalent cation can be rather easily replaced by fluctuating monovalent cations or by a water molecule, which are both less likely to be detected. Description of base stacking, H-bonding, and water binding can be considered as the most accurate part of the force field, since these interactions can be well captured by the point charge electrostatic model and the Lennard-Jones potential (77–79).
We evidenced another kind of hitherto unreported combined cation binding and hydration events, which are especially apparent in the catalytic pocket. Many of the long-residency water molecules present in the pocket participate in the Na+ first ligand shell while acting as structural bridges to the RNA. Thus, when the Na+ cations occupy the pocket, the pocket is filled by a complex network of water molecules structured by the ions and the solute. The binding times of water molecules in the first-shell of bound Na+ cations range up to ∼7 ns, which is almost two orders of magnitude longer than in the case of bulk Na+ cations. Therefore, Na+ plays an important role in structuring of water molecules at the active site in analogy to Mg2+ and could possibly participate in catalysis in the absence of Mg2+. Interestingly, the same conclusion can be drawn regarding water molecules participating in the second shell of the bound Mg2+ ions. These water molecules also form stable water bridges between the hydrated cation and solute, and their binding time is substantially enhanced compared to the second shell water molecules of bulk Mg2+.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
This work was supported by the Wellcome Trust International Senior Research Fellowship in Biomedical Science in Central Europe GR067507, grants GA203/05/0388 and GA203/05/0009, Grant Agency of the Czech Republic, grant 1QS500040581 by Grant Agency of the Academy of Sciences of the Czech Republic, by Research Center LC512, and research projects AVO Z5 004 0507, AVO Z4 055 0506, and MSM0021622413 by the Ministry of Education of the Czech Republic, by National Institutes of Health grant GM62357, including supplement S2 for acquisition of a computer cluster to N.G.W., and by a Margaret and Herman Sokol International Summer Research Fellowship, a NATO Science Fellowship, a Center for the Education of Women Sarah Winans Newman Scholarship, and an Eli Lilly Fellowship to Jana S.
References
- 1.Lai, M. M. 1995. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64:259–286. [DOI] [PubMed] [Google Scholar]
- 2.Perrotta, A. T., I. Shih, and M. D. Been. 1999. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 286:123–126. [DOI] [PubMed] [Google Scholar]
- 3.Nakano, S., D. M. Chadalavada, and P. C. Bevilacqua. 2000. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 287:1493–1497. [DOI] [PubMed] [Google Scholar]
- 4.Ferre-D'Amare, A. R., K. Zhou, and J. A. Doudna. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature. 395:567–574. [DOI] [PubMed] [Google Scholar]
- 5.Ferre-D'Amare, A. R., K. Zhou, and J. A. Doudna. 1998. A general module for RNA crystallization. J. Mol. Biol. 279:621–631. [DOI] [PubMed] [Google Scholar]
- 6.Ke, A., K. Zhou, F. Ding, J. H. Cate, and J. A. Doudna. 2004. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 429:201–205. [DOI] [PubMed] [Google Scholar]
- 7.Das, S. R., and J. A. Piccirilli. 2005. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 1:45–52. [DOI] [PubMed] [Google Scholar]
- 8.Nakano, S., D. J. Proctor, and P. C. Bevilacqua. 2001. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 40:12022–12038. [DOI] [PubMed] [Google Scholar]
- 9.Nakano, S., A. L. Cerrone, and P. C. Bevilacqua. 2003. Mechanistic characterization of the HDV genomic ribozyme: classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 42:2982–2994. [DOI] [PubMed] [Google Scholar]
- 10.Pyle, A. M. 2002. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 7:679–690. [DOI] [PubMed] [Google Scholar]
- 11.Hanna, R., and J. A. Doudna. 2000. Metal ions in ribozyme folding and catalysis. Curr. Opin. Chem. Biol. 4:166–170. [DOI] [PubMed] [Google Scholar]
- 12.Murray, J. B., A. A. Seyhan, N. G. Walter, J. M. Burke, and W. G. Scott. 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 5:587–595. [DOI] [PubMed] [Google Scholar]
- 13.Perrotta, A. T., and M. D. Been. 1990. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 18:6821–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami, J., P. K. Kumar, Y. A. Suh, F. Nishikawa, K. Kawakami, K. Taira, E. Ohtsuka, and S. Nishikawa. 1993. Identification of important bases in a single-stranded region (SSrC) of the hepatitis delta (delta) virus ribozyme. Eur. J. Biochem. 217:29–36. [DOI] [PubMed] [Google Scholar]
- 15.Ferre-D'Amare, A. R., and J. A. Doudna. 2000. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 295:541–556. [DOI] [PubMed] [Google Scholar]
- 16.Rangan, P., and S. A. Woodson. 2003. Structural requirement for Mg2+ binding in the group I intron core. J. Mol. Biol. 329:229–238. [DOI] [PubMed] [Google Scholar]
- 17.Draper, D. E., D. Grilley, and A. M. Soto. 2005. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34:221–243. [DOI] [PubMed] [Google Scholar]
- 18.Ennifar, E., P. Walter, and P. Dumas. 2003. A crystallographic study of the binding of 13 metal ions to two related RNA duplexes. Nucleic Acids Res. 32:2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auffinger, P., L. Bielecki, and E. Westhof. 2004. Anion binding to nucleic acids. Structure. 12:379–388. [DOI] [PubMed] [Google Scholar]
- 20.Reblova, K., N. Spackova, J. E. Sponer, J. Koca, and J. Sponer. 2003. Molecular dynamics simulations of RNA kissing-loop motifs reveal structural dynamics and formation of cation-binding pockets. Nucleic Acids Res. 31:6942–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano, S., and P. C. Bevilacqua. 2001. Proton inventory of the genomic HDV ribozyme in Mg(2+)-containing solutions. J. Am. Chem. Soc. 123:11333–11334. [DOI] [PubMed] [Google Scholar]
- 22.Shih, I. H., and M. D. Been. 1999. Ribozyme cleavage of a 2,5-phosphodiester linkage: mechanism and a restricted divalent metal-ion requirement. RNA. 5:1140–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shih, I. H., and M. D. Been. 2002. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 71:887–917. [DOI] [PubMed] [Google Scholar]
- 24.Westhof, E. 1988. Water: An integral part of nucleic acid structure. Annu. Rev. Biophys. Biophys. Chem. 17:125–144. [DOI] [PubMed] [Google Scholar]
- 25.Schneider, B., K. Patel, and H. M. Berman. 1998. Hydration of the phosphate group in double-helical DNA. Biophys. J. 75:2422–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auffinger, P., and E. Westhof. 2001. RNA solvation: A molecular dynamics simulation perspective. Biopolymers. 56:266–274. [DOI] [PubMed] [Google Scholar]
- 27.Schneider, C., M. Brandl, and J. Suhnel. 2001. Molecular dynamics simulation reveals conformational switching of water-mediated uracil-cytosine base-pairs in an RNA duplex. J. Mol. Biol. 305:659–667. [DOI] [PubMed] [Google Scholar]
- 28.Brandl, M., M. Meyer, and J. Suhnel. 2000. Water-mediated base pairs in RNA: A quantum-chemical study. J. Phys. Chem. A. 104:11177–11187. [Google Scholar]
- 29.Reblova, K., N. Spackova, J. Koca, N. B. Leontis, and J. Sponer. 2004. Long-residency hydration, cation binding, and dynamics of loop E/helix IV rRNA-L25 protein complex. Biophys. J. 87:3397–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csaszar, K., N. Spackova, R. Stefl, J. Sponer, and N. B. Leontis. 2001. Molecular dynamics of the frame-shifting pseudoknot from beet western yellows virus: the role of non-Watson-Crick base-pairing, ordered hydration, cation binding and base mutations on stability and unfolding. J. Mol. Biol. 313:1073–1091. [DOI] [PubMed] [Google Scholar]
- 31.Guo, J. X., and W. H. Gmeiner. 2001. Molecular dynamics simulation of the human U2B'' protein complex with U2 snRNA hairpin IV in aqueous solution. Biophys. J. 81:630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reblova, K., N. Spackova, R. Stefl, K. Csaszar, J. Koca, N. B. Leontis, and J. Sponer. 2003. Non-Watson-Crick basepairing and hydration in RNA motifs: molecular dynamics of 5S rRNA loop E. Biophys. J. 84:3564–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spackova, N., T. E. Cheatham, F. Ryjacek, F. Lankas, L. van Meervelt, P. Hobza, and J. Sponer. 2003. Molecular dynamics simulations and thermodynamics analysis of DNA-drug complexes. Minor groove binding between 4′,6-diamidino-2-phenylindole and DNA duplexes in solution. J. Am. Chem. Soc. 125:1759–1769. [DOI] [PubMed] [Google Scholar]
- 34.Razga, F., J. Koca, J. Sponer, and N. B. Leontis. 2005. Hinge-like motions in RNA kink-turns: The role of the second A-minor motif and nominally unpaired bases. Biophys. J. 88:3466–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Razga, F., N. Spackova, K. Reblova, J. Koca, N. B. Leontis, and J. Sponer. 2004. Ribosomal RNA kink-turn motif–a flexible molecular hinge. J. Biomol. Struct. Dyn. 22:183–194. [DOI] [PubMed] [Google Scholar]
- 36.Auffinger, P., L. Bielecki, and E. Westhof. 2004. Symmetric K+ and Mg2+ ion-binding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol. 335:555–571. [DOI] [PubMed] [Google Scholar]
- 37.Krasovska, M. V., J. Sefcikova, N. Spackova, J. Sponer, and N. G. Walter. 2005. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis delta virus as revealed by molecular dynamics simulations. J. Mol. Biol. 351:731–748. [DOI] [PubMed] [Google Scholar]
- 38.Accelrys Software, I. 1997. InsightII. San Diego, CA.
- 39.Gresh, N., J. E. Sponer, N. Spackova, J. Leszczynski, and J. Sponer. 2003. Theoretical study of binding of hydrated Zn(II) and Mg(II) cations to 5′-guanosine monophosphate. Toward polarizable molecular mechanics for DNA and RNA. J. Phys. Chem. B. 107:8669–8681. [Google Scholar]
- 40.Varnai, P., and K. Zakrzewska. 2004. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 32:4269–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wadkins, T. S., I. Shih, A. T. Perrotta, and M. D. Been. 2001. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol. 305:1045–1055. [DOI] [PubMed] [Google Scholar]
- 42.Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheathan III, J. Wang, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, R. V. Stanton, A. L. Cheng, J. J. Vincent, M. Crowley, V. Tsui, H. Gohlke, R. J. Radmer, Y. Duan, J. Pitera, I. Massova, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 2002. AMBER 7:2002. University of California, San Francisco, San Francisco, CA.
- 43.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- 44.Wang, J. M., P. Cieplak, and P. A. Kollman. 2000. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21:1049–1074. [Google Scholar]
- 45.Cheatham, T.E., P. Cieplak, and P.A. Kollman. 1999. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 16:845–862. [DOI] [PubMed] [Google Scholar]
- 46.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 47.Aqvist, J. 1990. Ion water interaction potentials derived from free-energy perturbation simulations. J. Phys. Chem. 94:8021–8024. [Google Scholar]
- 48.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- 49.Berendsen, H. J. C., J. P. M. Postma, W. F. Vangunsteren, A. Dinola, and J. R. Haak. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 50.DeLano, W. L. 2002. The PyMOL Molecular Graphics System.
- 51.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: Visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 52.Schneider, B., and H. M. Berman. 1995. Hydration of the DNA bases is local. Biophys. J. 69:2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McRee, D. E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156–165. [DOI] [PubMed] [Google Scholar]
- 54.Gilson, M. K., K. A. Sharp, and B. H. Honig. 1988. Calculating the electrostatic potential of molecules in solution—method and error assessment. J. Comput. Chem. 9:327–335. [Google Scholar]
- 55.Schneider, B., Z. Moravek, and H. M. Berman. 2004. RNA conformational classes. Nucleic Acids Res. 32:1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez, J. M., R. R. Pappalardo, and E. S. Marcos. 1999. First-principles ion-water interaction potentials for highly charged monatomic cations. Computer simulations of Al3+, Mg2+, and Be2+ in water. J. Am. Chem. Soc. 121:3175–3184. [Google Scholar]
- 57.Markham, G. D., J. P. Glusker, and C. W. Bock. 2002. The arrangement of first- and second-sphere water molecules in divalent magnesium complexes: Results from molecular orbital and density functional theory and from structural crystallography. J. Phys. Chem. B. 106:5118–5134. [Google Scholar]
- 58.Misra, V. K., and D. E. Draper. 1998. On the role of magnesium ions in RNA stability. Biopolymers. 48:113–135. [DOI] [PubMed] [Google Scholar]
- 59.Misra, V. K., and D. E. Draper. 2002. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 317:507–521. [DOI] [PubMed] [Google Scholar]
- 60.Burgess, J. M. 1988. Ions in Solution. Ellis Horwood, Chichester, UK.
- 61.Auffinger, P., L. Bielecki, and E. Westhof. 2003. The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations. Chem. Biol. 10:551–561. [DOI] [PubMed] [Google Scholar]
- 62.Tanner, N. K., S. Schaff, G. Thill, E. Petit-Koskas, A. M. Crain-Denoyelle, and E. Westhof. 1994. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr. Biol. 4:488–498. [DOI] [PubMed] [Google Scholar]
- 63.Leontis, N. B., J. Stombaugh, and E. Westhof. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 30:3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rueda, M., E. Cubero, C. A. Laughton, and M. Orozco. 2004. Exploring the counterion atmosphere around DNA: What can be learned from molecular dynamics simulations? Biophys. J. 87:800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matysiak, M., J. Wrzesinski, and J. Ciesiolka. 1999. Sequential folding of the genomic ribozyme of the hepatitis delta virus: structural analysis of RNA transcription intermediates. J. Mol. Biol. 291:283–294. [DOI] [PubMed] [Google Scholar]
- 66.Lafontaine, D. A., S. Ananvoranich, and J. P. Perreault. 1999. Presence of a coordinated metal ion in a trans-acting antigenomic delta ribozyme. Nucleic Acids Res. 27:3236–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeong, S., J. Sefcikova, R. A. Tinsley, D. Rueda, and N. G. Walter. 2003. Trans-acting hepatitis delta virus ribozyme: catalytic core and global structure are dependent on the 5′ substrate sequence. Biochemistry. 42:7727–7740. [DOI] [PubMed] [Google Scholar]
- 68.Harris, D. A., R. A. Tinsley, and N. G. Walter. 2004. Terbium-mediated footprinting probes a catalytic conformational switch in the antigenomic hepatitis delta virus ribozyme. J. Mol. Biol. 341:389–403. [DOI] [PubMed] [Google Scholar]
- 69.Pallan, P. S., W. S. Marshall, J. Harp, F. C. Jewett, Z. Wawrzak, B. A. Brown, A. Rich, and M. Egli. 2005. Crystal structure of a luteoviral RNA pseudoknot and model for a minimal ribosomal frameshifting motif. Biochemistry. 44:11315–11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yaremchuk, A., M. Tukalo, M. Grotli, and S. Cusack. 2001. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: Comparison with histidyl-tRNA synthetase. J. Mol. Biol. 309:989–1002. [DOI] [PubMed] [Google Scholar]
- 71.Nissen, P., J. A. Ippolito, N. Ban, P. B. Moore, and T. A. Steitz. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. USA. 98:4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sponer, J., H. A. Gabb, J. Leszczynski, and P. Hobza. 1997. Base-base and deoxyribose-base stacking interactions in B-DNA and Z-DNA: A quantum-chemical study. Biophys. J. 73:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barone, F., F. Lankas, N. Spackova, J. Sponer, P. Karran, M. Bignami, and F. Mazzei. 2005. Structural and dynamic effects of single 7-hydro-8-oxoguanine bases located in a frameshift target DNA sequence. Biophys. Chem. 118:31–41. [DOI] [PubMed] [Google Scholar]
- 74.Beveridge, D. L., G. Barreiro, K. S. Byun, D. A. Case, T. E. Cheatham, S. B. Dixit, E. Giudice, F. Lankas, R. Lavery, and J. H. Maddocks. 2004. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(C(p)G) steps. Biophys. J. 87:3799–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fadrna, E., N. Spackova, R. Stefl, J. Koca, T. E. Cheatham, and J. Sponer. 2004. Molecular dynamics simulations of guanine quadruplex loops: Advances and force field limitations. Biophys. J. 87:227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spackova, N., and J. Sponer. 2006. Molecular dynamics simulations of sarcin-ricin rRNA motif. Nucleic Acids Res. 34:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jurecka, P., J. Sponer, and P. Hobza. 2004. Potential energy surface of the cytosine dimer: MP2 complete basis set limit interaction energies, CCSD(T) correction term, and comparison with the AMBER force field. J. Phys. Chem. B. 108:5466–5471. [Google Scholar]
- 78.Sponer, J., P. Jurecka, and P. Hobza. 2004. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 126:10142–10151. [DOI] [PubMed] [Google Scholar]
- 79.Sponer, J. E., N. Spackova, J. Leszczynski, and J. Sponer. 2005. Principles of RNA base pairing: Structures and energies of the trans Watson-Crick/sugar edge base pairs. J. Phys. Chem. B. 109:11399–11410. [DOI] [PubMed] [Google Scholar]