Abstract
The structures of two mutants of the site-specific recombinase, γδ resolvase, that form activated tetramers have been determined. One, at 3.5-Å resolution, forms a synaptic intermediate of resolvase that is covalently linked to two cleaved DNAs, whereas the other is of an unliganded structure determined at 2.1-Å resolution. Comparisons of the four known tetrameric resolvase structures show that the subunits interact through the formation of a common core of four helices. The N-terminal halves of these helices superimpose well on each other, whereas the orientations of their C termini are more variable. The catalytic domains of resolvase in the unliganded structure are arranged asymmetrically, demonstrating that their positions can move substantially while preserving the four-helix core that forms the tetramer. These results suggest that the precleavage synaptic tetramer of γδ resolvase, whose structure is not known, may be formed by a similar four-helix core, but differ in the relative orientations of its catalytic and DNA-binding domains.
Keywords: site-specific recombination, serine recombinase, hyperactive mutant, cleaved complex, crystallography
Site-specific recombinases can be divided into two families that achieve strand exchange by fundamentally different mechanisms (1). The tyrosine recombinases, of which λ integrase is a prototypical member, catalyze the formation of Holliday junction intermediates (2). A series of structures of Cre bound to loxP show that recombination in this family requires only subtle movements of nucleic acid and protein and can occur in the context of a largely fixed protein scaffold (3, 4). In the members of the serine recombinase family, catalytic serines attack the phosphodiester backbone to generate covalently linked intermediates that contain double-strand breaks (5–8). The structure of such an intermediate (9), obtained by using a hyperactivated mutant of γδ resolvase, differs dramatically from that of an unactivated complex (10) and establishes that relatively large protein movements and interface rearrangements are required for recombination by the serine recombinase family. Here we explore which of these structural rearrangements can exist before the chemical step in the recombination pathway and which are specifically induced upon the formation of a covalent bond between protein and nucleic acid.
Wild-type γδ resolvase is a remarkably specific enzyme. Recombination by resolvase requires two 114-bp-long res sequences that are oriented to form direct repeats in negatively supercoiled DNA. These res sequences contain three sites, each of which binds a dimer of γδ resolvase. Cleavage takes place only at site I and requires activating signals from resolvase dimers bound to sites II and III (5). These activating signals are mediated by an interface that contains residues Arg-2 and Glu-56 (5, 11–13).
The numerous known crystal structures of wild-type γδ resolvase (10, 12, 14, 15), show that it is composed of three structural elements: an N-terminal catalytic domain (residues 1–101), a long α helix (the “E” helix, residues 102–137) and a C-terminal DNA binding domain (residues 138–183). The wild-type resolvase in all of these structures is dimeric; some of the contacts observed within the crystals correspond to dimer–dimer interactions that are presumed to occur in the context of a full synaptic complex containing res sequences in supercoiled DNA. Consistent with a lack of resolvase activity on DNA containing only site I, the catalytic Ser-10 residues in the structure of wild-type resolvase bound to a site I analog are >14 and 20 Å distant from the scissile phosphates (10).
Mutations that increase the activity and reduce the topological specificity of γδ and Tn3 resolvase have been isolated in a number of studies. For example, the mutation E124Q removes the requirement for supercoiled substrate (5, 16), whereas the triple-mutant G101S/E102Y/M103I recombines short linear substrates containing only site I (5, 17, 18). The additional mutations R2A and E56K increase the activity of the triple mutant, underscoring its independence from the effects of sites II and III. A chimera, in which residues 96–105 of resolvase were replaced by the homologous sequence from Hin, contains an alternate set of activating mutations and also can cleave linear DNA substrates containing just site I (19). Activating mutations that occur at positions corresponding to residues in the E helix of resolvase also have been found in the invertases Gin, Hin, and Cin (20–22). Taken together, these studies suggest that in the resolvase/invertase family, the activating conformational changes usually induced by signals from accessory sites also can be achieved through selected mutations in the E helix or in the loop linking it with the catalytic domain.
Using resolvase containing the activating mutations R2A/E56K/G101S/E102Y/M103I/E124Q, the structure of an intermediate along the recombination pathway in which a synaptic tetramer of resolvase was linked covalently through Ser-10 to the scissile phosphate of a site I analog recently was determined (9). Superimposing the resolvase subunits from this cleaved complex on those from dimer structures showed that activation involves a hinge motion in the loop connecting the catalytic domain and E helix, resulting in the subunit interfaces within the tetramer being very different from those seen in dimer structures. Thus, in the dimer structures, the E helix of a monomer docks into a groove formed by the catalytic domain and E helix of another monomer, whereas in the context of a tetramer, E helices form antiparallel pairs, and the groove of the dimer is replaced by a flat interface between subunits.
The presence of this flat, hydrophobic interface in the tetramer implies that strand exchange may be accomplished by subunit rotation. Recombination by γδ resolvase proceeds with a topological change equivalent to a 180° rotation during the exchange of half-sites at site I (23, 24). The previous structure of a dimer complexed with site I contained a grooved interface that made it difficult to understand how two such dimers could associate and achieve such a rotation. In contrast, the flat interface of the tetramer structure is compatible with the rotation of resolvase subunits and their accompanying half sites. Extensive hydrophobic interactions across this interface could prevent dissociation during the course of rotation (9).
Although the structure of the cleaved complex suggests a mechanism for strand exchange, a number of questions remain unanswered. For example, in this cleaved complex, the free 3′ OH moieties are 14–16 Å from the scissile phosphates to which they were linked previously. A substrate complex containing intact DNA and a tetramer of resolvase with Ser-10 residues poised for attack therefore must differ from this cleaved complex, although it is unclear what the precise differences are. Nor is it apparent, from the cleaved complex, how interactions with resolvase bound at accessory sites II and III trigger conformational changes in resolvase bound at site I. To explore the conformational plasticity of a synaptic tetramer, we determined the structure of a synaptic complex containing a cleaved intermediate with a distinct set of activating mutations and that of a tetramer in the absence of DNA substrate.
Results
Structure of a Resolvase:Hin Chimera.
To study the interactions that are mediated by the loop preceding the E helix of Tn3 resolvase, a very close homologue of γδ resolvase, Wenwieser et al. (19) replaced residues 96–105 of Tn3 and γδ resolvase with the corresponding residues of Hin. Surprisingly, the hybrid proteins were active on site I DNA substrates lacking sites II and III. The hyperactive properties of the γδ resolvase chimera made it an attractive target for the purpose of structural studies (19). The chimeric γδ resolvase protein containing the Hin sequence of residues 96–105 and the additional mutations R2A, E56K, and E124Q crystallized in the presence of a site I analog.
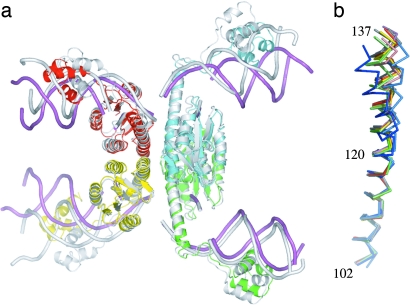
This chimeric protein complexed with site I DNA crystallized in spacegroup P21212 and yielded data to 3.5-Å resolution (Table 1). Its structure was determined by molecular replacement by using the coordinates of Protein Data Bank (PDB) ID code 1ZR4 (9) and contained a dimer in each asymmetric unit, because the resolvase tetramer lies on a crystallographic twofold axis (Fig. 1a). The structure has been refined to an Rcryst of 28.2% and Rfree of 32.3% (Table 1).
Table 1.
Crystallographic statistics
| Resolvase:Hin chimera | Activated apo resolvase | |
|---|---|---|
| Spacegroup | P21212 | P212121 |
| Resolution, Å | 50.0–3.5 | 50–2.1 |
| Rmerge,* % | 9.1 | 8.9 |
| I/σ† | 12.9 (1.2) | 15.7 (1.9) |
| Completeness, % | 96.5 (84.3) | 99.8 (100) |
| Unique reflections | 16,189 | 59,517 |
| Redundancy | 3.5 | 3.8 |
| Copies in AU | 2 | 4 |
| rms deviation bond length, Å | 0.008 | 0.007 |
| rms deviation bond angle, ° | 1.2 | 1.0 |
| Rcryst,‡ % | 28.2 (20–3.5 Å) | 21.9 |
| Rfree,‡ % | 32.3 (20–3.5 Å) | 25.5 |
| PDB ID | 2GM4 | 2GM5 |
*Rmerge is Σj|Ij−〈I〉|, where Ij is the intensity of an individual reflection and 〈I〉 is the mean intensity for multiply recorded reflections.
†I/σ is the mean of intensity divided by the SD. The values in brackets are for the high resolution shell (3.63–3.5 Å and 2.18–2.1 Å, respectively).
‡Rcryst is Σ‖Fo|−|Fc‖/Σ|Fo|, where Fo is an observed amplitude and Fc is a calculated amplitude; Rfree is the same statistic calculated over a subset of the data that have not been used for refinement.
Fig. 1.
Structural consequences of variable conformation within E helices. (a) The structure of the resolvase:Hin chimera (in color) superimposed by using Cα atoms 2–120 in all four chains on a tetramer with a different set of activating mutations (shown in white; PDB ID code 1ZR4). Differences between the structures are apparent at the C termini of the E helices and lead to different orientations for the DNA and DNA-binding domains. (b) Individual E helices (residues 102–137) superimposed by using their N-terminal Cα atoms (102–120) from dimeric resolvase (blue and cyan; PDB ID code 1GDT), the resolvase:Hin chimera (green and lime), and other tetrameric cleaved intermediate structures (yellow, red, magenta, and salmon, PDB ID code 1ZR4; gray and black, PDB ID code 1ZR2). The location of the Cα atom of residue 137 differs by up to 6 Å when only the tetrameric resolvases are considered and by 8 Å when the dimeric structure of wild-type resolvase bound to a site analog (PDB ID code 1GDT) is included as well. Figures in this paper were generated by using pymol (www.pymol.org).
The chimeric protein superimposes well on the previously determined activated resolvase structures and thus represents a similar reaction intermediate. As in these earlier structures, resolvase has cut the DNA to form double strand breaks and a covalent link between Ser-10 and the scissile phosphate. The rms deviation between Cα atoms 2–120 (excluding the loop residues 40–43) in the 1ZR4 tetramer (9) superimposed on the corresponding atoms in the chimera is 1.3 Å. Separate superpositions of Cα atoms 138–183 of the DNA binding domains yield rms deviation values between 0.3 and 0.8 Å. The differences between these tetramers are thus largely due to variations in residues at the C terminus of the E helix and can be seen by superimposing the E helices from all DNA bound resolvase structures (Fig. 1b).
The six mutated residues between 96 and 105 of the chimeric protein (G96S/S98D/D100S/G101S/E102A/K105R) must account, in large measure, for its increased activity. One way in which they could do so is by stabilizing the observed tetrameric form of resolvase relative to a dimeric form. Because the residues at positions 98 and 100 appear to occupy similar environments in either the dimer or tetramer, the mutations S98D and D100S presumably do not contribute substantially to activation. Because residues at positions 96 and 105 make distinctly different contacts in the dimer and tetramer structures, the conservative mutation K105R may lead to stabilization of the tetramer by promoting interactions with D84, and the change G96S might enhance interactions with M103. However, there are no independent data with which to assess whether either of these mutations alone are activating.
Such data exist for residues at position 101 and 102. In Tn3 resolvase, a D102A mutant is known to have a mildly activated phenotype, and the mutation G101S increases the activity of a D102Y mutant (18). The mutation G101S may contribute to activation both by formation of hydrogen bonds and through restricting the phi-psi angles a residue in this position can access (9). Consistent with the observation that mutation of residue 102 to alanine is much less activating than mutation to tyrosine (18), the alanine does not form part of an oligomeric interface, whereas the aromatic moiety of the tyrosine interacts with the E helix of an adjacent monomer in 1ZR4.
Structure of a Truncated, Activated Resolvase Mutant.
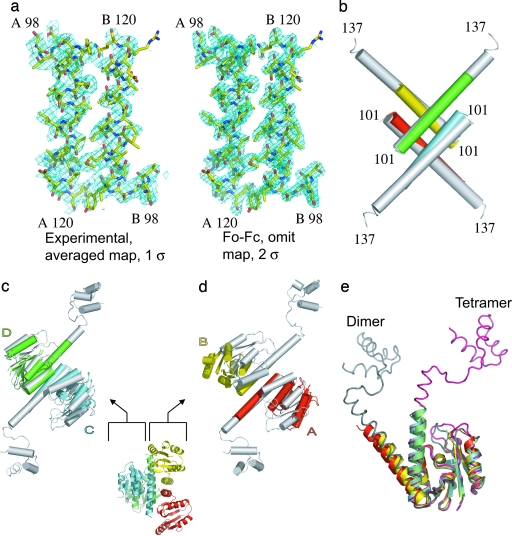
To ascertain what role, if any, is played by DNA in maintaining the conformation of the synaptic tetramer, we determined the structure of resolvase containing the mutations R2A/E56K/R68H/G101S/E102Y/M103I in the absence of the DNA-binding domain and DNA. Because the DNA-binding domain of unliganded resolvase is disordered, we cloned constructs containing only the catalytic domain and part of the E helix. A construct that contained resolvase residues 1–134 followed by a C-terminal hexa-histidine tag formed a tetramer of the expected molecular weight in solution and crystallized readily in an orthorhombic crystal form containing four monomers per asymmetric unit (Table 1). Initial attempts to solve this structure by using molecular replacement failed. However, data from a crystal containing selenomethionyl-labeled protein that diffracted to 2.1-Å resolution and multiwavelength anomalous dispersion phasing in conjunction with noncrystallographic averaging and solvent flattening produced interpretable maps (Fig. 2a). The electron density was generally of good quality but did not allow the placing of the C-terminal residues of the construct (presumably because of fraying of the ends of the E helices) or of residues in some mobile loops. With the addition of 147 waters, the structure was refined to an Rcryst of 21.9% (Rfree 25.5%).
Fig. 2.
The structure of the apo, activated, resolvase fragment. (a Left) The experimental electron density map averaged, solvent-flattened, and calculated at 3-Å resolution is shown. (a Right) A map made with residues 98–120 in chains A and B omitted by using the coefficients Fo − Fc and calculated to 2.1-Å resolution. Both maps cover the same two E helix segments. (b) The E helix cores of the apo (chain A, red; chain B, yellow; chain C, cyan; and chain D, lime) and cleaved complex structure (white; PDB ID code 1ZR4) superimpose well. The catalytic domains in chains C and D also superimpose well on the cleaved complex structure (c), whereas chains A and B do not because these domains have moved apart (d). (e) Superimposing the four monomers by using their catalytic domains shows that the hinge region between the catalytic domain and E helix can bend to generate a variety of orientations. Representative catalytic domains from a dimer structure (chain A from PDB ID code 1GDT is shown in gray) and from the cleaved intermediate also are shown (chain A from PDB ID code 1ZR4 is shown in pink).
Despite some striking similarities to the structures of activated resolvases linked to cleaved DNA, the apo structure is far more asymmetric. Four E helices, which pack with point group 222 symmetry, form the cores of both sets of structures and superimpose well on each other (Fig. 2b). The rms deviation between superposition of Cα atoms 104–120 of the four E helices in the fragment structure on the corresponding atoms of the cleaved complex structure 1ZR4 is 1.5 Å. Two of the catalytic domains in the fragment structure are oriented identically to and superimpose well on those of the resolvase from the cleaved complex structures; the rms deviation between them after superposition of all Cα atoms in chains C and D is 1.5 Å (Fig. 2c). As in the cleaved complex structures, this pair of monomers interact with each other both through an antiparallel association of E helices and through contacts at the ends of the D helices. In contrast, whereas monomers A and B maintain an antiparallel association of E helices, their catalytic domains have a different orientation relative to the E helices and, thus, do not contact each other because the axes of their D helices are separated by 25 Å (chains A and B; Fig. 2d). Indeed, despite the dramatic differences in quaternary structures of dimeric and tetrameric resolvase, the tertiary structures of chains A and B resemble those of dimeric resolvase more closely than those of cleaved intermediates (Fig. 2e).
The asymmetric arrangement of catalytic domains around the E helix core in the apo structure is achieved through two different conformations of the loop connecting the E helix to the catalytic domain. Separate superpositions of the four E helices (residues 104–120) and the four catalytic domains (residues 2–99) show that substantial differences in main-chain conformation occur only at residues 100 and 101 (Fig. 2e). Significantly, although the local environment around Ile-103 differs between the two pairs of monomers, Tyr-102 assumes a similar rotomer and packs against residues from an adjacent E helix in all four monomers. That the mutation E102Y appears to contribute more to hyperactivation than either G101S or M103I (18) may account for the apo fragment remaining tetrameric despite having two different loop and catalytic domain orientations.
DNA from the cleaved complex structure can be docked onto the structure of the fragment by using an alignment based on superposition of either the four E helices or individual catalytic domains. The former results in clashes between the catalytic domains in the fragment and the modeled DNA, whereas in the latter, the DNA half sites are related by implausibly acute angles. It therefore would seem that the precise arrangement of catalytic domains in the structure is unlikely to be observed in any synaptic species containing DNA. Rather, the two conformations observed in this fragment structure indicate that considerable variability in the orientation of the catalytic domains is possible within the confines of a tetrameric intermediate.
Discussion
A synaptic complex before cleavage of DNA must lie on the reaction path connecting two dimers to the cleaved intermediate tetramer. Although crystal forms containing intact DNA and activated resolvase mutants have been obtained, they diffract to low resolution, and a structure has not yet been determined. In the absence of such a structure, information on this intermediate is indirect and derives from two main sources: (i) disulfide cross-linking studies and (ii) extrapolations from existing structures. A simple question can be asked regarding the nature of this synaptic intermediate: Does it more closely resemble two associated, unactivated dimers or the cleaved complex tetramer? More specifically, are the E helices at its core arranged as in the tetramer?
Matching Structures, Engineered Disulfides, and Activity.
One approach to studying a complex machine is to restrict its motion and ask how its function is affected. By following this approach, disulfide bonds have been engineered in γδ resolvase, Tn3 resolvase, and Hin (refs. 9, 13, 19, 25, and 26; Fig. 3). For example, oxidation of either the M106C mutant of γδ resolvase or the D95C/A113C mutant of Tn3 resolvase results in disulfide bond formation that locks resolvase into conformations resembling its dimer form. Neither of these mutants, in their trans oxidized forms, supports double-strand cleavage or recombination activity (13, 19, 25). On the other hand, an intramolecular disulfide bond between residues 73 and 112 in Tn3 resolvase makes it hyperactive in DNA cleavage (19). These residues are in contact in the cleaved complex structures but not in the context of dimeric resolvase. Thus, these disulfide cross-linking studies appear to support the idea that the precleavage synaptic tetramer resembles the cleaved intermediate more than a pair of dimers.
Fig. 3.
Disulfide links can lock mutant resolvases into specific quaternary associations. Resolvase structures in ribbon form, with the E helix represented as a cylinder and DNA shown as a surface. The Cα positions of selected residues are shown as spheres. The DNA-bound dimer structure is taken from ref. 10. The resolvase mutant, M106C, can form disulfides readily in the context of a dimer but not when either cleaved-intermediate or activated apo tetramers are used as scaffolds. Conversely, of these three scaffolds, only the activated apo form appears appropriate for the formation of G96C-mediated disulfides. The cleaved complex scaffold appears compatible with the formation of four intramolecular T73C/S112C links, and the activated apo scaffold appears compatible with only two, consistent with the observation that the tertiary conformations of two of the monomers in the activated apo form resemble those of unactivated resolvase.
By introducing the additional change S94C to an activated Hin mutant, Johnson et al. (26) obtained cross-linking data that appear more difficult to reconcile with either the cleaved intermediate or dimer structures. The corresponding mutation in γδ resolvase, G96C, also readily forms disulfides in the presence of activating mutations, although the oxidation reaction does not proceed to completion (ref. 9; data not shown). These results are difficult to account for because the Cα atoms of residue 96 are separated by >20 Å in dimer structures and 18 Å in cleaved intermediate structures.
The cross-linking data therefore suggest the presence of additional conformations that differ from the known dimer and cleaved intermediate tetramer structures and are consistent with either one of the following possibilities. First, the disulfide-linked species might be an intermediate in the rotation of pairs of domains. To bring residue 96 into contact with itself across the flat interface would require a rotational motion of ≈70°. Second, the structure of the apo resolvase fragment suggests an alternative way in which the S94C cross-linking data can be explained. When this apo structure is superimposed on the cleaved complex structures as described above, the catalytic domains of monomers A and B in the two structures are oriented very differently. One consequence of these differences is that, in the apo structure, the Cα atoms of residue 96 in chains B and C are separated by <7 Å and those between chains A and D are within 9 Å of each other (Fig. 3). If this second explanation were true, then it would appear that at least some of the variation in the orientation of the catalytic domains with respect to the E helices in the apo structure also might occur in the context of DNA-bound complexes.
Structural Variation in the C-Terminal Halves of E Helices.
The orientation of the C-terminal halves of the E helices in the cleaved complex structures cannot be maintained in the context of a synaptic tetramer before cleavage. In these structures, the DNA half sites are separated and would need to move 15 Å toward each other before they could be covalently linked through the scissile phosphate. Because the E helix of resolvase from residues R119 onward binds to the minor groove of a DNA half site, movement of the cleaved ends of the DNA toward each other would require a concomitant movement of this half of the E helix.
Three lines of evidence suggest that the C-terminal halves of E helices are flexible. First, when the Cα atoms of residues 104–120 of the E helices taken from various cocrystal structures are superimposed, the C termini of these E helices are up to 8 Å apart (Fig. 1b). Second, disulfide cross-linked mutants γδ M106C or Tn3 D95C/A113C can function at sites II and III (12, 19). Although these disulfide links significantly constrain the orientation of the N-terminal halves of the E helices, the C-terminal halves nevertheless are able to interact with half sites that are separated by 10 bp in the case of site II and only 1 bp in site III. Third, in structures of resolvase not bound to DNA, the C termini of the E helices are disordered (12, 14, 15). One possible source of this disorder would be fraying of the C-terminal halves of these helices, which would be consistent with secondary structure predictions that indicate a sharp drop in helical propensity around residue 125 (27). The position of the N-terminal half of an E helix thus does not appear to lock its C-terminal half into a unique orientation or conformation.
Features of a Synaptic Tetramer Before DNA Cleavage.
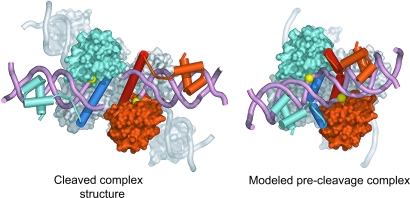
What structural features is a prechemistry synaptic complex likely to possess? The structures of resolvase in four crystal forms containing tetrameric complexes have been determined to date (1ZR2, 1ZR4, and the two presented here). All of them contain superimposable cores made of four E helices (residues 103 to ≈120). The results described here show that this core is formed in the presence of various activating mutations, in the presence of substantial movements of the catalytic domain, and even in the absence of DNA substrate. In the postcleavage complexes, the free 3′ OH moieties are ≈15 Å from the scissile phosphates, whereas in the precleavage synaptic complex, this distance must be reduced to that of a covalent bond. We therefore constructed a model to see whether, despite the fact that the DNA within it must be arranged in a substantially different way, the precleavage synaptic complex also could contain a similar E helical core.
In our modeling, we moved individual half sites of DNA, along with their associated DNA-binding domain, as rigid bodies relative to the catalytic core and N-terminal halves (residues 103–120) of the E helices (Fig. 4). The C-terminal halves of the E helices were treated as separate rigid bodies in this modeling exercise (residues 121–137), and their positions adjusted to maintain the connections between the core regions and the DNA-binding domains. Moving the N- and C-terminal halves of the E helices separately appeared reasonable because structural comparisons suggest that the positions and conformations of the C-terminal halves of E helices can vary substantially. To ensure the formation of covalently intact DNA, we also manually adjusted the positions of the central four base pairs of each site. Rearrangements of the catalytic domains relative to the E helix core of a tetramer are plausible and, in fact, are present in our truncated activated mutant. Nevertheless, to reduce the number of parameters in our models, we did not alter the positions of the catalytic domains. The positions of the catalytic domains thus match those observed in activated tetramers containing DNA and chains C and D in the apo-fragment structure. Although it is not yet clear whether cleavage of both DNA strands in site I is sequential or simultaneous, we have retained the pseudo 222 symmetry of the cleaved complex structures in the model and, consequently, all of the catalytic serines in the model remain within van der Waal’s contact of the scissile phosphates.
Fig. 4.
Comparison of the structure of the resolvase:Hin chimera cleaved complex (Left) with a model of the synaptic complex before DNA cleavage (Right). Two of the monomers in the tetramer, colored white, are shown in a surface representation. The other two monomers are colored light red and cyan, with their respective E helices shown in darker shades. The catalytic domains of these monomers also are shown as surfaces, whereas the E helices and DNA-binding domains are represented as cartoon traces. The positions of the scissile phosphates are indicated with yellow spheres. The modeled precleavage complex was made by introducing sharp kinks in the E helices and moving the DNA and DNA-binding domains of the cleaved complex closer together.
The model of a precleavage synaptic intermediate obtained in this manner contains two features of interest (Fig. 4). First, because the DNA from a left and right half site are joined in the model, it is substantially more compact than the cleaved complex. Although the DNA from a left and right half site in the cleaved complex lie largely within a single plane, the DNA in the model has moved substantially out of this plane, in a fashion similar to that seen in structures containing dimers of wild-type resolvase bound to DNA. This movement appears necessary for the close approach of a free 3′ hydroxyl end to a scissile phosphate. Second, the model suggests that the C-terminal half of an E helix may interact with the catalytic domain of an adjacent monomer, indicating the possibility of a novel interface in the precleavage synaptic tetramer. The modeling thus suggests that a synaptic tetramer before DNA cleavage may contain a core of E helices similar to those seen in cleaved complex structures but could differ from them both in terms of the orientation of the C-terminal halves of the E helices and also in the positions of the catalytic domains relative to the E helices.
Methods
The structure of the resolvase:Hin chimera cleaved DNA complex was determined by using molecular replacement software in the cns suite of programs with a tetrameric catalytic domain as a search model (28). Cycles of manual building by using the program o (29) and refinement with refmac (30) resulted in a model with a final Rcryst of 28.2% (Rfree of 32.3%). The model contains all protein residues except the N-terminal methionine of chain B, residues 40–43 in both protein chains, and all of the DNA except a terminal base pair in chains J and K.
The structure of the apo activated fragment of resolvase was determined by using multiwavelength anomalous dispersion (MAD) phasing. Selenomethionine positions were found by using the program suites solve and cns (28, 31). After MAD phasing with solve, electron density was averaged and solvent flattened with resolve (32) to yield a clearly interpretable map (Table 1 and Fig. 2a). Cycles of building in o and refinement with cns and refmac resulted in a structure with an Rcryst of 21.9% and an Rfree of 25.5% (28–30). The final model contains residues 2–38 and 44–128 in chain A; 2–11, 15–38, and 44–123 in chain B; 2–9, 15–39, and 43–125 in chain C; 2–9, 13–37, and 44–127 in chain D; and 147 water molecules.
Protein purification, crystallization, and stabilization methods are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank the staff at National Synchrotron Light Source beamlines X12B, X12C, and X25; the staff at Advanced Photon Source beamlines 19-ID and 8-BM; the Steitz laboratory for help with data collection and useful discussions; and the staff of the Center for Structural Biology core facility at Yale University. This work was funded by National Institutes of Health Grants GM 57510 (to T.A.S.) and GM 28470 (to N.D.F.G.).
Abbreviation
- PDB
Protein Data Bank.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2GM4 and 2GM5).
References
- 1.Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Mobile DNA II. Washington, DC: Am. Soc. Microbiol.; 2002. [Google Scholar]
- 2.Azaro A. M., Landy A. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 118–148. [Google Scholar]
- 3.Guo F., Gopaul D. N., Van Duyne G. D. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 4.Van Duyne G. D. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 93–117. [Google Scholar]
- 5.Grindley N. D. F. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 272–302. [Google Scholar]
- 6.Johnson R. C. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 230–271. [Google Scholar]
- 7.Reed R. R., Grindley N. D. F. Cell. 1981;25:721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- 8.Hatfull G. F., Grindley N. D. F. Proc. Natl. Acad. Sci. USA. 1986;83:5429–5433. doi: 10.1073/pnas.83.15.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W., Kamtekar S., Xiong Y., Sarkis G. J., Grindley N. D. F., Steitz T. A. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 10.Yang W., Steitz T. A. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 11.Hughes R. E., Hatfull G. F., Rice P. A., Steitz T. A. Cell. 1990;63:1331–1338. doi: 10.1016/0092-8674(90)90428-h. [DOI] [PubMed] [Google Scholar]
- 12.Rice P. A., Steitz T. A. EMBO J. 1994;13:1514–1524. doi: 10.2210/pdb1gdr/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murley L. L., Grindley N. D. F. Cell. 1998;95:553–562. doi: 10.1016/s0092-8674(00)81622-0. [DOI] [PubMed] [Google Scholar]
- 14.Sanderson M. R., Freemont P. S., Rice P. A., Goldman A., Hatfull G. F., Grindley N. D. F., Steitz T. A. Cell. 1990;63:1323–1329. doi: 10.1016/0092-8674(90)90427-g. [DOI] [PubMed] [Google Scholar]
- 15.Rice P. A., Steitz T. A. Structure (London) 1994;2:371–384. doi: 10.1016/s0969-2126(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 16.Arnold P. H., Blake D. G., Grindley N. D. F., Boocock M. R., Stark W. M. EMBO J. 1999;18:1407–1414. doi: 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkis G. J., Murley L. L., Leschziner, Boocock M. R., Stark W. M., Grindley N. D. F. Mol. Cell. 2001;8:623–631. doi: 10.1016/s1097-2765(01)00334-3. [DOI] [PubMed] [Google Scholar]
- 18.Burke M. E., Arnold P. H., He J., Wenwieser S. V. C. T., Rowland S. J., Boocock M. R., Stark W. M. Mol. Microb. 2004;51:937–948. doi: 10.1046/j.1365-2958.2003.03831.x. [DOI] [PubMed] [Google Scholar]
- 19.Wenwieser S. V. C. T. Ph.D. thesis. Glasgow, U.K.: University of Glasgow; 2001. [Google Scholar]
- 20.Klippel A., Cloppenborg K., Kahmann R. EMBO J. 1988;7:3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haykinson M. J., Johnson L. M., Soong J., Johnson R. C. Curr. Biol. 1996;6:163–177. doi: 10.1016/s0960-9822(02)00449-9. [DOI] [PubMed] [Google Scholar]
- 22.Haffter P., Bickle T. A. EMBO J. 1988;7:3991–3996. doi: 10.1002/j.1460-2075.1988.tb03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cozarrelli N. R., Krasnow M. A., Gerrard S. P., White J. H. Cold Spring Harbor Symp. Quant. Biol. 1984;49:383–400. doi: 10.1101/sqb.1984.049.01.045. [DOI] [PubMed] [Google Scholar]
- 24.Stark W. M., Sherratt D. J., Boocock M. R. Cell. 1989;58:779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- 25.Hughes R. E., Rice P. A., Steitz T. A., Grindley N. D. F. EMBO J. 1993;12:1447–1458. doi: 10.1002/j.1460-2075.1993.tb05788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhar G., Sanders E. R., Johnson R. C. Cell. 2004;119:33–45. doi: 10.1016/j.cell.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Leschziner A. E., Grindley N. D. F. Mol. Cell. 2003;12:775–781. doi: 10.1016/s1097-2765(03)00351-4. [DOI] [PubMed] [Google Scholar]
- 28.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger T. C. Acta Crystallogr. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.