Abstract
The major surface proteins of the parasitic protozoon Leishmania mexicana are anchored to the plasma membrane by glycosylphosphatidylinositol (GPI) anchors. We have cloned the L. mexicana GPI8 gene that encodes the catalytic component of the GPI:protein transamidase complex that adds GPI anchors to nascent cell surface proteins in the endoplasmic reticulum. Mutants lacking GPI8 (ΔGPI8) do not express detectable levels of GPI-anchored proteins and accumulate two putative protein–anchor precursors. However, the synthesis and cellular levels of other non–protein-linked GPIs, including lipophosphoglycan and a major class of free GPIs, are not affected in the ΔGPI8 mutant. Significantly, the ΔGPI8 mutant displays normal growth in liquid culture, is capable of differentiating into replicating amastigotes within macrophages in vitro, and is infective to mice. These data suggest that GPI-anchored surface proteins are not essential to L. mexicana for its entry into and survival within mammalian host cells in vitro or in vivo and provide further support for the notion that free GPIs are essential for parasite growth.
INTRODUCTION
Leishmania are protozoan parasites that cause a spectrum of diseases in humans. These parasites alternate between a flagellated promastigote stage that proliferates within the midgut of the sandfly vector and a nonmotile amastigote stage that invades mammalian macrophages, where they occupy the phagolysosome compartment. The cell surface of the promastigote stage is coated by a protective glycocalyx that comprises a number of glycosylphosphatidylinositol (GPI)-anchored glycoproteins, a complex GPI-anchored lipophosphoglycan (LPG), and a family of free GPIs (termed glycoinositolphospholipids [GIPLs]) (McConville and Ferguson, 1993; Beverley and Turco, 1998; Figure 1). Components in this glycoclayx are thought to be essential for parasite survival and infectivity in the diverse host (McConville and Ferguson, 1993; Beverley and Turco, 1998). The GPI-anchored glycoproteins include an abundant metalloproteinase, termed gp63 or leishmanolysin, and the promastigote surface antigen (PSA2)/gp46 family of glycoproteins (Medina-Acosta et al., 1989; Murray et al., 1989; Frommel et al., 1990; Lohman et al., 1990). Gp63 has been shown to be proteolytically active against a wide variety of different peptide substrates and has been reported to act as a ligand for macrophage receptors, either directly or after opsonization with complement, to protect the parasites from complement-mediated lysis and also to contribute to the pathology of lesion development (see Alexander and Russell, 1992; Joshi et al., 1998). However, the fact that these proteins are encoded by multicopy polymorphic genes (Button et al., 1989; Lohman et al., 1990; Symons et al., 1994) has hindered elucidation of their function by genetic analysis (Joshi et al., 1998). Moreover, surface expression of gp63 and PSA2 is dramatically down-regulated in the amastigote stage of some Leishmania species and variable within particular parasite populations of others (Bahr et al., 1993; Handman et al., 1995) such that the precise function of these parasite proteins in the mammalian host remains unclear.
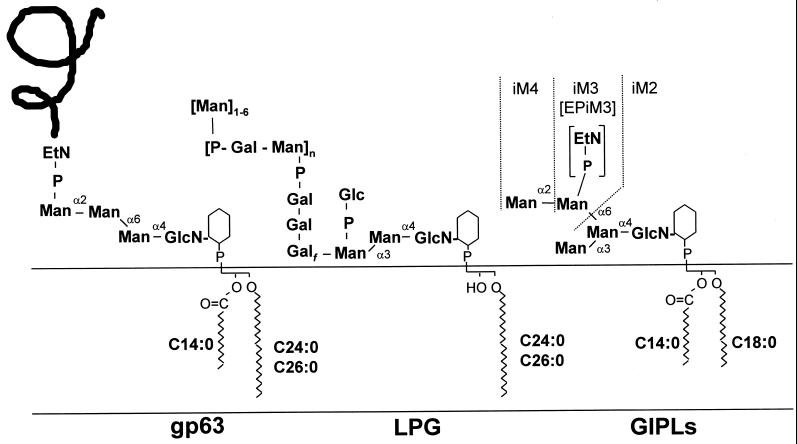
Figure 1.
Structures of the three distinct classes of GPIs synthesized by L. mexicana promastigotes. These glycolipids act as membrane anchors for the major surface glycoprotein (gp63) and an abundant surface lipophosphoglycan (LPG) as well as being the major class of free glycolipids (GIPLs) (McConville et al., 1993; Ralton and McConville, 1998). The nomenclature used for different GPI structures is as follows: M2–4 denotes GPIs with the structures Man2–4GlcN-PI; EP denotes the presence of an EtN-P cap or side chain; and the prefix i denotes the presence of an α1–3-linked mannose in the core structure.
Proteins that are destined to receive a GPI anchor are synthesized as precursor proteins with an N-terminal signal sequence that targets the protein to the endoplasmic reticulum (ER) and a C-terminal GPI attachment signal. The latter consists of three main domains: a C-terminal hydrophobic portion, a short hydrophilic spacer, and an attachment site (ω) (Caras et al., 1989; Udenfriend and Kodukula, 1995). This signal is recognized by the GPI:protein transamidase complex, which proteolytically cleaves the precursor protein at the ω amino acid and links the newly generated C terminus to the terminal ethanolamine phosphate (EtN-P) of a preassembled GPI anchor precursor having the structure EtN-P-Man3GlcN-phosphatidylinositol (PI) (Udenfriend and Kodukula, 1995). The transamidase is thought to form an intermediate with the precursor protein, resulting in an activated carbonyl group at the ω site (Maxwell et al., 1995b; Sharma et al., 1999). The nucleophilic ethanolamine component of the GPI anchor reacts with the active carbonyl, and the resultant GPI-anchored protein is then trafficked to the cell surface. This proposed reaction mechanism is supported by the finding that nucleophiles such as hydrazine can replace the GPI anchor in a cell-free assay system for GPI anchoring (Maxwell et al., 1995b; Sharma et al., 1999) and that the process is ATP and GTP independent (Mayor et al., 1991). Interestingly, the GPI signal sequence differs between species, and that of Trypanosoma brucei variable surface glycoprotein functioned poorly when expressed in mammalian cells (Moran and Caras, 1994). Mutagenesis studies have suggested that the sizes of the peptide binding pocket in the trypanosome and mammalian GPI:protein transamidases differ (Moran and Caras, 1994). There are hopes that these differences may be exploited in the design of new antiparasite agents.
The GPI:protein transamidase complex contains at least two protein components, Gaa1p and Gpi8p, which have recently been identified by analysis of several temperature-sensitive yeast mutants defective in the GPI-anchoring of proteins. Gaa1p is a lumenally oriented ER glycoprotein with several transmembrane domains (Hamburger et al., 1995), whereas Gpi8p is an ER–membrane glycoprotein with a large lumenal domain, a single C-terminal transmembrane domain, and a short cytoplasmic tail (Benghezal et al., 1996). Yeast mutants defective in the functioning of these individual proteins accumulate preassembled GPI anchors and have no GPI-anchored proteins. Homologues of both of these proteins have now been identified in humans (Yu et al., 1997; Hiroi et al., 1998). It is believed that Gpi8p is the catalytic subunit (Yu et al., 1997). This protein shares significant homology with a family of previously characterized cysteine proteinases, the asparaginyl endopeptidases known as legumains (Benghezal et al., 1996), which were originally identified in the seeds of leguminous plants (Ishii, 1994). Together these proteins have been categorized as family C13 of cysteine peptidases (Riezman and Conzelmann, 1998).
We report here the cloning of GPI8 from the parasitic protozoon Leishmania mexicana and the production of GPI8 null mutants by targeted gene replacement. Despite the finding that GPI-anchored proteins are no longer detectable on the cell surface of the null mutants, these parasites show normal growth in in vitro culture, are able to differentiate into amastigotes, and survive within macrophages in vitro and can establish an infection in mice. These results suggest that GPI-anchored proteins are not essential for the parasite's infectivity and survival in a mammalian host. In addition, we show that some of the protein anchor precursors in Leishmania contain an acyl modification on the inositol head group. These data suggest that the leishmanial pathway of protein anchor biosynthesis is more similar to that of the intensively studied African trypanosomes than was previously thought and provide another example of how different classes of GPI intermediates are differentially processed in these parasites.
MATERIALS AND METHODS
Growth of Parasites
Wild-type L. mexicana promastigotes (MNYC/BZ/62/M379) were maintained at 25°C in either modified Eagle's medium (HOMEM) or RPMI-1640 supplemented with 2 mM l-glutamine (Life Technologies, Gaithersburg, MD) containing 10% (vol/vol) heat-inactivated fetal calf serum. The required antibiotics were added as follows: ClonSat (Hans Knoll Institute, Jena, Germany) at 25 μg/ml, puromycin (Calbiochem, La Jolla, CA) at 10 μg/ml, and neomycin (G418, Geneticin; Life Technologies) at 25 μg/ml. Parasites were grown as amastigotes axenically in vitro using the method described previously (Bates et al., 1992).
Cloning of GPI8
Degenerate oligonucleotide primers (IUB group codes) were designed against the following three peptide sequences based on those of yeast, human, and Caenorhabditis elegans GPI8 proteins: 1) TNNWAVLV (OL76), 5′-ACS AAY AAY TGG GCN GTB CTY GT-3′; 2) RFWFNYRH (OL77), 5′-CGB TTY TGG TTY AAY TAY CGB CA-3′; and 3) (I/F)YMTGHGG (OL79), 5′-CCV CCR TGV CCS GTV AKR TAR CT-3′ (antisense).
Nested PCR was carried out with Taq Polymerase (Applied Biosystems, Foster City, CA) in a 20-μl reaction volume. In the first round, primers OL76 and OL79 were used with 100 ng of procyclic promastigote, metacyclic promastigote, and amastigote L. mexicana first-strand cDNA and genomic DNA using the following conditions: 1 cycle of 94°C for 4 min, 25 cycles of 94°C for 1 min, 42°C for 1 min and 72°C for 1 min, and 1 cycle of 72°C for 4 min. Two microliters of first-round PCR were then reamplified with primers OL77 and OL79 using the same conditions as for the first round. Reactions were electrophoresed on a 1% agarose gel, and the 362-bp PCR products were gel purified (Qiaquick gel extraction kit; Qiagen, Hilden, Germany) and cloned into pTAG vector (R & D Systems, Minneapolis, MN) for sequencing.
The full-length GPI8 was isolated by screening an L. mexicana λ DashII genomic library (Mottram et al., 1997) using the PCR product, labeled with α-32P, as a probe. Two λ clones were isolated, and three subclones containing the GPI8 gene were isolated and cloned into pBluescript (Stratagene, La Jolla, CA); a 2.1-kb SalI fragment (pGL185), a 2.4-kb XhoI fragment (pGL186), and a 4.5-kb XbaI fragment (pGL187). Portions of these subclones were sequenced to provide double-stranded sequence covering the GPI8 gene and ∼1 kb of 5′ and 3′ flanks. The nucleotide sequence of L. mexicana GPI8 is available in the European Molecular Biology Laboratory database under accession number AJ242865. Multiple sequence files produced using the program Pileup were used with the Internet analysis tool Boxshade version 3.21 (http://www.isrec.isb-sib.ch/software/BOX_form.html) to highlight amino acid identities and similarities.
Disruption of GPI8
Sequential rounds of transfection with SAT and PAC resistance knockout constructs were used to disrupt the GPI8 gene. These constructs were designed to replace precisely the GPI8 with each antibiotic resistance gene so that the endogenous processing signals would allow expression of the resistance genes. A 1.2-kb DNA fragment containing the 5′ flank of GPI8 was amplified from plasmid pGL186 with primers OL83 and OL84 and cloned into pCR script. A 900-bp DNA fragment containing the 3′ flank of GPI8 was amplified from plasmid pGL185 with primers OL85 and OL86 and cloned into pCR script. OL83 and OL84 were engineered with HindIII and SpeI sites, respectively, whereas OL85 and OL86 were engineered with BamHI and BglII sites, respectively. Primer sequences were as follows: OL83, 5′-GCCAAGCTTTGCTCAGATGACCGAGCCGGCGC-3′; OL84, 5′-GACTAGTAAACAGCCGGA-ACTGCACTAGCT-3′; OL85, 5′-CGCGGATCCCGTGTGGCATC-TACCTCCCTGCG-3′; and OL86, 5′GAAGATCTTTGCTCGTGATACGACGGCGTGG-3′.
The BamHI–BglII fragment containing the GPI8 5′ flank was cloned into BamHI–BglII-digested pGL207 and pGL51 (plasmids that contain the PAC and SAT genes, respectively (Brooks and J.C. Mottram, unpublished data). Subsequently the GPI8 3′ flank was used to replace the HindIII–SpeI fragment to give pGL236 (PAC) and pGL237 (SAT). The integration cassette was released by digestion with HindIII and BglII. The complete open reading frame (ORF) of GPI8 together with 1.1 kb of 5′ and 1.0 kb of 3′ flanking regions, respectively, is found in the λ clone on a 3.1-kb HincII fragment. This HincII fragment was cloned into the SmaI site of pXG (Ha et al., 1996) to produce plasmid clone pGL269.
Transfection of L. mexicana promastigotes was as described previously (Mottram et al., 1996). Briefly, for gene knockout experiments the cassettes were excised by digestion with HindIII and BglII, and the insert was gel purified using a Qiextract kit (Qiagen). Five to 10 μg of DNA were used for each transfection with 4 × 107 promastigotes. For introduction of the episome pGL269, DNA was prepared using a Qiagen Tip100 column as outlined by the manufacturer (Qiagen). Transfection used 10 μg of pGL269 DNA and 4 × 107 ΔGPI8 promastigotes. After electroporation, cells were allowed to recover in 10 ml of HOMEM for 24 h at 25°C, and then transfectants were selected on 1% agar/HOMEM plates containing appropriate antibiotics.
DNA Isolation and Southern Blot Analysis
Southern blotting using the hybridization membrane Hybond-N (Amersham, Little Chalfont, United Kingdom) was carried out as previously described (Mottram et al., 1993) with 5 μg of genomic DNA prepared by the method of Medina-Acosta and Cross (1993). Probes were labeled by a random priming using Prime It II (Stratagene) and purified using a NucTrap column (Stratagene).
Analysis of GPI-anchored Proteins
For Western blotting, 108 promastigotes were lysed on ice with 100 μl of 0.25% Triton X-100 before immediate addition of reducing Laemmli sample buffer. Samples were boiled for 5 min before fractionation by 10% SDS-PAGE. Proteins were electroblotted onto Hybond-C, and the membranes were incubated overnight with blocking buffer (10% horse serum and 5% nonfat milk powder in 20 mM Tris-HCl, pH 7.6, 300 mM NaCl, and 0.05% Tween 20) at 4°C. Anti-gp63 monoclonal antibodies were a gift from Dr. Robert McMaster (University of British Columbia, British Columbia, Canada). They were used at a 1:25 dilution in blocking buffer at room temperature for 90 min followed by washing in 20 mM Tris-HCl, pH 7.6, 300 mM NaCl, and 0.05% Tween 20. Affinity-purified anti-CRK1 antibodies (Mottram et al., 1993) were used at a 1:100 dilution. Secondary antibody (either anti-mouse or anti-rabbit immunoglobulin-horseradish peroxidase conjugate; Promega, Madison, WI) was used at a 1:5000 dilution. Enhanced chemiluminescence (Pierce, Rockford, IL) was used to detect antibody binding.
GPI-anchored proteins were metabolically labeled with [3H]EtN as described below. Proteins were extracted in boiling 1% SDS and solvent precipitated for analysis by 10% SDS-PAGE. Gels were incubated in En3Hance (New England Nuclear, Boston, MA) for detection of labeled protein by fluorography.
Characterization of EtN-P-containing GPIs
Wild-type and ΔGPI8 promastigotes were metabolically labeled with [3H]EtN (20 μCi/ml) in RPMI-1640 with 10% fetal calf serum for 15 h at 27°C. Alternatively, promastigotes were pulse labeled with [3H]mannose for 30 min, as previously described (Ralton and McConville, 1998). Metabolically labeled promastigotes were extracted in chloroform/methanol/water (1:2:0.8 vol/vol), and GPI lipids were recovered by biphasic partitioning between 1-butanol (200 μl) and water (200 μl) (Ralton and McConville, 1998). [3H]EtN-labeled GPIs were purified by high-performance TLC (HPTLC) on aluminum-backed Silica Gel60 (Merck, Darmstadt, Germany), developed in chloroform/methanol/ammonium hydroxide/ammonium acetate/water (180:140:9:9:23 vol/vol) (Ralton and McConville, 1998) and then reanalyzed by HPTLC in chloroform/methanol/1 M ammonium hydroxide (10:10:3 vol/vol) before or after various treatments. Digestion with Bacillus cereus PI-specific phospholipase C (PI-PLC) was performed in 50 mM triethanolamine-HCl buffer, pH 7.5, 10 mM EDTA, and 0.16% sodium deoxycholate for 15 h at 37°C. Jack bean α-mannosidase digestion was performed in 0.1 M sodium acetate buffer, pH 5.0, for 15 h at 37°C. Base hydrolysis was performed in either 0.1 M methanolic NaOH for 2 h at 37°C (to cleave all ester-linked fatty acids) or a mixture of NH4OH/methanol (1:1 vol/vol) for 2 h at 37°C (to preferentially cleave inositol acyl groups). The products of these treatments were recovered by passage down a column (0.5 ml) of octyl-Sepharose, equilibrated in 0.1 M sodium acetate and 5% 1-propanol, and eluted in 40% 1-propanol (Ralton and McConville, 1998). For characterization of [3H]Man-labeled Lipid X, this species was purified by HPTLC as described above and then treated with jack bean α-mannosidase in 0.1 M sodium acetate, pH 5.0, containing 0.2% taurodeoxycholic acid for 15 h at 37°C to hydrolyze comigrating GIPLs. The α-mannosidase-resistant Lipid X was then recovered by 1-butanol and water partitioning, and the glycan head group was obtained by hydrofluoric acid (HF) dephosphorylation (48% HF, 0°C, 48 h) followed by nitrous acid deamination and reduction with NaBH4 as previously described (Ralton and McConville, 1998). Partial acetolysis of the released glycan head group was also performed as previously described (Zawadzki et al., 1998).
Infections of Macrophages In Vitro and in Mice
Freshly culled BALB/c mice were injected intraperitoneally with 5 ml of ice-cold RPMI-1640 medium complete with 10 U/ml penicillin and 10 μg/ml streptomycin sulfate (Life Technologies). After 5 min of gentle massaging of the mouse abdomen, the RPMI-1640 that contained macrophages was harvested and kept on ice. Macrophages (2 × 105) were placed in each chamber of an eight-chamber Permanox slide (Labtek, Campbell, CA) and allowed to adhere overnight at 32°C under 5% CO2/95% air. Chambers were washed with RPMI-1640 to remove nonadhered cells before 2 × 105 stationary phase promastigotes were added in RPMI-1640 with 10% fetal calf serum and incubated at 32°C under 5% CO2/95% air. After 4 h, slides were again washed, and fresh RPMI-1640 with 10% fetal calf serum was added. Infection was allowed to proceed at 32°C under 5% CO2/95% air. Individual chambers were fixed with 100% methanol for 5 min at 8 h and 1, 3, and 7 d after addition of promastigotes. Slides were stained with modified Giemsa stain (Sigma, St. Louis, MO), diluted 20-fold with distilled water for 30 min, and viewed by light microscopy using a Zeiss (Jena, Germany) Axioplan microscope. Infections of mice involved percutaneous inoculation of 5 × 106 stationary phase promastigotes into the shaven rumps of BALB/c mice. Amastigotes were isolated from lesions after 8 mo of infection and reinoculated into BALB/c mice (2.5 × 107 per mouse). The subcutaneous lesions produced were monitored over 4 mo.
RESULTS
Cloning and Characterization of the L. mexicana GPI8
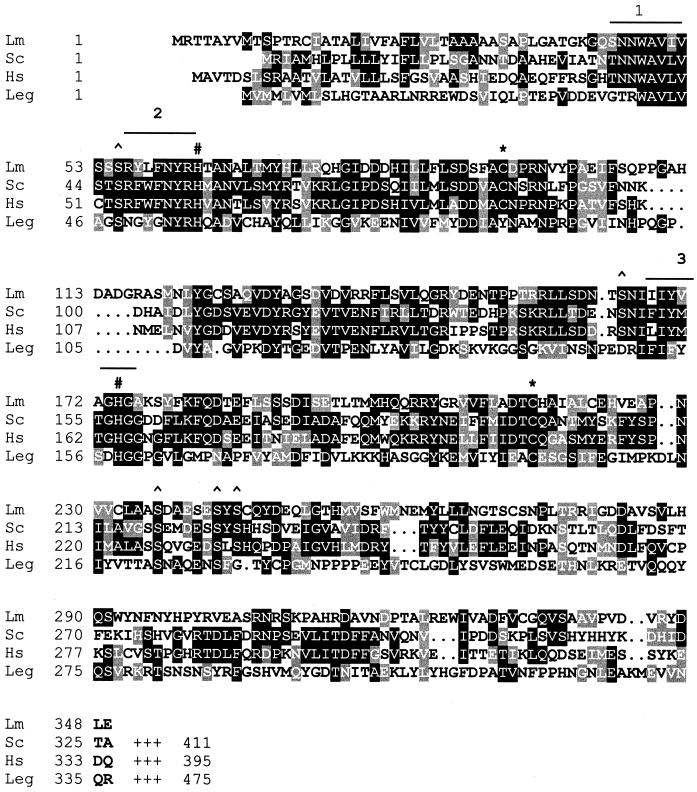
Degenerate oligonucleotide primers, based on sequences of similarity between the GPI8 sequences from Saccharomyces cerevisiae, human, and C. elegans, were used to amplify regions of L. mexicana cDNA or genomic DNA. A 362-bp PCR product was identified and cloned from cDNA produced from three life cycle stages of L. mexicana: procyclic promastigotes, metacyclic promastigotes, and amastigotes. Three identical PCR products were identified that showed encoded protein domains with significant sequence identity to human and yeast GPI8 proteins. The full-length L. mexicana GPI8 gene was subsequently isolated from a λ genomic library and sequenced. The L. mexicana GPI8 encodes a protein of 349 amino acids with a predicted size of 38 kDa. It has 31% sequence identity to yeast and human GPI8s and 19% identity to the plant asparaginyl endopeptidase legumain. A hydropathy analysis showed that the L. mexicana GPI8 has a similar overall profile to that of the yeast homologue, with an N-terminal hydrophobic domain that is likely to be a signal sequence directing entry into the ER. It lacks, however, a C-terminal hydrophobic domain found in yeast and human GPI8s that is predicted to be a transmembrane helix (Benghezal et al., 1996). The GPI:protein transamidases belong to family C13 of the cysteine peptidases (Riezman and Conzelmann, 1998) and so are likely to possess the classical catalytic dyad residues, cysteine and histidine, that mediate activity. It has been shown that GPI:protein transamidase activity is susceptible to sulfydryl alkylating agents, implying that the protein has an essential cysteine (Sharma et al., 1999). Two cysteine (Cys94 and Cys216, L. mexicana GPI8 numbering) and two histidine (His63 and His174) residues are conserved among leishmanial, yeast, and human GPI8 (Figure 2). Both histidine residues are also conserved in legumain, whereas only one of the cysteine residues is conserved across all C13 family members (Cys216). Thus this residue is the prime candidate for the active site cysteine. Three serine residues are also conserved between the three GPI8 sequences and legumain (Ser55, Ser236, and Ser242), and two more (Ser165 and Ser244) are conserved between the three GPI8 proteins but are absent from legumain.
Figure 2.
GPI8 protein sequence of L. mexicana (Lm; this study) aligned with S. cerevisiae (Sc; SwissProt, accession number P39012), human (Hs; Q92643), and C. ensiformis legumain (Leg; P49046). Gaps (.) were introduced to help the alignment; the C-terminal ends of Sc, Hs, and Leg have been omitted. Amino acid identities and similarities are highlighted. The highly conserved regions used to design degenerate oligonucleotide primers to isolate the L. mexicana GPI8 are overlined and numbered. Conserved cysteine (∗), histidine (#) and serine (^) residues are indicated. L. mexicana GPI8 has three potential N-glycosylation sites at positions N163, N268, and N307, none of which is conserved with yeast GPI8 that is heavily glycosylated (Benghezal et al., 1996).
Genomic Organization of GPI8 and Production of GPI8 Null Mutants
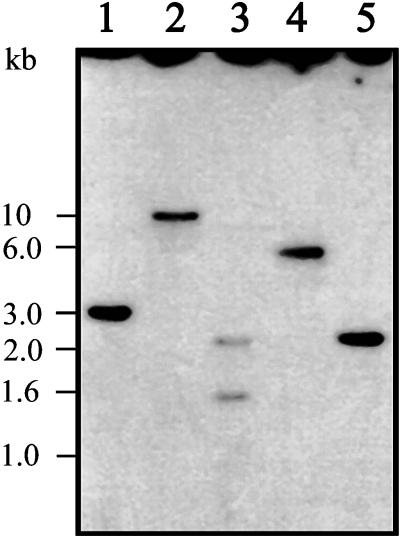
Southern blot analysis revealed that L. mexicana GPI8 is a single-copy gene (Figure 3). A single hybridizing fragment was detected with HincII, HindIII, XbaI, and XhoI digests, whereas two DNA fragments were detected with a PstI digest because of the presence of a PstI site in the GPI8 gene (Figure 4A). The sizes of the HincII (3.1 kb; Figure 3, lane 1) and XhoI (2.4 kb; Figure 3, lane 5) fragments correlate precisely with the 3.3 kb of sequence data obtained for the GPI8 locus.
Figure 3.
Southern blot. Restriction digests of L. mexicana wild-type genomic DNA (5 μg) using HincII (lane 1), HindIII (lane 2), PstI (lane 3), XbaI (lane 4), and XhoI (lane 5) were electrophoresed on a 0.8% agarose gel and blotted onto a nylon membrane. The blot was probed with a radiolabeled 1.1-kb XbaI–XhoI fragment containing the GPI8 ORF.
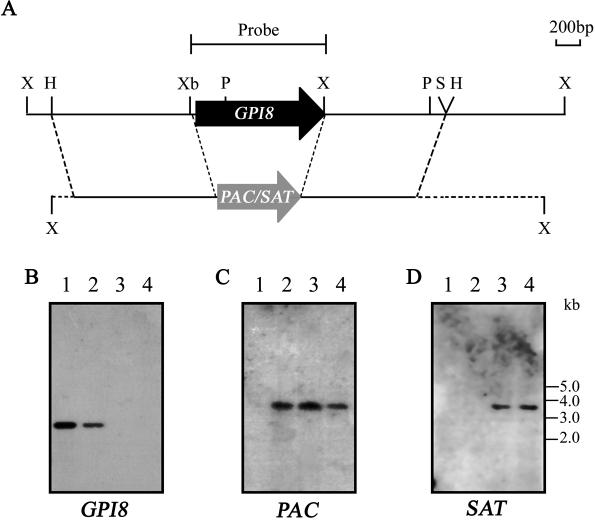
Figure 4.
Southern blot analysis of GPI8 mutants. (A) Map of GPI8 locus and predicted recombination events. X, XhoI; H, HincII; Xb, XbaI; P, PstI; S/H, SalI and HincII. Note the loss of the XhoI site within GPI8 after recombination. (B) Five micrograms of genomic DNA from wild-type L. mexicana (lane 1), W1238 heterozygote mutant (lane 2), W1234 ΔGPI8 clone 1 (lane 3), and W1236 ΔGPI8 clone 2 (lane 4) were digested with XhoI and fixed to a nylon membrane. The blot was probed with a radiolabeled 1.1-kb XbaI–XhoI fragment containing the GPI8 ORF. The same blot was stripped and reprobed with radiolabeled PAC (C) and SAT (D) probes.
To investigate the role of GPI8 in L. mexicana, sequential rounds of targeted gene replacement were performed using SAT and PAC antibiotic resistance genes flanked by GPI8 5′ and 3′ sequence to replace GPI8 (Figure 4A). A heterozygote clone (W1238, GPI8::PAC) in which one allele of GPI8 had been removed by integration with PAC was subjected to a second round of transfection with the SAT-containing cassette, and clones resistant to both antibiotics (ΔGPI8 clones W1234 and W1236) were analyzed by Southern blotting to confirm that they were indeed GPI8 deficient and had undergone the predicted genomic rearrangements. XhoI-digested genomic DNA from wild-type L. mexicana, W1238 (GPI8::PAC, clone 1), W1234 (ΔGPI8, clone 1), and W1236 (ΔGPI8 clone 2) was probed with a 1-kb XbaI–XhoI fragment containing the GPI8 ORF. A 2.4-kb XhoI fragment was detected in wild-type L. mexicana corresponding to the two GPI8 alleles (Figure 4B, lane 1). One GPI8 allele remained in the W1238 heterozygote (lane 2); however, the GPI8 gene could not be detected in either of the ΔGPI8 mutants (lanes 3 and 4). To confirm that SAT and PAC had integrated into the correct locus, the same blot was stripped and reprobed with the drug resistance genes (Figure 4, C and D). As predicted, the PAC probe hybridized to a band of 3.7 kb with the W1238 heterozygote (Figure 4C, lane 2) and both ΔGPI8 clones (lanes 3 and 4) but not wild-type parasites (lane 1). The SAT probe hybridized to a 3.6-kb band in the ΔGPI8 clones (Figure 4C, lanes 3 and 4), which was absent from the wild-type and W1238 heterozygote (lanes 1 and 2). These results, together with further Southern blot analysis with a 5′ flank probe (our unpublished results), indicate that both alleles of GPI8 had been replaced with SAT and PAC and that the mutants are GPI8 deficient. GPI8 was cloned into the pXG episomal expression vector and introduced into ΔGPI8 clones 1 and 2 by electroporation to give G418-resistant cell lines W1235 ΔGPI8[pXG-GPI8] and W1237 ΔGPI8[pXG-GPI8].
ΔGPI8 Mutants Lack GPI-anchored Proteins
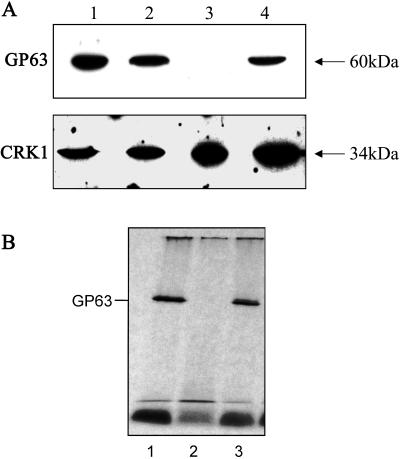
The major GPI-anchored surface protein of Leishmania promastigotes is the metalloproteinase gp63 (Schneider et al., 1990). Immunofluorescence was carried out with a monoclonal antibody directed against L. mexicana gp63 on promastigotes of wild-type L. mexicana, ΔGPI8, and ΔGPI8[pXG-GPI8]. A strong signal was detected with wild-type L. mexicana, but no gp63 was detected with ΔGPI8 (our unpublished results). Levels of surface expression of gp63 were restored to wild-type levels in the cell line expressing an episomal copy of GPI8. The presence of gp63 in cell lysates from these cell lines was investigated by Western blotting with anti-gp63 antiserum (Figure 5A). An ∼60-kDa protein was detected by immunoblotting with wild-type L. mexicana (lane 1) and the heterozygote (lane 2) but was absent from ΔGPI8 (lane 3). gp63 was restored in the ΔGPI8[pXG-GPI8] cell line (lane 4). Equal loading of cell lysates was confirmed with an antiserum against the CRK1 protein kinase (Mottram et al., 1993). Long-term exposure of the blot revealed that ΔGPI8 contained a small amount of protein recognized by the anti-gp63 antibody (our unpublished results). It had a similar mass to gp63 itself and could either have been undegraded precursor protein or possibly the isoenzyme characteristically present in the lysosomes of amastigotes, which is not GPI anchored (Medina-Acosta et al., 1989; Ilg et al., 1993). The residual gp63 protein detected was not GPI anchored, because no [3H]ethanolamine-labeled gp63 could be detected in the ΔGPI8 mutant (Figure 5B). Interestingly, even after a very-long-term (3 mo) exposure of the autoradiograph no other GPI-anchored proteins could be detected in this strain of L. mexicana.
Figure 5.
Analysis of gp63 expression. (A) Western blotting. Lysates of wild-type (lane 1), W1238 GPI8 heterozygote (lane 2), W1234 ΔGPI8 (lane 3), and W1235 ΔGPI8[pXG-GPI8] (lane 4) promastigotes were probed with an anti-gp63 mouse monoclonal antibody or anti-CRK1 kinase antibodies as a control for protein loading. (B) Metabolic labeling. Promastigotes were metabolically labeled to steady state with [3H]EtN, and labeled GPI:proteins were detected after separation by SDS-PAGE. Lysates of wild-type (lane 1), W1234 ΔGPI8 (lane 2), and W1235 ΔGPI8[pXG-GPI8] (lane 3) promastigotes are shown.
ΔGPI8 Mutants Accumulate a Novel Inositol-acylated GPI Species
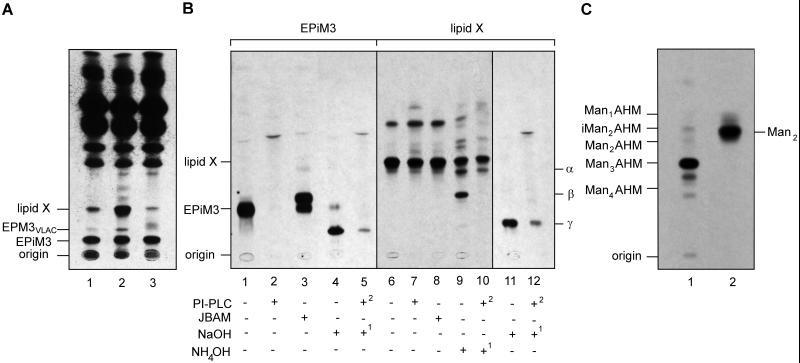
Wild-type parasites and ΔGPI8 and ΔGPI8[pXG-GPI8] mutants were metabolically labeled to steady state with [3H]EtN to investigate whether disruption of protein anchoring resulted in the accumulation of any EtN-P-containing protein–anchor precursors. Two [3H]EtN-labeled species were present in fivefold higher levels in ΔGPI8 compared with wild-type cells (Figure 6A, compare lanes 1 and 2). We have previously shown that one of these bands, EPM3VLAC, is likely to be the major protein GPI anchor precursor in L. mexicana and has the structure EtN-P-Man3GlcN-PI (Ralton and McConville, 1998). The second species, termed Lipid X, represents a novel species that has not been previously characterized. Interestingly, levels of both EPM3VLAC and Lipid X were decreased in ΔGPI8[pXG-GPI8] compared with the wild-type parasites, suggesting that both species may be used at a higher rate in cells overexpressing this component of the transamidase (Figure 6A, lane 3). The cellular levels of other EtN-labeled phospholipids, as well as EPiM3, the major EtN-P-containing free GPI (see Figure 1 for structure), were essentially unchanged in all three cell lines (Figure 6A, lanes 1–3).
Figure 6.
Accumulation of protein–anchor intermediates in GPI8 mutants. (A) HPTLC analysis of [3H]EtN-labeled lipids in wild-type (lane 1), W1234 ΔGPI8 (lane 2), and W1235 ΔGPI8[pXG-GPI8] (lane 3) promastigotes. The elution positions of the major EtN-P-modified GIPL, EPiM3, and a previously characterized protein anchor precursor, EPM3VLAC, are indicated (VLAC refers to GPIs with very long [C24:0/C26:0] alkyl chains in the lipid moiety). Lipid X represents a second putative protein anchor precursor that accumulates in ΔGPI8. (B) [3H]EtN-labeled EPiM3 and Lipid X were purified by HPTLC and treated with PI-PLC, jack bean α-mannosidase (JBAM), and either strong (NaOH) or mild base (NH4OH), as indicated. Numbers refer to the order in which each treatment was performed. The products were recovered by 1-butanol partitioning and analyzed by HPTLC. The base treatments generated several novel species (α, β, and γ) that may represent the products of acyl migration or deacylation (discussed in RESULTS). (C) HPTLC analysis of the neutral glycan head group of Lipid X. Lipid X was metabolically labeled with [3H]mannose and purified by HPTLC. The neutral glycan obtained after HF hydrolysis and nitrous acid deamination and reduction was analyzed by HPTLC before (lane 1) or after (lane 2) acetolysis. The migration positions of standard glycans, Man1–4AHM (glycan head groups derived from Man1–4GlcN-PI), and Manα1–2Man (Man2 derived from [3H]mannose-labeled Man4AHM) are indicated.
To characterize Lipid X further, the [3H]EtN-labeled species was purified by HPTLC and subjected to a series of enzymatic and chemical treatments. Lipid X was resistant to both PI-specific phospholipase C and jack bean α-mannosidase digestions (Figure 6B, lanes 6–8). In contrast, EPiM3 was completely digested by PI-PLC and was partially converted to a faster HPTLC migrating species after jack bean α-mannosidase digestion, consistent with the removal of the α1–3-linked mannose side chain (Figures 1 and 6B, lanes 1–3). Treatment of Lipid-X with methanolic-NaOH using conditions that should remove all ester-linked acyl groups generated a new species with a slower HPTLC mobility (species γ; Figure 6B, lane 11) that was sensitive to PI-PLC digestion (Figure 6B, lane 12). This species migrated with a slightly faster HPTLC mobility than the base-generated lyso-EPiM3, which will retain a single C18:0 alkyl chain (Figures 1 and 6B, lanes 4 and 5). When Lipid X was subjected to milder base hydrolysis, using conditions that preferentially cleave the inositol-linked acyl chains rather than glycerolipids, two new species with intermediate HPTLC mobilities were generated (species α and β; Figure 6B, lane 9). Species α had a slightly slower HPTLC mobility than Lipid X and remained resistant to PI-PLC, whereas species β migrated with the same HPTLC mobility as the putative protein anchor precursor EPM3VLAC and was sensitive to PI-PLC digestion (Figure 6B, lane 10). Collectively these results suggested that Lipid X corresponds to a GPI with two base-sensitive acyl chains and a single base-resistant (alkyl) chain. The resistance of Lipid X and species α to PI-PLC suggested that both of these lipids retained a base-labile constituent on the inositol head group. Species α (which is present in small amounts in untreated samples) may be generated by base-catalyzed migration of an acyl group around the inositol ring. Species β most likely corresponded to Lipid X minus the inositol acyl modification (rendering it susceptible to PI-PLC), whereas species γ corresponded to Lipid X minus both the inositol acyl modification and the sn-2 fatty acyl chain in the PI glycerolipid.
To define further the structure of the glycan head group of Lipid X, ΔGPI8 was metabolically labeled with [3H]mannose, and a species with the same HPTLC mobility as Lipid X was purified by HPTLC and exhaustive α-mannosidase digestion to remove comigrating free GPIs (the latter species are not modified with an EtN-P and are therefore digested by α-mannosidase [Ralton and McConville, 1998]). The neutral glycan head group of Lipid X obtained by hydrogen fluoride dephosphorylation, nitrous acid deamination, and reduction (which cleaves the PI lipid moiety and converts the terminal GlcN to 2,5-anhydromannitol [AHM]) comigrated with authentic Man3AHM on HPTLC (Figure 6C, lane 1). Partial acetolysis of this glycan, which is expected to cleave the 1–6 linkage in the conserved Man3GlcN backbone of the gp63 anchor (Schneider et al., 1990), generated a faster migrating species, which comigrated with the disaccharide Manα1–2Man (Figure 6C, lane 2). This is the expected product if most of the [3H]mannose label was incorporated into the two outer mannose residues. These data suggested that Lipid X contained the same Man3GlcN glycan core as the gp63 GPI anchor (Figure 1). Moreover, based on the HPTLC mobilities of the deacylated Lipid-X species it is likely that this species had the same 1-O-alkyl(C24:0/C26:0)-2-O-acyl-PI lipid moiety as EPM3VLAC and the mature protein-linked GPIs (Schneider et al., 1990; Ralton and McConville, 1998). However, this species is unusual in containing an inositol acyl modification that has not been identified on any other leishmanial GPIs.
Cellular Levels of Non–Protein-linked GPIs Are Unchanged in the ΔGPI8 Mutant
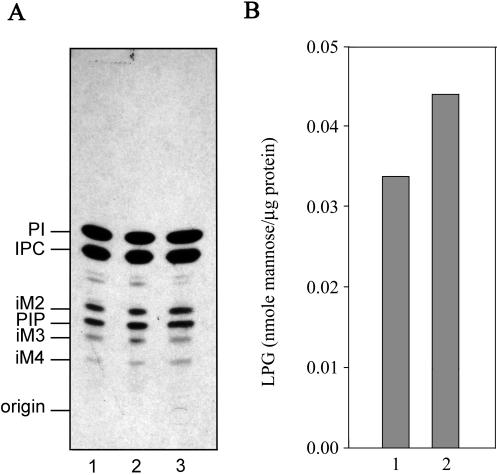
To determine whether the cellular levels of non–protein-linked GPIs were altered in the ΔGPI8 mutants, cells were labeled to steady state with [3H]inositol, and total inositol lipids were analyzed by HPTLC. [3H]Inositol is incorporated into the major phospholipids PI and inositolphosphoceramide, as well as into the predominant free GPI species iM2, iM3, and iM4 (see Figure 1 for structures). The levels of these free GPIs, relative to the levels of PI, were essentially the same in wild-type parasites, ΔGPI8, and ΔGPI8[pXG-GPI8] (Figure 7A). Deletion of GPI8 also had no effect on the levels of the major GPI-anchored macromolecule LPG (Figure 7B). Thus an accumulation of protein–anchor precursors does not reduce the rate of synthesis of non–protein linked GPIs.
Figure 7.
Cellular levels of LPG and protein-free GPIs are not affected by deletion of GPI8. (A) Cells were metabolically labeled to steady state with [3H]myoinositol, and total inositol lipids were analyzed by HPTLC. Inositol lipids from wild-type (lane 1), W1234 ΔGPI8 (lane 2), and W1235 ΔGPI8[pXG-GPI8] (lane 3) promastigotes are shown. PI, phosphatidylinositol; IPC, inositolphosphoceramide; PIP, phosphatidylinositolphosphate; iM2, iM3, and iM4, the major free GPIs containing glycan head groups with two (iM2), three (iM3), and four (iM4) mannose residues. (B) LPG was extracted from delipidated cell pellets and purified by octyl-Sepharose chromatography. Cellular levels (given as nanomoles of mannose per microgram of cellular protein) in wild-type (lane 1) and W1234 ΔGPI8 (lane 2) promastigotes were quantitated by GC-MS.
ΔGPI8 Infect and Replicate in Macrophages and Mice
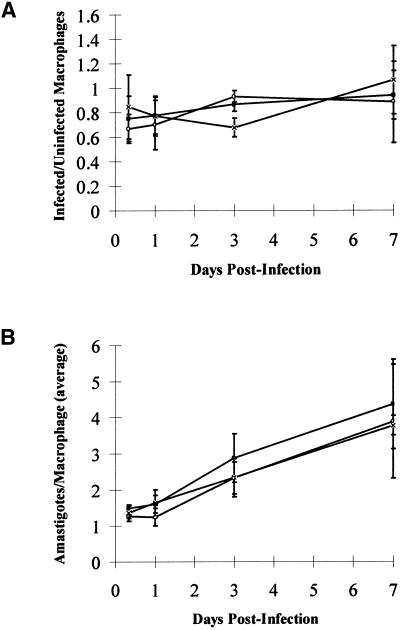
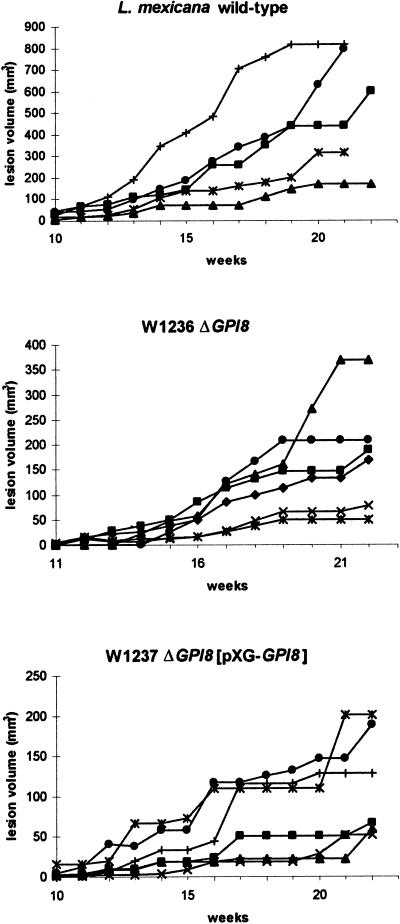
Promastigotes of the two ΔGPI8 clones were found to grow at the same rate in liquid culture as wild-type L. mexicana promastigotes and could differentiate into amastigote-like forms in vitro. To test whether the mutants were capable of surviving in mammalian cells, invasion experiments were performed with macrophages in vitro (Figure 8). The mouse macrophages were allowed to adhere to slides before a 1:1 exposure to stationary-phase promastigotes of wild-type L. mexicana, ΔGPI8, and ΔGPI8[pXG-GPI8]. Samples were taken at 8 h and 1, 3, and 7 d after infection. ΔGPI8 and ΔGPI8[pXG-GPI8] mutants were found to infect macrophages in vitro to wild-type levels (Figure 8A). In addition, the numbers of amastigotes per infected macrophage increased over the 7-d period to a similar extent for each of the three cell lines (Figure 8B). This shows that macrophage entry by promastigotes, differentiation into amastigotes, and survival and replication within the macrophage phagolysosome is not dependent on GPI-anchored proteins. ΔGPI8 and ΔGPI8[pXG-GPI8] mutants were inoculated into BALB/c mice, and after 8 mo amastigotes were isolated from lesions for inoculation into six BALB/c mice. The size of the subsequent lesions was monitored over a 4-mo period (Figure 9). All animals infected with ΔGPI8 and ΔGPI8[pXG-GPI8] produced large cutaneous lesions ranging in volume from 50 to 350 mm3 after 22 wk. In general, wild-type L. mexicana amastigotes produced larger lesions (100–800 mm3 after 22 wk) that formed slightly more rapidly than those caused by the mutant parasites. These data show that amastigote replication and survival in mice is not dependent on GPI-anchored proteins.
Figure 8.
Macrophage infectivity assays. Peritoneal exudate cells from BALB/c mice were infected 1:1 with stationary phase promastigotes of wild-type L. mexicana (×), W1234 ΔGPI8 (▪), and W1235 ΔGPI8[pXG-GPI8] (○). Samples were taken over a 7-d period, and the number of intracellular parasites was counted after fixation and Giemsa staining. More than 100 macrophages were counted for each time point, and the results are expressed in terms of the ratio of infected to uninfected macrophages (A) and the number of amastigotes per infected macrophage (B). The data are means and SDs from several experiments.
Figure 9.
Infectivity to mice. BALB/c mice were inoculated with lesion amastigotes of wild-type L. mexicana, W1236 ΔGPI8, and W1237 ΔGPI8[pXG-GPI8], and the subsequent lesion sizes were monitored over 22 wk. Data from individual mice in each group are shown to highlight the variation in lesion size between them.
DISCUSSION
Several lines of evidence point to the L. mexicana GPI8 being a functional homologue of yeast Gpi8p and a component of the GPI:protein transamidase complex. First, there is the high level of sequence identity among the predicted yeast, human, and L. mexicana GPI8 proteins, including the conservation of cysteine and histidine residues that potentially have key involvement in the catalytic mechanism. Second, deletion of GPI8 from L. mexicana led to the accumulation of the putative protein–anchor precursor EPM3VLAC (Ralton and McConville, 1998). Third, ΔGPI8 mutants are deficient in GPI-anchored gp63, the major surface protein of L. mexicana. Interestingly, [3H]ethanolamine labeling experiments (Figure 5B) confirmed the presence of GPI-anchored gp63 in wild-type L. mexicana but failed to detect other GPI-anchored proteins that have been reported to be present at low abundance on the surface of other Leishmania species (Murray et al., 1989; Lohman et al., 1990). It remains a formal, albeit unlikely, possibility that a transamidase structurally unrelated to GPI8 carries out the GPI:protein transamidase reaction for low-abundance GPI-anchored proteins. However, the buildup of the protein:anchor precursor and the loss of GPI-anchored gp63 in the ΔGPI8 mutant, together with the finding that GPI8 is a single-copy gene (as found in yeast), argues strongly that GPI8 is an essential component of the single GPI:protein transamidase in Leishmania.
Sequence comparisons suggest that the L. mexicana GPI8 protein is smaller than the S. cerevisiae and human homologues (38 vs. 46 kDa) and that it lacks the C-terminal hydrophobic domain that is thought to be involved in anchoring these other GPI8 proteins to the lumenal leaflet of the ER (Benghezal et al., 1996; Yu et al., 1997). Attachment of L. mexicana GPI8 to the ER membrane, where the protein will be able to interact with the preformed GPI anchor, may thus require one or more other integral membrane proteins. A possible candidate is Gaa1p, an integral membrane protein that appears to an essential component of the GPI:transamidation complex of yeast (Hamburger et al., 1995). This type of variation between ER isoenzymes involved in GPI biosynthesis is not unprecedented. Two classes of dolichol-phosphate-mannose synthase have been identified in different species (Colussi et al., 1997). The human and fission yeast enzymes lack a C-terminal hydrophobic domain, whereas the trypanosome and budding yeast enzymes contain one. The two classes of this enzyme, however, are functionally equivalent, because both the human and S. cerevisiae genes can complement a lethal null mutant in fission yeast (Colussi et al., 1997).
Leishmania promastigotes synthesize a number of GPI-anchored proteins that are minor, but significant, components in the well-defined glycocalyx that covers the promastigote surface. The other major components in this glycocalyx are LPG and the family of free GPIs (GIPLs) that are likely to form a densely packed layer immediately above the plasma membrane (McConville and Ferguson, 1993). Although a variety of in vitro studies have suggested that these GPI-anchored proteins fulfill a number of functions in different developmental stages of the parasite, the relative significance of these proteins for parasite infectivity and survival in the insect or mammalian hosts has been unclear. Previous attempts to obtain parasites that are defective in one or more of these proteins by targeted gene deletion have been hampered by the fact that most of the proteins are encoded by multigene families. Interestingly, targeted deletion of six of the seven L. major gp63 genes did not affect growth in vitro or prevent the formation of disease in mice (Joshi et al., 1998). An alternative approach to obtaining Leishmania parasites lacking GPI-anchored proteins exploited the finding that ectopic expression of T. brucei GPI-PLC in Leishmania results in the selective degradation of protein anchor GPI precursors (Mensa-Wilmot et al., 1994; Ilgoutz et al., 1999b; Mensa-Wilmot et al., 1999). However, this approach does not result in the complete removal of all GPI-anchored proteins and may also lead to the partial degradation of non–protein-linked GPIs (Mensa-Wilmot et al., 1999) and, therefore, is unable to provide definitive data on the specific functions of GPI-anchored proteins. In our study, we have eliminated the expression on the parasite surface of the major GPI-anchored glycoprotein gp63. Although expression of the much less abundant PSA2/gp46 could not be detected even in wild-type L. mexicana, it is likely that the GPI-anchoring and therefore surface expression of these glycoproteins are also prevented in the ΔGPI8 mutant.
The absence of GPI-anchored proteins had no effect on the growth of the parasite in liquid culture or its ability to infect and survive in macrophages in vitro. Importantly, the ΔGPI8 mutant was also able to establish an infection in mice. These data suggest that GPI-anchored proteins are not essential for growth or infectivity to mammals. It remains to be determined whether they play a key role for the parasite in the insect host. In contrast to our findings with the ΔGPI8 mutant, we have recently shown that the gene encoding dolichol-phosphate mannose synthase, a key enzyme in GPI biosynthesis, is essential for growth of L. mexicana promastigotes (Ilgoutz et al., 1999b). Taken together with the results described here, it is likely that this enzyme is essential because of a requirement for continued synthesis of non–protein-linked free GPIs in the promastigote stage.
Although all three classes of GPI synthesized by Leishmania contain the same core, they each contain distinct glycan head group modifications and lipid compositions (Figure 1). Moreover, the synthesis of the protein and LPG anchors, but not the GIPLs, is developmentally regulated (McConville and Blackwell, 1991). These findings suggest that the regulation of GPI biosynthesis in Leishmania is complex and may require the presence of distinct enzyme complexes and/or some degree of compartmentalization of these pathways (Ilgoutz et al., 1999a). The finding in this study that a subpool of mature protein anchor precursors becomes inositol acylated provides another example of how this class of GPI intermediates is processed differently from the GIPL or LPG anchor intermediates. The inositol acylated GPI intermediate (Lipid X) has been overlooked in previous analyses (McConville et al., 1993; Ralton and McConville, 1998), because it is present in low levels in wild-type cells and comigrates in HPTLC analysis with one of the major GIPL species. However, the involvement of Lipid X in the GPI protein anchor pathway is now strongly suggested by the structural characterization of this species and the finding that both Lipid X and the protein anchor precursor EPM3VLAC are specifically accumulated in ΔGPI8 mutant cells. Inositol acylation of GPI anchor intermediates usually involves the addition of a saturated fatty acid to the 2-hydroxyl of the inositol residue and appears to occur in all eukaryotes (Ferguson 1999; Tiede et al., 1999; McConville and Menon, 2000). However, the timing, nature of the acyl donor, and function of this modification may vary between higher and lower eukaryotes (Güther and Ferguson, 1995; Smith et al., 1997). For example, in animal cells and yeast, inositol acylation occurs at an early stage in GPI biosynthesis (after de-N-acetylation of GlcN-PI), whereas in African trypanosomes it occurs later in the pathway (Güther and Ferguson, 1995). Inositol acylation in Leishmania appears to be analogous to the situation in African trypanosomes, because we have found no evidence for the inositol acylation of early GPI intermediates. Moreover, as in the trypanosomes, there is no evidence that these inositol acylated GPI species are added to protein (Schneider et al., 1990). In trypanosomes inositol acylation of Man3GlcN-PI is thought 1) to be a prerequisite for addition of the EtN-P head group and for subsequent fatty acid remodeling reactions of latter intermediates and 2) to play a role in regulating the catabolism of excess GPI intermediates (Güther and Ferguson, 1995; Milne et al., 1999). Pulse–chase labeling experiments suggest that Lipid X may also be part of a catabolic pathway in Leishmania (J.L. Zawadzki and M.J. McConville, unpublished data). Whatever its function, this type of modification appears to be highly selective for protein anchor precursors, because it is not found on the more abundant and structurally similar Man3GlcN-PI intermediates in GIPL biosynthesis (Ralton and McConville, 1998). It is possible that the GPI inositol acyltransferase only recognizes GPI molecular species with very long alkyl chains (C24:0/C26:0) that are selectively incorporated into the protein and LPG anchors (Ralton and McConville, 1998). Alternatively, the acyltransferase may only have access to this pool of GPIs if intermediates from different pathways are present in different membranes or have different membrane topologies. Given the distinct properties and probable function of inositol acylation in trypanosomes compared with animal cells, it has been proposed that the inositol acyltransferase may be a target for new anti-trypanosome therapeutics (Smith et al., 1999). However, the fact that this modification is restricted to nonessential protein–anchor precursors in Leishmania suggests that therapeutics such as these would not be useful against these parasites.
The lack of detectable gp63 in the ΔGPI8 mutants suggests that the newly synthesized gp63 is either rapidly degraded or secreted. We have been unable to detect any secretion of gp63 from ΔGPI8, suggesting that this protein is being degraded intracellularly. In contrast, depletion of intracellular levels of GPI protein anchor precursors by ectopic expression of T. brucei GPI-PLC in L. major resulted in the secretion of a hydrophilic form of gp63 (Mensa-Wilmot et al., 1994). Similarly, cleavage of the signal peptide without anchor addition has been reported in both mammalian and trypanosomal in vitro systems and resulted in the release of processed, but GPI-free, protein (Maxwell et al., 1995a; Ramalingam et al., 1996; Sharma et al., 1999). However, in the absence of the proteolytic processing event (as would be the situation in ΔGPI8), the protein is targeted for degradation. Interestingly, mutation of the putative ω residue of gp63 (Asn577) resulted in the secretion of the protein (McGwire and Chang, 1996). It now seems probable that this mutated protein was proteolytically processed but not modified with a GPI anchor.
The GPI:protein transamidase of yeast is essential (Benghezal et al., 1996). It is also likely to be crucial for growth of parasitic protozoa that synthesize GPI:glycoprotein coats, such as the African trypanosomes and Plasmodium falciparum (McConville and Ferguson, 1993; Ferguson, 1999). As such, selective inhibitors of fungal or protozoan GPI8 may have therapeutic potential against a variety of pathogens. Despite the fact that some caution is required in interpreting the results from genetic manipulation to validate drug targets in parasitic protozoa (Barrett et al., 1999), the finding that ΔGPI8 mutants infected mice provides convincing evidence that the GPI:protein transamidase is not essential for Leishmania proliferation in mammals; therefore, inhibitors of the protein will not have clinically useful antileishmanial activity. However, the viability of the GPI8 null mutants of Leishmania, together with the ability to reexpress mutated forms of GPI8 in these parasites, makes Leishmania an excellent model system for analyzing the structure and function of the transamidase complex.
ACKNOWLEDGMENTS
We thank Dr. Robert McMaster and Dr. David Russell for anti-gp63 antibodies. This work was supported by the Medical Research Council (United Kingdom). J.D.H. was supported by a University of Glasgow postgraduate scholarship. J.C.M. is a Medical Research Council Senior Research Fellow. M.J.M. is an Australian National Health and Medical Research Council Principal Research Fellow.
REFERENCES
- Alexander J, Russell DG. The interaction of Leishmania species with macrophages. Adv Parasitol. 1992;31:175–254. doi: 10.1016/s0065-308x(08)60022-6. [DOI] [PubMed] [Google Scholar]
- Bahr V, Stierhof Y-D, Ilg T, Demar M, Quinten M, Overath P. Expression of lipophosphoglycan, high-molecular weight phosphoglycan and glycoprotein 63 in promastigotes and amastigotes of Leishmania mexicana. Mol Biochem Parasitol. 1993;58:107–122. doi: 10.1016/0166-6851(93)90095-f. [DOI] [PubMed] [Google Scholar]
- Barrett MP, Mottram JC, Coombs GH. Recent advances in identifying and validating drug targets in trypanosomes and leishmanias. Trends Microbiol. 1999;7:82–88. doi: 10.1016/s0966-842x(98)01433-4. [DOI] [PubMed] [Google Scholar]
- Bates PA, Robertson CD, Tetley L, Coombs GH. Axenic cultivation and characterization of Leishmania mexicana amastigote-like forms. Parasitology. 1992;105:193–202. doi: 10.1017/s0031182000074102. [DOI] [PubMed] [Google Scholar]
- Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 1996;15:6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Beverley SM, Turco SJ. Lipophosphoglycan (LPG) and the identification of virulence genes in the protozoan parasite Leishmania. Trends Microbiol. 1998;6:35–40. doi: 10.1016/S0966-842X(97)01180-3. [DOI] [PubMed] [Google Scholar]
- Button LL, Russell DG, Klein HL, Medina-Acosta E, Karess RE, McMaster WR. Genes encoding the major surface glycoprotein in leishmania are tandemly linked at a single chromosomal locus and are constitutively transcribed. Mol Biochem Parasitol. 1989;32:271–283. doi: 10.1016/0166-6851(89)90076-5. [DOI] [PubMed] [Google Scholar]
- Caras IW, Weddell GN, Williams SR. Analysis of the signal for attachment of a glycophospholipid membrane anchor. J Cell Biol. 1989;108:1387–1396. doi: 10.1083/jcb.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi PA, Taron CH, Mack JC, Orlean P. Human and Saccharomyces cerevisiae dolichol phosphate mannose synthases represent two classes of the enzyme, but both function in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1997;94:7873–7878. doi: 10.1073/pnas.94.15.7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MAJ. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- Frommel TO, Button LL, Fujikura Y, McMaster WR. The major surface glycoprotein (gp63) is present in both life stages of Leishmania. Mol Biochem Parasitol. 1990;38:25–32. doi: 10.1016/0166-6851(90)90201-v. [DOI] [PubMed] [Google Scholar]
- Güther MLS, Ferguson MAJ. The role of inositol acylation and inositol deacylation in GPI biosynthesis in Trypanosoma brucei. EMBO J. 1995;14:3080–3093. doi: 10.1002/j.1460-2075.1995.tb07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha DS, Schwarz JK, Turco SJ, Beverley SM. Use of the green fluorescent protein as a marker in transfected Leishmania. Mol Biochem Parasitol. 1996;77:57–64. doi: 10.1016/0166-6851(96)02580-7. [DOI] [PubMed] [Google Scholar]
- Hamburger D, Egerton M, Riezman H. Yeast gaa1p is required for attachment of a completed gpi anchor onto proteins. J Cell Biol. 1995;129:629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handman E, Osborn AH, Symons F, Van Driel R, Cappai R. The Leishmania promastigote surface antigen 2 complex is differentially expressed during the parasite life cycle. Mol Biochem Parasitol. 1995;74:189–200. doi: 10.1016/0166-6851(95)02500-6. [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Komuro I, Chen R, Hosoda T, Mizuno T, Kudoh S, Georgescu SP, Medof ME, Yazaki Y. Molecular cloning of human homolog of yeast GAA1 which is required for attachment of glycosylphosphatidylinositols to proteins. FEBS Lett. 1998;421:252–258. doi: 10.1016/s0014-5793(97)01576-7. [DOI] [PubMed] [Google Scholar]
- Ilg T, Harbecke D, Overath P. The lysosomal gp63-related protein in Leishmania mexicana amastigotes is a soluble metalloproteinase with an acidic pH optimum. FEBS Lett. 1993;327:103–107. doi: 10.1016/0014-5793(93)81049-6. [DOI] [PubMed] [Google Scholar]
- Ilgoutz SC, Mullin KA, Southwell BR, McConville MJ. Glycosylphosphatidylinositol biosynthetic enzymes are localized to a stable tubular subcompartment of the endoplasmic reticulum in Leishmania mexicana. EMBO J. 1999a;18:3643–3654. doi: 10.1093/emboj/18.13.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgoutz SC, Zawadzki J, Ralton JE, McConville MJ. Evidence that free GPI glycolipids are essential for growth of Leishmania mexicana. EMBO J. 1999b;18:2746–2755. doi: 10.1093/emboj/18.10.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S. Legumain—asparaginyl endopeptidase. Methods Enzymol. 1994;244:604–615. doi: 10.1016/0076-6879(94)44044-1. [DOI] [PubMed] [Google Scholar]
- Joshi PB, Sacks DL, Modi G, McMaster WR. Targeted gene deletion of Leishmania major genes encoding developmental stage-specific leishmanolysin (GP63) Mol Microbiol. 1998;27:519–530. doi: 10.1046/j.1365-2958.1998.00689.x. [DOI] [PubMed] [Google Scholar]
- Lohman KL, Langer PJ, McMahon-Pratt D. Molecular cloning and characterization of the immunologically protective surface glycoprotein gp46/m-2 of Leishmania amazonensis. Proc Natl Acad Sci USA. 1990;87:8393–8397. doi: 10.1073/pnas.87.21.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell SE, Ramalingam S, Gerber LD, Brink L, Udenfriend S. An active carbonyl formed during glycosylphosphatidylinositol addition to a protein is evidence of catalysis by a transamidase. J Biol Chem. 1995a;270:19576–19582. doi: 10.1074/jbc.270.33.19576. [DOI] [PubMed] [Google Scholar]
- Maxwell SE, Ramalingam S, Gerber LD, Udenfriend S. Cleavage without anchor addition accompanies the processing of a nascent protein to its glycosylphosphatidylinositol-anchored form. Proc Natl Acad Sci USA. 1995b;92:1550–1554. doi: 10.1073/pnas.92.5.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Menon AK, Cross GAM. Transfer of glycosyl-phosphatidylinositol membrane anchors to polypeptide acceptors in a cell free system. J Cell Biol. 1991;114:61–71. doi: 10.1083/jcb.114.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville MJ, Blackwell JM. Developmental regulation of the glycosylphosphatidylinositols of Leishmania donovani: characterization of the promastigote and amastigote glycolipids. J Biol Chem. 1991;266:15170–15179. [PubMed] [Google Scholar]
- McConville MJ, Collidge TAC, Ferguson MAJ, Schneider P. The glycoinositol phospholipids of Leishmania mexicana promastigotes. Evidence for the presence of three distinct pathways of glycolipid biosynthesis. J Biol Chem. 1993;268:15595–15604. [PubMed] [Google Scholar]
- McConville MJ, Ferguson MAJ. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville, M.J., and Menon, A.K. (2000). Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids. Mol. Membr. Biol. (in press). [DOI] [PubMed]
- McGwire BS, Chang KP. Post-translational regulation of a Leishmania HEXXH metalloprotease (gp63)—the effects of site-specific mutagenesis of catalytic, zinc binding, N-glycosylation, and glycosyl phosphatidylinositol addition sites on N-terminal end cleavage, intracellular stability, and extracellular exit. J Biol Chem. 1996;271:7903–7909. doi: 10.1074/jbc.271.14.7903. [DOI] [PubMed] [Google Scholar]
- Medina-Acosta E, Cross GAM. Rapid isolation of DNA from trypanosomatid protozoa using a simple mini-prep procedure. Mol Biochem Parasitol. 1993;59:327–329. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- Medina-Acosta E, Karess RE, Schwartz H, Russell DG. The promastigote surface protease (gp63) of Leishmania is expressed but differentially processed and localized in the amastigote stage. Mol Biochem Parasitol. 1989;37:263–273. doi: 10.1016/0166-6851(89)90158-8. [DOI] [PubMed] [Google Scholar]
- Mensa-Wilmot K, Garg N, McGuire BS, Lu HG, Zhong L, Armah DA, LeBowitz JH, Chang K-P. Roles of free GPIs in amastigotes of Leishmania. Mol Biochem Parasitol. 1999;99:103–116. doi: 10.1016/s0166-6851(99)00003-1. [DOI] [PubMed] [Google Scholar]
- Mensa-Wilmot K, LeBowitz JH, Chang K-P, Al-Qahtani A, McGwire BS, Tucker S, Morris JC. A glycosylphosphatidylinositol (GPI)-negative phenotype produced in Leishmania major by GPI phospholipase C from Trypanosoma brucei: topography of two GPI pathways. J Cell Biol. 1994;124:935–947. doi: 10.1083/jcb.124.6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne KG, Ferguson MAJ, Englund PT. A novel glycosylphosphatidylinositol in African trypanosomes—a possible catabolic intermediate. J Biol Chem. 1999;274:1465–1471. doi: 10.1074/jbc.274.3.1465. [DOI] [PubMed] [Google Scholar]
- Moran P, Caras IW. Requirements for glycosylphosphatidylinositol attachment are similar but not identical in mammalian-cells and parasitic protozoa. J Cell Biol. 1994;125:333–343. doi: 10.1083/jcb.125.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram JC, Frame MJ, Brooks DR, Tetley L, Hutchison JE, Souza AE, Coombs GH. The multiple cpb cysteine proteinase genes of Leishmania mexicana encode isoenzymes which differ in their stage-regulation and substrate preferences. J Biol Chem. 1997;272:14285–14293. doi: 10.1074/jbc.272.22.14285. [DOI] [PubMed] [Google Scholar]
- Mottram JC, Kinnaird J, Shiels BR, Tait A, Barry JD. A novel CDC2-related protein kinase from Leishmania mexicana, lmmCRK1, is post-translationally regulated during the life-cycle. J Biol Chem. 1993;268:21044–21051. [PubMed] [Google Scholar]
- Mottram JC, Souza AE, Hutchison JE, Carter R, Frame MJ, Coombs GH. Evidence from disruption of the lmcpb gene array of Leishmania mexicana that cysteine proteinases are virulence factors. Proc Natl Acad Sci USA. 1996;93:6008–6013. doi: 10.1073/pnas.93.12.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Spithill TW, Handman E. The PSA-2 glycoprotein complex of Leishmania major is a glycosylphosphatidylinositol-linked promastigote surface antigen. J Immunol. 1989;143:4221–4226. [PubMed] [Google Scholar]
- Ralton JE, McConville MJ. Delineation of three pathways of glycosylphosphatidylinositol biosynthesis in Leishmania mexicana—precursors from different pathways are assembled on distinct pools of phosphatidylinositol and undergo fatty acid remodeling. J Biol Chem. 1998;273:4245–4257. doi: 10.1074/jbc.273.7.4245. [DOI] [PubMed] [Google Scholar]
- Ramalingam S, Maxwell SE, Medof ME, Chen R, Gerber LD, Udenfriend S. COOH-terminal processing of nascent polypeptides by the glycosylphosphatidylinositol transamidase in the presence of hydrazine is governed by the same parameters as glycosylphosphatidylinositol addition. Proc Natl Acad Sci USA. 1996;93:7528–7533. doi: 10.1073/pnas.93.15.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H, Conzelmann A. Glycosylphosphatidylinositol:protein transamidase. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. London: Academic Press; 1998. pp. 756–759. [Google Scholar]
- Schneider P, Ferguson MAJ, McConville MJ, Mehlert A, Homans SW, Bordier C. Structure of the glycosylphosphatidylinositol membrane anchor of the Leishmania major promastigote surface protease. J Biol Chem. 1990;265:16955–16964. [PubMed] [Google Scholar]
- Sharma DK, Vidugiriene J, Bangs JD, Menon AK. A cell-free assay for glycosylphosphatidylinositol anchoring in African trypanosomes. J Biol Chem. 1999;274:16479–16486. doi: 10.1074/jbc.274.23.16479. [DOI] [PubMed] [Google Scholar]
- Smith, T.K., Sharma, D.K., Crossman, T., Brimacombe, J.S., and Ferguson, M.A.J. (1999). Selective inhibition of the glycosylphosphatidylinositol biosynthetic pathway of Trypanosoma brucei. EMBO J. 5922–5930. [DOI] [PMC free article] [PubMed]
- Smith TK, Sharma DK, Crossman A, Dix A, Brimacombe JS, Ferguson MA. Parasite and mammalian GPI biosynthetic pathways can be distinguished using synthetic substrate analogues. EMBO J. 1997;16:6667–6675. doi: 10.1093/emboj/16.22.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons FM, Murray PJ, Ji H, Simpson RJ, Osborn AH, Cappai R, Handman E. Characterization of a polymorphic family of integral membrane proteins in promastigotes of different Leishmania species. Mol Biochem Parasitol. 1994;67:103–113. doi: 10.1016/0166-6851(94)90100-7. [DOI] [PubMed] [Google Scholar]
- Tiede A, Bastisch I, Schubert J, Orlean P, Schmidt RE. Biosynthesis of glycosylphosphatidylinositols in mammals and unicellular microbes. Biol Chem. 1999;380:503–523. doi: 10.1515/BC.1999.066. [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane-proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- Yu JL, Nagarajan S, Knez JJ, Udenfriend S, Chen R, Medof ME. The affected gene underlying the class K glycosylphosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc Natl Acad Sci USA. 1997;94:12580–12585. doi: 10.1073/pnas.94.23.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki J, Scholz C, Currie G, Coombs GH, McConville MJ. The glycoinositolphospholipids from Leishmania panamensis contain unusual glycan and lipid moieties. J Mol Biol. 1998;282:287–299. doi: 10.1006/jmbi.1998.2014. [DOI] [PubMed] [Google Scholar]