Abstract
Saccharomyces cerevisiae cnm67Δ cells lack the spindle pole body (SPB) outer plaque, the main attachment site for astral (cytoplasmic) microtubules, leading to frequent nuclear segregation failure. We monitored dynamics of green fluorescent protein–labeled nuclei and microtubules over several cell cycles. Early nuclear migration steps such as nuclear positioning and spindle orientation were slightly affected, but late phases such as rapid oscillations and insertion of the anaphase nucleus into the bud neck were mostly absent. Analyzes of microtubule dynamics revealed normal behavior of the nuclear spindle but frequent detachment of astral microtubules after SPB separation. Concomitantly, Spc72 protein, the cytoplasmic anchor for the γ-tubulin complex, was partially lost from the SPB region with dynamics similar to those observed for microtubules. We postulate that in cnm67Δ cells Spc72–γ-tubulin complex-capped astral microtubules are released from the half-bridge upon SPB separation but fail to be anchored to the cytoplasmic side of the SPB because of the absence of an outer plaque. However, successful nuclear segregation in cnm67Δ cells can still be achieved by elongation forces of spindles that were correctly oriented before astral microtubule detachment by action of Kip3/Kar3 motors. Interestingly, the first nuclear segregation in newborn diploid cells never fails, even though astral microtubule detachment occurs.
INTRODUCTION
During mitosis, precise positioning and migration of the nucleus as well as orientation of the spindle are crucial events for the successful propagation of the genetic material. In the genetically tractable budding yeast Saccharomyces cerevisiae, the dividing nucleus has to penetrate a small channel (bud neck) between mother and daughter cell. The migration of the nucleus to the bud neck and orientation of the mitotic spindle along the mother–daughter axis depend on dynamic forces that act on the nucleus via astral (cytoplasmic) microtubules (Huffaker et al., 1988; Sullivan and Huffaker, 1992; Carminati and Stearns, 1997; Shaw et al., 1997). Microtubules are nucleated by the γ-tubulin complex (Sobel and Snyder, 1995; Marschall et al., 1996), which is anchored to the spindle pole body (SPB) (Geissler et al., 1996; Spang et al., 1996b; Knop et al., 1997; Knop and Schiebel, 1998; Pereira et al., 1999), the yeast microtubule-organizing center. The SPB consists of three main, electron-dense layers: the inner, central, and outer plaques. The inner plaque serves as the attachment site for intranuclear microtubules, the outer plaque is the major structure for binding of astral microtubules (Byers, 1981; Rose et al., 1993; Winey and Byers, 1993; Kilmartin, 1994; Snyder, 1994; Bullitt et al., 1997).
Only recently research focused on another SPB substructure that is also capable of organizing astral microtubules: the half-bridge or, upon its duplication, the bridge (Brachat et al., 1998; Adams and Kilmartin, 1999; O'Toole et al., 1999; Pereira et al., 1999), which had already been described by Byers and Goetsch (1975). It lies adjacent to the central plaque within the nuclear envelope and initiates microtubules during the early part of the cell cycle before SPB duplication and continuing until SPB separation. For the rest of the cell cycle microtubules are organized by the SPB outer plaque (Byers and Goetsch, 1975). The cell cycle–dependent change of microtubule attachment to distinct SPB substructures is concomitant with different phases of nuclear migration. The nucleus is moved from an apparently random position in the mother cell to a region close to the mother–daughter boundary, the bud neck (nuclear positioning). The nucleus then rapidly elongates along the mother–bud axis at the onset of anaphase (fast spindle elongation). Proper orientation of the elongated nucleus and the mitotic spindle (spindle orientation) at this stage depends on the action of astral microtubules (Huffaker et al., 1988; Sullivan and Huffaker, 1992). In wild-type cells fast elongation of the anaphase nucleus is accompanied by its movement into the bud (spindle insertion). This phase is followed by rapid oscillations, with subsequent slow spindle elongation and final division of the nucleus, as concluded from in vivo video microscopy observations (Yeh et al., 1995).
The observation of mutant phenotypes such as spindle misorientation and nuclear division within the mother cell have identified several factors that contribute to precise nuclear migration in S. cerevisiae. Among them are cytoskeletal components and proteins that are thought to support the microtubule-based motor dynein (Magdolen et al., 1988; Haarer et al., 1990; Palmer et al., 1992; Eshel et al., 1993; Li et al., 1993; Clark and Meyer, 1994; McMillan and Tatchell, 1994; Muhua et al., 1994; Yeh et al., 1995). Several members of the kinesin class of microtubule-based motor proteins have also been associated with roles in nuclear migration and spindle orientation (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller et al., 1998), as well as two suggested microtubule–cortex mediators (Farkasovsky and Kuntzel, 1995; Miller and Rose, 1998).
We recently characterized Cnm67, a novel component of the SPB outer plaque, which is required for the formation of this structure (Brachat et al., 1998, Wigge et al., 1998). In cells lacking Cnm67, the SPB outer plaque was not detectable by electron microscopy, indicating that the main attachment site for astral microtubules was defective. Nevertheless, cnm67Δ cells were still able to organize long astral microtubules. By electron microscopy, Brachat et al. (1998) could show that in cnm67Δ mutants astral microtubules exclusively emanated from the bridge or half-bridge throughout the cell cycle. Therefore, cnm67Δ mutants were forced to rely solely on half-bridge- and bridge-organized microtubules for nuclear migration. These cells showed impaired spindle orientation and nuclear migration, leading to the accumulation of bi- and multinucleate cells (Brachat et al., 1998).
In this study, we investigate the underlying mechanism for both failed and successful nuclear migrations in cells lacking the SPB outer plaques. We analyze nuclear movements as well as astral and spindle microtubule dynamics during mitotic divisions in a large number of wild-type and cnm67Δ mutant cells by in vivo time-lapse microscopy. In addition, using a series of double mutants, we address the question of which astral microtubule motors and cortex interactions are required for the remaining successful nuclear migrations in this mutants. Our results not only allow us to explain the changes in nuclear dynamics in cnm67Δ cells but also point to a general cell cycle–dependent switch in astral microtubule organization.
MATERIALS AND METHODS
Strains, Media, and Yeast Transformation
Yeast strains used in this study are summarized in Table 1. Yeast media were prepared as described by Guthrie and Fink (1991). The yeast transformation procedure was based on the protocol by Schiestl and Gietz (1989). After the heat shock step, cells were pelleted and resuspended in 5 ml of YPD and incubated for 2 h at 30°C. Cells were again pelleted, resuspended in 1 ml of distilled H2O (dH2O), and plated on selective YPD-G418 medium (200 mg/l geneticin). The Escherichia coli strain XL1-blue (Bullock et al., 1987) was used to propagate plasmids.
Table 1.
Yeast strains used in this work
| Name | Genotype | Source or reference |
|---|---|---|
| FY 1679 | MATα/a ura3-52Δ1/ura3-52Δ1 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ200/HIS3 | B. Dujon |
| CEN.PK2 | MATα/a ura3-52Δ1/ura3-52Δ1 trp1-289/trp1-289 leu2-3, 112/leu2-3,112 his3Δ1/his3Δ1 | K. D. Entian |
| DHY6 | MATα/a HHF2::GFP-KanMX6/HHF2 ura3-52Δ1/ura3-52Δ1 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ 200/HIS3 | This study |
| DHY14 | MATα/a HHF2::GFP-KanMX6/HHF2 cnm67Δ1::KanMX4/cnm67Δ1::KanMX4 ura3-52Δ1/ura3-52Δ1 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ200/HIS3 | This study |
| DHY31 | MATα HHF2::GFP-His3MX6 ura3-52Δ1 TRP1 leu2Δ1 his3Δ200 | This study |
| DHY48 | MATα HHF2::GFP-His3MX6 cnm67Δ1::KanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY49 | MATa HHF2::GFP-His3MX6 cnm67Δ1::KanMX4 ura3-52Δ1 TRP1 LEU2 | This study |
| DHY57 | MATα HHF2::GFP-His3MX6 kip2Δ1::KanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY58 | MATα HHF2::GFP-His3MX6 kip2Δ1::KanMX4 cnm67Δ1::KanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY75 | MATα HHF2::GFP-His3MX6 kip3Δ1::kanMX4 ura3-52Δ1 trp1Δ63 leu2Δ1 | This study |
| DHY77 | MATa HHF2::GFP-His3MX6 kip3Δ1::KanMX4 cnm67Δ1::KanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY79 | MATα HHF2::GFP-His3MX6 kar3Δ1::KanMX4 cnm67Δ1::KanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY80 | MATa HHF2::GFP-His3MX6 kar3Δ1::KanMX4 ura3-52Δ1 TRP1 LEU2 | This study |
| DHY108 | MATa HHF2::GFP-His3MX6 dhc1Δ1::KanMX4 cnm67Δ1::KanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY109 | MATa HHF2::GFP-His3MX6 dhc1Δ1::KanMX4 ura3-52Δ1 TRP1 LEU2 | This study |
| DHY111 | MATα HHF2::GFP-His3MX6 yeb1Δ1::KanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY112 | MATa HHF2::GFP-His3MX6 she1Δ1::kanMX4 cnm67Δ1::kanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY113 | MATa HHF2::GFP-His3MX6 kar9Δ1::KanMX4 cnm67Δ1::KanMX4 ura3-52Δ1 trp1Δ63 leu2Δ1 | This study |
| DHY121 | MATα HHF2::GFP-His3MX6 kar9Δ1::KanMX4 ura3-52Δ1 trp1Δ63 leu2Δ1 HIS3?? | This study |
| DHY133 | MATα/a HHF2::GFP-His3 MX6/HHF2 bud9Δ1::KanMX4/bud9Δ1::KanMX4 cnm67Δ1::KanMX4/cnm67Δ1:kanMX4 ura3-52Δ1/ura3-52Δ1 TRP1/TRP1 LEU2/leu2Δ1 | This study |
| DHY134 | MATa HHF2::GFP-His3MX6 yeb1Δ1::kanMX4 cnm67Δ1::kanMX4 ura3-52Δ1 TRP1 leu2Δ1 | This study |
| DHY143 | MATα/a HHF2::GFP-His3 MX6/HHF2 bud8Δ1::KanMX4/bud8Δ1::KanMX4 cnm67Δ1::KanMX4/cnm67Δ1::KanMX4 ura3-52Δ1/ura3-52Δ1 TRP1/trp1Δ63 LEU2/leu2Δ1 | This study |
| DHY154 | MATα HHF2::GFP-His3MX6 num1Δ1::KanMX4 ura3-52Δ1 trp1Δ63 LEU2 | This study |
| DHY165 | MATα HHF2::GFP-His3MX6 num1Δ1::KanMX4 cnm67Δ1::kanMX4 ura3-52Δ1 trp1Δ63 LEU2 | This study |
| DHY179 | MATα cnm67Δ1::KanMX4 SPC72::GFP-His3MX4 ura3-52Δ1 trp1-289 leu2-3,112 his3Δ1 | This study |
| DHY194 | MATα ura3-52Δ1::GFP-TUB1-URA3(pAFS125) trp1Δ63 leu2Δ1 his3Δ200 | This study |
| DHY192 | MATα cln3-1::k1TRP1 HHF2::GFP-kanMX6 ura3-52Δ1 trp1Δ63 leu2Δ1 his3Δ200 | This study |
| DHY201 | MATα cnm67Δ1::k1TRP1 ura3-52Δ1::GFP-TUB1-URA3(pAFS125) trp1Δ63 leu2Δ1 his3Δ200 | This study |
| DHY209 | MATα/a ura3-52Δ1::GFP-TUB1-URA3(pAFS125)/ura3-52Δ1::GFP-TUB1-URA3(pAFS125) | This study |
DNA Manipulations and Strain Constructions
Standard DNA manipulations were performed as described by Sambrook et al. (1989). We applied a PCR-based method to construct gene deletion cassettes that were used in yeast transformations (Wach et al., 1994). DNA of E. coli plasmids pFA6-KanMX4 (Wach et al., 1994), pFA6-HIS3MX6, pFA6-GFP-KanMX6, pFA6-GFP-HIS3MX6 (Wach et al., 1997), and pYM3-klTRP1 (Knop et al., 1999) served as template for preparative PCR reactions. Correct genomic integration of the corresponding construct was verified by analytical PCR (Huxley et al., 1990; Wach et al., 1994). Yeast strains were grown on YPD-G418 (200 mg/l geneticin) to select for transformants that had integrated kanMX4 or GFP-kanMX6 deletion cassettes. Growth on SD plates lacking histidine and tryptophane selected for GFP-HIS3MX6, HIS3MX6, or klTRP1 integration.
For gene deletions we followed the EUROFAN guidelines (A. Wach, A. Brachat, and P. Philippsen [1996]. Guidelines for EUROFAN B0 program ORF deletants, plasmid tools, basic functional analyses, available at www.mips.biochem.mpg.de/proj/eurofan/index.html) for gene replacement in S. cerevisiae. The cnm67Δ1::kanMX mutant strain was constructed as published by Brachat et al. (1998). We used the following oligonucleotide pairs for generation of the kanMX4, His3MX6, klTRP1 deletion cassettes with flanking homologies to the target genes: dhc1Δ1, 5′-TACTCGTTCAGAGCTTAAATTGGAAAGTACGTCAAAACGTTTTTTAGGCAGGTCGACGGATCCCCGGG-3′ and 5′-TTTGAACCTGTTCTATACAATTTTGTATATCATCTTTTAG-TTTGA-TGACTACATCGATGAATTCGAGCTCG-3′; kip2Δ1, 5′-AGA-ACACT-TGATAAAATTCTTACCATAATACCACCATTGATGCGTACGCTGCAGGTCGAC-3′ and 5′-AACAATCCGCACAAGGAA-AAAAGCACCCGAGATCTGGGACCCCATCGATGAATTCGAGCT-CG-3′; kip3Δ1, 5′-CTTGAGTTTTCTTTCCAGCTGTATACTATTGACACTAACATGCGTACGCTGCAG-GTCGAC-3′ and 5′-AATGCT-GGCGGAAAGAAGTTATATTCGATAGTTTACGTAGGACATCGATG-AATTCGAGCTCG-3′; kar3Δ1, 5′-AAATAAAGGACTTAGAGAAATTCTGGCAACTATTAAACTATGCGTACGCTG-CAGGTCGAC-3′ and 5′-GTCTCTGTCATTTGTCAAAGGAGTGAAAAAGACCAGA-AAAGGCATCGATGAATTCGAGCTCG-3′; kar9Δ1, 5′-GTCTGTACAAGCCTTAAAGATTTCAGTAGCACTGCCATGCGTACGCTGCA-GGTCGAC-3′ and 5′-TATAAAAATGTATAACTATACAGTTTTAGGTTAGTATCACATCGATGAATTCGAGCTC-3′; num1Δ1, 5′-ATGTCCCACAACAACAGGCATAAAAAGAATAACGATAAAGACAGCGGTCGACGGATCCCCGGG-3′ and 5′-CTATCGTAAATTGCCAAATGATCGGCTTTGTGGTACTCTATGTCTTACATCGATGAATTCGAGCTCG-3′; bud8Δ1, 5′-CCAATATCCTCTTTCTACTTGAGAATTTTTTCGATTCTACATGAAGTGGTCGACGGATCCCCG-GG3′ and 5′-AGTTTTTTATTTTTTATCCTATTTGATGAATGAT-ACAGTTTCTTATTCCATCGATGAATTCGAGCTCG-3′; she1Δ1, 5-AATTCTAAAACACAAAAAAACAAAAAAAATCCTATAACCAGTTCTCCCGCGGTCGACGGATCCCCGGG-3′ and 5′-AGGATATA-TGTATATATACATATATACATATATGGGCGTATATTTACTTTGC-ATCGATGAATTCGAGCTCG-3′; yeb1Δ1, 5′-AGGCAGACTCAAA-AGCAAGGATAATATTCCACCAAATCAGGGACGAAGGGTCGA-CGGATCCCCGGG-3′ and 5′-ATAATACATATTCGAAAACAAT-ACTGCTTTTTAGTTCTCAACTTAAAACATCGATGAATTCGAGC-TCG-3′; cln3-1, 5′-ATTCTACCGTTCACAAGAATTGGAAACCA-TGTACAATACTATCTTTGCTCAGTAAAAAGTGGAACGATCATT-CA-3′ and 5′-GTGATGCAGAGTCTGGGGTCGTCATTGAACTTGA-A-GTCTTGTAAGGGGAACATCGATGAATTCGAGCTCG-3′; and cln3Δ1, 5′-AAGACACTGATTTGATACGCTTTCTGTACGATGGCC-ATATTGAAGGATACGCTTCGTACGCTGCAGGTCG-3′ and 5′ATTTATTTGTTGTTAAATGCATTTTTTTTTTGTCGTTTCAGCGAG-TTTTCCATCGATGAATTCGAGCTCG-3′.
C-terminal fusion of the S65T variant of green fluorescent protein (GFP) to Hhf2 (histone H4) was performed as described by Wach et al. (1997). This label was used in one copy in diploid strains. Growth rate and morphology of these strains were indistinguishable from those of wild type. A slight growth rate reduction could be observed in haploid strains. The haploid strains carrying the HHF2::GFPHis3MX6 label in addition to other genetic alterations were obtained by crossing DHY31 with the corresponding strains that were histidine prototrophic followed by sporulation and tetrad dissection. Selected strains that showed Hhf2-GFP fluorescence were prototrophic for histidine. We did not determine in each case whether these strains carried in addition the S. cerevisiae wild-type HIS3 allele or the mutant HIS3Δ200 allele.
We integrated a GFP (N-terminal fusion)–labeled copy of the TUB1 gene under control of the endogenous promoter into the URA3 locus as described by Straight et al. (1997) using plasmid pAFS125. Spindle and astral microtubules were clearly observable under the fluorescence microscope upon successful transformation. The suitability of this label for in vivo studies has already been demonstrated (Straight et al., 1997).
C-terminal fusion of the S65T variant of GFP to Spc72 was performed as described by Wach et al. (1997). The label was shown to be functional because it fully complemented the deletion phenotype and was generated by the use of the following oligonucleotide pair: SPC72-GFP, 5′-AGAGTGACTGAGTGTTACATTAAATATATTTATATATAAACGTATATATCATCGATGAATTCGAGCTCG-3′ and 5′-GAGTCATTGAGATCGAAACTTTTCAACCTATCAATCAACA-ATCCCGGTCGACGGATCCCCGGG-3′.
The cln3-1 mutant was generated by deleting the last 76 codons of the CLN3 gene and introducing a stop codon, thereby recreating the original cln3-1 mutation (Cross, 1988). To allow for correct termination of transcription we fused the terminator region of the HA tag of pYM3-klTRP1 (Knop et al., 1999) after the stop codon.
Generation of double mutants was achieved either by crossing of the single mutants followed by sporulation and dissection of the four-spored asci or by serial gene deletion using the kanMX4/His3MX6 with the klTRP1 cassette. Spore colonies were checked for GFP fluorescence visually and for presence of the corresponding deletion by analytical PCR.
Video Microscopy Setup
The video microscopy setup consisted of an Axioplan 2 microscope (Carl Zeiss, Feldbach, Switzerland), equipped with a 75W/XBO epifluorescence illumination source, a motorized stage, a Plan-Neofluar 100×, 1.3 oil PH3 objective, and a 2× camera magnification lens. HiQ FITC, DAPI, and TRITC filter sets were used (Chroma Technology, Brattleboro, VT). Because the spectrum of an XBO lamp shows a considerable emission in the infrared range, we doubled the normal heat absorption and reflection filters in the microscope to protect the specimen. Fluorescence excitation was controlled by a shutter controller in combination with a MAC2000 shutter system (Ludl controller MAC 2000; Ludl Electronics, Hawthorne, NY). We used a TE/CCD-1000PB back-illuminated cooled charge-coupled device camera mounted on the primary port and used in combination with the ST-133 controller (Princeton Instruments, Trenton, NJ).
Fluorescence Microscopy Procedures and Techniques
For in vivo time-lapse studies Hhf2-GFP– or GFP-Tub1–labeled strains were grown in YPD medium to early logarithmic phase. One milliliter of the culture was transferred to a reaction tube, pelleted, and resuspended in 1 ml of fresh YPD. Three microliters were then placed on top of the agarose surface of a ground well microscopy slide (Huber, Reinach, Switzerland). This had been prepared by placing a drop of hot growth medium (SD complete medium plus 1.7% agarose) onto the slide and covering it with a coverslip. To enrich the growth medium with oxygen, it had been vigorously vortexed before. As soon as the medium was solid, the coverslip was carefully pushed sideward, resulting in a very smooth agarose surface. Cells were placed on top and covered by another coverslip that was sealed to allow for long investigation times without liquid loss. Steady growth rates were observed until the whole layer was covered with a thin layer of cells up to 3 d later.
For DAPI staining, yeast cells were grown in YPD to early log phase. Approximately 5 × 107 cells were pelleted, resuspended in 50 μl of dH2O, and fixed for 5 min by addition of 1 ml of 70% ethanol. One microliter of a 1 mg/ml DAPI stock solution was added, and the suspension was incubated for 1 min at room temperature. Cells were subsequently washed twice in dH2O and mounted on a poly-l-lysine–treated slide for microscopy.
For visualization of bud and birth scars, cells were grown in YPD to early log phase. Approximately 5 × 107 cells were pelleted, washed with 1 ml of dH2O, resuspended in 100 μl of a 1 mg/ml calcofluor (fluorescence brightener; Sigma-Aldrich, Deisenhofen, Germany) solution, and incubated for 5 min at room temperature. Cells were washed in 1 ml of dH2O, pelleted, and resuspended in 50 μl of dH2O. Approximately 4 μl were mounted on a poly-l-lysine–treated slide for microscopy.
Membranes were stained using 1,1-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate (DiI; Fluka, Buchs, Switzerland). Cells were grown in YPD to early log phase. Approximately 5 × 107 cells were pelleted, washed three times in 1 ml of dH2O, and resuspended in 0.5 ml of dH2O, and 1.5 μl of a 1 mg/ml DiI solution were added. After 5 min of incubation at room temperature, cells were washed twice in 1 ml of dH2O and finally resuspended in 50 μl of dH2O. Approximately 4 μl were mounted on a poly-l-lysine-treated slide for microscopy.
Image Acquisition and Picture Processing
The microscope, camera, and fluorescence shutter were controlled by Metamorph 3.51 software (Universal Imaging, West Chester, PA). General Hhf2-GFP acquisition settings were 1-min interval time, 0.1-s exposure time, 3% illumination transmission/, one z-axis plane, and no binning. General GFP-Tub1 acquisition settings were 2-min interval time, 0.4-s exposure time, 50% illumination transmission, three z-axis planes spaced by 0.8 μm, and 2 × 2 binning. Using these conditions cells showed steady growth for >72 h. Nuclear and microtubule dynamics of individual cells could be tracked for more than eight divisions. Acquisition settings for the short GFP-Tub1 or Spc72-GFP studies were 15-s interval time, 1-s exposure time, 100% illumination transmission, three z-axis planes spaced by 0.8 μm, and no binning. The temperature of immersion oil on the microscopy slide near the sample was ∼24°C. The z-axis stacks were merged into one plane using the “stack arithmetic: maximum” command of Metamorph. The stored images were then scaled and converted to 8-bit files. A red look-up table was assigned to the phase-contrast image, and a green look-up table was assigned for the fluorescence image. The phase-contrast and fluorescence 8-bit planes were then overlaid using the built-in “Overlay” command with the default balance. For time-lapse analysis we then assembled the picture files to a movie in QuickTime format (Apple Computer, Cupertino, CA) with a frame rate of 10 frames per second using the Premiere 4.2 program (Adobe Systems Europe, Edinburgh, Scotland).
RESULTS
Nuclear Segregation in cnm67Δ Cells Depends on Correctly Positioned Nuclei and Oriented Spindles
The dynamic behavior of nuclear migration was assessed by in vivo time-lapse fluorescence microscopy of cells carrying a bright nuclear Hhf2-GFP label (GFP fused to histone H4), which we had previously introduced (Wach et al., 1997). This label colocalized with the conventional DAPI staining of nuclei. We developed a mounting procedure (see MATERIALS AND METHODS) that supported cell growth on microscopy slides over many generations. Using this procedure the analysis of nuclear dynamics in a large number of consecutive cell cycles became possible.
We compared distinct steps of nuclear migration between diploid wild-type and cnm67Δ mutant cells: nuclear positioning (movement of the nucleus to the bud neck before anaphase), nuclear orientation during anaphase (alignment of the elongating nucleus along the mother–bud axis, equivalent to spindle orientation; see below), insertion of the anaphase nucleus into the bud neck (equivalent to spindle insertion), fast and slow nuclear elongation (equivalent to fast and slow spindle elongation, as described by Yeh et al., 1995), separation into two nuclei, and postanaphase movements. Each of 120 observed wild-type cell cycles displayed these nuclear dynamic steps in a very similar mode with the exception of occasionally delayed mitoses (Movie 1). A representative example is shown in Figure 1, which also indicates that completion of cell separation can be monitored by rotational movement of the daughter relative to the mother cell. Time-lapse studies of 325 cnm67Δ mutant cell cycles revealed characteristic alterations in some nuclear migration phases, leading to bi- and multinucleate cells within a few generations (Movie 2), with representative examples shown in Figure 2. We first describe our analyses of nuclear dynamics in single nucleate cells; the fate of multinucleate cells and empty buds will be discussed in later sections.
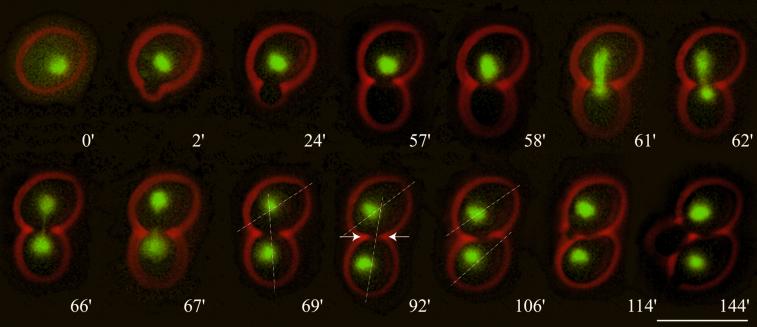
Figure 1.
Nuclear migration dynamics of diploid Hhf2-GFP–labeled wild-type cells observed by in vivo time-lapse microscopy. (0′) At onset of bud emergence the nucleus is positioned randomly. (2′–57′) During the positioning phase the nucleus moves to the bud neck region. (58′) The nucleus starts to elongate, marking the onset of anaphase B. (58′–62′) In a rapid process the nucleus becomes bar shaped, elongates along the mother–bud axis, and starts to penetrate the bud neck. This corresponds to spindle orientation, elongation, and penetration. (62′–67′) The labeled chromosomal material transforms into an hourglass shape. This phase is typically accompanied by bending and rapid oscillations of the nucleus within the bud neck. This can be seen by the fast change in position of the elongated nucleus. (69′) Final separation of chromosomal masses. At this time point the separated chromosomal masses show slow coordinated movements indicating that the spindle is still intact. (92′) Onset of cytokinesis can be observed by a slight change of the orientation of the longitudinal axis of the daughter cell relative to that of the mother and by a constriction of the bud neck (indicated with arrows). (106′) A sudden change of the orientation of the daughter cell relative to that of the mother indicates completion of cell separation. (114′) Next bud emergence of the mother cell. (144′) First distal bud emergence of the daughter cell. Bar, 10 μm. Movie 1: Nuclear dynamics as seen in Hhf2-GFP–labeled wild-type cells followed for >10 h. Two delayed mitoses can be seen in the second half of the time-lapse sequence. Acquisition interval, 1 min; movie speed, 10 frames per second = 10 min/s. One z-axis plane fluorescence image was acquired.
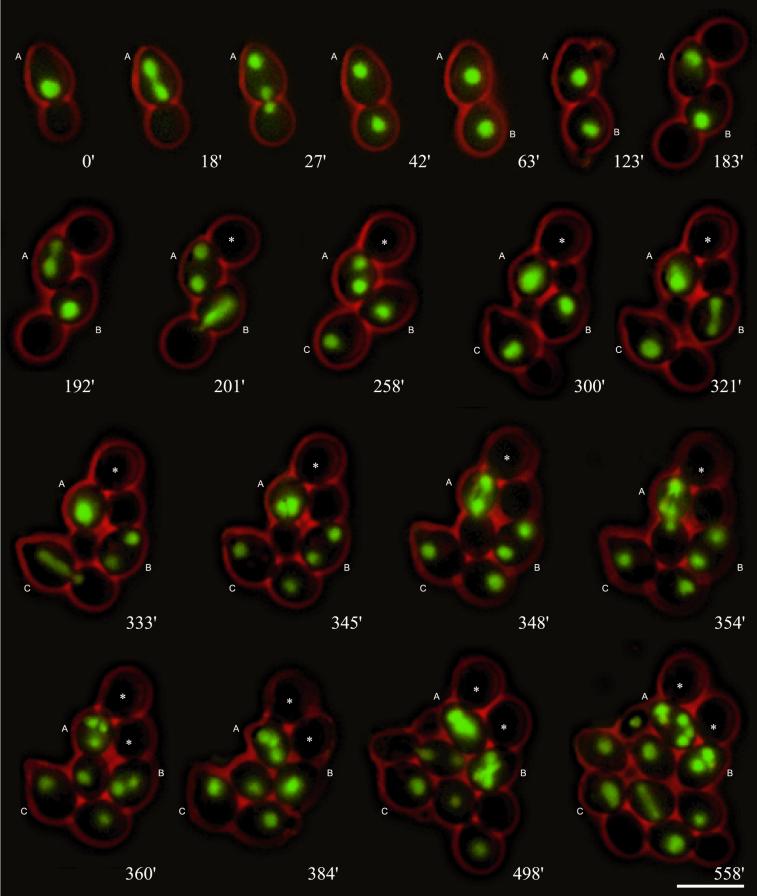
Figure 2.
Nuclear migration dynamics of diploid cnm67Δ cells. Mitotic cell cycles were observed by in vivo time-lapse fluorescence microscopy. Nuclear positioning of the preanaphase nucleus to the bud neck is usually similar to wild type (e.g., 0′, cell A). Unlike in wild type, spindle elongation during anaphase is mostly restricted to the mother cell (e.g., 18′, cell A). Elongation of the anaphase spindle usually occurs along the mother–bud axis (e.g., 18′, cell A) and is normally followed by a successful nuclear movement into the bud (e.g., 27′ and 42′, cell A). In case of spindle misalignment (192′, cell A), the subsequent nuclear migration often fails (201′–345′, cell A) leading to a binucleate cell with an anucleate bud. Nuclear divisions in binucleated cells occur simultaneously (354′, cell A, and 498′, cell B). After successful transit of a nucleus into the bud, separation can be observed by a change of the position of the bud relative to the mother cell during cytokinesis (e.g., 42′ and 63′, cell A). Anucleate buds (marked with asterisks) are not separated from the mother cell (cell A from 300′ and cell B from 360′). Bar, 10 μm. Movie 2: Nuclear dynamics as seen in Hhf2-GFP–labeled cnm67Δ cells followed for 8 h. Acquisition interval, 1 min; movie speed, 10 frames per second = 10 min/s. One z-axis plane fluorescence image was acquired.
A quantitative evaluation revealed that correct nuclear positioning of the preanaphase nucleus was accomplished in 73% of cnm67Δ mutant cells and that 75% of all mutant cells elongated the nucleus along the mother–daughter axis. Only 13.5% of cnm67Δ cells showed early insertion of the anaphase nucleus into the bud neck, whereas this was always the case for wild-type cells. Hence elongation of the anaphase nucleus was usually limited to the mother cell in cnm67Δ mutants. Rapid oscillations and bending of the elongated nucleus typical for anaphase in wild-type cells were completely absent in the mutant, indicating weakened forces acting on the nuclei at this cell cycle stage.
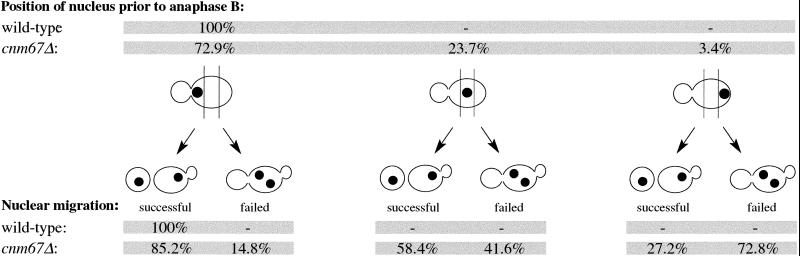
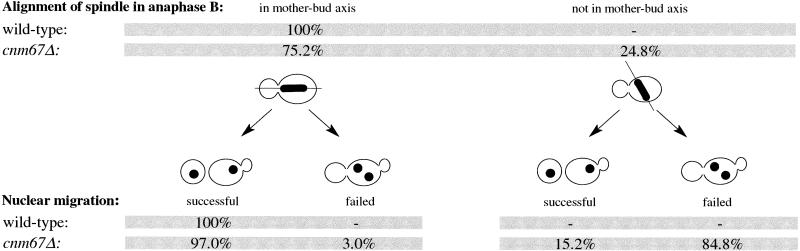
The analyses of pedigrees also allowed us to answer the question of which degree failed nuclear positioning or spindle misorientation influenced distribution of nuclei between mother and daughter cells. If the preanaphase nucleus was positioned close to the bud neck, successful nuclear migration was achieved in most cnm67Δ cells, although it was often delayed. A failure in nuclear positioning to the bud neck resulted in a high rate of nuclear mis-segregation leading to binucleate mother cells and anucleate buds. Data are summarized in Table 2. If the spindle had been aligned along the mother–bud axis, almost all mutant cells showed a successful nuclear migration. If the spindle had been misaligned, only few cells succeeded in directing a nucleus into the daughter cells. Data are summarized in Table 3. In conclusion, successful nuclear migration in cnm67Δ cells was dependent on the correct execution of early nuclear migration steps such as nuclear positioning and spindle orientation.
Table 2.
Dependency of nuclear migration success on nuclear positioning before anaphase B in wild-type and cnm67Δ cells
The relation between nuclear positioning and subsequent nuclear migration success was analyzed by time-lapse fluorescence microscopy of diploid Hhf2-GFP–labeled cells. In all cases the last time point before nuclear elongation was taken into account for the depicted nuclear positioning. Nuclear migration was counted as failed when the nucleus had not entered the bud at the time the cell started a new cell cycle with new bud emergence. Later entry of a nucleus into a bud from a previous cell cycle was not observed. (wild-type, n = 120; cnm67Δ, n = 325).
Table 3.
Dependency of nuclear migration success on spindle orientation in wild-type and cnm67Δ cells
The relation between spindle orientation and subsequent nuclear migration success was analyzed by time-lapse fluorescent microscopy of diploid Hhf2-GFP–labeled cells. Spindle orientation was counted as aligned along the mother–bud axis when the elongated nucleus pointed into the bud neck. Nuclear migration was counted as failed when the nucleus had not entered the bud at the time the cell started a new cell cycle with new bud emergence. Later entry of a nucleus into a bud from a previous cell cycle was not observed (wild-type, n = 120; cnm67Δ, n = 319).
Astral Microtubules Detach in cnm67Δ Cells after SPB Duplication
Nuclear migration is achieved by forces acting on the nucleus via astral microtubules (Huffaker et al., 1988; Sullivan and Huffaker, 1992; Carminati and Stearns, 1997; Shaw et al., 1997). Therefore, it was important to investigate whether the observed nuclear segregation phenotype in cnm67Δ could be explained by changes in astral microtubule dynamics. Using a GFP-Tub1 fusion (Straight et al., 1997), we constructed diploid wild-type and cnm67Δ cells with fluorescently labeled microtubules. We monitored astral microtubules by time-lapse studies over several generations (Movies 3 and 4) and additionally by acquisition of still images of live cells from log phase cultures (Figures 3 and 4). Morphology and dynamics of the astral microtubule arrays early in the cell cycle (G1 and early S phase) were similar in wild-type and cnm67Δ mutant cells (Movies 3 and 4; Figures 3A and 4A). For this phase we could confirm in wild-type and mutant cells the previously described search-and-capture mechanism of astral microtubules making contact with the nascent bud, followed by interaction of microtubule bundles with the bud cortex and movement of the nucleus toward the bud neck (Shaw et al., 1997).
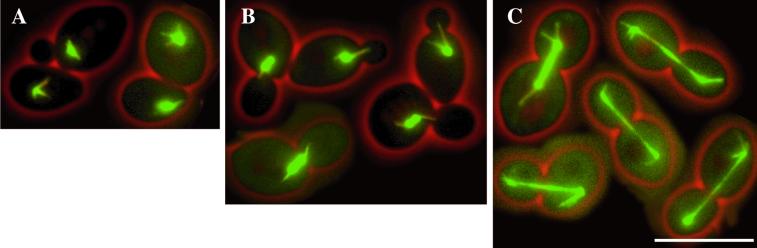
Figure 3.
Microtubule morphology in different cell cycle stages in wild-type cells. (A) Early in the cell cycle an aster of astral microtubules is visible, eventually interacting with the cortex. (B) Later in the cell cycle the spindle poles separate, and a short, thick spindle is formed. Bundles of astral microtubules interact with the bud, position the nucleus, and orient the spindle. (C) Anaphase, elongation of the spindle. Short, thick bundles of astral microtubules interact with the cortex. The pictures represent merged still images of four z-axis planes of selected GFP-Tub1–labeled wild-type cells. Bar, 10 μm. Movie 3: Microtubule dynamics as seen in GFP-Tub1–labeled wild-type cells followed for >7 h. Acquisition interval, 2 min; movie speed, 10 frames per second = 20 min/s. Three z-axis plane fluorescence images were acquired and merged.
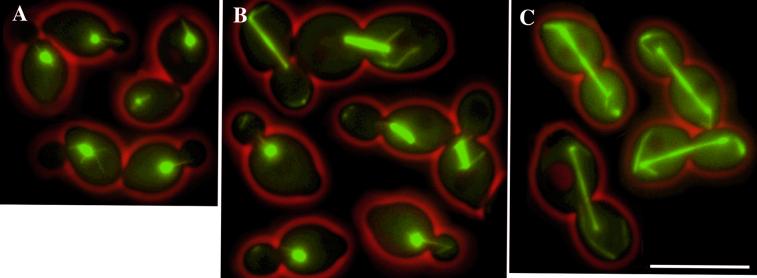
Figure 4.
Microtubule morphology in different cell cycle stages in cnm67Δ cells. (A) Early in the cell cycle the aster of astral microtubules is similar to wild type. (B) Upon spindle formation bundles of microtubules detached from the SPB appear in the buds. (C) At anaphase bundles of detached cytoplasmic microtubules are located in the bud and the mother cell. Some SPBs organize newly nucleated cytoplasmic microtubules. The pictures represent merged still images of four z-axis planes of selected GFP-Tub1–labeled cnm67Δ cells. Bar, 10 μm. Movie 4: Microtubule dynamics as seen in GFP-Tub1–labeled cnm67Δ cells followed for 6 h. Acquisition interval, 2 min; movie speed, 10 frames per second = 20 min/s. Three z-axis plane fluorescence images were acquired and merged.
After the onset of SPB separation and the formation of a thick, short spindle, microtubules detached from the SPB in all 56 analyzed cnm67Δ cells (Movie 4; Figure 4, B and C). In 47% of the cases this was accompanied by a slight movement of the nucleus away from the bud neck. The microtubules that were detached from the SPB still interacted with the bud–cortex in a manner that had been described as a “sweeping mechanism” (Carminati and Stearns, 1997). We observed that the tip of a microtubule touched the cortex, moved back and forth, and started to align along the bud cortex. This resulted in the translocation of the entire microtubule into the bud. Detached astral microtubules in the bud appeared to be very stable. Detachment of microtubules occurred from both SPBs, but we observed a five times higher detachment frequency from the SPB destined for the bud. The formation of new astral microtubules at the SPBs could be observed in all cnm67Δ cells, which detached again in 87% of the cells. A higher resolution of this detachment process, 15 s instead of 2 min, can be viewed in Movies 5 and 6, with 12 representative frames of Movie 5 depicted in Figure 5. In wild-type cells we never observed detachment of astral microtubules, and such an event has not been reported in previous studies focusing on astral microtubule dynamics (Carminati and Stearns, 1997; Shaw et al., 1997).
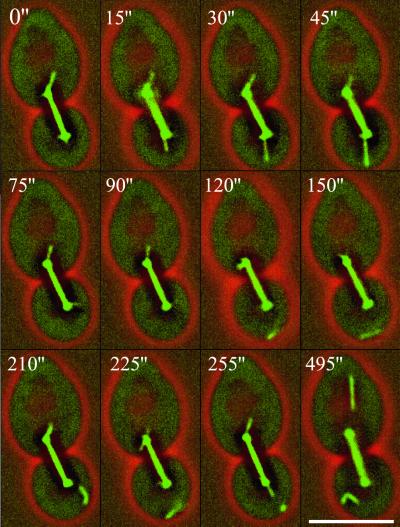
Figure 5.
Microtubule detachment as seen in short time-lapse sequences in cnm67Δ cells. One astral microtubule bundle is formed at the SPB located in the bud and contacts the bud–cortex (0"–45"). Upon contact with the cortex the bundle rapidly changes its orientation (45" and 75"). Suddenly the bundle is no longer visible, although the spindle is perfectly in focus (90"). The microtubule bundle is detected again but does not show any connections to the SPB and is moving along the cortex (120"–225"). New microtubules at the SPB in the bud are formed (255"). Although the spindle is clearly captured by this z-image series, both SPBs are devoid of astral microtubules, indicating new detachment (495"). This is supported by two detached microtubule bundles that are localized in the bud and one in the mother cell. Bar, 5 μm. Movies 5 and 6: Microtubule detachment as seen in GFP-Tub1–labeled cnm67Δ cells followed for 14 and 22 min, respectively. Acquisition interval, 15 s; movie speed, 10 frames per second = 2.5 min/s. Three z-axis plane fluorescence images were acquired and merged.
The ends of astral microtubules at the SPB are capped by the γ-tubulin binding complex, which is anchored by Spc72 to the half-bridge or bridge and the SPB outer plaque (Knop and Schiebel, 1998, Pereira et al., 1999). In wild-type cells, an Spc72-GFP fusion protein exclusively localizes to the SPBs (Soues and Adams, 1998). We monitored the localization of an Spc72-GFP fusion protein in a total of 100 cnm67Δ cells. In G1 cells Spc72-GFP exclusively localized to the SPB as in small budded cells with just separated SPBs (Figure 6A). After further separation of the SPBs, we detected additional spots of Spc72-GFP clearly separated from the nuclear periphery (Figure 6B). In time-lapse experiments we observed that these new Spc72-GFP spots detached from the SPBs, displayed straight movement into the bud, and moved along the bud cortex with kinetics very similar to that described above for detached microtubules. One representative example of Spc72-GFP loss observed at high time resolution is shown in Movie 7.
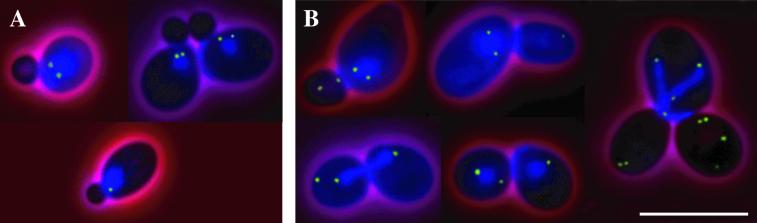
Figure 6.
Spc72-GFP and DNA localization in cnm67Δ cells. (A) In small budded cells before or after SPB separation, the Spc72-GFP signal localizes to bright spots close to the nuclear periphery corresponding to the SPBs. (B) During later cell cycle stages most Spc72-GFP still localizes to the nuclear periphery, but spots of Spc72-GFP signal can be detected in the bud not associated with the nuclear periphery. This is obvious in a binucleate anaphase cell with two buds (right panel). The larger bud from the previous cell cycle does not contain DNA (consistent with two anaphase nuclei in the mother cell), but Spc72-GFP can be detected in this bud. Also, the new bud already shows Spc72-GFP spots not associated with DNA. The DNA was stained with DAPI. These still images consist of five FITC z-axis planes (green), one DAPI plane (blue), and one phase-contrast image (red) merged into one plane. Bar, 10 μm. Movie 7: γ-Tubulin complex detachment as seen in Spc72-GFP–labeled cnm67Δ cells followed for 15 min. Acquisition interval, 15 s; movie speed, 10 frames per second = 2.5 min/s. Three z-axis plane fluorescence images were acquired and merged.
Our observations suggest that microtubules including Spc72 and thus the γ-tubulin complex detach from the SPBs after SPB separation. The presence of the γ-tubulin complex at one end of detached microtubules is also supported by the observed stability of these microtubules, as apparent in Movies 4–6. The observed time point of detachment, after the onset of SPB separation, allows the conclusion to be drawn that astral microtubule-directed forces on the nucleus are most likely absent or at least severely impaired in later phases of cell cycles in cnm67Δ cells.
Synthetic Effects in cnm67Δ Double Mutants
Dynein and three kinesin-related proteins have been identified in S. cerevisiae as motor proteins exerting forces on the nucleus via astral microtubules (Meluh and Rose, 1990; Eshel et al., 1993; Li et al., 1993; Carminati and Stearns, 1997; Cottingham et al., 1997, DeZwaan et al., 1997; Saunders et al., 1997; Miller et al., 1998). To determine which of these motor proteins is essential or important for the observed remaining nuclear segregation in cells lacking the SPB outer plaque, we deleted genes encoding these motors in a cnm67Δ background. None of the double deletions was lethal, indicating that, as in wild-type cells, none of the four motors is essential. As a measure of a change in nuclear migration failure in the double mutants compared with the cnm67Δ single mutant, we determined the frequency of bi- and multinucleate cells in log phase cultures and investigated growth on solid media. Data are summarized in Table 4.
Table 4.
Nuclear segregation failure and growth of wild-type, cnm67Δ single, and double mutants
| Strain | Sum of bi- and multinucleate cells (%) | Increase in bi- and multinucleate cells relative to cnm67Δ (%) | Growth of cnm67Δ double mutants relative to cnm67Δ |
|---|---|---|---|
| Wild type | 0.8 | ||
| cnm67Δ | 33.0 | Slow growth | |
| Microtubule motors | |||
| dhc1Δ | 9 | ||
| dhc1Δ cnm67Δ | 30.9 | −6.4 | Unchanged |
| kip3Δ | 3.0 | ||
| kip3Δ cnm67Δ | 39.5 | +19.7 | Much reduced |
| kar3Δ | 3.0 | ||
| kar3Δ cnm67Δ | 41.0 | +24.2 | Much reduced |
| kip2Δ | 5.0 | ||
| kip2Δ cnm67Δ | 25.0 | −24.2 | Improved |
| Microtubule-cortex interaction mediators | |||
| kar9Δ | 6.0 | ||
| kar9Δ cnm67Δ | 37.5 | +13.6 | Reduced |
| num1Δ | 4.0 | ||
| num1Δ cnm67Δ | 25.5 | −22.7 | Improved |
The number of Hhf2-GFP–labeled nuclei was determined in haploid mutant strains in exponential growth phase. More than 800 cells were counted for each strain. For comparison of growth the corresponding single and double mutants were streaked on the same plate containing solid full medium. The plates were incubated at 30 and 20°C for 72 h. cnm67Δ cells showed a clear slow growth phenotype; all other single mutants showed only slightly reduced growth compared with wild-type.
Loss of Dhc1, the large subunit of dynein, in a cnm67Δ background only caused a marginal change in the frequency of bi- and multinucleate cells, and we did not observe differences in growth compared with the cnm67Δ single mutant. Initial time-lapse analyzes of dhc1Δ cnm67Δ cells did not point toward differences in nuclear migration or frequency of microtubule detachment compared with cnm67Δ cells (our unpublished results). This result is on one hand expected because dynein acts late in nuclear migration (Carminati and Stearns, 1997; Cottingham and Hoyt, 1997) after the time point of astral microtubule detachment. On the other hand, it also shows that the process of detachment is dynein independent. Loss of the kinesin-related proteins Kip3 or Kar3 in a cnm67Δ mutant background increased the frequency of bi- and multinucleate cells by 20 and 24%, respectively, and led to severely reduced growth compared with the cnm67Δ single mutant. This enhanced failure rate in nuclear migration is not unexpected, because Kip3 and Kar3 have been suggested to function early in the cell cycle in nuclear positioning and spindle orientation (Cottingham and Hoyt, 1997; DeZwaan et al., 1997). Indeed, in initial time-lapse studies of kar3Δ cnm67Δ cells, we observed a high portion of cells that failed to localize the nucleus near the bud neck before anaphase B and that did not correctly align the spindle (our unpublished results). In addition, these movies showed that astral microtubule detachment was similar to the single mutant. An interesting case is the kinesin-related protein Kip2, which was described as counteracting force to Kip3 and also Dhc1 with the potential of moving the nucleus from the bud neck back into the mother cell (Cottingham and Hoyt, 1997; Huyett et al., 1998). Not unexpectedly, loss of Kip2 in the cnm67Δ background resulted in a significant improvement of nuclear migration (24% less bi- and multinucleate cells) and to improved growth compared with the cnm67Δ single mutant.
To generate forces on the nucleus, the distal ends of astral microtubules have to interact with the bud cortex or the mother cell cortex, respectively (Carminati and Stearns, 1997). Kar9 has been suggested to mediate astral microtubule–cortex interactions in the bud and Num1 in the mother cell (Farkasovsky and Kuntzel, 1995; Miller and Rose, 1998). This implies that loss of Kar9 may impair pulling forces acting on the nucleus toward the bud, and loss of Num1 may impair those acting in the opposite direction. We deleted KAR9 or NUM1 in a cnm67Δ background and determined the frequency of bi- and multinucleate cells as well as growth on solid media. Data are summarized in Table 4, revealing for kar9Δ cnm67Δ cells an increase in bi- and multinucleate cells and reduced growth compared with cnm67Δ. In contrast, num1Δ cnm67Δ mutants showed improved nuclear migration (23% less bi- and multinucleate cells) and concomitantly improved growth very similar to those in kip2Δ cnm67Δ cells.
Spindle Elongation Provides the Pushing Force for Nuclear Transfer in cnm67Δ Cells
In cnm67Δ cells transit of a nucleus through the bud neck mostly occurred after detachment of astral microtubules. This raised the question of how the forces for bud neck penetration were established. Analysis of our time-lapse data revealed that, unlike in wild-type, initial spindle elongation in cnm67Δ mutants was mostly restricted to mother cells (Movies 2 and 4). Only when the spindle was as long as the diameter of the mother cell and continuing to elongate could we observe transit of one SPB through the bud neck. Successful insertion of a nucleus during this process was dependent on the correct orientation of the elongating spindle, as depicted in Table 3. The potential of spindle dynamics to generate pushing forces became apparent in cases in which the spindle was misaligned: it continued to elongate despite spatial constrictions by the mother cell, resulting in bent spindles. We observed bent spindles aligned along the cortex with >1.5 times the length of the mother cell diameter (Movie 4). The dependence of successful nuclear segregation on the pushing forces of elongating spindles was supported by the observation that in 56 cnm67Δ divisions we never observed transit of a nucleus through the bud neck after spindle disassembly.
We also addressed the question of whether spindle dynamics are influenced by loss of the Cnm67 protein. We measured spindle dynamics by two methods: directly in GFP-Tub1–labeled cells and indirectly by the kinetics of chromatin separation as visualized by Hhf2-GFP. In Hhf2-GFP–labeled cells the round-shaped nucleus became bar shaped during spindle elongation (Figure 1, 61 min) and elongated further, adapting a typical hourglass-like shape (Figure 1, 66 min) before the chromosomal masses were focused into two spherical structures with very similar GFP intensities (Figure 1, 69 min). We compared the length of these distinct elongated nuclear shapes and the dynamics of their formation in wild-type and cnm67Δ cells (Table 5). In both strains we measured very similar parameters. In addition, from the onset of anaphase to final spindle disassembly (apparent in GFP-Tub1–labeled cells of Movies 3 and 4), we measured similar time intervals and maximal spindle lengths in both strains.
Table 5.
Dynamics of chromatin separation and spindle elongation in diploid wild-type and cnm67Δ cells
| Formation of
bar-shaped nucleus (Hhf2-GFP label)
|
Formation of
hourglass nucleus (Hhf2-GFP label)
|
Spindle
elongation (GFP-Tub1 label)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Maximal length (μm) | Time required (min) | No. of cells measured | Maximal length (μm) | Total time required (min) | No. of cells measured | Maximal length (μm) | Total time required (min) | No. of cells measured | |
| Wild-type | 5.5 (ς = 0.17) | 4.2 (ς = 0.5) | 30 | 7.9 (ς = 0.2) | 11 (ς = 2.1) | 30 | 9.47 (ς = 0.54) | 23.8 (ς = 2.4) | 50 |
| cnm67Δ | 5.4 (ς = 0.19) | 3.8 (ς = 0.4) | 30 | 8.1 (ς = 0.2) | 10 (ς = 2.7) | 30 | 9.56 (ς = 0.56) | 26.2 (ς = 2.8) | 50 |
The average fluorescent diameter of the round-shaped Hhf2-GFP–labeled nuclei was 2.5 μm (ς = 0.16; n = 50); the average-length of the short GFP-Tub 1 labeled spindle prior to elongation was 1.9 μm (ς = 0.14; n = 50). The last frame of a time-lapse sequence showing a round-shaped nucleus or unelongated spindle before anaphase was taken as time 0 min.
In summary, our data support the notion that unaltered spindle elongation dynamics contribute the main force for transit of one nucleus through the bud neck in cnm67Δ cells.
Consequences of Failed Nuclear Migrations
cnm67Δ cells that failed to direct one nucleus into the growing bud did not show a permanent growth arrest but initiated a new cell cycle, as seen by new bud emergence, SPB duplication, and spindle assembly. Multiple nuclei in cnm67Δ cells underwent simultaneous mitotic divisions in the next cell cycle (Figure 2, 348 min; Movies 2 and 4). This led to a large number of cells that carried more than two nuclei, as depicted in Table 4. Transfer of more than one nucleus into the new bud was not observed in cnm67Δ cells (n = 325). Interestingly, deposition of a nucleus into a still attached anucleate bud of a previous cell cycle was also not detected (n = 325). In addition, we detected no transfer of detached microtubules or Spc72-GFP into a bud of a previous cell cycle. cnm67Δ cells with >12 nuclei were observed giving rise to single nucleate daughter cells that followed mutant characteristics described above.
We were also able to follow cell separation in wild-type and cnm67Δ cells, which was apparent by a sudden change in the position of the bud relative to that of the mother cell (Figures 1 and 2; Movies 1–4). In wild-type and cnm67Δ cells separation of nucleate buds promptly occurred ∼30 min after entry of the nucleus into the bud. However, we never observed separation of anucleate buds in cnm67Δ cells (Figure 2, 192–558 min, cell A, 354–558 min cell B, Movie 2 and 4). The process of cytokinesis and separation of new daughter cells did not change the attachment of anucleate buds generated in previous cell cycles as concluded from unchanged positions of these buds relative to the mother cell (Figure 2, 354–384 min cell A, 498–558 min, cell B, Movies 2 and 4). Our observations point toward a link between successful nuclear migration and cell separation in cnm67Δ mutants.
The First Nuclear Segregation of a Diploid cnm67Δ Daughter Cell Is Always Successful
One striking observation during analyzes of consecutive cell cycles revealed that the first nuclear migration of a newborn daughter cell was always successful (n = 768) (Figure 2, cells B and C; Movies 2 and 4). To our knowledge this phenomenon has not been described previously. It did not depend on 1) the number of nuclei in the mother, 2) astral microtubule detachment, or 3) the age of the mother that had generated a daughter cell.
We investigated the involvement of other known mother–daughter-specific differences such as bud site selection or cell size in generating the observed mother–daughter-specific division characteristic in cnm67Δ cells. It was shown that the first bud emergence of a diploid daughter cell usually occurs distally from the birth scar, whereas later mother cell divisions follow a bipolar pattern (Freifelder, 1960; Chant and Pringle, 1995). The distal pole of a daughter cell is usually the region where the nucleus is positioned by the spindle elongation process during the previous cell cycle. Hence a distal first bud emergence might facilitate nuclear positioning in cnm67Δ cells. We investigated whether the high fidelity of first daughter nuclear migration events decreased in cells with a first proximal bud emergence. Mutation of BUD8 was previously shown to lead to a proximal budding pattern in diploid cells (Zahner et al., 1996). We deleted both BUD8 alleles of a diploid homozygous cnm67Δ mutant. Of 67 first proximal daughter divisions analyzed by time-lapse microscopy, we never observed nuclear segregation failure. In addition, growth on solid medium and the number of multinucleate cells in log phase cultures did not suggest a worsening of the nuclear migration defect (our unpublished results). Thus the high fidelity of nuclear migration during the first division of a diploid daughter cell was not directly linked to distal first bud emergence.
Another general mother–daughter difference consisted of the size of the cells. We tested whether the high fidelity in first nuclear segregations of daughter cells was linked to their smaller cell size. We deleted the CLN3 gene in a cnm67Δ background. It has been shown that lack of this cyclin delays START (Nash et al., 1988). Therefore, daughter cells have more time to grow and reach larger cell sizes before they enter the first cell cycle. Measuring the size of daughter cells in the cnm67Δ cln3Δ background indeed revealed larger-daughter cell size at the time of the first bud emergence (6.7 μm; ς = 0.42; n = 25) compared with cnm67Δ daughter cells (5.3 μm; ς = 0.44; n = 30). Although cnm67Δ cln3Δ daughter cells were even bigger than cnm67Δ mother cells (5.6 μm; ς = 0.42; n = 30), all 42 first daughter cell divisions observed by time-lapse microscopy were successful. Growth on solid medium and the number of bi- and multinucleate cells in log phase cultures was similar to cnm67Δ single mutants (our unpublished results). In addition, we created a cln3-1 mutant (Cross, 1988) in a cnm67Δ deletion background. This truncated form of the Cln3 protein lacks the C-terminal destruction box, and the high cellular levels of Cln3 protein promoted early START (Cross, 1988), leading to a smaller daughter cell size at the moment of the first bud emergence (4.9 μm; ς = 0.42; n = 25). Although daughter cells still showed a successful first division (n = 48), nuclear migration in mother cells failed with the normal cnm67Δ frequency, although cln3-1 cnm67Δ mother cells were smaller (5.4 μm; ς = 0.44; n = 25) than cnm67Δ daughter cells. Our time-lapse data were consistent with the nuclear segregation defect as determined for log phase cultures and the growth on solid medium (our unpublished results). Our observations did not point to a direct link between cell size and the site of the first bud emergence or the high fidelity of first nuclear migrations in daughter cells.
DISCUSSION
In this study we investigated the underlying mechanism for failed and successful nuclear migration in cells lacking the SPB outer plaques. Our observation of nuclear and astral microtubule dynamics suggested that the main and primary reason for most phenotypic changes in nuclear migration in cnm67Δ cells was a cell cycle–dependent detachment of astral microtubules from the SPB after onset of SPB separation. This led to a decrease or lack of forces acting on the nucleus at a critical phase of the cell cycle, resulting in a failure of early insertion of anaphase spindles into the bud necks, complete lack of rapid spindle oscillations, spindle misorientations, and, consequently, failed nuclear migrations. Successful nuclear migration in cnm67Δ cells was achieved only when the preanaphase nucleus was positioned near the bud neck and the spindle was correctly aligned early in the cell cycle before astral microtubule detachment. In such cells, the elongating spindle often still pointed into the bud when astral microtubule detachment was observed. Further elongation of the spindle forced the insertion of one SPB into the bud, which led to successful nuclear migration. This remaining ability to properly orient the spindle in a substantial number of cnm67Δ cells is essential for viability. Cells that are unable to orient the spindle because of complete lack of astral microtubules, as in tub2-104 mutants (Huffaker et al., 1988) or cnm67Δ kar1Δ mutants (Pereira et al., 1999), are inviable despite the presence of a functional spindle.
Astral microtubule detachment in cnm67Δ cells started at a cell cycle stage at which a major astral microtubule reorganization occurs, as already described in the pioneering work by Byers and Goetsch (1975): in wild-type cells with duplicated but unseparated SPBs, long astral microtubules emanate from the bridge, and after SPB separation astral microtubules are organized by the SPB outer plaque. Astral microtubules are anchored to these distinct SPB substructures via Spc72 (Knop and Schiebel, 1998; Pereira et al., 1999) that binds the γ-tubulin complex, which is necessary for nucleation of microtubules (Sobel and Snyder, 1995; Marshall et al., 1996; Knop and Schiebel, 1998; Soues and Adams, 1998). The half-bridge–specific protein Kar1 (Spang et al., 1995) was shown to bind Spc72 and is thus viewed as the anchor site for cytoplasmic microtubules at the half-bridge (Pereira et al., 1999). The Spc72-interacting protein at the SPB outer plaque is not yet known, although localization of Spc72 to the outer plaque has been shown (Soues and Adams, 1998). We observed that upon SPB separation in cnm67Δ cells astral microtubules detached concomitantly with Spc72-GFP. This suggests that intact microtubules, still carrying γ-tubulin caps plus Spc72, detach from the SPB. In addition, astral microtubule detachment was not blocked or decreased in cnm67Δ double mutants deleted for astral microtubule motors. These observations imply a release of intact astral microtubules rather than rupture or breakage exerted by external forces. In the context of our observations and the reported cell cycle–dependent change in astral microtubule organization (Byers and Goetsch, 1975), it is conceivable that astral microtubules switch upon SPB separation from the half-bridge to the SPB outer plaque, where they are organized for the rest of the cell cycle. In cnm67Δ cells half-bridge–released microtubules fail to be anchored to the region of the missing outer plaque and detach.
Switching of astral microtubules might by accomplished by altered affinity of Spc72 to the half-bridge or the outer plaque. Consistent with the idea of Spc72 being involved in switching of microtubules, Pereira et al. (1999) could show that in cells with an Spc72-Kar1 fusion astral microtubules were restricted to the half-bridge throughout the cell cycle and that accumulation of binucleated cells did not occur even in the absence of the Cnm67 protein. Apparently, astral microtubules that are stably anchored at the half-bridge throughout the cell cycle are able to fulfill all mitotic functions, which raises the intriguing question about the biological relevance of the cell cycle–dependent change in astral microtubule organization in S. cerevisiae.
Long-term time-lapse investigations allowed pedigree analysis of cnm67Δ cells. This revealed a successful first mitosis of all diploid, single-nucleate daughter cells, whereas later divisions of the same cells often failed. We observed the same characteristic in a different SPB mutant also affecting nuclear migration (our unpublished results). This points toward a general mother–daughter difference in nuclear migration rather than a cnm67Δ-specific characteristic. One known general mother–daughter difference consists of the identity of S. cerevisiae mother and daughter cells because of the asymmetric distribution of the transcriptional regulator Ash1, a repressor of the HO endonuclease gene (Bobola et al., 1996). She1 is a myosin involved in establishing the asymmetric distribution of Ash1 between mother and daughter cell (Bobola et al., 1996). We tested whether She1 was also involved in transport of daughter cell–specific products that influence nuclear migration. Loss of She1 in a cnm67Δ mutant background, however, did not alter the daughter fidelity phenotype of cnm67Δ mutants (our unpublished results). Another general mother–daughter difference concerns cell size. Daughter cells are smaller than mother cells at the time of their first bud emergence. Smaller cell size might facilitate force generation by spindle elongation, increase the chance of penetrating the bud neck because of smaller cell-surface, and facilitate astral microtubule-cortex interactions. However, we observed no effect on nuclear migration by altering the size of cnm67Δ daughter and mother cells. A third general difference between mother and daughter cells consists of bud site selection. Daughter cells produce the first bud from the distal pole, whereas mother cells have a bipolar bud site selection (Freifelder, 1960, Chant and Pringle, 1995). Distal bud site selection may facilitate nuclear positioning, because spindle elongation of the previous cell cycle deposits the nucleus in the daughter cell near the region where the new bud is produced. But the high fidelity of first daughter divisions was not directly linked to a general first distal bud site selection in daughter cells, as shown in homozygous diploid bud8Δ cnm67Δ mutants in this study. Changes of these three parameters, cell size, mating-type asymmetry, and first distal bud site selection, did not affect the high fidelity in nuclear migration of daughter cells. Our observations with cnm67Δ are reminiscent of the bud1Δ, budΔ2, and bud5Δ mutant phenotypes (Chant and Herskowitz, 1991). In mutants deleted for any of those genes the budding pattern is randomized except for the first division of a daughter cell, which maintains a first distal bud emergence. It appears conceivable that during bud emergence, bud growth, and later cytokinesis, cortical landmarks or cytoskeletal arrangements are positioned in the daughter by the mother cell, which by default are used by the daughter cell during its first cell cycle. Similarly, maternal effects have been described in other organisms, e.g., during the oogenesis in Drosophila in which mRNA gradients are established, defining anterior and posterior poles (Nusslein-Vollhard, 1996). Other ways of generating mother–daughter cell asymmetries can be generated by genomic imprinting and have been proposed to exist in yeast (Jiang and Stillman, 1996).
Our time-lapse studies of cnm67Δ mutants indicated that cytokinesis depends on successful nuclear migration. The time required from entry of the nucleus into the bud until completion of cytokinesis was the same for wild-type and mutants. This indicated that the cell separation process itself was not affected by Cnm67 loss. The observed lack of separation of anucleate buds in cnm67Δ cells suggests that the nucleus carries a cytokinesis-triggering signal. Recently, a putative cytokinesis checkpoint involving the Yeb1/Bim1 protein was postulated (Schwartz et al., 1997; Muhua et al., 1998). Loss of Yeb1 in the act5Δ nuclear migration mutant allowed separation of anucleate buds (Muhua et al., 1998). When we introduced the yeb1Δ deletion in a cnm67Δ background, we also saw separation of anucleate buds in time-lapse studies (our unpublished results). Although separation of anucleate buds in cnm67Δ yeb1Δ cells did not occur in all cases, we were able to visualize septa separating empty buds from the mother cells by staining with DiI (our unpublished results). Such septa were not observed in cnm67Δ single mutants. This also suggests that a nucleus in the bud is not per se a requirement for cytokinesis. Observations of the ndc1 mutant showed that the transfer of a defective SPB without any chromosomal material can be sufficient to trigger cytokinesis (Thomas and Botstein, 1986). This finding and our observations that detached γ-tubulin-capped microtubules did not trigger cell separation suggest that the cytokinesis-triggering signal may be associated with entrance of one (incomplete) SPB into the bud.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jürgen Wendland for helpful discussions, Florian Schärer for help setting up the microscope, and Tom Bickle for critical reading of the manuscript. We also thank A.F. Straight for the pAFS125 plasmid. This work was supported by grants from the University of Basel and the Swiss Federal Office for Education and Science (grants 95.0191-12 and 97.0537-1).
Footnotes
Online version of this article contains video material for Figures 1–7. Online version available at www.molbiolcell.org.
REFERENCES
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachat A, Kilmartin JV, Wach A, Philippsen P. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Bullock WO, Fernandez JM, Short JM. XLl-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. Biotechniques. 1987;5:376–378. [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 59–96. [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Herskowitz I. Genetic control of bud-site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;7:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SW, Meyer DI. ACT3: a putative centractin homologue in S. cerevisiae is required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin related KIP3 of Saccharomyces cerevisiae is required for a distinct step of nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11186. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J Cell Biol. 1995;131:1003–1014. doi: 10.1083/jcb.131.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. Bud formation in Saccharomyces cerevisiae. J Bacteriol. 1960;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin JV, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:14–15. [PubMed] [Google Scholar]
- Haarer BK, Lillie SH, Adams AE, Magdolen V, Bandlow W, Brown SS. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- Huyett A, Kahana J, Silver P, Zeng X, Saunders WS. The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J Cell Sci. 1998;111:295–301. doi: 10.1242/jcs.111.3.295. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Stillman DJ. Epigenetic effects on yeast trascription by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV. Genetic and biochemical approaches to spindle function and chromosome segregation in eukaryotic microorganisms. Curr Opin Cell Biol. 1994;6:50–54. doi: 10.1016/0955-0674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdolen V, Oechsner U, Muller G, Bandlow W. The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol Cell Biol. 1988;8:5108–5115. doi: 10.1128/mcb.8.12.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. Kar3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Frisen L, Wallack DL, Loayza D, Gammie AE, Rose MD. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell. 1998;8:2051–2068. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L, Adames NR, Murphy MD, Shields CR, Cooper JA. A cytokinesis checkpoint requiring the yeast homologue of an APC-binding protein. Nature. 1998;4:487–491. doi: 10.1038/31014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA. A yeast actin related homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- Nash R, Tokiwa G, Anand S, Stojcic C, Hazlett M, Erickson K, Futcher B. Cloning and partial characterization of the WHI1 (cln3) gene of S. cerevisiae. Yeast. 1988;4:51–62. [Google Scholar]
- Nusslein-Vollhard C. Gradients that organize embryo development. Sci Am. 1996;2:54–55. doi: 10.1038/scientificamerican0896-54. , 58–61. [DOI] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Grueneberg U, Knop M, Schiebel E. Interaction of the yeast γ-tubulin complex-binding protein Spc72 with Kar1p is essential for microtubule function during karyogamy. EMBO J. 1999;15:4180–4195. doi: 10.1093/emboj/18.15.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Biggins S, Satterwhite LL. Unraveling the tangled web at the microtubule-organizing center. Curr Opin Cell Biol. 1993;5:105–115. doi: 10.1016/s0955-0674(05)80015-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schwartz K, Richards K, Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol Biol Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The spindle pole body of yeast. Chromosoma. 1994;103:369–380. doi: 10.1007/BF00362281. [DOI] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soues S, Adams IR. SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J Cell Sci. 1998;111:2809–2818. doi: 10.1242/jcs.111.18.2809. [DOI] [PubMed] [Google Scholar]
- Spang A, Courtney I, Grein K, Matzner M, Schiebel E. The Cdc31p-binding protein Kar1 is a component of the half bridge of the yeast spindle pole body. J Cell Biol. 1995;134:429–441. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. Gamma-tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtu-bules and is required for mitotic spindle formation. J Cell Biol. 1996b;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living yeast: anaphase A but not metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986;44:65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud-site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.