Abstract
Mitotic segregation of chromosomes requires spindle pole functions for microtubule nucleation, minus end organization, and regulation of dynamics. γ-Tubulin is essential for nucleation, and we now extend its role to these latter processes. We have characterized a mutation in γ-tubulin that results in cold-sensitive mitotic arrest with an elongated bipolar spindle but impaired anaphase A. At 30°C cytoplasmic microtubule arrays are abnormal and bundle into single larger arrays. Three-dimensional time-lapse video microscopy reveals that microtubule dynamics are altered. Localization of the mutant γ-tubulin is like the wild-type protein. Prediction of γ-tubulin structure indicates that non-α/β-tubulin protein–protein interactions could be affected. The kinesin-like protein (klp) Pkl1p localizes to the spindle poles and spindle and is essential for viability of the γ-tubulin mutant and in multicopy for normal cell morphology at 30°C. Localization and function of Pkl1p in the mutant appear unaltered, consistent with a redundant function for this protein in wild type. Our data indicate a broader role for γ-tubulin at spindle poles in regulating aspects of microtubule dynamics and organization. We propose that Pkl1p rescues an impaired function of γ-tubulin that involves non-tubulin protein–protein interactions, presumably with a second motor, MAP, or MTOC component.
INTRODUCTION
γ-Tubulin is central to mitotic spindle formation and is a ubiquitous component of MTOCs (Gueth-Hallonet et al., 1993; McDonald et al., 1993; Muresan et al., 1993; Palacios et al., 1993; Rizzolo and Joshi, 1993; Liu et al., 1994; for review, see Oakley, 1994, and references therein). Genetic studies in fungi and Drosophila indicate that the protein is essential and required for spindle function (Oakley et al., 1989, 1990; Horio et al., 1991; Stearns et al. 1991; Sobel and Snyder, 1995; Sunkel et al., 1995; Spang et al., 1996; Martin et al., 1997). A direct role for γ-tubulin in microtubule nucleation itself has been shown by antibody inhibition or depletion of γ-tubulin (Joshi et al., 1992; Felix et al., 1994), in vitro using a purified γ-tubulin-containing ring complex (Zheng et al., 1995) and genetically (Masuda and Shibata, 1996; Marschall et al., 1996; Spang et al., 1996; Martin et al., 1997). Surprisingly, cells depleted of γ-tubulin still nucleate cytoplasmic microtubules, albeit with abnormal length and number (Sobel and Snyder, 1995; Spang et al., 1996).
γ-Tubulin itself is expected to help form the link between microtubules and the centrosome or spindle pole body (SPB). Its sequence is similar to α- and β-tubulins (Oakley, 1994), and the structure is predicted to be analogous (Downing and Nogales, 1998; Nogales et al., 1998). Consistent with this view, γ-tubulin binds microtubules in vitro (Melki et al., 1993; Raff et al., 1993; Stearns and Kirschner, 1994) at their minus ends (Li and Joshi, 1995; Zheng et al., 1995). Two γ-tubulin-interacting MTOC proteins, Spc97p and Spc98p, were originally characterized in Saccharomyces cerevisiae (Geissler et al., 1996; Knop et al., 1997; Knop and Schiebel, 1997; Pereira et al., 1998; Wigge et al., 1998), and sequence and functional homologues are present in metazoan centrosomes (Moritz et al., 1995; Zheng et al., 1995; Martin et al., 1998; Murphy et al., 1998; Tassin et al., 1998; Oegema et al., 1999). Human γ-tubulin can replace the endogenous protein in fission yeast (Horio and Oakley, 1994), suggesting that key aspects of γ-tubulin function are broadly conserved.
Microtubule motors play important roles in spindle assembly and dynamics. A trimolecular complex, composed of cytoplasmic dynein, dynactin, and NuMA, is required for focusing spindle minus ends in higher eukaryotes (Heald et al., 1996; for review, see Compton, 1998). In its absence, nucleation continues, but spindle poles lack organization, and chromosome segregation is impaired. In the fungus Nectria hematococca, dynein may play a similar role for astral microtubules (Inoue et al., 1998a,b). In yeast, dynein affects nuclear positioning (Plamann et al., 1994; for review see Stearns, 1997; Shaw et al., 1997; Kahana et al., 1998; Yamamoto et al., 1999). Members of the Ncdp/Kar3p, BimCp, and MCAKp kinesin-like protein (klp) families affect centrosome integrity (Endow et al., 1994a; Endow and Komma, 1996), microtubule number and length (Endow et al., 1994b; Pidoux et al., 1996; Walczak et al., 1996; Saunders et al., 1997; Huyett et al., 1998; Desai et al., 1999), or the balance of forces for spindle assembly and function (Hagan and Yanagida, 1992; Gaglio et al., 1996; Pidoux et al. 1996; Huyett et al., 1998). In addition, Ncdp has been shown to be required for proper localization of γ-tubulin to the meiosis II spindle (Endow and Komma, 1998).
The Schizosaccharomyces pombe klp Pkl1p localizes to the SPB and spindle. It is nonessential in wild type; however, moderate overexpression of Pkl1p results in spindle shrinkage or collapse (Pidoux et al., 1996). To identify functional overlap with Pkl1p, we isolated mutations conferring dependence on pkl1 for viability. Two mutants recovered suggest that Pkl1p and a subset of γ-tubulin functions are closely linked. In this report we characterize one of these mutants, an allele of γ-tubulin. Mutation of a single conserved residue allows microtubule nucleation but impairs chromosome segregation and has dramatic effects on cytoplasmic microtubule arrays. Our analysis identifies a novel role for γ-tubulin at the MTOC in regulating microtubule organization and dynamics.
MATERIALS AND METHODS
Genetics, Media, and General Yeast Strains and Plasmids
Standard genetic procedures, lithium acetate transformation, and rich (YEAC–yeast extract + adenine + casamino acids) or minimal media (MSA or EMM) have been described (Moreno et al., 1991; Egel et al., 1994). Adenine, leucine, or uracil (75 μg/ml each) was supplemented as necessary. Color selection used 25 μg/ml adenine. Drug sensitivity was on YEAC- or EMM-supplemented plates containing 10 μg/ml thiabendazole (Sigma, St. Louis, MO) by spotting increasing serial dilutions of strains.
The following strains and plasmids were kind gifts: mad2− strain (h− ade6-M210, leu1-32, ura4-D18, mad2::ura4+) from Dr. Shelly Sazar (Verna and Marrs McLean Departments of Biochemistry and Cell Biology, Baylor College of Medicine, Houston, TX; He et al., 1997); strain YY105 (h90 leu1-32, ura4-D18, lys1+::[GFP-atb2]) and plasmids pGFPatb2 (Ding et al., 1998) and pD817 (green fluorescent protein [GFP]-cytochrome P450 reductase, ars1, LEU2; Tange et al., 1998) from Dr. Da-Qiao Ding and Dr. Yasushi Hiraoka (Structural Biology Section, Kansai Advance Research Center, Communications Research Laboratory, Kobe, Japan); strain EG367 (h90 mei1-102, his2, swi2–3,ade6-M210) used to test dominance of the mutations in SL1 and SL2, from Dr. Richard Egel (Department of Genetics, University of Copenhagen, Copenhagen, Denmark; Egel, 1973; Kohli et al., 1977); tubulin plasmids pHutubG, pNda2, and pAtb2 from Dr. Satoru Uzawa (Department of Molecular and Cell Biology, University of California, Berkeley, CA; Toda et al., 1984); pTS446, containing genomic S. pombe gtb1 from Dr. Robert Jeng and Dr. Tim Stearns (Department of Biology, Stanford University, Stanford, CA); and pREP3 from Dr. Kinsey Maundrell (Department of Molecular Microbiology, Glaxo Institute for Molecular Biology, Geneva, Switzerland; Maundrell, 1993).
Wild-type S. pombe 972 h− and FY392 (h− ade6-210, leu1-32, his3-D1, ura4-D18) and tubulin strains h+ nda3-KM311, h− nda2-340, h− nda2-890, and h− nda2-167 were from laboratory stocks. An h+ nda2-340 strain was constructed by crossing with 972 h+ leu1-32. pJPgtb1 (S. pombe gtb1 in pREP3) was constructed as follows: pTS446 contains S. pombe gtb1 in pUR19, with 2.0-kb upstream and 154-bp downstream sequences. A PstI–SacI S. pombe gtb1 fragment from pTS446 was cloned into the same pREP3 sites, removing the nmt1 promoter and terminator. pGFP-gtb1 was constructed in three steps: pEGFP (Clontech, Palo Alto, CA) was used to produce a PCR fragment of enhanced GFP (EGFP) with SpeI, PstI, and NcoI flanking sites, digested with SpeI–PstI and cloned into pJPgtb1 at these sites. The oligonucleotides were (restriction sites underlined; Operon Technologies, Emeryville, CA) 5′-TGCAAATCACACTAGTACCATGGTGAGCAAGGGCGAGG-3′ and 5′-GTATGCGATCTGCAGTAGAGTCGCGGCCGCTTTA-3′. A PCR fragment containing 3′ gtb1 and a linker (KLGGRQ), inserted between the last coding amino acid in gtb1 and the initiator methionine of EGFP, was generated to fuse the proteins in frame by inserting the SpeI–NcoI fragment. The oligonucleotides used for PCR were 5′-CGAACCACCGTGTTAGTGGACTTATGCTTGCAAATCACACTAGTATTGCCTCTGT-3′ and 5′-GCATAGCTCCATGGCTTGTCGACCGCCAAGCTTAAGAGATAAATAATT-GGGATCTTCAC-3′. The fusion in frame was confirmed by sequencing. pGFPgtb1-PL301 was constructed in two steps. A PCR fragment covering the 3′ half of gtb1-PL301 was generated using SL1 chromosomal DNA and cloned into pCRII (TA cloning kit; Invitrogen, Carlsbad, CA). Internal SpeI–Bsu36I sites were used to move the gtb1-PL301 sequence from pCRII into pGFP-gtb1, replacing the wild-type sequence. Replacement was confirmed by sequencing. Plasmids pGFPatb2, pGFPgtb1, and pGFPgtb-PL301 were integrated into the appropriate strains at the ars1 locus using the unique mluI site.
Synthetic Lethality Strains, Plasmids, and Screen
The protocol for synthetic lethality, plasmid shuffle strain MP18 (h− ade1-D25, ade6-M210, leu1-32, ura4-D18) and pNPT/ADE1 (ars1, adh1-neoR, ade1+ in pUC119) were gifts from Dr. Michael J. Moser and Dr. Trisha Davis (Department of Biochemistry, University of Washington, Seattle, WA). The screen was initiated by Dr. A. Pidoux (Medical Research Council Human Genetics Unit, Edinburgh, United Kingdom) when in the laboratory of Dr. W.Z. Cande and completed by Dr. J. L. Paluh. A strain replacing 1.2 kb of pkl1+ with ura4+ (Pidoux et al., 1996) was used to make strain ZC94 (h− ade1-D25, ade6-M210, leu1-32, ura4-D18, pkl1-D12::ura4+). pUR19-gpkl1 (Pidoux et al., 1996) and pKSpkl1, a 6.7-kb PstI–KpnI genomic fragment containing pkl1 cloned in Bluescript (Stratagene, La Jolla, CA), were used to construct pNPT/ADE1/pkl1. A BglII–SacI fragment from pUR19-gpkl1 was cloned into BamHI–SacI sites of pNPT/ADE1, and a SacI fragment from pKSpkl1, containing the 5′ end of pkl1+, was added. Strain ZC94 bearing pNPT/ADE1/pkl1 was mutagenized with ethylmethane sufonate, and 20,000 gene equivalents were screened for plasmid dependence. Fifty solid red or mostly red colonies were picked and restreaked. Sixteen isolates sectored to white and were discarded, and thirty-four remaining were transformed with pSMpkl1 (Pidoux et al., 1996) to confirm pkl1 dependence. Three were chosen for back-crossing (four rounds) and characterization. Two isolates (SL1 and SL2) exhibited conditional growth and are described here: SL1 (h− ade1-D25, ade6-M210, leu1–32, ura4-D18, pkl1-D12::ura4+, gtb1-PL301, [pNPT/ADE1/pkl1]) and SL2 (h− ade1-D25, ade6-M210, leu1–32, ura4-D18, pkl1-D12::ura4+, slp2, [pNPT/ADE1/pkl1]). The undefined locus in SL2 has been designated slp2 for “synthetic lethality with pkl.” Strains SL1sc (h− ade6-M210, leu1-32, ura4-D18, gtb1-PL301) and SL2sc (h− ade6-M210, leu1-32, ura4-D18, slp2) are wild-type for pkl1 (native genomic single copy) and were generated by crossing with strain h+ ade6-M210, leu1-32, ura4-D18.
Chromosomal DNA Isolation, PCR, and Sequence Analysis
Chromosomal DNA was isolated using glass beads (Moreno et al., 1991), digested with 100 μg/ml Proteinase K (Amresco, Solon, OH) for 30 min and stored in ethanol. PCR was performed using the Boehringer Expand long PCR kit (Boehringer Mannheim, Indianapolis, IN). PCR fragments were gel isolated on 1.0% agarose in 67 mM Tris-HCl, pH 8.3, 67 mM borate, 1 mM EDTA buffer, purified using QIAEX resin (gel extraction kit; Promega, Madison WI), phenol extracted, and ethanol precipitated. Direct sequencing of PCR products on both strands was done at the University of California Berkeley Sequencing Facility and always compared with DNA from 972 h− cells. The previously undetermined mutation in nda3-KM311 is amino acid 93 (GGA to GAA) Gly to Glu. Sequences were obtained from the S. pombe Sequencing Group at Sanger Center (Wellcome Trust Genome Campus, Cambridge, United Kingdom; http://www.sanger.ac.uk).
Transmission Electron Microscopy (TEM) and Cell Fixation and Immunofluorescence
Cells for TEM were grown to early log phase in appropriate media and kept circulating at the desired temperature until immediately before harvesting, high-pressure freezing, and embedding (McDonald, 1999). Serial sectioning was done at 40–60 nm thickness using a Reichert Ultracut E (Reichert Jung, Vienna, Austria). Sections were picked up on grids and stained with uranyl acetate and lead citrate. Cells were imaged on a JEOL (Peabody, MA) 100 CX electron microscope operating at 80 kV at the University of California Berkeley Electron Microscope Laboratory.
Immunofluorescence was as described (Hagan and Hyams, 1988). Cells were grown in YEAC or supplemented EMM liquid media at the appropriate temperature to early log phase and harvested on Whatman (Hillsboro, OR) GF/C glass microfiber 25-mm filters for fixation in methanol at −80°C for 30 min or fixed in 0.2% glutaraldehyde and 2.6% paraformaldehyde (Electron Microscopy Sciences, Ft. Washington, PA). Results obtained were similar using both methods. The TAT1 anti-tubulin antibody was a gift from Dr. Keith Gull (University of Manchester, Manchester, United Kingdom; Woods et al., 1989) and was used at 1:25 dilution. Fluorescein or Texas Red goat anti-mouse secondary antibodies (EY Laboratories, San Mateo, CA) were used at 1:200 dilution. DNA staining was with Hoechst (Sigma). Microsocopy and image processing are described below.
Three-dimensional (3D) and Time-lapse Video Microscopy of Living Cells Using GFP Fusion Proteins
Microtubules in living cells were visualized using GFPatb2 (see Genetics, Media, and General Yeast Strains and Plasmids). Cells were grown on YEAC- or EMM-supplemented plates and transferred to the same liquid media or grown with high aeration in low volume (1–5 ml) for enhanced visualization of GFP. Microscopy was performed on a Zeiss (Thornwood, NY) Axiovert S100 fluorescence microscope with an oil immersion 100× NEOFLUAR lens, numerical aperture 1.3, at room temperature of 21 or 25°C. Coverslips were coated with poly-l-lysine or 0.1% polyethyleneimine and sealed with 3% agarose. 3D optical data sets were collected using a SenSys charge-coupled device (CCD) camera equipped with automated fine focus, at a Z spacing of 0.2 to 0.4 μm using ISEE software (Inovision, Durham, NC). Wild-type haploid S. pombe is 3.5 μm wide and increases in length from 7 to 15 μm (Johnson et al., 1989). Ten to 30 Z-sections at 0.2 or 0.4 μm were typically collected depending on the application and cells. Raw image stacks were imported into Deltavision software (Applied Precision Incorporated, Issaquah, WA) (Chen et al., 1995) and processed using constrained iterative (15 rounds) deconvolution. Deconvolved images were saved as tagged image format files and transferred to Adobe Photoshop 4.0 (Adobe Systems, San Jose, CA) and Canvas 5.0 (Deneba Software, Miami, FL) for compiling image plates.
For movies 13 serial Z-section images at 0.4 μm spacing were taken. Fifteen seconds were typically needed to capture each Z-stack of images. Time intervals between Z-stacks were 15–45 s. Short exposure times and neutral density filters were used to avoid photobleaching. Time recordings were limited to 10–20 min, although recordings up to 4 h were possible with few or single Z-stacks. Figure 5 movie conditions used are indicated below and correspond to these parameters (exposure time, neutral density filter [NDF], number of cycle iterations, time interval between completion of stacks, and total time): Figure 5C, 0.2 s, 0.6 NDF, 28, 60 s, and 35 min; Figure 5D, 0.2 s, 0.3 NDF, 20, 15 s, and 10 min; Figure 5E, 0.2 s, 0.3 NDF, 40, 15 s, and 20 min; Figure 5F, 0.3 s, no NDF, 20, 15 s, and 10 min; Figure 5G, 0.8 s, 0.3 NDF, 15, 15 s, and 7.5 min; and Figure 5H, 0.3 s, no NDF, 20, 15 s, and 10 min.
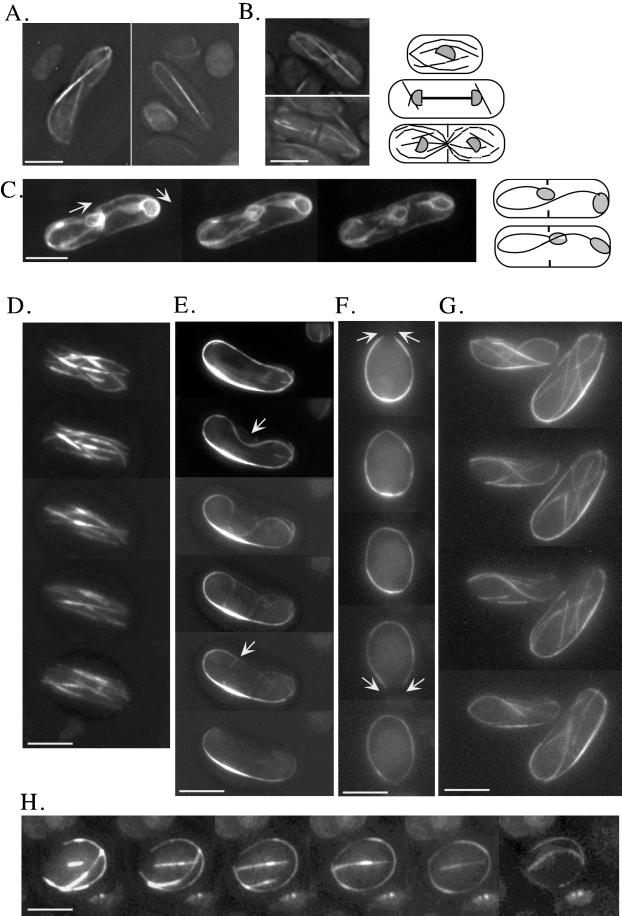
Figure 5.
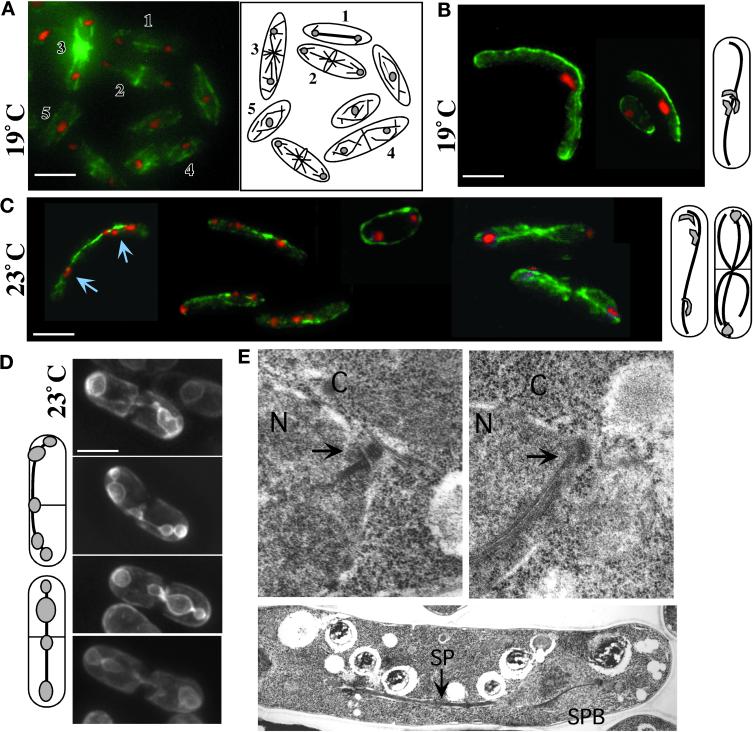
Time-lapse video microscopy of microtubule dynamics in living wild-type and gtb1-PL301 cells (for details see MATERIALS AND METHODS). Microtubules were visualized in gtb1-PL301 cells, with only the native copy of pkl1 present, using a GFP-α-tubulin fusion protein. Images shown are 3D projections of all microtubules. Projections are generated from a series of images taken through the cell at a Z-spacing of 0.2–0.4 μm. Representative examples show the γ-tubulin mutant of spindles at 19°C (A) and retained spindles with postanaphase microtubule arrays at 23°C (B) viewed using GFP-α-tubulin. The diagrams to the right depict normal wild-type patterns of microtubule dynamics in G2, anaphase B, and cytokinesis (for review, see Hagan, 1998). (C–H) 3D time-lapse video microscopy of spindle and microtubule behavior in the mutant. Selected frames are shown. Entire movies are available. (C) GFP-NADPH cytochrome P450 reductase is used as a nuclear envelope marker to confirm that spindle elongation can curve around the cell ends. Arrows indicate the result of pushing forces from spindle elongation. The diagrams at the right show the beginning and end stages. (D–H) 3D time-lapse video microscopy of microtubules in wild-type (D) and the mutant (E–H). See RESULTS for details. Bar, 5 μm.
γ-Tubulin Structural Model
Sequence identity between S. pombe and human γ-tubulin is 71.6% (Horio and Oakley, 1994), well above that between any tubulin and FtsZ. Given the striking similarity between the structures of FtsZ and that of α- and β-tubulin, γ-tubulin is expected to greatly resemble α- and β-tubulin. The structure of γ-tubulin was approximated using the atomic model of mammalian α-tubulin obtained by electron crystallography. A model was created using the program O by substituting residues not conserved between mammalian α-tubulin and γ-tubulin. Insertions and deletions were modeled to minimize disruption of the starting model.
RESULTS
A γ-Tubulin Mutation Confers Dependence on Kinesin-like Pkl1p for Cell Viability
Pkl1p is one of two mitotic klps characterized in fission yeast (Hagan and Yanagida, 1990; Pidoux et al., 1996; for review, see Su and Yanagida, 1997). Its overexpression affects spindle structure and dynamics, suggesting that it may share similar functions with the homologous S. cerevisiae KAR3p for tubulin dimer removal at microtubule minus ends (Endow et al., 1994b; Saunders et al., 1997). Because Pkl1p is nonessential in wild-type cells, we performed a synthetic lethality screen to identify proteins that overlap functionally (Figure 1; see MATERIALS AND METHODS). Genomic mutations were generated that are lethal in the absence of pkl1, resulting in plasmid dependence that can be monitored by a color assay. Two mutant strains were recovered that shared similar phenotypes (Figure 1, SL1 and SL2). Genetic analysis indicates that these mutations are recessive and unlinked (see MATERIALS AND METHODS).
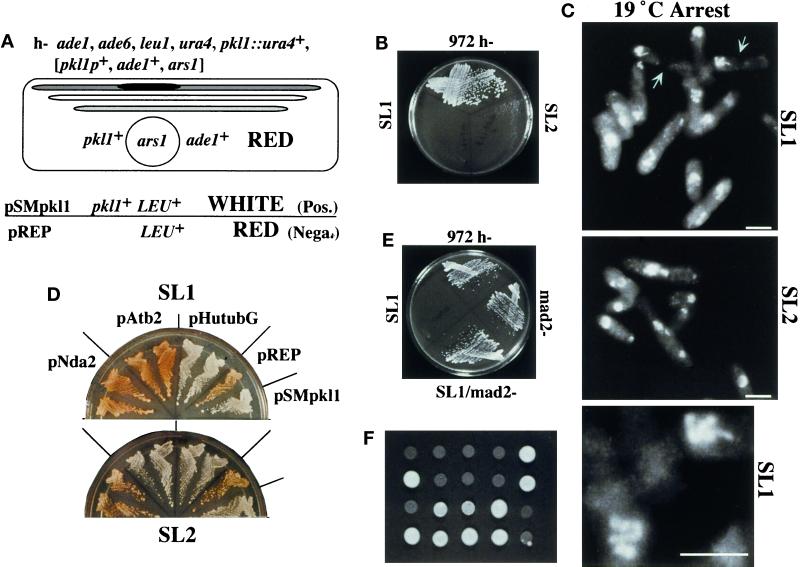
Figure 1.
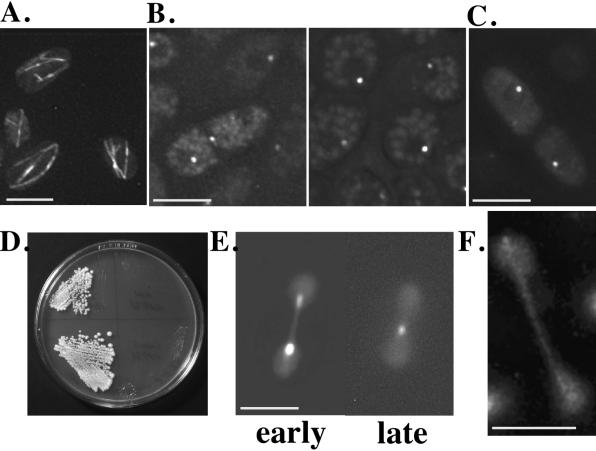
Two mutations genetically link Pkl1p and γ-tubulin. (A) Synthetic lethality strategy with strain genotype (see MATERIALS AND METHODS for details). The pkl1 gene is deleted in the genome (bars represent three chromosomes; dark is the pkl1 locus) and replaced by a plasmid (circle) carrying wild-type pkl1+ and ade1+. Red and white screening uses ade1 and ade6 mutations (Moser et al., 1995). Ade6− cells appear red on low-adenine plates, whereas [ade1−, ade6−] cells are white. Mutations that cause plasmid dependence result in red cells, because ade1− is complemented. S. cerevisiae LEU2+ complements S. pombe leu1− and is a genetic marker in lieu of ade1+ on control and test plasmids. Control plasmids: (negative) pREP [LEU+, no pkl1]; (positive) pSMpkl1 [LEU+, pkl1+]. (B) Cold-sensitive growth of SL1 and SL2 synthetic lethal mutants at 19°C compared with wild-type S. pombe 972 h−. (C) Photograph of condensed chromosomes in SL1 and SL2 cells at 19°C, stained with Hoechst. Microtubule and Hoechst combined images acquired by CCD camera are shown for SL1 only in Figure 2. The bottom image is an enlargement of chromosomes in SL1. Bar, 5 μm. SL1 cells arrest in mitosis and only rarely proceed to cytokinesis. (Arrows in the upper panel point to a septum in two cells. DNA is off center.) (D) SL1 and SL2 cells, transformed with plasmid controls (pREP and pSMpkl1) and tubulin genes (see this section of RESULTS), were streaked onto selective plates (EMM, no leucine) containing low adenine. Duplicate transformants for each tubulin gene are shown. Red indicates that the original plasmid [pkl1+, ade1+, ars1] is still present and required for growth. White cells have lost the original plasmid. Labels for streaks in SL2 are as shown for SL1. (E) Disruption of the checkpoint gene mad2 in SL1 cells with single-copy pkl1 removes the cold-sensitive mitotic block. (F) Tetrads tested for growth at 19°C. SL1 with single-copy pkl1 was crossed to strain h+ nda2-340, which carries a cold-sensitive microtubule-destabilizing mutation in α-tubulin (see last section of RESULTS). Only the parental genotypes remained cold sensitive.
Both SL1 and SL2 mutants were cold sensitive for growth at 19°C (Figure 1B) and arrested with highly condensed chromosomes indicative of a block in mitosis (Figure 1C). The Hoechst-stained pattern of the chromosomes, for SL1 cells in particular, was reminiscent of that of a well-characterized cold-sensitive mutation in β-tubulin, nda3-KM311 (Umesono et al., 1983; for review, see Fantes, 1989). This prompted us to test the possibility that SL1 and SL2 cells might contain mutations in a tubulin gene. Using plasmid shuffle, S. pombe α-tubulin (two genes, atb2 and nda2) or γ-tubulin (gtb1) and human γ-tubulin (tubG) genes were tested (Figure 1D). Human γ-tubulin has previously been shown to efficiently complement an S. pombe γ-tubulin deletion (Horio and Oakley, 1994). Interestingly, for both mutants γ-tubulin was able to substitute for pkl1 (only human tubG shown). For SL2 cells, atb2 also substituted for pkl1, whereas the essential α-tubulin nda2 did not. This may suggest a specific advantage for atb2-encoded α-tubulin in these cells. In S. pombe, as in S. cerevisiae, even one additional copy of β-tubulin is lethal (Hiraoka et al., 1984; Burke et al., 1989; Katz et al., 1990; Javerzat et al., 1996). Linkage to the β-tubulin locus was tested by crossing SL1 and SL2 cells to the mutant nda3-KM311. Random spore and tetrad analysis revealed that neither mutation was linked to β-tubulin (our unpublished data).
Chromosomal DNA was isolated from SL1, SL2, and wild-type 972 cells for PCR and direct sequencing of genes (see MATERIALS AND METHODS). SL1 contained a single point mutation in gtb1, encoding γ-tubulin, resulting in an amino acid change at position 301 from proline, conserved at this position in metazoan and fungal γ-tubulins, to leucine (gtb1-PL301; see Figure 3A). Genetic crossing and sequence analysis (see MATERIALS AND METHODS) confirmed that this gtb1 mutation is linked to the cold-sensitive growth arrest and synthetic lethality with pkl1. No changes from wild type were present in SL2 cells for any of the tubulin genes, or for pkl1, and the identity of the gene that carries the mutation remains under investigation. Thus, SL1 is a mutation in γ-tubulin, whereas the mutation in SL2 remains unknown.
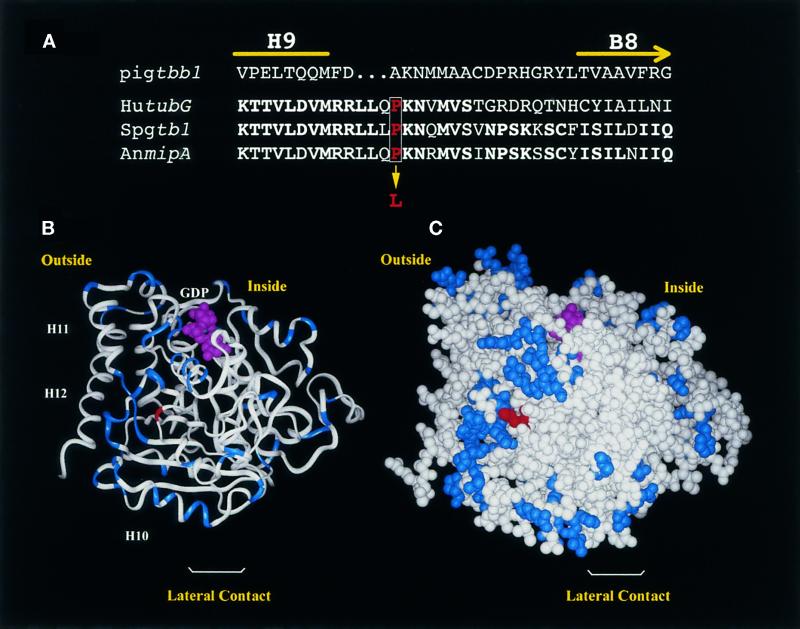
Figure 3.
Predicted structure of γ-tubulin showing the position of the SL1 mutation. (A) Partial sequence alignment of pig β-tubulin (Pigtbb1) and γ-tubulin from human (hutubG), S. pombe (Spgtb1), and A. nidulans (AnmipA) showing amino acids from H9 through B8. This corresponds to amino acids 298–324 in Spgtb1. Identical amino acids are in bold. The proline (red type, boxed) that is mutated in SL1 to a leucine is indicated. (B and C) Predicted 3D structure of γ-tubulin, obtained by replacing differing amino acids in pig α-tubulin with the corresponding γ-tubulin amino acids. Residues that are similar in α- or β-tubulin but that have a totally different character in γ-tubulin (e.g., small vs. large or hydrophobic vs. charged) are highlighted in blue. The PL301 mutation is shown in red. (B) Ribbon diagram. The nucleotide and the last C-terminal helices (H10, H11, and H12) are indicated. Klps have been reported to bind at H11 and H12 of β-tubulin. (C) CPK representation. The orientation is such that, if attached to a microtubule end, the viewer will be facing a site of lateral interaction. The inside and outside of the microtubule are indicated.
Pkl1 Copy Number Influences the Severity of the γ-Tubulin Phenotype
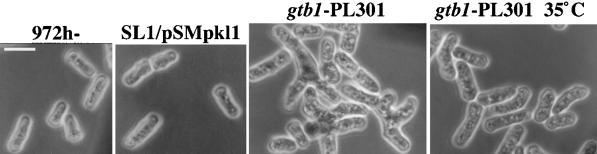
For the synthetic lethality screen pkl1 is present on a multicopy plasmid. To analyze the phenotype of the γ-tubulin mutation alone, we tested whether the native genomic copy of pkl1 itself was sufficient for viability. At 19°C mitotic arrest was similar in SL1 cells with single or multiple copies of pkl1. However, at 30°C, near wild-type cell morphology required multicopy pkl1 (Figure 2, compare first three panels). When only the single native copy of pkl1 was present (gtb1-PL301), cells grew poorly versus wild-type (972 h−) or mutant cells that had multiple copies of pkl1 (SL1/pSMpkl1), and bent and branched cells were observed (Figure 2, third panel). By raising the growth temperature to 35°C, near normal cell morphology was restored to the mutant with single-copy pkl1, although bent cells were still present (Figure 2, last panel).
Figure 2.
Lowering pkl1 copy number exacerbates the gtb1-PL301 phenotype. Phase microscopy of wild-type cells (972 h−) or SL1 cells with pkl1+ on a multicopy plasmid (SL1/pSMPkl1) is compared with SL1 cells with only the native copy of pkl1+ present (gtb1-PL301) grown at either 30°C (first three panels) or 35°C (last panel). Bent and branched cells were observed only when the gene for Pkl1p was not present in multicopy. Bar, 5 μm.
The γ-Tubulin Mutation Lies in a Domain Expected to Form Non-Tubulin Protein–Protein Interactions
The structure of α/β-tubulin dimers was recently determined by electron crystallography (Nogales et al., 1998). Its alignment with a lower-resolution 3D density map of tubulin now creates a detailed view of the microtubule lattice that provides information on lateral contacts between protofilaments and longitudinal contacts between tubulin dimers (Nogales et al., 1999). γ-Tubulin is ∼30% identical to α- and β-polymer tubulins, and its 3D structure can be approximated by replacing differing amino acids in the sequence of α-tubulin with the corresponding residues for γ-tubulin (Figure 3, B and C; see MATERIALS AND METHODS). Two helices positioned on the outer face of polymer tubulins that are predicted to form part of the site for binding klps (for review, see Mandelkow and Hoenger, 1999) are retained in the γ-tubulin sequence.
By predicting the structure of γ-tubulin, we localized the gtb1-PL301 mutation to the surface of the protein between helix 9 (H9) and β-sheet 8 (B8) (Figure 3, B and C). The mutated proline lies in a loop in γ-tubulin that has variability in length and sequence across species. However, comparison of γ-tubulin sequences from several organisms revealed that proline is the predominant residue at this position. This includes γ-tubulin from human, Xenopus, Drosophila, the fungi Aspergillus nidulans and Neurospora crassa, and the alga Chlamydomonas. Threonine was also found at this position in Zea mays, Arabidopsis, and Tetrahymena. The flowering fern Anemia phyllitidis has an alanine at this position. Proline 301 is not far from residues in the M-loop, between B7 to H9, that are thought to compose the central elements in the lateral interaction surface of polymer tubulins. In the 3D model this proline lies in a region of the most significant differences with respect to α- and β-tubulins. The clustering of these γ-tubulin-specific residues at one surface of the protein suggests a specialized face for protein–protein interaction. Importantly, the site of the mutation does not correspond to any of the sites of tubulin–tubulin interactions in microtubules.
Chromosome Segregation Fails in gtb1-PL301 Cells Despite Normal Formation of a Bipolar Spindle
To determine whether a spindle was present in the γ-tubulin mutant at 19°C, we performed immunofluorescence microscopy of microtubules on cells carrying only the single native copy of pkl1 (Figure 4, B and C) or multiple copies of pkl1 (results similar to single-copy pkl1; our unpublished data) using TAT1 antibody against tubulin (see MATERIALS AND METHODS). We found that a spindle was present that had elongated to the cell ends (Figure 4B) or was sometimes hyperelongated so that it curved back around from the cell end toward the cell center (Figures 4C, middle cell, and 5, A, right, and C). Hyperelongating spindles sometimes extended to twice the cell length unless blocked by formation of the septum.
Figure 4.
Impaired chromosome segregation in gtb1-PL301 cells despite formation and elongation of a bipolar spindle. Immunofluorescence microscopy of microtubules (A–C), GFP nuclear membrane visualization (D), and TEM (E) are shown. Wild-type 972 h− cells are shown in A and E, top left. In A a field of wild-type cells shows different microtubule arrays: anaphase B spindle (1), telophase and central ring formation (2), postanaphase arrays (3), cytokinesis (4), and G2 (5). gtb1-PL301 cells with only the native copy of pkl1 present (B–E, right and lower) were grown at either 19 or 23°C as indicated. Microtubules (green) were visualized using TAT1 mAb. DNA staining (red) was with Hoechst. The cells shown in A–D are 3D projections of microtubules present throughout the cell taken at 0.2 μm spacing using an automated fine focus and CCD camera (see MATERIALS AND METHODS). Diagrams to the right of B and C and left of D indicate the predominant phenotype of the mutant observed at that temperature. Chromosome behavior is different at 19°C (B) versus 23°C (C and D). At 19°C chromosomes generally remained aggregated and nonsegregated (also see Figure 1C), whereas at 23°C chromosomes segregated to the poles or trailed along the spindle. Arrows in C indicate trailing chromsomes. (D) The cytochrome P-450 NADPH reductase first 80 amino acids fused to GFP (see MATERIALS AND METHODS) was used as a nuclear membrane marker in the mutant. In S. pombe the nuclear envelope collapses around the spindle, and mis-segregating chromosomes are evident as bumps along its length. (E) TEM of an anaphase SPB (arrows) in wild-type (left) or gtb1-PL301 mutant (right) cells (upper images). Lower image, a spindle in gtb1-PL301. Cells were grown at 23°C. N, nucleus; C, cytoplasm; SP, spindle. Bar (in A–D), 5 μm. Magnification for TEM images, 19,080× for SPB images; 7590× for the spindle.
The phenotype of gtb1-PL301 cells is different at 23 versus 19°C. Chromosomes no longer are aggregated at 23°C but instead have either segregated to the poles (Figure 4C, right) or are spread out along the spindle (Figure 4C, left, arrows). Trailing chromosomes are typical of cells known to be defective in attachment to or maintenance of chromosomes on the spindle. These phenotypes are even more evident using GFP-NADPH cytochrome P450 reductase to visualize the nuclear envelope surrounding the spindle and chromosomes (Figure 4D; Tange et al., 1998; see MATERIALS AND METHODS). During mitosis in many fungi the nuclear envelope does not break down but instead persists and remains closely associated with these structures as anaphase proceeds. The cell in Figure 4D, upper panel, has near normal nuclear envelope morphology during mitosis; the bulk of chromosomes have segregated near the poles and spindle length (as judged by nuclear envelope morphology around the spindle) is only slightly longer than expected in wild-type cells. In the last three cells trailing chromosomes are seen as bumps in the nuclear envelope. At least 30% of cells at this temperature (and all shown in Figure 4D) progress from mitosis to cytokinesis while retaining the spindle. In wild-type cells the spindle is no longer present during this stage of cytokinesis.
To determine whether obvious structural alterations in the SPB or spindle were present in the mutant, we performed transmission electron microscopy using high-pressure freezing and freeze substitution on gtb1-PL301 (single-copy pkl1) and wild-type cells (see MATERIALS AND METHODS). SPB structures of wild-type (Figure 4E, upper left) and gtb1-PL301 (Figure 4E, upper right) cells or spindle structure in gtb1-PL301 (Figure 4E, lower) versus wild-type cells (our unpublished data) were similar. The SPB in the mutant was not larger in size, and the number of microtubules associated with the SPB appeared normal. However, it is still possible that subtle structural changes could be missed without extensive serial section analysis of the spindle and SPB.
Mad2 Is Required for the Mitotic Block, Suggesting Impaired Chromosome Attachment to Kinetochore Microtubules
Our analysis of the gtb1-PL301 mitotic arrest phenotype by immunofluorescence indicated that a bipolar spindle formed and elongated without chromosome segregation occurring. To test whether chromosome attachment to the spindle was altered, signaling Mad2p-sensitive mitotic arrest, we analyzed [gtb1-PL301, mad2−] progeny for growth at 19°C on plates (Figure 1E; He et al., 1997; see MATERIALS AND METHODS). The Mad2p spindle checkpoint protein monitors chromosome attachment and microtubule tension and blocks mitosis when these are inadequate (Waters et al., 1998, 1999). Poor attachment of chromosomes to kinetochores (not all microtubules attached at all kinetochores) would be expected to result in greater viability than the complete absence of chromosome attachment. In the mad2− background, microscopic analysis of cells indicated that chromosomes decondensed, and a significant number of cells progressed through mitosis and cytokinesis. Hoechst staining indicated that some cell death from cut DNA and chromsome mis-segregation was occurring in the population (our unpublished data).
Altered Microtubule Dynamics and Organization in gtb1-PL301 Cells
TEM analysis of the spindle and SPB in the mutant was similar to wild type, suggesting that microtubule dynamics instead might be altered. We used a GFP-α-tubulin fusion protein (GFP-atb2; Ding et al., 1998) to visualize microtubules in living S. pombe cells. The microtubules we visualized in this manner accurately reflected those arrays, as observed by immunofluorescence of tubulin in fixed cells, as was noted previously (compare Figures 4 and 5; Ding et al., 1998). We integrated GFP-atb2 into gtb1-PL301 cells that had only the native copy of pkl1 present. Microscopy was performed using a motorized fine focus drive and a CCD camera to take serial Z-section images of living cells (see MATERIALS AND METHODS). In this manner, images of the microtubule arrays present throughout the entire cell were captured as a 3D projection (Figure 5, A and B, gtb1-PL301 cells). The dynamic behavior of microtubules and the reorganization of microtubules into larger bundles was monitored over time by capturing and observing stacks of images at selected intervals (see submitted videos; 3D clips from the movies are shown in Figure 5, D, wild-type cells, and E–H, gtb1-PL301 cells). Because acquiring each complete 3D stack of images takes 10–15 s, rate measurements on the same cells are not possible. Nonetheless, the 3D time-lapse images of microtubules are invaluable for monitoring microtubule organization and dynamic behavior within the entire cell as it occurs in wild type versus the mutant.
In gtb1-PL301 cells grown at 19 or 23°C, the spindle extended the entire length of the cell (Figure 5A, left) or was curved around cell ends (Figure 5A, right). In each case the bright zone of microtubule overlap remained constant at one-half to one-third of the cell length, consistent with excessive polymerization versus depolymerization as the cause for increased length. At 23°C cells frequently progressed to cytokinesis while retaining the mitotic spindle (Figure 5B). In wild-type cells spindle microtubules are normally depolymerized before cytokinesis is complete. New microtubule arrays emanating from the cell central MTOCs were often abnormal in appearance and number when the spindle failed to depolymerize (Figure 5B, upper cell). Astral microtubules were also affected at these temperatures and could be abnormally long (Figure 5, A left cell, and B, lower cell). Variation in astral microtubule number and length have been observed previously in fixed cells among the phenotypes present when γ-tubulin is depleted (for review, see Martin et al., 1997, discussion). To confirm spindle hyperelongation in the mutant, GFP-NADPH cytochrome P450 reductase was again used (Figure 5C; also refer back to Figure 4D) to visualize the nuclear envelope surrounding the spindle. Three time-lapse images from a movie (see videos) are shown. Although the chromosomes appear to have segregated to the poles, and cytokinesis is proceeding, the spindle continues to extend in length. In the cell shown the spindle has curved around the cell end and back toward the cell middle. Spindle hyperelongation was also observed when chromosomes trailed along the spindle (our unpublished data) and could be twice the cell length. Thus, dependent on the temperature, cells either arrested in mitosis or proceeded to cytokinesis with unsuccessful spindle microtubule depolymerization.
Cytoplasmic microtubules in the mutant, at temperatures that did not result in mitotic arrest, were also abnormal. 3D time-lapse video microscopy of these microtubule arrays at 30°C revealed altered organization and dynamics. In wild-type S. pombe, cytoplasmic microtubules consist of six to eight microtubule bundles that extend between the cell ends and form a basket around the nucleus (Figure 5D). Two to three microtubules are likely to be present in each bundle and are believed to be of mixed polarity (for review, see Hagan, 1998). Cytoplasmic microtubule arrays in gtb1-PL301 cells at 30°C with multiple copies of pkl1 present (our unpublished data) are similar to wild-type arrays (Figure 5D). However, when only the native copy of pkl1 was present (Figure 5, E–H), a single larger microtubule bundle was predominant and positioned at the cell periphery. In some cells this was the only microtubule array present (Figure 5, E and F), whereas in other cells an inner array of microtubules was also observed (Figure 5, G and H). Although catastrophe was observed in these bundles, it appeared limited to a region near the very ends of microtubules, because overall microtubule length did not significantly diminish and in fact increased. However, it is impossible to resolve depolymerization of a single microtubule when it is within a bundle.
The dynamic behavior observed for the cytoplasmic microtubule bundles is best illustrated by the cells shown in Figure 5, E and F. In Figure 5E either microtubule ends appear unable to depolymerize, or depolymerization is outpaced by polymerization. The microtubule bundle behaves as a contiguous structure and as a whole begins to invaginate (top arrow) away from the cell edge. This continues until a portion of the invaginating bundle is near perpendicular to its original position and snaps, presumably from tension (bottom arrow). Despite the breaking of the microtubules, the majority of this bundle remains intact. In the cell in Figure 5F, microtubules in a ring move in opposite directions to each other, as if sliding over each other. In the top panel, microtubule ends come together (upper arrows), cross each other, and then appear to flatten down onto the bundle (middle image in the series). A seamless circle of microtubules is momentarily observed, before further time points reveal a new opening in the circular array (lower arrows), and the process repeats as these ends come together. The common feature of all of the altered cytoplasmic microtubule arrays was a propensity to bundle into a single larger array and the stability of these arrays.
Cellular Localization of GFP-gtb1-PL301 Is Similar to Wild Type
To determine whether localization of the γ-tubulin mutant was similar to the wild-type protein, we constructed a functional genomic GFP-γ-tubulin fusion protein under control of its native promoter for visualization of γ-tubulin in living cells (GFPgtb1p; see MATERIALS AND METHODS). GFPgtb1p was able to substitute effectively for wild-type γ-tubulin in plasmid shuffle of SL1 cells and in a deletion strain of γ-tubulin (see MATERIALS AND METHODS; our unpublished data). Mutant gtb1-PL301 cells that carry GFPgtb1p no longer arrested at 19°C, and normal G2 microtubule arrays were observed (Figure 6A). The cellular location of the fusion protein (Figure 6B) corresponded to that observed for wild-type γ-tubulin by antibody localization (Hagan, 1998).
Figure 6.
Mutant γ-tubulin and Pkl1p localization and function were examined using GFP fusion proteins. (A) G2 arrays at 19°C in gtb1-PL301 cells with single-copy pkl1 carrying the GFPgtb1p (wild-type) fusion protein. (B) Left, GFPgtb1p localization at the two SPB MTOCs and at the cell equatorial MTOC. The central location is a transient stage and not always readily detected. Right, G2 cells with single SPB dots. (C) GFPgtb1-PL301 (mutant protein) integrated in gtb1-PL301 cells with single-copy pkl1. (D) Streaked cells of wild-type (left) and mutant gtb1-PL301 (right) carrying multiple copies (upper) or the integrated GFPgtb1-PL301 construct (lower) grown at 19°C. (E and F) GFPpkl1p localization in the γ-tubulin mutant and effect of overexpression on spindle dynamics. (E) Two time points showing spindle shrinkage by GFPpkl1p (left, early; right, late). Fluorescence staining of the spindle and poles is from GFPpkl1p; background fluorescence (dumbbell shape) shows the general outline of the nucleus. (F) Pkl1p localization to the spindle and poles is less evident in late anaphase as reported for wild-type cells. Bars, 5 μm.
To observe localization of the mutant γ-tubulin protein, we constructed a GFPgtb1-PL301 fusion protein (see MATERIALS AND METHODS), and transformed the plasmid into gtb1-PL301 cells that had only the native pkl1 gene present. Whether integrated (Figure 6C) or on a multicopy plasmid (our unpublished data), GFPgtb1-PL301p showed similar localization as wild type, with similar intensity of fluorescence. We also observed that the presence of multicopy plasmid GFP-gtb1-PL301 did not prevent mitotic arrest at 19°C (Figure 6D) and did not alter the phenotype.
Pkl1p Localization and Function Are Unaffected by the γ-Tubulin Mutation
The PL301 mutation is predicted to lie on a lateral surface of γ-tubulin and to affect nontubulin protein interactions. We examined whether Pkl1p localization or activity were altered in the γ-tubulin mutant using the fusion protein GFPpkl1p. This fusion protein is able to replace Pkl1p in plasmid shuffle with the γ-tubulin mutant (our unpublished data) and shows similar localization as with antibody (Pidoux et al., 1996). We found no change in localization or behavior of GFPpkl1p in the mutant versus wild-type cells. The protein was seen along the spindle and at the poles, and multicopy GFPpkl1p was able to cause spindle shrinkage in the mutant cells (Figure 6E). As in wild-type cells, GFPpkl1p staining in the γ-tubulin mutant (25°C shown) was less prominant in late anaphase spindles (Figure 6F; Pidoux et al., 1996).
Stable Microtubules in gtb1-PL301 and β-Tubulin benA33 Cells Share Similarities
γ-Tubulin was discovered as a suppressor of A. nidulans benA33, a mutation in β-tubulin that resulted in mitotic arrest with a spindle present. Because of the similarity in the mitotic arrest phenotypes and the involvement of γ-tubulin, we tested gtb1-PL301 cells (single-copy pkl1) for known features relating to benA33 microtubules. Oakley et al. (1987) showed that a destabilizing mutation in α-tubulin was capable of rescuing the benA33 phenotype; however, microtubule-destabilizing drugs were only partially effective (Oakley and Morris, 1981). We combined the gtb1-PL301 mutation with the α-tubulin microtubule-destabilizing mutation nda2-340. Tetrads analyzed from this cross (Figure 1E) indicated that the conditional growth arrest at 19°C present in the parental strains is not seen in the combined genetic background. Growth in the double mutant was similar to that in wild type. As in benA33 cells, gtb1-PL301 cells were also found to be thiabendazole resistant. Cell growth in the presence of 5–200 μg/ml thiabendazole (described in MATERIALS AND METHODS) did not prevent growth arrest at 19°C, although microtubules were depolymerized at the higher levels. Therefore, as with benA33, not all methods of destabilizing microtubules relieve the mitotic arrest of gtb1-PL301.
DISCUSSION
γ-Tubulin is a specialized tubulin isoform that is a component of MTOCs and required for nucleation of spindle microtubules (Oakley and Oakley, 1989; see INTRODUCTION). We propose an additional essential role for γ-tubulin at the MTOC in regulating microtubule dynamics and organization for spindle segregation of chromosomes and the establishment of normal cytoplasmic microtubule arrays. This role appears to be genetically separable from microtubule nucleation and not the result of mislocalization of mutant γ-tubulin. That human γ-tubulin was able to rescue all aspects of the phenotype suggests a conserved role for γ-tubulin in these processes.
Befitting the phenotype, the γ-tubulin mutation was recovered in a synthetic lethality screen with the Ncdp/Kar3p family klp, Pkl1p. This klp localizes to the spindle and poles and forces spindle shrinkage or collapse when overexpressed (Pidoux et al., 1996). Thus the γ-tubulin mutation stabilizes spindle microtubules, whereas Pkl1p appears to have opposite functions. Pkl1p was required for viability of the γ-tubulin mutant and needed in multicopy to restore near wild-type morphology. Its localization and function appear unaltered, suggesting that Pkl1p is needed to rescue a lost redundant function. By predicting the 3D structure of γ-tubulin, we have localized the mutation and present possible models for the requirement of Pkl1.
Anaphase A Is Impaired in the gtb1-PL301 Mutant and Is Mad2p Dependent
The essential process impaired in the γ-tubulin PL301 mutant is the ability to effectively segregate chromosomes. A bipolar spindle is formed, and chromosomes are condensed and grouped, but anaphase A is blocked. The spindle checkpoint protein Mad2p senses both attachment and tension of kinetochore microtubules, and Mad2p removal occurs as kinetochore microtubules accumulate (Waters et al., 1999). In S. pombe, as in mammalian cells, multiple microtubules attach to each kinetochore (Ding et al., 1993), and Mad2p regulates the spindle checkpoint (He et al., 1997). The extent of kinetochore microtubule attachment to chromosomes in the mutant at 19°C was unclear. At elevated temperatures segregation did occur; however, trailing chromosomes in many cells indicated that chromosome attachment was not always effectively maintained. Deletion of the spindle checkpoint gene mad2 removed the cold-sensitive mitotic arrest without severely reducing cell viability. This favors the view that microtubule attachment to kinetochores is incomplete or lacks tension, rather than being altogether absent. In budding yeast S. cerevisiae, a variety of lesions, including spindle pole-associated events, are able to activate the spindle checkpoint (Hardwick et al., 1999). It is possible in the mutant that Mad2p, in addition to sensing problems at the kinetochore, is also detecting a problem at the spindle poles relating to the γ-tubulin functions we have proposed.
We attribute the failure of appropriate kinetochore attachment to the increased microtubule stability and impaired dynamics in the mutant. That cold temperatures exacerbate the phenotype is consistent with previous studies demonstrating that two classes of microtubules are especially stable at cold temperatures, kinetochore microtubules (Brinkley and Cartwright, 1975; Rieder, 1981) and microtubules of the midbody (Salmon et al., 1976; Mullins and McIntosh, 1982). Stabilization of spindle microtubules against depolymerization could prevent effective search and capture of kinetochores (for review, see Desai et al., 1997) and block timely removal of the spindle and may have contributed to spindle hyperelongation.
The γ-Tubulin Mutation Alters Cytoplasmic Microtubule Arrays
Growth at 30°C does not result in mitotic arrest of gtb1-PL301 cells. However, without multicopy pkl1 present, cytoplasmic microtubule arrays are combined into a few bundles or a single large bundle. 3D time-lapse video microscopy using GFP-α-tubulin showed that a single microtubule or a subset of microtubules often extruded briefly a short distance from the large bundle end. Extruded microtubules were dynamic and underwent growth and catastrophe, but the length of the large bundle itself was maintained or increased in size. This suggests that microtubule polymerization is outpacing depolymerization.
In S. pombe, new cell equatorial MTOCs become active for nucleation late in mitosis and along with SPB MTOCs contribute to the microtubule arrays of daughter cells (for review, see Su and Yanagida, 1997; Hagan, 1998). Multicopy pkl1 is not able to alleviate mitotic arrest in the mutant; however, it does restore normal cytoplasmic microtubule arrays and cell morphology. Because there is no evidence that Pkl1p has cytoplasmic functions, we expect that excess Pkl1p may improve the transition in microtubule arrays from mitosis to cytokinesis, perhaps by helping remove the spindle. In the mutant, retention of the spindle during cytokinesis resulted in a prominent knot of microtubules that impaired formation of these arrays. Abnormal astral microtubules may have contributed to the spindle hyperelongation phenotype that was also observed. The extended length or absence of astral microtubules may have interfered with any cortical sensing of spindle length for triggering depolymerization of spindle microtubules.
Pkl1p Helps Regulate a Subset of γ-Tubulin Functions
The γ-tubulin mutation gtb1-PL301 is synthetically lethal with loss of Pkl1p. Another mutant isolated in the same screen, SL2, is suppressible by multicopy γ-tubulin and arrests with defects in chromosome segregation. Both mutations suggest that Pkl1p function may be closely linked with that of γ-tubulin in regulating microtubule dynamics for anaphase A. Another member of this family of klps, Drosophila Ncdp, is required for proper localization of γ-tubulin during meiosis II in vivo (Endow and Komma, 1998).
Pkl1p may or may not interact directly with γ-tubulin to rescue the mutant phenotype. However, we favor a mechanism requiring Pkl1p binding to γ-tubulin based on the localization of Pkl1p, involvement of γ-tubulin with SL2, and our preliminary evidence that Pkl1p antibody can coimmunoprecipitate γ-tubulin (J.L. Paluh and W.Z. Cande, unpublished results). The localization of Pkl1p and its ability to affect spindle dynamics appear normal in the γ-tubulin mutant. Thus, if Pkl1 binds to γ-tubulin, it may rescue or restore an altered protein interaction at another site on γ-tubulin, closer to the site of the mutation. This view makes sense because Pkl1p is not essential in wild type, and impaired Pkl1p function should be tolerated. Because multicopy pkl1 allows normal cytoplasmic arrays, the mutation is unlikely to enhance Pkl1p binding or function. If Pkl1p does bind to γ-tubulin, this interaction could help tether or organize microtubules at the MTOC, because Pkl1p can bundle microtubules in vitro (Pidoux et al., 1996). Alternatively, such an interaction could be required for tubulin dimer removal by Pkl1p. It is not known whether yeast kinetochore microtubules undergo flux and, if so, what contribution it has to anaphase A. Although the kinetochore is the primary site of microtubule disassembly in somatic cells for anaphase A, flux coupled to depolymerization of microtubule minus ends is the predominant mechanism in Xenopus (Desai et al., 1998, and references therein). Further investigation will be needed to decipher the exact mechanism underlying Pkl1p rescue of this subset of γ-tubulin functions.
We predicted the 3D structure of γ-tubulin (Figure 3) to allow us to approximate the location of the gtb1-PL301 mutation and to better understand the mechanism behind the phenotype. In Figure 3, B and C, we have highlighted residues that are absolutely conserved in α- and β-tubulins but different in γ-tubulins. The PL301 mutation is not positioned near known sites of tubulin–tubulin contact and so is not expected to affect microtubules directly. It lies at the edge of a divergent region on the γ-tubulin surface, which is flanked on the opposite side by two helices. The similarly positioned helices in microtubules, helices 11 and 12, have been identified as primary sites of interaction with klp motor domains (for review, see Mandelkow and Hoenger, 1999). The importance of a divergent face on γ-tubulin is unknown. Still, the 3D model is expected to provide a useful tool for analyzing other γ-tubulin mutations as well as non-tubulin protein interactions.
Two obvious candidates for proteins whose functions may be altered in gtb1-PL301 are the conserved γ-tubulin binding MTOC components. Sequence homologues to the S. cerevisiae Spc97p and Spc98p proteins (Knop and Schiebel, 1997) are present in the S. pombe database. Another protein that helps organize spindle poles in metazoans and astral microtubules in fungi is dynein. How dynein interacts with microtubules or other proteins to accomplish this is not known. Although loss of dynein is synthetically lethal with loss of Kar3p in S. cerevisiae or loss of BimCp in A. nidulans, two klps with known mitotic functions (Saunders et al., 1995; DeZwaan et al., 1997; Efimov and Morris, 1998), it is not synthetically lethal with loss of Pkl1p (J.L. Paluh and W.Z. Cande, unpublished results). However, it is still possible that dynein may influence organization of astral or interphase arrays in S. pombe.
The ability of γ-tubulin to associate with specialized classes of proteins, including MTOC components, microtubules, and motor proteins, is practical. It would provide this central MTOC component with the capability to intimately regulate microtubule minus end dynamics and organization as well as nucleation. Our results clearly indicate that γ-tubulin itself can dramatically affect microtubule dynamics and organization. With this broader view of γ-tubulin function, the challenge will be to decipher the protein interactions that mediate both important roles and how these interactions are coordinated.
Phenotypic Similarities between Spindle Phenotypes of S. pombe gtb1-PL301 and A. nidulans benA33
γ-Tubulin was originally identified as an extragenic suppressor of a mutation in β-tubulin, A. nidulans benA33, that resulted in mitotic arrest with a stable bipolar spindle (Oakley and Oakley, 1989; Burns, 1995). Its discovery in this manner has remained intriguing, because spindle nucleation obviously occurred and because α-tubulin is now known to be unequivocally positioned as the ultimate subunit at the minus end of the microtubule (Mitchison, 1993; Fan et al., 1996; Nogales et al., 1999). An end-on interaction between γ-tubulin and β-tubulin therefore is unlikely. It has been postulated that the role of γ-tubulin in suppression of the benA33 phenotype might not relate to microtubule nucleation but instead to some as yet undefined role for γ-tubulin (Burns, 1995). Although the mechanism of γ-tubulin suppression of benA33 mitotic arrest is unknown (Oakley and Morris, 1981; Jung et al., 1998), striking similarities are present with the gtb1-PL301 phenotype. Both results clearly argue for a broader view of γ-tubulin function.
Supplementary Material
ACKNOWLEDGMENTS
We authors are indebted to several laboratories for supplying yeast strains, plasmids, and antibodies (see MATERIALS AND METHODS), Dr. Satoru Uzawa for assistance with time-lapse video microscopy and lively discussions on S. pombe mitosis, Pete Carlton for help with Deltavision, Defne Yarar who helped construct the GFPgtb1p fusion protein, and Reena Zalpuri for advice on TEM. We also thank Dr. Frank W. Pfrieger, Dr. Rebecca Heald, and Cande laboratory members, particularly Carrie Cowan and So-Ching Brazer, for helpful suggestions and comments concerning this work. J.L.P. is supported by a fellowship from the National Institutes of Health (NIH), grants 5 F32 GM-19145-02 and GM-23238; E.N. is supported by NIH grant GM-51487; B.R.O. is supported by NIH grant GM31837.
Abbreviations used:
- B
β-sheet
- CCD
charge-coupled device
- EGFP
enhanced green fluorescent protein
- GFP
green fluorescent protein
- H
helix
- klp
kinesin-like protein
- NDF
neutral density filter
- SPB
spindle pole body
- 3D
three dimensional
- TEM
transmission electron microscopy
Footnotes
Online version of this article contains video material for Figure 5. Online version available at www.molbiolcell.org.
REFERENCES
- Brinkley BR, Cartwright J., Jr Cold-labile and cold-stable microtubules in the mitotic spindle of mammalian cells. Ann NY Acad Sci, 1975;253:428–439. doi: 10.1111/j.1749-6632.1975.tb19218.x. [DOI] [PubMed] [Google Scholar]
- Burke D, Gasdaska P, Hartwell L. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns RG. Analysis of the γ-tubulin sequences: implications for the functional properties of γ-tubulin. J Cell Sci. 1995;108:2123–2130. doi: 10.1242/jcs.108.6.2123. [DOI] [PubMed] [Google Scholar]
- Chen H, Swedlow JR, Grote MA, Sedat JW, Agard DA. The collection, processing and display of digital three- dimensional images of biological specimens. In: Pawley J, editor. Handbook of Biological Confocal Microscopy. New York: Plenum Press; 1995. pp. 197–210. [Google Scholar]
- Compton DA. Focusing on spindle poles. J Cell Sci. 1998;111:1477–1481. doi: 10.1242/jcs.111.11.1477. [DOI] [PubMed] [Google Scholar]
- Desai A, Maddox PS, Mitchison TJ, Salmon ED. Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J Cell Biol. 1998;141:703–713. doi: 10.1083/jcb.141.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TA. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. KinI kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111:701–712. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing KH, Nogales E. Tubulin and microtubule structure. Curr Opin Cell Biol. 1998;10:16–22. doi: 10.1016/s0955-0674(98)80082-3. [DOI] [PubMed] [Google Scholar]
- Efimov VP, Morris NR. A screen for dynein synthetic lethals in Aspergillus nidulans identifies spindle assembly checkpoint genes and other genes involved in mitosis. Genetics. 1998;149:101–116. doi: 10.1093/genetics/149.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. Commitment to meiosis in fission yeast. Mol Gen Genet. 1973;121:277–284. [Google Scholar]
- Egel R, Willer M, Kjaerulff S, Davey J, Nielsen O. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast. 1994;10:1347–1354. doi: 10.1002/yea.320101012. [DOI] [PubMed] [Google Scholar]
- Endow SA, Chandra R, Komma DJ, Yamamoto AH, Salmon ED. Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J Cell Biol. 1994a;107:859–867. doi: 10.1242/jcs.107.4.859. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast KAR3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994b;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Komma DJ. Centrosome and spindle function of the Drosophila Ncd microtubule motor visualized in live embryos using Ncd-GFP fusion proteins. J Cell Sci. 1996;109:2429–2442. doi: 10.1242/jcs.109.10.2429. [DOI] [PubMed] [Google Scholar]
- Endow SA, Komma DJ. Assembly and dynamics of an anastral:astral spindle: the meiosis II spindle of Drosophila oocytes. J Cell Sci. 1998;111:2487–2495. doi: 10.1242/jcs.111.17.2487. [DOI] [PubMed] [Google Scholar]
- Fan J, Griffiths AD, Lockhart A, Cross RA, Amos LA. Microtubule minus ends can be labeled with a phage display antibody specific to alpha-tubulin. J Mol Biol. 1996;259:325–330. doi: 10.1006/jmbi.1996.0322. [DOI] [PubMed] [Google Scholar]
- Fantes PA. Cell cycle controls. In: Nasim A, Young P, Johnson BF, editors. Molecular Biology of the Fission Yeast. San Diego: Acdemic Press; 1989. pp. 127–204. [Google Scholar]
- Felix M-A, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of γ-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gueth-Hallonet C, Antony C, Aghion J, Santa-Maria A, Lajoie-Mazenc I, Wright M, Maro B. γ-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci. 1993;105:157–166. doi: 10.1242/jcs.105.1.157. [DOI] [PubMed] [Google Scholar]
- Hagan IM. The fission yeast microtubule cytoskeleton. J Cell Sci. 1998;111:1603–1612. doi: 10.1242/jcs.111.12.1603. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 1990;347:563–566. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Li R, Mistrot C, Chen R-H, Dann P, Rudner A, Murray AW. Lesions in many different spindle components activate the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics. 1999;152:509–518. doi: 10.1093/genetics/152.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The nda3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Horio T, Oakley BR. Human γ-tubulin functions in fission yeast. J Cell Biol. 1994;126:1465–1473. doi: 10.1083/jcb.126.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast γ-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Huyett A, Kahana J, Silver P, Zeng X, Saunders WS. The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J Cell Sci. 1998;111:295–301. doi: 10.1242/jcs.111.3.295. [DOI] [PubMed] [Google Scholar]
- Inoue S, Turgeon BG, Yoder OC, Aist JR. Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J Cell Sci. 1998a;111:1555–1566. doi: 10.1242/jcs.111.11.1555. [DOI] [PubMed] [Google Scholar]
- Inoue S, Yoder OC, Turgeon BG, Aist JR. A cytoplasmic dynein required for mitotic aster formation in vivo. J Cell Sci. 1998b;111:2607–2614. doi: 10.1242/jcs.111.17.2607. [DOI] [PubMed] [Google Scholar]
- Javerzat JP, Cranston G, Allshire RC. Fission yeast genes which disrupt mitotic chromosome segregation when overexpressed. Nucleic Acids Res. 1996;24:4676–4683. doi: 10.1093/nar/24.23.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BF, Miyata M, Miyata H. Morphogenesis of fission yeasts. In: Nasim A, Young P, Johnson BF, editors. Molecular Biology of the Fission Yeast. San Diego: Academic Press; 1989. pp. 331–366. [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. γ-Tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Jung MK, May GS, Oakley BR. Mitosis in wild type and β-tubulin mutant strains of Aspergillus nidulans. Fungal Genet Biol. 1998;24:146–160. doi: 10.1006/fgbi.1998.1057. [DOI] [PubMed] [Google Scholar]
- Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, Silver PA. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol Biol Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli j, Hottinger H, Munz P, Strauss A, Thuriaux P. Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics. 1977;87:471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Joshi HC. γ-Tubulin is a minus-end specific microtubule binding protein. J Cell Biol. 1995;131:207–214. doi: 10.1083/jcb.131.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP. γ-Tubulin in Arabidopsis: gene sequence, immunoblot, and immunofluorescence studies. Plant Cell. 1994;6:303–314. doi: 10.1105/tpc.6.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E, Hoenger A. Structures of kinesin and kinesin-microtubule interactions. Curr Opin Cell Biol. 1999;11:34–44. doi: 10.1016/s0955-0674(99)80005-2. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MA, Osmani SA, Oakley BR. The role of γ-tubulin in mitotic spindle formation and cell cycle progression in Aspergillus nidulans. J Cell Sci. 1997;110:623–633. doi: 10.1242/jcs.110.5.623. [DOI] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a γ-tubulin-associated protein with an essential role in γ-tubulin ring complex (γTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Shibata T. Role of γ-tubulin in mitosis-specific microtubule nucleation from the Schizosaccharomyces pombe spindle pole body. J Cell Sci. 1996;109:165–177. doi: 10.1242/jcs.109.1.165. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- McDonald AR, Liu B, Joshi HC, Palevitz BA. γ-Tubulin is associated with a cortical-microtubule-organizing zone in the developing guard cells of Allium cepa L. Planta. 1993;191:357–361. doi: 10.1007/BF00195693. [DOI] [PubMed] [Google Scholar]
- McDonald K. High pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol Biol. 1999;117:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- Melki R, Vainberg IE, Chow RL, Cowan NJ. Chaperonin-mediated folding of vertebrate actin-related protein and γ-tubulin. J Cell Biol. 1993;122:1301–1310. doi: 10.1083/jcb.122.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. Localization of an exchangeable GTP binding site at the plus end of microtubules. Science. 1993;261:1044–1047. doi: 10.1126/science.8102497. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moser MJ, Lee SY, Klevit RE, Davis TN. Ca2+ binding to calmodulin and its role in Schizosaccharomyces pombe as revealed by mutagenesis and NMR spectroscopy. J Biol Chem. 1995;270:20643–20652. doi: 10.1074/jbc.270.35.20643. [DOI] [PubMed] [Google Scholar]
- Mullins JM, McIntosh JR. Isolation and initial characterization of the mammalian midbody. J Cell Biol. 1982;94:654–661. doi: 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Joshi HC, Besharse J. γ-Tubulin in differentiated cell types: localization in the vicinity of basal bodies in retinal photoreceptors and ciliated epithelia. J Cell Sci. 1993;104:1229–1237. doi: 10.1242/jcs.104.4.1229. [DOI] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian γ-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the α/β tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Oakley BR. γ-Tubulin. In: Hyam JS, Lloyd CW, editors. Microtubules. New York: John Wiley and Sons; 1994. pp. 33–45. [Google Scholar]
- Oakley BR, Morris NR. A β-tubulin mutation in Aspergillus nidulans that blocks microtubule function without blocking assembly. Cell. 1981;24:837–845. doi: 10.1016/0092-8674(81)90109-4. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Rinehart JE. Conditionally lethal tubA α-tubulin mutations in Aspergillus nidulans. Mol Gen Genet. 1987;208:135–144. doi: 10.1007/BF00330434. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. γ-Tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of γ-tubulin, a new member of the tubulin superfamily encoded by the mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios M, Joshi HC, Simerly D, Schatten G. Dynamic reorganization of γ-tubulin during mouse fertilization and early development. J Cell Sci. 1993;104:383–389. doi: 10.1242/jcs.104.2.383. [DOI] [PubMed] [Google Scholar]
- Pereira G, Knop M, Schiebel E. Spc98p directs the yeast γ-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol Biol Cell. 1998;9:775–793. doi: 10.1091/mbc.9.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, LeDizet M, Cande WZ. Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol Biol Cell. 1996;7:1639–1655. doi: 10.1091/mbc.7.10.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW, Kellogg DR, Alberts BM. Drosophila γ-tubulin is part of a complex containing two previously identified centrosomal MAPs. J Cell Biol. 1993;121:823–835. doi: 10.1083/jcb.121.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- Rizzolo LJ, Joshi HC. Apical orientation of the microtubule organizing center and associated γ-tubulin during the polarization of the retinal pigment epithelium in vivo. Dev Biol. 1993;157:147–156. doi: 10.1006/dbio.1993.1119. [DOI] [PubMed] [Google Scholar]
- Salmon ED, Goode D, Maugel TK, Bonar DB. Pressure-induced depolymerization of spindle microtubules. III. Differential stability in HeLa cells. J Cell Biol. 1976;69:443–454. doi: 10.1083/jcb.69.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent γ-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle. J Cell Biol. 1997;138:957–960. doi: 10.1083/jcb.138.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Su SSY, Yanagida M. Mitosis and cytokinesis in the fission yeast, Schizosaccharomyces pombe. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 765–825. [Google Scholar]
- Sunkel CE, Gomes R, Sampaio P, Perdigao J, Gonzalez C. γ-Tubulin is required for the structure and function of the microtubule organizing center in Drosophila neuroblasts. EMBO J. 1995;14:28–36. doi: 10.1002/j.1460-2075.1995.tb06972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange Y, Horio T, Shimanuki M, Ding D-Q, Hiraoka Y, Niwa O. A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J Cell Biol. 1998;140:247–258. doi: 10.1083/jcb.140.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Celati C, Moudjou M, Bornens M. Characterization of the human homologue of the yeast spc98p and its association with γ-tubulin. J Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Adachi Y, Hiraoka Y, Yanagida M. Identification of the pleiotropic cell division cycle gene nda2 as one of two different α-tubulin genes in Schizosaccharomyces pombe. Cell. 1984;37:233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Umesono K, Toda T, Hayashi S, Yanagida M. Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J Mol Biol. 1983;168:271–284. doi: 10.1016/s0022-2836(83)80018-7. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Gorbsky GJ, Salmon ED, Niklas RB. Mad2 binding by phosphorylated kinetochores links error detection and checkpoint action in mitosis. Curr Biol. 1999;9:649–652. doi: 10.1016/s0960-9822(99)80287-5. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.