Abstract
Unusual amino acids such as β-methoxytyrosine (β-MeOTyr), allo-Thr and allo-Ile were derivatized with Nα-(2,4-dinitro-5-fluorophenyl)-l-alaninamide (FDAA), 2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl isothiocyanate (GITC), (S)-N-(4-nitrophenoxycarbonyl)phenylalanine methoxyethyl ester (S-NIFE), or o-phthalaldehyde/isobutyryl-l-cysteine (OPA-IBLC), and then separated via reversed phase high performance chromatography followed by UV and electrospray ionization mass spectrometry detection. FDAA generally showed the highest enantioselectivity but the lowest sensitivity among the chiral derivatizing agents (CDAs) investigated. The detection limit of FDAA-derivatized amino acids was in the low picomolar range. Although the enantioselectivity of FDAA derivatives was generally quite high, its selectivity among β-MeOTyr isomers was poor. The best separation of β-MeOTyr stereoisomers was achieved with S-NIFE. Due to the complex relationships between the investigated CDAs, stereochemical analyses using a combination of two or more of the CDAs gave the most reliable results for a given separation problem. In general, the methods described are selective and reliable, and are being applied to the analysis of unusual amino acids as they occur in marine peptides.
Keywords: chirality, Marfey's reagent, GITC, S-NIFE, OPA-IBLC, hydrolysis, amino acid, liquid chromatography, electrospray-mass spectrometry
1. Introduction
In recent years, considerable effort has been made to elucidate the structures of novel and pharmacologically active marine natural products, in particular, peptides and depsipeptides (peptides that contain at least one ester linkage) [1-4]. One main focus in marine biomedical research is currently the purification, characterization and total synthesis of marine peptides with high potential as anti-cancer and anti-HIV lead compounds [2, 5-7]. The cyclic depsipeptide Callipeltin A, which was isolated from the New Caledonian Lithistida sponge Callipelta sp., is one of the more prominent examples [7]. The structural elucidation of Callipeltin A was soon followed by the discovery of Callipeltin B and C [8] and other cyclic depsipeptides such as Papuamides A-D, which were isolated from the Lithistida sponges Theonella mirabilis and T. swinhoei [6].
Marine peptides and depsipeptides not only contain members of the 22 known proteinogenic L-amino acids [9-11] but they often contain D- and allo-configurated amino acid isomers and various other non-proteinogenic amino acids [1-2, 6-8]. Some of the unusual components identified in marine peptides include amino acid residues such as β-methoxy-tyrosine (β-MeOTyr), homoproline (homoPro), and numerous methylated amino acids [1-2, 6-8].
Interest in the synthesis of marine peptides has been driven by the diverse pharmacological activities of these compounds [2, 12-14]. Efforts to synthesize the callipeltins and papuamides are underway in several laboratories and the synthesis of several amino acid components of these compounds have already been reported [14-18]. These syntheses are demanding, because enantiomerically pure unusual amino acids need to be prepared prior to the assembly of the parent peptide. Syntheses are complicated by the fact that the enantiomeric excess of synthetic amino acids is often less than 100%, even when asymmetric synthetic routes are applied. Successful syntheses also rely on the correct stereochemical assignment of all of the amino acids in the naturally occurring peptides.
However, the structural elucidation of complex marine peptides, which occur in low abundance, is challenging and mistaken stereochemical assignments can occur [19]. While NMR studies can often establish the structure of the individual amino acid residues in a peptide, assignment of the configuration is generally achieved through HPLC or GCMS methodologies. These techniques rely on acid hydrolysis of the parent peptide, derivatization of the resulting amino acids and comparison with appropriate amino acid standards. The development of sensitive methods that allow the identification of the amino acid residues and the assignment of their absolute configuration is therefore essential. MS investigations of marine peptides can be done in a manner similar to classical composition analysis of peptides and proteins, namely by acid hydrolysis and subsequent separation of the resulting amino acid mixture by direct or indirect chromatography [20]. In trace analysis, indirect liquid chromatography (LC) that uses highly sensitive labels is often preferred over direct chromatography, which uses expensive short-lived optically active stationary phases. It is also preferred over indirect gas chromatography (GC), which frequently requires more difficult derivatizations to prepare volatile derivatives, even though some elegant methods have been described for GC [40]. For LC, both the amino group and the carboxylic acid group can be derivatized to enhance chromatographic properties and detector response of the analyte. Labeling of the amino group is much more common than labeling the carboxylic acid group. Thus more derivatization agents are available for the amino group. Among the chiral derivatizing agents (CDAs) that have been used for indirect chromatography, Marfey's reagent Nα-(2,4-dinitro-5-fluorophenyl)-l-alaninamide (FDAA) [21] has been most widely used for assigning the stereochemistry of amino acids in trace amounts [22-31]. Other CDAs include 2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl isothiocyanate (GITC) [30-33], (S)-N-(4-nitrophenoxycarbonyl)phenylalanine methoxyethyl ester (S-NIFE) [34-36, 43] and o-phthalaldehyde/isobutyryl-l-cysteine (OPA-IBLC) [22, 37].
In this paper, the separation and stereochemical assignment of a variety of unusual amino acids by reversed phase high performance liquid chromatography-electrospray ionization mass spectrometry (RPHPLC-ESI-MS) is described. The 34 amino acids investigated included uncommon amino acids such as β-methoxytyrosine (β-MeOTyr), allo-configured and/or N-methylated forms of threonine (Thr), isoleucine (Ile) and phenylalanine (Phe). Indirect chromatography was carried out after precolumn derivatization with FDAA, GITC, S-NIFE, and OPA-IBLC separately, and subsequent separation of the resulting diastereomers of each amino acid monitored by UV and MS. The chromatographic separation of the unusual amino acids could generally be achieved in a single HPLC run. Nevertheless, for certain amino acids that might coelute if the conditions were to be changed and thus, to increase confidence in the results, it is recommended that more than one CDA should be used to confirm the assignment of unusual amino acids. The strengths and limitations of each CDA are discussed in detail.
2. Experimental
2.1 Chemicals and reagents
Glacial acetic acid (99%), triethylamine, FDAA, GITC, OPA and available amino acid standards were purchased from Sigma (St. Louis, MO). S-NIFE was obtained from Peptisyntha (Brussels, Belgium), IBLC was from Calbiochem-Novabiochem (San Diego, CA). N-MeThr 5-8 were synthesized as previously described [6]. Boc protected N-MeLeu and N-MeIle standards were obtained from BACHEM Bioscience Inc., King of Prussia, PA. N-MeLeu 23-24 and N-MeIle 25-28 were generated from their Boc protected standards by deprotection with TFA. β-MeOTyr 31-34 were synthesized at the Department of Chemistry, Purdue University, West Lafayette, Indiana. A detailed chemical characterization of these compounds will be described in a separate paper. Acetonitrile and water were HPLC gradient grade quality and supplied by Fisher (Pittsburgh, PA). All other chemicals and reagents were of the highest available purity and were used without further purification.
2.2. Preparation of the solutions
Aqueous solutions of amino acids were prepared at a concentration of 1 mg/ml. If necessary, methanol was added to dissolve precipitating amino acids. Separate solutions were prepared for each of the CDAs: FDAA, GITC and S-NIFE were dissolved in acetone to give the respective 1% solutions. OPA was prepared as a 30 mM solution in methanol, and IBLC (90 mM) was dissolved in water. Triethylamine (6%) was used as the basic buffer (pH = 11.9) and acetic acid (5%) was used to quench the reactions.
2.3. Derivatization with FDAA, GITC, S-NIFE or OPA-IBLC
To a 1 μl (1μg/μl) aliquot of an aqueous solution of the amino acid or amino acid mixture, 10 μl of 6% triethylamine and 10 μl of a solution of CDA were added and allowed to react at rt for 10 min (GITC), 20 min (S-NIFE, OPA) or at 50°C for 1h (FDAA). In case of the OPA-IBLC derivatization, 10 μl of the IBLC solution was immediately added. The reaction mixture was diluted with 10 μl of 5% acetic acid and an aliquot (20 μl) was analyzed by LC-MS.
2.4. Liquid chromatography – mass spectrometry system (LC-MS)
Derivatized amino acids were separated and analyzed on an HP1100 LC-MSD (Agilent Technologies, Palo Alto, CA) consisting of a binary pump, a degasser, an autosampler, a DAD detector, an 1100 MSD and a ChemStation. A ZorbaxSB column (C18 150 × 2.1 mm, Agilent, Palo Alto, CA) equipped with an Opti-Guard column (C18 15 mm × 1 mm, Bodman, Baltimore, MD) was used for the separation of the derivatized amino acids. The column temperature was maintained at 50 °C and flow rate was 250 μl/min. Mobile phase A was 5% acetic acid pH 2.6 and B was acetonitrile containing 10% methanol. Linear gradients started with 5% mobile phase B and reached 50% over 50 min. To wash the column, the concentration of the mobile phase B was increased to 100% over 5 min and maintained for 5 min (in case of OPA-IBLC for 15 min) at 100% B. The diode array detector was monitored at 340 nm (FDAA derivatives), 254 nm (GITC and S-NIFE derivatives), and 280 nm (OPA-IBLC derivatives), respectively. Whenever necessary, UV spectra from 300 to 600 nm were also recorded. To avoid contamination of the mass spectrometer, the eluent was directed into the mass spectrometer after a 4 or 5 minute delay, depending on the reagent. The molecular mass of the samples was determined using the HP MSD mass spectrometer in positive ion mode. Data were acquired from m/z 300 to m/z 1000 every four seconds.
3. Results and Discussion
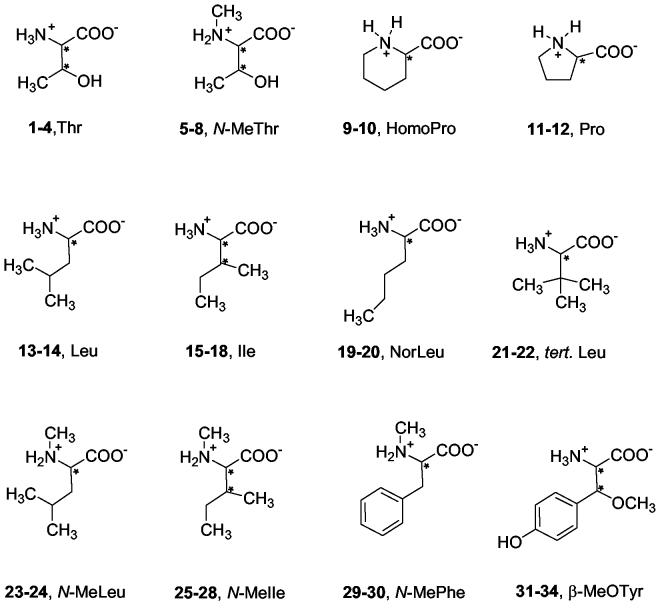
In this study, chiral amino acids 1-34 were investigated (Fig. 1). All amino acids were reacted under basic conditions with the respective CDAs to form the corresponding diastereomers (Fig. 2). Subsequently, derivatized amino acids were separated on a reversed phase column and analyzed by UV and ESI-MS. The major advantages of using a mass spectrometric detection are the increased sensitivity and selectivity that allows the identification and characterizations of unknowns. The mass spectrometer can thus be viewed as an added dimension to the first-dimensional chromatographic separation. Detection by both UV and MS facilitated the assignment of the peaks, especially when amino acids and possible side products of different m/z co-eluted and could therefore not be distinguished with UV detection only.
Figure 1.
Amino acids 1-34 were investigated in this study. 1 = L-Threonine (L-Thr), 2 = D-Threonine (D-Thr), 3 = L-allo-Threonine (L-allo-Thr), 4 = D-allo-Threonine (D-allo-Thr), 5 = L-N-Methyl-Threonine (L-N-MeThr), 6 = D-N-Methyl-Threonine (D-N-MeThr), 7 = L-allo-N-Methyl-Threonine (L-allo-N-MeThr), 8 = D-allo-N-Methyl-Threonine (D-allo-N-MeThr), 9 = L-homo-Proline = L-piperidine-2-carboxylic acid (L-homoPro), 10 = D-homo-Proline = D-piperidine-2-carboxylic acid (DhomoPro), 11 = L-Proline (L-Pro), 12 = D-Proline (D-Pro), 13 = L-Leucine (L-Leu), 14 = D-Leucine (D-Leu), 15 = L-Isoleucine (L-Ile), 16 = D-Isoleucine (D-Ile), 17 = L-allo-Isoleucine (L-allo-Ile), 18 = D-allo-Isoleucine (D-allo-Ile), 19 = L-Norleucine (L-NorLeu), 20 = D-Norleucine (D-NorLeu), 21 = L-tert.-Leucine (L-t.-Leu), 22 = D-tert.--Leucine (D-t.-Leu), 23 = L-N-Methyl-Leucine (L-N-MeLeu), 24 = D-N-Methyl-Leucine (D-N-MeLeu), 25 = L-N-Methyl-Isoleucine (L-N-MeIle), 26 = D-N-Methyl-Isoleucine (D-N-MeIle), 27 = L-allo-N-Methyl-Isoleucine (L-allo-N-MeIle), 28 = D-allo-N-Methyl-Isoleucine (D-allo-N-MeIle), 29 = L-N-Methyl-Phenylalanine (L-N-MePhe), 30 = D-N-Methyl-Phenylalanine (D-N-MePhe), 31 = L-threo-β-Methyoxytyrosine (L-threo-β-MeOTyr; 2R,3S), 32 = D-threo-β-Methyoxytyrosine (D-threo-β-MeOTyr; 2S,3R), 33 = L-erythro-β-Methyoxytyrosine (L-erythro-β-MeOTyr; 2R,3R), 34 = D-erythro-β-Methyoxytyrosine (D-erythro-β-MeOTyr; 2S,3S). * Indicates chiral centers.
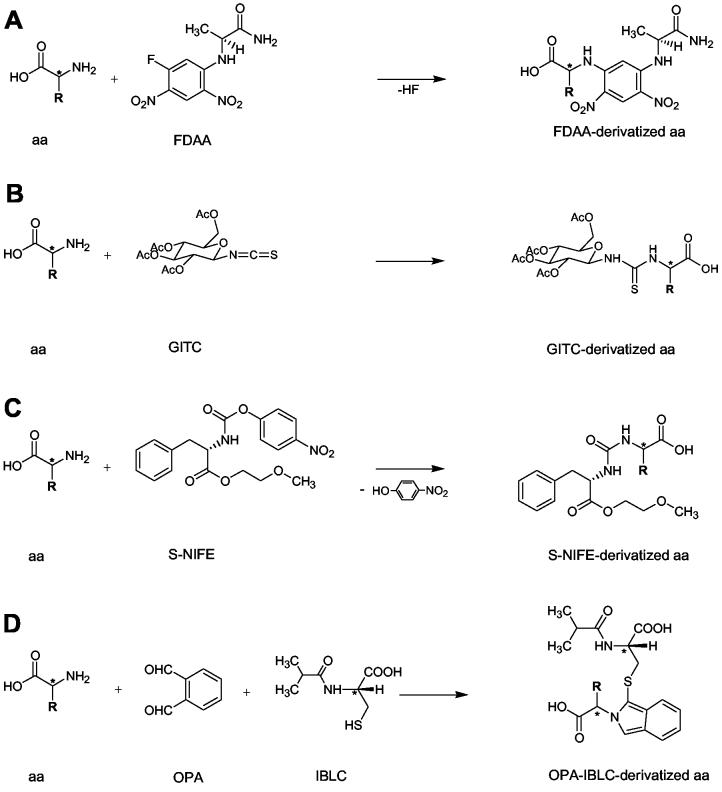
Figure 2.
Reaction schemes for the chiral derivatizing agents FDAA (A), GITC (B), S-NIFE (C) and OPA-IBLC (D). Reaction of the CDA with the amino group of the amino acid yields the respective diastereomers. For limitations and exceptions see text.
Chromatographic separation of amino acids derivatized with FDAA
The reaction scheme of FDAA with amino acids is outlined in Fig. 2A. Nucleophilic substitution of FDAA by the amino group of the amino acid yields the respective diastereomers. Besides the reaction of primary amines, other nucleophiles such as thiols and aromatic alcohols can also react with FDAA, leading to single and double substitutions in the case of Lys, His, Cys and Tyr [23-24, 26-28, 42]. The resulting 2,4-dinitrophenyl-l-alanine amide (DNP) derivatives show strong UV absorbance at 340 nm, which facilitates their identification in the chromatograms [21]. Because of the stability and high enantioselectivity of these DNP derivatives, FDAA has been widely used for stereochemical studies and some modifications to the original procedure have been introduced [23-31]. For our purposes, we needed to adjust the method. For instance, the originally proposed, chromatographically optimized phosphate buffer system is not compatible with online-coupled mass spectrometry and thus has been replaced with acetic acid. Using acetic acid instead of the alternatively suggested TFA [26-27], which is known to greatly reduce the signal intensity in ESI-MS, resulted in approximately a fourfold sensitivity increase. Therefore, we did not need to replace FDAA by FDLA (Nα-(2,4-Dinitro-5-fluorophenyl)-l-leucinamide; not commercially available), which was reported to show greater sensitivity [27]. The addition of 10% methanol to the acetonitrile phase was found to be beneficial for peak shape and resolution.
The retention times and separation factors of all investigated DNP-derivatized amino acids are given in Table 1. The most hydrophilic amino acid of this series was DNP-l-allo-Thr with a retention time of 19.01 min and the most hydrophobic one was di-DNP-l-threo-β-MeOTyr with a retention time of 51.05 min. All other amino acids were distinguishable in a single HPLC run. This result is in good agreement with earlier studies, where high enantioselectivity for DNP-derivatives has been reported [21-31]. Only hydrophilic hydroxy and acidic amino acids have been reported to have poor resolution [26]. As shown in Table 1, DNP-derivatives of non-aromatic amino acids show the highest enantioselectivity when compared to GITC (Table 2), S-NIFE (Table 3) and OPA-IBLC-derivatized amino acids (Table 4), with a few exceptions including N-MeThr and allo-N-MeIle. In these cases, S-NIFE derivatives were considerably better resolved (Table 3). DNP-derivatives of aromatic amino acids showed lower enantioselectivity than non-aromatic ones. Here again, S-NIFE derivatives were better resolved (Tables 1 and 3). As already noted [26], DNP derivatives of N-methyl amino acids generally have longer retention times than unmethylated amino acids and show decreased resolution.
Table 1.
Retention times (tRi), unadjusted relative retention (rG) and separation factors of amino acids derivatized with FDAA. As internal standard, an DNP-derivative with an m/z = 348 was chosen, which eluted around 38 min. Conditions: Mobile Phase A: 5% acetic acid, mobile phase B: acetontirile containing 10 % (v/v) of methanol, linear gradient from 5 to 50% B in 50 min, C18 reversed phase column (150 × 2.1 mm); 250 μl/min.
| Amino acid | [M+H]+ | Elution order | tRi(L)in min | tRi(D)in min | rG(L)1 | rGl(D)1 | rG(L)/rG(D) |
|---|---|---|---|---|---|---|---|
| Thr | 372 | L < D | 19.41 | 24.85 | 0.511 | 0.654 | 0.781 |
| allo-Thr | 372 | L < D | 19.01 | 22.38 | 0.506 | 0.588 | 0.861 |
| N-MeThr | 386 | L < D | 21.17 | 22.93 | 0.556 | 0.602 | 0.924 |
| allo-N-MeThr | 386 | L < D | 25.60 | 26.81 | 0.673 | 0.705 | 0.955 |
| Homo-Pro | 382 | D < L | 35.10 | 33.20 | 0.929 | 0.875 | 1.062 |
| Pro | 368 | L < D | 26.00 | 27.93 | 0.683 | 0.734 | 0.931 |
| Leu | 384 | L < D | 38.18 | 43.80 | 1.008 | 1.150 | 0.877 |
| Ile | 384 | L < D | 37.15 | 43.18 | 0.976 | 1.135 | 0.860 |
| allo-Ile | 384 | L < D | 37.31 | 43.32 | 0.980 | 1.138 | 0.861 |
| NorLeu | 384 | L < D | 38.37 | 44.23 | 0.985 | 1.165 | 0.845 |
| t.-Leu | 384 | L < D | 36.93 | 42.68 | 0.970 | 1.121 | 0.865 |
| N-MeLeu | 398 | L < D | 41.03 | 44.70 | 1.079 | 1.175 | 0.918 |
| N-MeIle | 398 | L < D | 40.81 | 42.81 | 1.074 | 1.127 | 0.953 |
| allo-N-MeIle | 398 | L < D | 41.32 | 44.73 | 1.086 | 1.173 | 0.926 |
| N-MePhe | 432 | L < D | 38.93 | 39.42 | 1.024 | 1.037 | 0.987 |
| erythro-β-MeOTyr | 463 | D < L | 24.81 | 23.75 | 0.653 | 0.627 | 1.041 |
| threo-β-MeOTyr | 463 | L < D | 23.97 | 24.83 | 0.631 | 0.655 | 0.963 |
| di-erythro-β-MeOTyr | 716 | D < L | 45.36 | 45.05 | 1.194 | 1.189 | 1.004 |
| di-threo-β-MeOTyr | 716 | D < L | 51.05 | 47.19 | 1.344 | 1.246 | 1.079 |
The Unadjusted Relative Retention rG was calculated as tRi/tR(St).
Table 2.
Retention times (tRi), unadjusted relative retention (rG) and separation factors of amino acids derivatized with GITC. As internal standard, a GITC-derivative with an m/z = 463 was chosen, which was eluting around 38 min. Chromatographic conditions are described under Table 1.
| Amino acid | [M+H]+ | Elution order | tRi(L)in min | tRi(D)in min | rG(L)1 | rGl(D)1 | rG(L)/rG(D) |
|---|---|---|---|---|---|---|---|
| Thr | 509 | L < D | 25.20 | 27.28 | 0.659 | 0.713 | 0.924 |
| allo-Thr | 509 | L < D | 24.75 | 25.33 | 0.644 | 0.663 | 0.971 |
| N-MeThr | 523 | L < D | 25.20 | 27.45 | 0.676 | 0.737 | 0.917 |
| allo-N-MeThr | 523 | D < L | 26.42 | 26.33 | 0.709 | 0.705 | 1.006 |
| Homo-Pro | 519 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Pro | 505 | L < D | 25.79 | 27.75 | 0.677 | 0.729 | 0.929 |
| Leu | 521 | L < D | 37.95 | 39.74 | 0.993 | 1.043 | 0.952 |
| Ile | 521 | L < D | 37.53 | 39.51 | 0.983 | 1.037 | 0.948 |
| allo-Ile | 521 | L < D | 36.66 | 38.72 | 0.980 | 1.035 | 0.947 |
| NorLeu | 521 | L < D | 38.60 | 40.53 | 1.016 | 1.066 | 0.953 |
| t.but.-Leu | 521 | L < D | 35.89 | 38.33 | 0.947 | 1.011 | 0.937 |
| N-MeLeu | 535 | L < D | 40.01 | 42.14 | 1.070 | 1.126 | 0.950 |
| N-MeIle | 535 | L < D | 39.04 | 40.49 | 1.047 | 1.078 | 0.971 |
| allo-N-MeIle | 535 | L < D | 39.60 | 42.02 | 1.058 | 1.121 | 0.944 |
| N-MePhe | 569 | L < D | 41.00 | 42.88 | 1.076 | 1.125 | 0.956 |
| erythro-β-MeOTyr | 601 | L < D | 29.97 | 30.89 | 0.794 | 0.816 | 0.973 |
| threo-β-MeOTyr | 601 | D < L | 31.70 | 30.47 | 0.839 | 0.806 | 1.041 |
The Unadjusted Relative Retention rG was calculated as tRi/tR(St).
Table 3.
Retention times (tRi), unadjusted relative retention (rG) and separation factors of amino acids derivatized with S-NIFE. As internal standard, the S-NIFE-derivative with an m/z = 473 was chosen, which was eluting around 45 min. Chromatographic conditions are described under Table 1.
| Amino acid | [M+H]+ | Elution order | tRi(L)in min | tRi(D)in min | rG(L)1 | rG(D)1 | rG(L)/rG(D) |
|---|---|---|---|---|---|---|---|
| Thr | 369 | L < D | 22.53 | 24.44 | 0.503 | 0.546 | 0.921 |
| allo-Thr | 369 | L < D | 22.45 | 23.40 | 0.501 | 0.523 | 0.958 |
| N-MeThr | 383 | L < D | 24.58 | 25.40 | 0.518 | 0.566 | 0.915 |
| allo-N-MeThr | 383 | L < D | 25.12 | 25.73 | 0.560 | 0.574 | 0.976 |
| Homo-Pro | 379 | L < D | 32.75 | 33.92 | 0.733 | 0.759 | 0.966 |
| Pro | 365 | L < D | 27.55 | 28.79 | 0.616 | 0.644 | 0.957 |
| Leu | 381 | L < D | 35.75 | 39.16 | 0.799 | 0.875 | 0.913 |
| Ile | 381 | L < D | 34.93 | 38.93 | 0.781 | 0.871 | 0.897 |
| allo-Ile | 381 | L < D | 35.01 | 39.04 | 0.783 | 0.871 | 0.899 |
| NorLeu | 381 | L < D | 36.67 | 39.84 | 0.821 | 0.891 | 0.921 |
| t.but.-Leu | 381 | L < D | 34.00 | 38.05 | 0.762 | 0.853 | 0.893 |
| N-MeLeu | 395 | L < D | 40.91 | 43.33 | 0.902 | 0.955 | 0.945 |
| N-MeIle | 395 | L < D | 40.00 | 41.98 | 0.894 | 0.939 | 0.952 |
| allo-N-MeIle | 395 | L < D | 40.48 | 42.65 | 0.770 | 0.941 | 0.818 |
| N-MePhe | 429 | L < D | 42.15 | 43.41 | 0.929 | 0.956 | 0.972 |
| erythro-β-MeOTyr | 461 | L < D | 21.80 | 26.51 | 0.526 | 0.591 | 0.890 |
| threo-β-MeOTyr | 461 | L < D | 26.23 | 28.69 | 0.586 | 0.640 | 0.916 |
The Unadjusted Relative Retention rG was calculated as tRi/tR(St).
Table 4.
Retention times (tRi), unadjusted relative retention (rG) and separation factors of amino acids derivatized with OPA-IBLC. As internal standard, an OPA-IBLC-derivative with an m/z = 481 was chosen, which was eluting around 48 min. Chromatographic conditions are described under Table 1.
| Amino acid | [M+H]+ | Elution order | tRi(L)in min | tRi(D)in min | rG(L)1 | rG(D)1 | rG(L)/rG(D) |
|---|---|---|---|---|---|---|---|
| Thr | 409 | L < D | 32.23 | 32.82 | 0.679 | 0.692 | 0.981 |
| allo-Thr | 409 | L < D | 33.92 | 34.93 | 0.715 | 0.736 | 0.971 |
| Leu | 421 | L < D | 48.83 | 50.07 | 1.029 | 1.051 | 0.979 |
| Ile | 421 | L < D | 47.95 | 49.55 | 1.013 | 1.047 | 0.968 |
| allo-Ile | 421 | L < D | 48.20 | 49.55 | 1.019 | 1.047 | 0.973 |
| NorLeu | 421 | L < D | 48.14 | 49.43 | 1.024 | 1.051 | 0.974 |
| t.but.-Leu | 421 | L < D | 48.80 | 49.08 | 1.038 | 1.045 | 0.993 |
| erythro-β-MeOTyr | 501 | D < L | 39.64 | 38.30 | 0.822 | 0.782 | 1.051 |
| threo-β-MeOTyr | 501 | L < D | 37.30 | 41.06 | 0.773 | 0.828 | 0.934 |
The Unadjusted Relative Retention rG was calculated as tRi/tR(St).
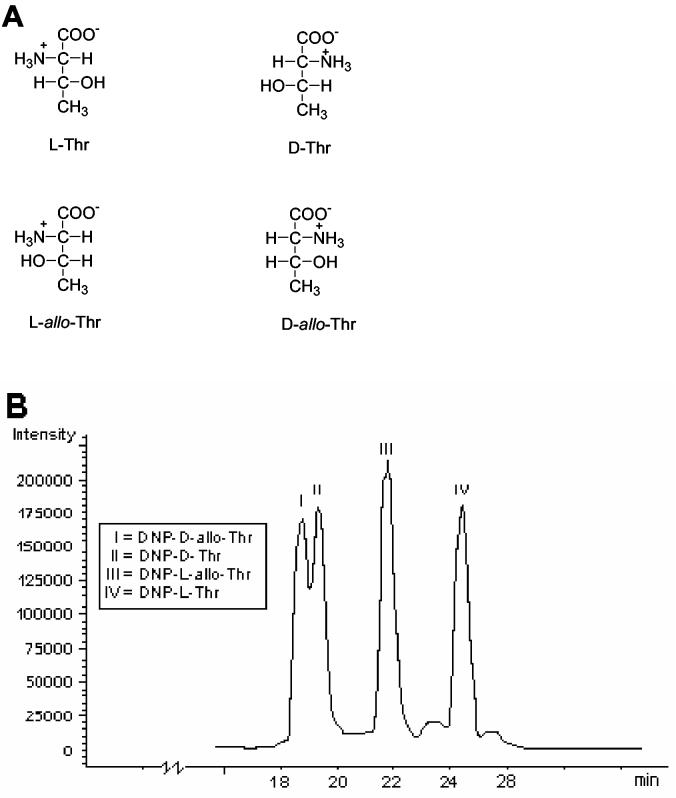
A major concern in natural product analysis is the resolution of stereoisomers, especially those that are difficult to distinguish by other techniques such as NMR. With the high enantioselectivity shown in Table 1, FDAA seems to be the best CDA for most non-aromatic amino acids. Since some amino acids such as Thr, Ile and β-MeOTyr contain two chiral centers, they can occur as four different stereoisomers. The configurational isomers of Thr (as shown in Fig. 3A) are l-Thr, which has the absolute configuration 2S,3R, d-Thr (2R,3S), l-allo Thr (2S,3S) and d-allo Thr (2R,3R). When investigated as their DNP-derivatives, the separation shown in Fig. 3B was obtained. Even though d-Thr and d-allo-Thr do not show baseline separation, they can be unambiguously assigned. While the resolution of the d,l-Thr and d,l-allo-Thr derivatives was satisfactory (Fig. 3B), the resolution of the l-allo-Ile/l-Ile and d-allo-Ile/d-Ile derivatives was not sufficient (Table 1 and 5). As apparent from Table 5, mono-DNP-l-erythro-β-MeOTyr (2R,3R) and mono-DNP-d-threo-β-MeOTyr (2S,3R) and their counterpart mono-DNP-l-threo-β-MeOTyr (2R,3S) and mono-DNP-d-erythro-β-MeOTyr (2S,3S) were not resolved. The poor separation of the mono-substituted β-MeO-Tyr was improved when di-substituted DNP-derivatives were used, but here the resolution of the d- and l-threo derivative remains critical (Table 5).
Figure 3.
The four stereoisomers of Thr (A). Separation of the Thr isomers 1-4 as their DNP-derivatives. DNP-D-allo-Thr I eluted first, followed by DNP-D-Thr II, DNP-L-allo-Thr III and DNP-L-Thr IV. Conditions: Mobile Phase A: 5% acetic acid, mobile phase B: acetontirile containing 10 % (v/v) of methanol, linear gradient from 5 to 50% B in 50 min, followed by a washing phase at 100% B; C18 reversed phase column (150 × 2.1 mm); 250 μl/min (B).
Table 5.
Comparison of the separation factors of allo- and threo-configurated diasteromeric amino acids. Separation was considered insufficient when a value of 1.000 ± 0.009 was achieved.
| L-allo/L-threo | L-allo/D-threo | L-allo/D-allo | L-threo/D-threo | L-threo/D-allo | D-allo/D-threo | |
|---|---|---|---|---|---|---|
| FDAA | ||||||
| Thr | 0.990 | 0.774 | 0.861 | 0.781 | 0.869 | 0.899 |
| N-MeThr | 1.210 | 1.118 | 0.955 | 0.924 | 0.789 | 1.171 |
| Ile | 1.004 | 0.863 | 0.861 | 0.860 | 0.858 | 1.003 |
| N-MeIle | 1.011 | 0.964 | 0.926 | 0.953 | 0.916 | 1.041 |
| β-MeOTyr1 | 0.966 | 1.006 | 0.963 | 1.041 | 0.997 | 1.045 |
| β-MeOTyr1 | 1.126 | 1.130 | 1.079 | 1.004 | 0.958 | 1.048 |
| GITC | ||||||
| Thr | 0.977 | 0.903 | 0.971 | 0.924 | 0.994 | 0.930 |
| N-MeThr | 1.049 | 0.962 | 1.006 | 0.917 | 0.959 | 0.957 |
| Ile | 0.997 | 0.945 | 0.947 | 0.948 | 0.950 | 0.998 |
| N-MeIle | 1.011 | 0.981 | 0.944 | 0.971 | 0.934 | 1.040 |
| β-MeOTyr1 | 1.057 | 1.028 | 1.041 | 0.973 | 0.985 | 0.988 |
| S-NIFE | ||||||
| Thr | 0.996 | 0.918 | 0.958 | 0.921 | 0.962 | 0.958 |
| N-MeThr | 1.081 | 0.989 | 0.976 | 0.915 | 0.902 | 1.014 |
| Ile | 1.003 | 0.899 | 0.899 | 0.897 | 0.897 | 1.000 |
| N-MeIle | 0.861 | 0.820 | 0.818 | 0.952 | 0.950 | 1.002 |
| β-MeOTyr1 | 1.114 | 0.992 | 0.916 | 0.890 | 0.822 | 1.083 |
| OPA-IBLC | ||||||
| Thr | 1.053 | 1.033 | 0.971 | 0.981 | 0.923 | 1.060 |
| Ile | 1.006 | 0.973 | 0.973 | 0.968 | 0.968 | 1.000 |
| β-MeOTyr1 | 0.940 | 0.988 | 0.934 | 1.051 | 0.993 | 1.059 |
The configuration L-allo refers to L-erythro in case of β-MeOTyr.
It has generally been noted that at higher retention times (>10 min) DNP-l-amino acids eluted before DNP-d-amino acids with a few exceptions reported so far [26-28, 44]. With our method, we have also found the reversed order of elution for the diastereomeric pairs of homoPro, mono-DNP-erythro-, di-DNP-erythro and di-DNP-threo-β-MeOTyr, whereas the threo-configurated β-MeOTyr eluted in the expected order (Table 1). Our results clearly indicate that one can not deduce the stereochemistry only on the basis of its elution order without additional supportive information. As a result of these experiments, we advise caution when non-empirically interpreting data without the appropriate standards. Our data clearly show that the proposed non-empirical method [26-27] may result in incorrect assignments since some amino acids showing this unusual behavior with the d-isomer eluted before the l-isomer.
The limit of detection (signal to noise ratio 2:1) determined with Gly standards at various concentrations was in the low picomolar range and was thus sensitive enough to investigate both synthetic standards and amino acids derived from marine natural products. The high stability of DNP derivatives was regarded as an advantage, whereas the relatively long incubation time for reaction may be regarded as a disadvantage.
Chromatographic separation of amino acids derivatized with GITC
In contrast to DNP-derivatives, the diasteromeric thiourea derivatives formed when amino acids were reacted with GITC (Fig. 2) do not show a visible chromophore and were therefore monitored at 254 nm. GITC also reacts more specifically with primary amines so double substitutions were only observed for Lys, but not for any other nucleophile [32-33]. GITC reacted with Pro but not with homoPro, which could be attributed to steric hindrance of this cyclic amine.
One advantage of GITC derivatization was the short incubation time of 10 min at rt, however, the instability of GITC derivatives can be regarded as a disadvantage. While FDAA derivatives are stable over several days, GITC derivatives need to be analyzed as soon as possible after their preparation and should not be stored longer than 24 hrs. As the originally proposed method was not designed for mass spectrometry coupling, it was adjusted in a manner similar to the FDAA method described above so that it was amenable to LC-MS analysis. Using this adjusted method, all of the investigated l-isomers eluted before their corresponding d-isomers, with the exception of allo-N-MeThr and threo-β-MeOTyr. This was in good agreement with earlier studies, where both proteinogenic and other uncommon amino acids have been investigated [32-33]. Exceptions from this rule have also been reported for a number of more rigid α-methylated amino acids [29]. Table 2 summarizes the retention times and separation factors of all investigated amino acids that were derivatized with GITC.
The most hydrophilic GITC-derivatized amino acid was l-allo-Thr with a retention time of 24.75 min and the most hydrophobic one was d-N-MePhe with a retention time of 42.14 min. Considering the fact that GITC does not form a bis-adduct with β-MeOTyr, the overall elution profile of the GITC-derivatives was similar to that of the DNP-derivatives. Derivatives of N-methyl amino acids showed longer retention times than the unmethylated parent amino acids due to increased hydrophobicity. The separation factors were similar to those of the unmethylated amino acids. This contrasted with the DNP-amino acids where a loss of chromatographic resolution was observed.
Even though the enantioselectivity of GITC-derivatized amino acids was generally lower than that of DNP-derivatives, the separation of the β-MeOTyr stereoisomers was better accomplished with GITC (Table 2, 5). The isomeric pairs l-Ile/l-allo-Ile, d-Ile/d-allo-Ile, l-allo-Thr/d-allo-Thr and d- and l-allo-MeThr coeluted when derivatized with GITC (Table 2 and 5). The GITC-tag showed an approximately four times higher response by MS than FDAA and was thus four times more sensitive when using MS detection.
Chromatographic separation of amino acids derivatized with S-NIFE
S-NIFE forms diasteromeric urea derivatives with amino acids by nucleophilic substitution of the carbamate and release of 4-nitrophenol as shown in Fig. 2C. S-NIFE derivatives were also monitored at 254 nm. Improved enantioselectivity has been described for those CDAs that have four or five bonds between their chiral centers [34]. This ideal distance is present in GITC (four bonds), S-NIFE (four bonds), and OPA-IBLC (five bonds) whereas FDAA has six bonds (Fig. 2).
S-NIFE reacted with all amino acids investigated including homoPro. S-NIFE reacts twice with lysine (α and ω-NH2), Cys and Tyr [34]. S-NIFE derivatives are easily formed within 15-20 minutes and are, in contrast to GITC-derivatized amino acids, stable over several days. In addition to 4-nitrophenol (Fig. 2C), phenylalanine methoxyethyl ester and N,N'-bis(methoxyethyl 3-phenylpropionate-2-yl)urea are formed as side products, and the latter product was used as an internal standard with a m/z 473 [M+H]+ eluted around 45 min.
For S-NIFE, d-isomers eluted without exception after their corresponding l-isomers. The retention times and separation factors of the S-NIFE derivatives are summarized in Table 3. The elution profile of S-NIFE derivatized amino acids was very similar to that of the GITC derivatized amino acids. As with GITC, the most hydrophilic S-NIFE derivative was l-allo-Thr (Rt = 22.45), and the most hydrophobic, d-N-MePhe (Rt = 43.41). There are some important distinctions though. In most cases, the enantioselectivity of S-NIFE derivatives was considerably better than that of GITC-derivatives but inferior to that of DNP-derivatives. As described for β-alkyl substituted amino acids [36], the enantioselecitivity of aromatic amino acids such as β-MeOTyr was extremely good. However, the separation of diasteromeric d-allo-MeOTyr and l-threo-MeOTyr is borderline with S-NIFE (Table 5). Baseline separation was not accomplished.
N-Methyl substituted amino acids eluted after their unmethylated parents with comparable resolution. Despite the better enantioselectivity of S-NIFE-derivatives, the separation of l-allo-Thr/l-Thr is not as good as the separation of those GITC-derivatives (Table 5). l-allo-Ile/l-Ile and d-allo-Ile/d-Ile and d-allo-MeIle/d-MeIle isomers were not resolved from each other under the conditions used, whereas l-allo-MeIle and l-MeIle isomers were separated (Table 5). The limit of detection was approximately 50 pmol on column, which was approximately two times more sensitive than the corresponding DNP-derivatives.
Chromatographic separation of amino acids derivatized with OPA-IBLC
As outlined in Fig. 2D, o-phthaldialdehyde (OPA) and the sulfhydryl group of IBLC react rapidly with primary amines to form the corresponding diastereomeric isoindole derivatives that have been exploited because of their intense fluorescence. Chiral sulfhydryl groups such as IBLC have been introduced to enable the separation of chiral amines. Secondary amines do not react [37]. There has been some discrepancy in the literature about the stability of OPA/sulfhydryl substituted amino acids. A rapid decrease of fluorescence intensity described for Gly and Lys derivatives [37] has recently been attributed to the transformation of kinetically formed mono-substituted derivatives into a thermodynamically stable di-substituted derivative [38, 39]. Isoindoles that carry an N-CH2-R group such as Gly (NH2-CH2-COOH), β-Ala (NH2-(CH2)2COOH), GABA (NH2-(CH2)3-COOH), Lys (NH2-(CH2)4CHNH2-COOH), are especially prone to form adducts that contain both an isoindole and a benzofuran moiety [38, 39].
The reported high enantioselectivity of the OPA-IBLC-derivatized amino acids made us investigate its applicability to our selection of unusual amino acids. For fluorescence detection, an alkaline mobile phase that ultimately destroys the stationary phase has been recommended for optimum sensitivity and chromatographic resolution [45-46]. The leaking of column material into the mass spectrometric detector would destroy the detector rather quickly and thus, we adjusted the LC-method so that it was suitable for online MS detection. l-Thr eluted with the shortest retention time (32.23 min) and d-Leu eluted at 50.07 min as the last peak of the investigated series. As with the other methods, derivatized l-amino acids generally eluted before their corresponding d-amino acids. An exception was the elution order of erythro-β-MeOTyr isomers.
Among the CDAs tested, OPA-IBLC-derivatized amino acids showed the least enantioselectivity for the amino acids examined (Table 4). However, enantioselectivity was sufficient in all cases to distinguish the d- and l-isomers. Since OPA-IBLC does not react with secondary amines, this CDA was not applicable to the N-methylated amino acids and imino acids investigated in this series and was thus of limited use. All Thr and allo-Thr derivatives were well separated from each other (Table 5). The l-allo-Ile/l-Ile and d-allo-Ile/d-Ile isomers that were generally found difficult to distinguish, were not effectively resolved with this method.
The resolution of OPA-IBLC-derivatives of β-MeOTyr was considerably better than that of their DNP- or GITC-derivatives, but here the separation of the L-erythro/D-allo pair was marginal.
The OPA-IBLC label showed approximately the same response as GITC in the mass selective detection mode and was thus four times more sensitive than FDAA. The limited applicability to primary amines only, the poorer resolution and the transformation of mono- into disubsituted OPA-IBLC derivatives was disadvantageous whereas the good separation of Thr and β-MeOTyr isomers was advantageous for OPA-IBLC.
Conclusion
The LC-ESI-MS methods described can be applied for the separation and identification of isomers of proteinogenic and unusual amino acids, especially those that are of high interest in biomedical research as components of marine peptides. In fact, it already has been successfully applied to naturally occurring marine peptides [41]. Separations were done in acetic acid-acetonitrile/methanol phases. Both UV and ESI-MS detection were used to corroborate the identity of the detected derivatives. DNP-derivatives have generally shown the highest enantioselectivity for non-aromatic amino acids followed by S-NIFE, GITC- and OPA-IBLC derivatives. For aromatic amino acids, S-NIFE derivatives showed the highest enantioselectivity, followed by derivatives of OPA-IBLC, GITC and FDAA. At the same time, DNP-derivatives have shown the lowest sensitivity, while S-NIFE provided approximately twice the signal intensity, and OPA-IBLC and GITC were ca. four times more sensitive. The limit of detection of DNP-derivatives was in the low picomolar range, which was found to be more than sufficient for the characterization of μg amounts of natural products. The limit of detection can be further improved with selected ion monitoring.
The current study has also shown the limitations of the CDAs investigated in terms of their separation power between diastereomers such as D-allo-Ile and D-Ile, which were not completely resolved with any of the CDAs. All of the other isomers investigated can be resolved either with one or two chromatographic separations. Due to the complex relationships of isomeric amino acids, especially with regard to the coexistence of common and uncommon amino acids, it is generally recommended to use more than one CDA for the structural assignment. In light of an increasing number of exceptions of the expected elution order of DNP-amino acids (L<D at retention times above 10 min), we also recommend the use of authentic standards to corroborate the assignments. This certainly improves confidence in the assignment and helps to avoid unexpected misassignments as have happened in the past [19].
References
- 1.Fusetani N, Matsunaga S. Chem. Rev. 1993;93:1793–1806. [Google Scholar]
- 2.Ballard CE, Yu H, Wang B. Current Med. Chem. 2002;9:471–498. doi: 10.2174/0929867023371049. [DOI] [PubMed] [Google Scholar]
- 3.Bewley CA, Faulkner DJ. Angew. Chem. Int. Ed. 1998;37:2162–2178. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2162::AID-ANIE2162>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Moore RE. J. Ind. Microbiol. 1996;16:134–143. doi: 10.1007/BF01570074. [DOI] [PubMed] [Google Scholar]
- 5.Kerr RG, Kerr SS. Exp. Opin. Ther. Patents. 1999;9:1207–1222. [Google Scholar]
- 6.Ford PW, Gustafson KR, McKee TC, Shigematsu N, Maurizi LK, Pannell LK, de Silva ED, Lassota P, Allen TM, Van Soest R, Andersen RJ, Boyd MR. J. Am. Chem. Soc. 1999;121:5899–5909. [Google Scholar]
- 7.Zampella A, D'Auria MV, Paloma LG, Casapullo A, Minale L, Debitus C, Henin Y. J. Am. Chem. Soc. 1996;118:6202–6209. [Google Scholar]
- 8.D'Auria MV, Zampella A, Paloma LG, Minale L, Debitus C, Roussakis C, Le Bert V. Tetrahedron. 1996;52:9589–9596. [Google Scholar]
- 9.Atkins JF, Gesteland R. Science. 2002;296:1409–1410. doi: 10.1126/science.1073339. [DOI] [PubMed] [Google Scholar]
- 10.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan G, James CM, Krzycki JA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 12.Liang B, Carroll PJ, Joullie MM. Org. Lett. 2000;2:4157–4160. doi: 10.1021/ol006679t. [DOI] [PubMed] [Google Scholar]
- 13.Rinehart KL. Med. Res. Rev. 2000;20:1–27. doi: 10.1002/(sici)1098-1128(200001)20:1<1::aid-med1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 14.Acevedo CM, Kogut EF, Lipton MA. Tetrahedron. 2001;57:6353–6359. [Google Scholar]
- 15.Okamoto N, Hara O, Makino K, Hamada Y. J. Org. Chem. 2002;67:9210–9215. doi: 10.1021/jo0258352. [DOI] [PubMed] [Google Scholar]
- 16.Thoen JC, Morales-Ramos AI, Lipton MA. Org. Lett. 2002;4:4455–4458. doi: 10.1021/ol0269852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto N, Hara O, Makino K, Hamada Y. Tetrahedron Asymmetry. 2001;12:1353–1358. [Google Scholar]
- 18.Guerlavais V, Carroll PJ, Joullie MM. Tetrahedron Asymmetry. 2002;13:657–680. [Google Scholar]
- 19.Zampella A, Randazzo A, Borbone N, Luciani S, Trevisis L, Debitus C, D'Auria MV. Tetrahedron Lett. 2002;43:6163–6166. [Google Scholar]
- 20.Fountoulakis M, Lahm HW. J. Chromatogr. A. 1998;826:109–134. doi: 10.1016/s0021-9673(98)00721-3. [DOI] [PubMed] [Google Scholar]
- 21.Marfey P. Carlsberg Res. Commun. 1984;49:591–596. [Google Scholar]
- 22.Toyo'oka T. Modern Derivatization Methods for Separation Sciences. Wiley; Chichester, New York: 1999. [Google Scholar]
- 23.Kochhar S, Christen P. Anal. Biochem. 1989;178:17–21. doi: 10.1016/0003-2697(89)90348-5. [DOI] [PubMed] [Google Scholar]
- 24.Brückner H, Gah C. J. Chromatogr. 1991;555:81–95. [Google Scholar]
- 25.Krishnamurthy T. J. Am. Soc. Mass Spectrom. 1994;5:724–730. doi: 10.1016/1044-0305(94)80004-9. [DOI] [PubMed] [Google Scholar]
- 26.Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada K-I. Anal. Chem. 1997;69:3346–3352. [Google Scholar]
- 27.Fujii K, Ikai Y, Oka H, Suzuki M, Harada K-I. Anal. Chem. 1997;69:5146–5151. [Google Scholar]
- 28.Harada K-I, Matsui A, Shimizu Y, Ikemoto R, Fujii K. J. Chromatogr. A. 2001;921:187–195. doi: 10.1016/s0021-9673(01)00884-6. [DOI] [PubMed] [Google Scholar]
- 29.Goodlett DR, Abuf PA, Savage PA, Kowlaski KS, Mukherjee TK, Tolan JW, Corkum N, Goldstein G, Crowther JB. J. Chromatogr. A. 1995;101:233–244. doi: 10.1016/0021-9673(95)00352-n. [DOI] [PubMed] [Google Scholar]
- 30.Peter A, Lazar L, Fülöp F, Armstrong DW. J. Chromatogr. A. 2001;926:229–238. doi: 10.1016/s0021-9673(01)01078-0. [DOI] [PubMed] [Google Scholar]
- 31.Peter A, Olajos E, Casimir R, Tourwe D, Broxterman QB, Kaptein B, Armstrong DW. J. Chromatogr. 2000;871:105–113. doi: 10.1016/s0021-9673(99)00889-4. [DOI] [PubMed] [Google Scholar]
- 32.Nimura N, Toyama A, Kinoshita T. J. Chromatogr. 1984;316:547–552. [Google Scholar]
- 33.Tian Z, Hrinyo-Pavlina T, Roeske RW, Rao PN. J. Chromatogr. 1991;541:297–302. [Google Scholar]
- 34.Peter A, Vekes E, Török G. Chromatographia. 2000;52:821–826. [Google Scholar]
- 35.Peter A, Vekes E, Toth G, Tourwe D, Borremans F. J. Chromatogr. A. 2002;948:283–294. doi: 10.1016/s0021-9673(01)01475-3. [DOI] [PubMed] [Google Scholar]
- 36.Vekes E, Török G, Péter A, Sápi J, Tourwé D. J. Chromatogr. A. 2002;949:125–139. doi: 10.1016/s0021-9673(01)01455-8. [DOI] [PubMed] [Google Scholar]
- 37.Brückner H, Haasmann S, Langer M, Westhauser T, Wittner R, Godel HJ. J. Chromatograph. A. 1994;666:259–273. [Google Scholar]
- 38.Molnar-Perl I, Vasanits A. J. Chromatogr. A. 1999;835:73–91. doi: 10.1016/s0021-9673(98)00977-7. [DOI] [PubMed] [Google Scholar]
- 39.Mengerink Y, Toth D, Csampai A, Molnar-Perl I. J. Chromatogr. A. 2002;949:99–124. doi: 10.1016/s0021-9673(01)01282-1. [DOI] [PubMed] [Google Scholar]
- 40.König WA. Journal of HRC & CC. 1982;5:588–595. [Google Scholar]
- 41.Davies-Coleman MT, Dzeha TM, Gray CA, Hess S, Pannell LK, Hendricks DT, Arendse CE. J. Nat. Prod. 2003;66:712–715. doi: 10.1021/np030014t. [DOI] [PubMed] [Google Scholar]
- 42.B'Hymer C, Montes-Bayon M, Caruso JA. J. Sep. Sci. 2003;26:7–19. [Google Scholar]
- 43.Peter A, Vekes E. E, Armstrong DW, Tourwe D. Chromatographia. 2002;56:S41–S47. [Google Scholar]
- 44.Adamson JG, Hoang T, Crivici A, Lajoie GA. Anal. Biochem. 1992;202:210–214. doi: 10.1016/0003-2697(92)90229-z. [DOI] [PubMed] [Google Scholar]
- 45.Michael G, Henrion G. GIT Fachz. Lab. 1995;9:769–773. [Google Scholar]
- 46.Molnar-Perl I. J. Chromatogr. A. 2001;913:283–302. doi: 10.1016/s0021-9673(00)01200-0. [DOI] [PubMed] [Google Scholar]