Abstract
It is thought that the H19 imprinting control region (ICR) directs the silencing of the maternally inherited Igf2 allele through a CTCF-dependent chromatin insulator. The ICR has been shown to interact physically with a silencer region in Igf2, differentially methylated region (DMR)1, but the role of CTCF in this chromatin loop and whether it restricts the physical access of distal enhancers to Igf2 is not known. We performed systematic chromosome conformation capture analyses in the Igf2/H19 region over >160 kb, identifying sequences that interact physically with the distal enhancers and the ICR. We found that, on the paternal chromosome, enhancers interact with the Igf2 promoters but that, on the maternal allele, this is prevented by CTCF binding within the H19 ICR. CTCF binding in the maternal ICR regulates its interaction with matrix attachment region (MAR)3 and DMR1 at Igf2, thus forming a tight loop around the maternal Igf2 locus, which may contribute to its silencing. Mutation of CTCF binding sites in the H19 ICR leads to loss of CTCF binding and de novo methylation of a CTCF target site within Igf2 DMR1, showing that CTCF can coordinate regional epigenetic marks. This systematic chromosome conformation capture analysis of an imprinting cluster reveals that CTCF has a critical role in the epigenetic regulation of higher-order chromatin structure and gene silencing over considerable distances in the genome.
Keywords: genomic imprinting, insulators, chromosome biology
A variety of control elements, such as enhancers, silencers, and chromatin insulators, are thought to establish and maintain domains of gene expression or repression in the genome. Insulators, for example, are needed to keep genes in silent domains, away from the influence of neighboring enhancers. The prototypical insulator from the chicken β-globin locus contains binding sites for the 11-zinc-finger protein CTCF, and binding of CTCF is necessary for insulator function, not only in the chicken but also in many mammalian insulators (1–4). How CTCF confers chromatin insulation is not clear, but recent work shows that DNA-bound CTCF molecules can dimerize in a binding site-specific fashion (5) and that CTCF binds nucleophosmin and may be tethered to the nuclear matrix (6). CTCF emerges, therefore, as a potential mediator of long-range interactions that are involved in establishing repressed domains.
Regional coordination of gene expression and repression is found in many imprinted gene clusters in the mammalian genome and is regulated by imprinting centers or imprinting control regions (ICRs) (7–10). A well characterized cluster is that containing the paternally expressed Igf2 and maternally expressed H19. Both genes share enhancers, and the ICR is located in the 5′ flank of the H19 gene (11). The deletion of this ICR results in biallelic expression of both Igf2 and H19, demonstrating a role of the ICR to repress the maternal Igf2 allele, which is located >80 kb away (12). The mechanism underlying this function has been proposed to involve a CTCF-dependent chromatin insulator located within the ICR (13–16), which is continuously required for Igf2 repression in somatic cells (17). However, the Igf2 gene itself also contains differentially methylated regions (DMRs), with DMR1 being a methylation-sensitive silencer (18, 19) and DMR2 being a methylation sensitive activator (20). Additional enhancer sequences located 5′ of the ICR (21, 22) suggest that a simple insulator function of the ICR is not sufficient to explain all aspects of maternal silencing of Igf2.
In addition to their insulator function, the CTCF target sites within the H19 ICR confer protection against de novo methylation in somatic cells (5, 23, 24), preimplantation conceptuses, embryonic stem cells, and oocytes (25, 26). Furthermore, maternal deletion of the H19 ICR leads to de novo methylation at DMR1 and DMR2 in Igf2, showing regional coordination of epigenetic modifications by the ICR, which was proposed to occur by higher-order chromatin conformations (27). By applying the chromosome conformation capture (3C) method (28), it was indeed demonstrated that in neonatal liver the H19 ICR interacts with the DMR1 at the Igf2 locus only on the maternal chromosome (29). Here we have carried out a systematic 3C analysis across the entire Igf2/H19 domain and have examined the effect of CTCF binding to the ICR on higher-order chromatin conformations and the epigenotype of the locus.
Results
Parent-of-Origin-Dependent Interaction Between the H19 Enhancers and the Chromatin Fiber Within the Igf2/H19 Domain.
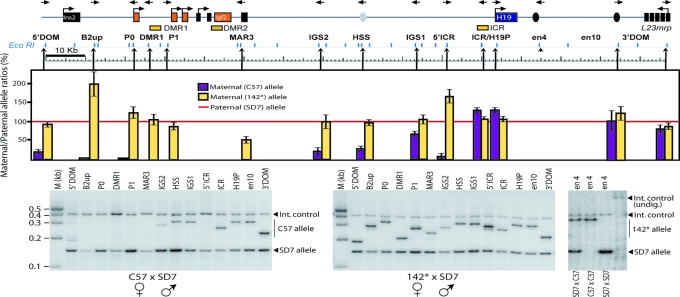
We used the 3C technique (28, 30) to detect long-range interactions within the Igf2/H19 expression domain. Briefly, digestion with the restriction enzyme of crosslinked, native chromatin can demonstrate the physical proximity of restriction fragments from remote locations if they were close together at the time of crosslinking. After ligation under very dilute DNA concentration, the ligated fragments can be detected using PCR with primers from the remote locations. We used the EcoRI restriction enzyme to study the crosslinking frequency of the endodermal enhancer region vs. the entire Igf2/H19 chromatin domain, including: 12.9 kb upstream of Ins2 (5′DOM); 6.4 kb upstream of B2 repeat region (B2UP); placental-specific promoter Po (Po); DMR1; P1 (just upstream of P1) or P1/DMR2; MAR3; intergenic sequence region (IGS)2 upstream of conserved DNase1 hypersensitive region (HSS); IGS1 downstream of HSS; 9 kb upstream of ICR (5′ICR); ICR; H19 promoter (H19P); enhancer-conserved sequence 10 (en10) corresponds to mesodermal enhancers (31); and within the second intron of the L23mrp gene (3′DOM) (Fig. 1). Fig. 6, which is published as supporting information on the PNAS web site, shows the absence of allelic bias in the digestion of crosslinked chromatin DNA, Fig. 7, which is published as supporting information on the PNAS web site, defines the conditions of crosslinking (crosslinking with 1% or 2% formaldehyde gave comparable results), and Fig. 8, which is published as supporting information on the PNAS web site, demonstrates the linearity of the amplification protocols used in this study. The 3C analyses were performed on neonatal liver cell nuclei of C57 × SD7, SD7 × C57, and 142* × SD7 crosses. The SD7 mice harbor the distal chromosome 7 imprinting region of Mus spretus origin in a Mus musculus domesticus background (32). The 142* allele, which does not interact with CTCF in vivo, was created by changing the sequence GTGG to ATAT in three of the four CTCF target sites within the CGCG(T/G)GGTGGCAG core motif of the H19 ICR (23).
Fig. 1.
Parent-of-origin-specific patterns of physical proximity between the H19 endodermal enhancer and the Igf2/H19 domain in neonatal liver. H19, Igf2, and the flanking Ins2 and L23mrp genes are indicated by squares. The orientation of the primers used is represented by the direction of the arrows at the top of the diagram. EcoRI restriction enzyme sites that are present all along the locus are shown below the locus diagram, whereas arrowheads pointing up show the location of the 3C primer with reference to the corresponding EcoRI restriction site. The hot-stop PCR analysis of the proximity between en4 and the entire Igf2/H19 domain was performed by comparing relative crosslinking frequencies between the maternal en4 allele and the rest of the locus after normalization of the wild-type paternal SD7 allele frequencies to 100% (red line). Also see Fig. 9 for direct comparison of frequencies of interactions. 3C analysis and allelic bias were corrected for as described in Supporting Materials and Methods. The rightmost image exemplifies a hot-stop PCR analysis of 3C samples, which were digested with KpnI to identify the SD7 allele. The completeness of the KpnI digestion was verified by incorporating a fragment covering the KpnI site, which is specific for the SD7 allele, as an internal digestion control. See Materials and Methods and Supporting Materials and Methods for additional information.
Using the endodermal H19 enhancer (en4) region as a common platform, we measured the relative frequency of proximity along the entire Igf2/H19 domain. Because all PCR protocols for Figs. 1 and 2 were identical, the PCR efficiency relative to the internal control (Ercc3) (33) gives an estimate of the actual amplification efficiency in relation to the internal control. Fig. 9, which is published as supporting information on the PNAS web site, shows that the frequency of interactions is largely correlated with the proximity of the sequences involved with reduced interactions with more distal elements. To discriminate between the parental alleles represented in the 3C products, we used a KpnI site, which is specific for the SD7 allele of the en4 region. After corrections for any allelic bias in the PCR conditions (see Fig. 10, which is published as supporting information on the PNAS web site), the signal obtained from the wild-type maternal allele is shown as cerise bars, in comparison with that from the paternal allele, which was normalized to 100% (red line in Fig. 1). The en4 region interacted equally with both parental alleles from the L23mrp gene up to the H19 ICR (Fig. 1). The equal representation of maternal- and paternal-specific en4–H19P interactions suggests that the repression of the paternal H19 gene by promoter methylation does not lead to physical exclusion of the enhancer from the silenced H19 allele.
In contrast to the silent H19 gene, there was no enhancer access to the maternal copy of the Igf2 gene, whereas the paternal copy was accessible (Fig. 1). Enhancer access on the maternal allele was severely but not completely blocked at the ICR. There was still some interaction between en4 and IGS1 and to a lesser extent with hypersensitive sites (34) and IGS2. Further 5′ of these regions, however, there was no enhancer access to the maternal allele at all. These observations do not support a simple model in which the maternal ICR blocks all physical access across the insulator.
We next asked to what extent CTCF binding to the maternal ICR was responsible for restricting the access of the H19 enhancers by using a maternally inherited mutant ICR allele that does not bind CTCF (142*, yellow bars in Fig. 1). The whole of the Igf2 region, including the promoters, was now accessible to the en4 enhancers. Hence, CTCF target sites in the H19 ICR prevent direct communication between the H19 enhancers and the Igf2 promoters on the maternal chromosome, thus presumably leading to the failure of the maternal Igf2 gene to be transcriptionally activated. To understand better how the ICR prevented this communication, we next focused on interactions between the ICR and the chromatin fiber of the entire Igf2/H19 domain.
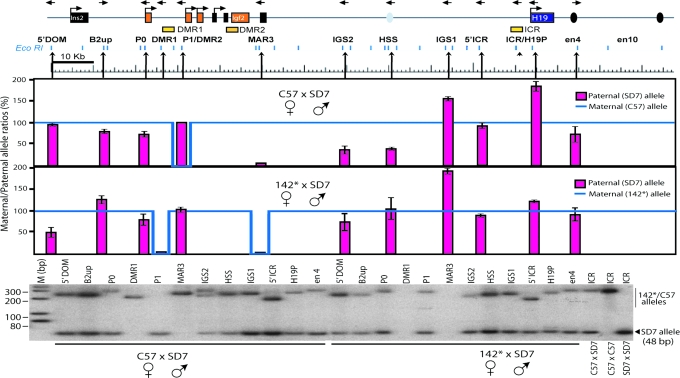
The H19 ICR Interacts with Igf2 Cis-Regulatory Elements in a Parent-of-Origin-Specific Manner.
As in the first experiment, EcoRI-digested nuclei of neonatal liver were used for 3C, but now the constant PCR primer was in the ICR, and all other primers were in the same locations as described above (Fig. 2). As for the enhancer–chromatin fiber interaction, albeit less marked, the ICR interactions reflect the proximity of the sequences involved with reduced interactions with more distal elements (Fig. 11, which is published as supporting information on the PNAS web site). A FauI polymorphic restriction site 61 bp downstream of the EcoRI restriction site within the ICR of specifically the SD7 allele was used to discriminate between parental alleles. After corrections for allelic bias in the PCR amplification steps (Fig. 12, which is published as supporting information on the PNAS web site), Fig. 2 displays the signal of the paternal allele (red bars) in relation to that of the maternal allele (blue lines) normalized to 100%.
Fig. 2.
Analysis of parent-of-origin-specific patterns of physical proximity between ICR and the Igf2/H19 domain in neonatal liver. The 3C analysis was performed by comparing relative crosslinking frequencies between the fixed ICR of the maternal allele and the rest of the locus after normalizing the maternal allele frequencies to 100% (blue line). Also see Fig. 11 for direct comparison of frequencies of interactions. Because of a lack of signal for the maternal allele in some instances, the blue line makes a dip, as indicated. 3C analysis and allelic bias were corrected for as described in Fig. 1 (see Fig. 12). The bottommost image exemplifies a hot-stop PCR analysis of 3C samples, which were digested with FauI to identify the SD7 allele. See Material and Methods for additional information.
This experiment shows that the ICR is in close physical proximity to all regions examined in the entire Igf2/H19 domain on both parental alleles, with three significant exceptions. First, the ICR is excluded from contact with P1/DMR2 specifically on the maternal chromosome and in close contact with P1/DMR2 only on the paternal chromosome. Second, the ICR is in close contact with DMR1 exclusively on the maternal chromosome. These results corroborate the findings by Murrell et al. (29). Third, and most interestingly, on the 3′ side of Igf2, the ICR is in contact with MAR3 exclusively on the maternal chromosome. Therefore, on the maternal allele, the Igf2 gene is located in a tight pocket made of contacts between the ICR, DMR1, and MAR3, which we suggest excludes the gene (P1/DMR2) from interactions with the enhancers.
Significantly, maternal transmission of the 142* allele (with the CTCF binding site mutations) led to a loss of the maternal interactions among the ICR, DMR1, and MAR3 and a gain of interaction between the ICR and P1/DMR2 (Fig. 2), showing that CTCF binding is the key regulator of the tight pocket between the ICR, DMR1, and MAR3.
The H19 ICR–CTCF Complex Controls the Epigenetic Status at Igf2 DMR1/2.
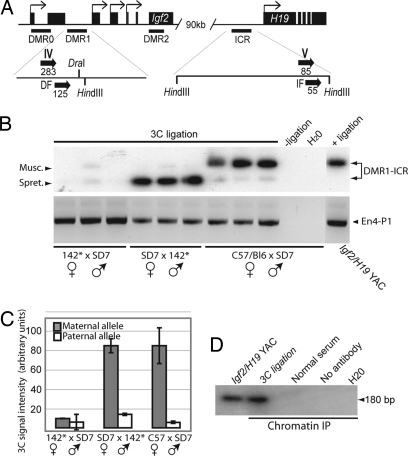
To understand the DMR1–ICR interaction in more detail, we performed 3C analysis using HindIII-digested chromatin (Fig. 3A). Fig. 3 B and C confirms that only the maternal DMR1 allele is engaged in the interaction with the H19 ICR and that this maternal interaction was lost when the mutated H19 ICR allele was inherited maternally. To demonstrate that CTCF is actually present in this complex, we combined the 3C analysis with chromatin immunopurification (ChIP), a method termed ChIP-loop assay (35). Formaldehyde crosslinked chromatin from livers of wild-type mice was digested with HindIII in the initial 3C step as outlined above and were immunopurified by a CTCF antibody (13–15). The purified DNA–protein complex was ligated and amplified for 3C analysis as above. Fig. 3D shows that the expected PCR product diagnostic of the H19 ICR–Igf2 DMR1 complex was specifically present in the ChIP material but absent in controls, including ChIP samples obtained with control serum. This result documents that CTCF is part of a complex that includes the Igf2 DMR1 and H19 ICR in close physical proximity.
Fig. 3.
CTCF target sites control the interaction between the H19 ICR and Igf2 DMR1 regions. (A) Schematic map of the Igf2 and H19 loci. The Igf2 DMR1 and H19 ICR domains are expanded to show the locations of 3C primers (marked with roman numerals and thick arrows to indicate their directions). The numbers indicate their distance from the HindIII sites. Primers IV/V span a polymorphic restriction site for DraI specific to the SD7 allele. (B) Three independent samples from each cross were subjected to the 3C assay. The PCR products were digested with DraI and subjected to Southern blot hybridization analysis to verify specificity of the amplified DNA fragments. The amplification of HindIII-digested and ligated yeast artificial chromosome DNA covering the entire Igf2/H19 domain was used as a positive control. (C) The intensity of the ICR–DMR1 bands of the image in B was calculated to normalize against variation in the 3C assay. The same result was obtained when the PCR products were quantified from ethidium bromide-stained agarose gels (data not shown). (D) CTCF is present in the ICR–DMR1 complex as determined by 3C analysis of CTCF–DNA complexes that had undergone ChIP. See Materials and Methods for additional information.
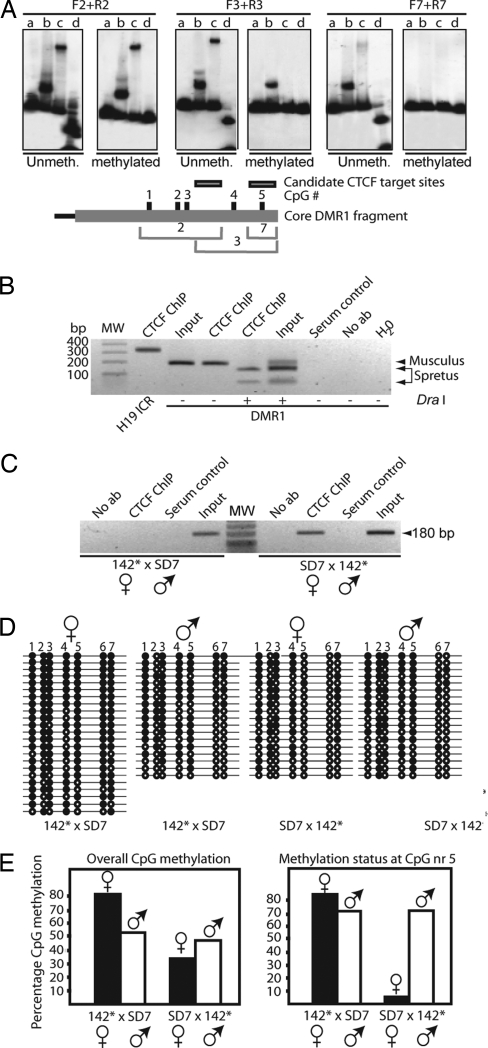
Our earlier demonstration that different CTCF–DNA complexes can interact physically with each other (5) prompted us to examine the possibility that the ICR–DMR1 complex involved CTCF target sites on both sides of the loop. EMSA revealed that only two overlapping DNA fragments in DMR1, 2 and 3 in Fig. 13 Upper, which is published as supporting information on the PNAS web site, displayed an ability to significantly interact in vitro with CTCF. Interestingly, the CTCF-positive fragments cover five of the seven CpGs analyzed in the bisulfite analysis for differential methylation of Igf2 DMR1 (see below). We therefore determined whether the CTCF binding to DMR1 was methylation-sensitive. Fig. 4A shows that only one DNA fragment, which encompasses CpG nr 5 (see below), interacted with CTCF in a methylation-sensitive manner.
Fig. 4.
ICR–DMR1, CTCF, and long-range epigenetic coordination. (A) (Upper) Effects of in vitro CpG methylation on CTCF binding to positive DNA fragments 2, 3, and 7 (the last fragment covers only CpG site nr 5; see Fig. 13). Digestion of unmethylated/methylated probe with the methylation-sensitive enzyme HpaII is depicted in lane d for each panel. Three pair of panels with both unmethylated and methylated [32P]DNA probes and in vitro-translated proteins are depicted (lanes are marked as in Fig. 13 Upper). (Lower) Shown is a map of core DMR1 fragment with CpG numbers and the positions of the overlapping DNA fragments. (B and C) CTCF interacts with the Igf2 DMR1 in vivo. ChIP analysis of neonatal liver derived from a cross between C57BL/6 and SD7 (B) or from 142* × SD7 and SD7 × 142* crosses (C). The DraI polymorphism specific for the M. spretus allele of the DMR1 in the recombinant SD7 mouse strain was exploited (see Fig. 3A). (D) Bisulfite sequencing data of DMR1 in neonatal liver. Filled and open circles represent methylated and unmethylated CpGs, respectively. Maternal and paternal alleles were distinguished using the DraI polymorphism. (E) Summary of the overall bisulfite data or, specifically, the fifth CpG site, given in percentage of CpG methylation.
Next, we examined the possibility that DMR1 interacted with CTCF in vivo by using ChIP assays. Fig. 4B shows that CTCF indeed interacts specifically with the maternal DMR1 allele. Strikingly, CTCF binding to the maternal DMR1 allele was lost when the mutated H19 ICR allele was inherited maternally. This result suggests that in vivo CTCF is recruited to DMR1 through the physical interaction between the H19 ICR and DMR1.
Given that CTCF binds to the maternal DMR1 allele in vivo dependent on CTCF binding sites within the H19 ICR, it was possible that this CTCF binding conferred protection against DNA methylation. We therefore carried out methylation analysis of DMR1, comparing paternal (control) to maternal inheritance of the CTCF binding mutant. With paternal inheritance of the 142* allele (control), the maternal DMR1 allele was comparatively less methylated than the paternal one (Fig. 4E), as described for the wild-type situation (27). However, when the mutant H19 ICR allele was maternally inherited, there was a significant overall increase of methylation of the maternal DMR1 allele. Particularly striking was the change at CpG nr 5, for which we showed methylation-dependent CTCF binding in vitro. In the control cross, the paternal allele was highly methylated, whereas the maternal allele was hypomethylated, but both alleles were hypermethylated when the mutant ICR was maternally derived. Clearly, removal of CTCF binding to the unmethylated CpG Nr 5 in DMR1 had removed the protection against de novo methylation. In addition to DMR1, we found that the CTCF binding sites in the ICR prevent de novo methylation of the maternally inherited DMR2 allele (Fig. 14, which is published as supporting information on the PNAS web site).
Discussion
We have carried out a systematic 3C analysis in an imprinting cluster. Although we have taken care to perform this study as quantitatively as possible, it is important to point out that all major conclusions are based on observing qualitative interactions. The significant conclusions from this work first include the observation that the distal enhancers, which are required both for Igf2 and H19 transcription, cannot access the maternal Igf2 allele but can do so with the paternal copy. Second, on the maternal allele, the H19 ICR is in close physical contact with DMR1 and MAR3, which flank the Igf2 gene. This conformation sequesters the maternal copy of Igf2 into a small loop of silent chromatin (Fig. 5). Presumably, the structure of this loop is what ultimately restricts the access of the enhancers in neonatal liver. The analysis of the access of the endodermal enhancers to the parental alleles in the Igf2/H19 region showed that the region as a whole is accessible to the enhancers on the paternal chromosome. This observation makes necessary the revision of the simple loop model proposed for the paternal chromosome (29) to a more compact model in which most parts of the chromatin in the H19/Igf2 region are in contact with the enhancers, perhaps located in an active chromatin hub or transcription factory (36). Whether such contact is static or alternatively involves the enhancers dynamically scanning the whole region is not known. By contrast, on the maternal allele, the enhancers are excluded from accessing large parts of the Igf2/H19 region, including, crucially, the promoters of Igf2. Thus, the repressed states of H19 (promoter methylation, enhancer access) and Igf2 (no promoter methylation, lack of enhancer access) are clearly established by fundamentally different epigenetic mechanisms.
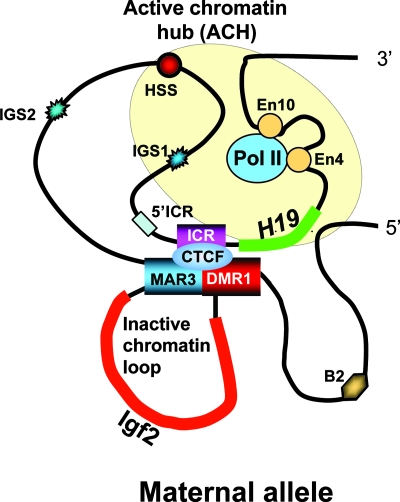
Fig. 5.
Model showing contacts established within the maternal allele at the H19 locus in neonatal liver. The model suggests a mechanism of H19 ICR function that takes into account active chromatin hubs (ACH) (40) and the repressing IDM–DMR–MAR3 complex on the maternal chromosome. This model is based on results from neonatal liver only and may not apply to other tissues. Additional work will be required (see Discussion) to determine the exact roles of DMR1 and MAR3 in liver, because complete deletion of DMR1 results in reexpression of the maternal Igf2 allele only in mesodermal tissues.
It was somewhat surprising, however, that enhancer access was not completely blocked to all areas located 5′ of the unmethylated ICR. This region, which encompasses at least 25 kb, includes a previously described hypersensitive site that can be found on both parental alleles (34). Interestingly, these hypersensitive sites have enhancer functions in both imprinted and nonimprinted tissues (22). Taken together with the HUC enhancers situated just upstream of the H19 ICR (21) and the suggestion of enhancers upstream of Igf2 (37), it is a formal possibility that the ICR’s insulator function collaborates in vivo with its function in establishing the ICR–MAR3–DMR1 complex and that this collaboration leads to complete repression of the maternal Igf2 allele. In addition, it is possible that insulators and enhancers in vivo are in a dynamic equilibrium, depending on the strength of both elements, and that physical restriction of access is not necessarily absolute (38).
An important component of how enhancer access is restricted away from the maternal Igf2 locus is presumably the pocket structure formed by physical contact between the ICR, DMR1, and MAR3. At least in the instance of the ICR–DMR1 complex, this interaction survives mitosis, suggesting that it is part of an epigenetic memory (39). It is important to realize that our analysis and therefore the model arising from it (Fig. 5) is exclusively based on neonatal liver, the major Igf2 expressing tissue in which the endoderm enhancers are active. Deletion or point mutation of DMR1 in liver reactivates the maternal Igf2 allele only marginally (18), whereas deletion of DMR1 in mesodermal tissues results in substantial reactivation (19). These observations suggest that there may be considerable differences of higher-order chromatin organization between embryonic lineages and tissues. In particular, in mesodermal tissues, the DMR1 may play a more prominent role than MAR3, whereas release from repression of Igf2 in liver may need deletion of MAR3 or deletion simultaneously of DMR1 and MAR3.
Both enhancer restriction and the ICR–DMR1–MAR3 structure clearly depend on CTCF binding to the maternal ICR. In the absence of CTCF binding, the higher-order chromatin structure of the region reverts from a maternal to a paternal pattern. CTCF binds to the maternal DMR1 in vivo, and, intriguingly, this binding requires intact CTCF binding sites in the ICR. Thus, whether CTCF binding to ICR and DMR1 directly mediates the contact (perhaps through formation of homodimers) or whether other proteins bound to DMR1 would make contact with CTCF on the ICR is not clear. When CTCF binding to DMR1 is abolished by CTCF binding-site mutations in the ICR, the maternal DMR1 becomes de novo methylated, particularly in the CpG that is part of a methylation-sensitive CTCF binding site. This finding is consistent with CTCF binding protecting against de novo methylation; once CTCF is removed, the binding site becomes methylated and can no longer bind CTCF (23, 24). We conclude that CTCF target sites of one region (the H19 ICR) can coordinate epigenetic marks at other regions in cis, located a long distance away.
Our results have provided a systematic view of higher-order chromatin structure in an imprinting cluster and have identified CTCF binding to the H19 ICR as a key component of this structure. The beauty of this system is that the parent-specific chromatin structure of the whole locus can be regulated epigenetically by one germ-line DMR, the paternally methylated ICR, through CTCF binding to its unmethylated maternal allele. It will be interesting to see whether similar principles apply to other imprinted or epigenetically regulated regions in which CTCF is implicated. Moreover, a number of CTCF-dependent chromatin insulators have recently been identified in the genome, many of them located outside of imprinted regions (3). It will be important to establish whether the organization of higher-order chromatin structure in cis is a general property of these elements.
Materials and Methods
3C Assay.
Neonatal mouse liver cells were dispersed by immediate mashing through a 70-mm nylon cell strainer into Dulbecco’s modified Eagle’s medium. The 3C assays were done essentially as described in refs. 29 and 40. The linear range of amplification was determined for the liver samples by serial dilution. The PCR products were subjected to Southern blot hybridization or hot-stop PCR ([α-32P]-ATP-labeled en4-R and ICR-R primer, respectively) (41) analysis, and the results were quantified by using multigauge version 2.2 PhosphorImager system (Fuji). All information about primers and PCR conditions are summarized in Table 1, which is published as supporting information on the PNAS web site. Consult the Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for a complete description of how 3C digestion, ligation, and PCR were controlled, and how quantitative values were normalized. Briefly, we normalized for 3C efficiency between experiments by using the unrelated gene locus Ercc3 for ligation and PCR bias of each primer pair with a Igf2-H19 yeast artificial chromosome and for parental PCR bias between M. m. domesticus and M. spretus alleles by mixing the 3C PCR products of the two strains in defined ratios. Each experiment was done on three different liver specimens from each mouse cross that is described in the study. Each liver sample was processed for 3C analysis three times.
ChIP-Loop Assay.
Formaldehyde-crosslinked chromatin was subjected to a ChIP-loop assay (35). Briefly, DNA–protein complexes were digested with HindIII, precleared for 4 h with protein G4 Fast Flow Sepharose beads (Amersham Pharmacia Biosciences), and then incubated with mouse monoclonal CTCF antibody (BD Biosciences, Franklin Lakes, NJ) overnight. After incubation with protein G4 Fast Flow Sepharose beads (and washing the complex four times), the beads were suspended in ligation buffer and subjected to the 3C analysis as described above.
Bisulfite Sequencing Analyses.
Genomic DNA was isolated from 1-day postpartum liver tissues (Promega). Approximately 1 μg of EcoRI-digested DNA was subjected to bisulfite treatment (42) and PCR amplification (27). The resulting PCR products were gel-purified (Qiagen, Hilden, Germany) and ligated into pCR2.1 (Invitrogen). The sequencing analyses were performed with the BigDye Terminator cycle sequencing kit (Applied Biosystems).
Nuclear Extracts and in Vitro Transcription–Translation.
Full-length human CTCF and the 11ZF CTCF-binding domain were in vitro translated from pET-7.1 and pET-11ZF, respectively (43), by using the TnT reticulocyte lysate-coupled in vitro transcription–translation system (Promega).
EMSA and in Vitro CpG Methylation.
The in vitro analysis of CTCF binding sites and CpG methylation effects was performed as described in ref. 15. A detailed account of the primer sequences is presented in Supporting Materials and Methods and Table 1.
Supplementary Material
Acknowledgments
We thank Drs. G. Smits and W. de Laat for helpful discussions. This work was supported by the Swedish Science Research Council (R.O.), the Swedish Cancer Research Foundation (R.O.), the Swedish Pediatric Cancer Foundation (R.O.), the Lundberg Foundation (R.O.), intramural research funding (to V.L.), the Wellcome Trust (A.M.), Cancer Research U.K. (W.R.), the European Union (W.R.), and the Biotechnology and Biological Sciences Research Council (W.R.).
Abbreviations
- 3C
chromosome conformation capture
- ChIP
chromatin immunopurification
- DMR
differentially methylated region
- en4
endodermal H19 enhancer
- ICR
imprinting control region
- IGS
intergenic sequence region
- MAR
matrix attachment region.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bell A. C., West A. G., Felsenfeld G. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 2.Bell A., West A., Felsenfeld G. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay R., Yu W.-Q., Whitehead J., Xu J.-W., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Quitschke W., Chernukhin I., et al. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohlsson R., Renkawitz R., Lobanenkov V. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 5.Pant V., Kurukuti S., Pugacheva E., Shamsuddin S., Mariano P., Renkawitz R., Klenova E., Lobanenkov V., Ohlsson R. Mol. Cell. Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yusufzai T., Tagami H., Nakatani Y., Felsenfeld G. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 7.Verona R., Mann M., Bartolomei M. Annu. Rev. Cell Dev. Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 8.Ohlsson R., Paldi A., Graves J. M. Trends Genet. 2001;17:136–141. doi: 10.1016/s0168-9525(00)02211-3. [DOI] [PubMed] [Google Scholar]
- 9.Sleutels F., Barlow D. Adv. Genet. 2002;46:119–163. doi: 10.1016/s0065-2660(02)46006-3. [DOI] [PubMed] [Google Scholar]
- 10.Reik W., Walter J. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 11.Leighton P., Saam J., Ingram R., Stewart C., Tilghman S. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 12.Thorvaldsen J. L., Duran K. L., Bartolomei M. S. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., Tilghman S. M. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 14.Bell A. C., Felsenfeld G. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 15.Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C.-F., Wolffe A., Ohlsson R., Lobanenkov V. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 16.Holmgren C., Kanduri K., Dell G., Ward A., Mukhopadhya R., Kanduri M., Lobanenkov V., Ohlsson R. Curr. Biol. 2001;11:1128–1130. doi: 10.1016/s0960-9822(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 17.Kaffer C. R., Srivastava M., Park K. Y., Ives E., Hsieh S., Batlle J., Grinberg A., Huang S. P., Pfeifer K. Genes Dev. 2000;14:1908–1919. [PMC free article] [PubMed] [Google Scholar]
- 18.Eden S., Constancia M., Hashimshony T., Dean W., Goldstein B., Johnson A., Keshet I., Reik W., Cedar H. EMBO J. 2001;20:3518–3525. doi: 10.1093/emboj/20.13.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constancia M., Dean W., Lopes S., Moore T., Kelsey G., Reik W. Nat. Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 20.Murrell A., Heeson S., Bowden L., Constancia M., Dean W., Kelsey G., Reik W. EMBO Rep. 2001;2:1101–1106. doi: 10.1093/embo-reports/kve248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drewell R., Arney K., Arima T., Barton S., Brenton J., Surani M. Development. 2002;129:1205–1213. doi: 10.1242/dev.129.5.1205. Cambridge, U.K. [DOI] [PubMed] [Google Scholar]
- 22.Charambolous M., Menheniott T., Bennet W., Kelly S., Dell G., Dandolo L., Ward A. Dev. Biol. 2004;271:488–497. doi: 10.1016/j.ydbio.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 23.Pant V., Mariano P., Kanduri C., Mattsson A., Lobanenkov V., Heuchel R., Ohlsson R. Genes Dev. 2003;17:586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoenherr C., Levorse J., Tilghman S. Nat. Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 25.Rand E., Ben-Porath I., Keshet I., Cedar H. Curr. Biol. 2004;14:1007–1012. doi: 10.1016/j.cub.2004.05.041. [DOI] [PubMed] [Google Scholar]
- 26.Fedoriw A., Stein P., Svoboda P., Schultz R., Bartolomei M. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 27.Lopes S., Lewis A., Hajkova P., Dean W., Oswald J., Forne T., Murrell A., Constancia M., Bartolomei M., Walter J., Reik W. Hum. Mol. Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- 28.Dekker J., Rippe K., Dekker M., Kleckner N. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 29.Murrell A., Heeson S., Reik W. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 30.Dekker J. Nat. Methods. 2005;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 31.Davies K., Bowden L., Smith P., Dean W., Hill D., Furuumi H., Sasaki H., Cattanach B., Reik W. Development. 2002;129:1657–1668. doi: 10.1242/dev.129.7.1657. Cambridge, U.K. [DOI] [PubMed] [Google Scholar]
- 32.Dean W., Bowden L., Aitchison A., Klose J., Meneses J., Reik W., Feil R. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. Cambridge, U.K. [DOI] [PubMed] [Google Scholar]
- 33.Tolhuis B., Palstra R., Splinter E., Grosveld F., de Laat W. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 34.Ainscough J., Koide T., Tada M., Barton S., Surani M. Development. 1997;124:3621–3632. doi: 10.1242/dev.124.18.3621. Cambridge, U.K. [DOI] [PubMed] [Google Scholar]
- 35.Horike S.-I., Cai S., Miyano M., Cheng J.-F., Kohwi-Shigematsu T. Nat. Genet. 2004;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 36.Osborne C., Chakalova L., Brown K., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J., Lopes S., Reik W., Fraser P. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 37.Ainscough J., Dandolo L., Surani M. Mech. Dev. 2000;91:365–368. doi: 10.1016/s0925-4773(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 38.Fourel G., Magdinier F., Gilson E. Bioassays. 2004;26:523–532. doi: 10.1002/bies.20028. [DOI] [PubMed] [Google Scholar]
- 39.Burke L., Zhang R., Bartkuhn M., Tiwari V., Tavoosidana G., Kurukuti S., Weth C., Leers J., Galjart N., Ohlsson R., Renkawitz R. EMBO J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Laat W., Grosveld F. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 41.Uejima H., Lee M., Cui H., Feinberg A. Nat. Genet. 2000;25:375–376. doi: 10.1038/78040. [DOI] [PubMed] [Google Scholar]
- 42.Kerjean A., Vieillefond A., Thiounn N., Sibony M., Jeanpierre M., Jouannet P. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.21.e106. E106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Awad T., Bigler J., Ulmer J., Hu Y., Moore J., Lutz M., Neiman P., Collins S., Renkawitz R., Lobanenkov V., Filippova G. J. Biol. Chem. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.