Abstract
Many genes encoding aminoacyl-tRNA synthetases and other amino acid–related products in Gram-positive bacteria, including important pathogens, are regulated through interaction of unacylated tRNA with the 5′-untranslated region (5′-UTR) of the mRNA. Each gene regulated by this mechanism responds specifically to the cognate tRNA, and specificity is determined by pairing of the anticodon of the tRNA with a codon sequence in the “Specifier Loop” of the 5′-UTR. For the 5′-UTR to function in gene regulation, the mRNA folding interactions must be sufficiently stable to present the codon sequence for productive binding to the anticodon of the matching tRNA. A model bimolecular system was developed in which the interaction between two half molecules (“Common” and “Specifier”) would reconstitute the Specifier Loop region of the 5′-UTR of the Bacillus subtilis glyQS gene, encoding GlyRS mRNA. Gel mobility shift analysis and fluorescence spectroscopy yielded experimental K ds of 27.6 ± 1.0 μM and 10.5 ± 0.7 μM, respectively, for complex formation between Common and Specifier half molecules. The reconstituted 5′-UTR of the glyQS mRNA bound the anticodon stem and loop of tRNAGly (ASLGlyGCC) specifically and with a significant affinity (K d = 20.2 ± 1.4 μM). Thus, the bimolecular 5′-UTR and ASLGlyGCC models mimic the RNA–RNA interaction required for T box gene regulation in vivo.
Keywords: gene regulation, Gram-positive bacteria, tRNA, RNA experimental model
INTRODUCTION
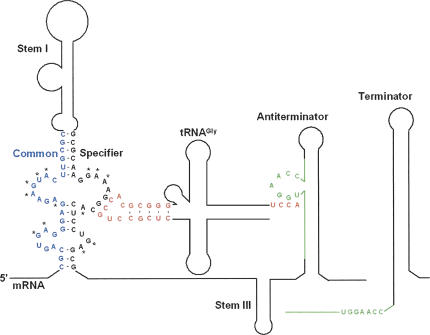
The T box transcription termination control system is commonly used in many Gram-positive bacteria, including pathogenic species, to regulate expression of genes encoding aminoacyl-tRNA synthetases (aaRS), amino acid biosynthetic enzymes, and transporter proteins (Grundy and Henkin 1993, 2003). Each gene in this family is induced in response to a decrease in aminoacylation of the cognate tRNA. Genes in the T box family contain a 5′-untranslated region (5′-UTR) ranging from 200 to 300 nucleotides (nt) in length. The 5′-UTRs exhibit a complex pattern of conserved sequence and structural elements, and generally include an intrinsic transcriptional terminator (Fig. 1). Formation of the terminator helix results in premature termination of transcription and repression of transcription of the downstream coding sequence, while sequestration of the 5′ side of the terminator helix into a competing antiterminator structure results in read-through of the termination site and continued transcription (Fig. 1). Folding of the nascent RNA into the antiterminator form depends on interaction of the nascent RNA with the cognate unacylated tRNA (Fig. 1). The specificity of tRNA recognition is directed by pairing of the tRNA anticodon with a triplet sequence, designated the “Specifier Sequence,” within the Stem I element of the 5′-UTR (Grundy and Henkin 1993). The terminator helix is more stable than the antiterminator, so that preventing termination requires stabilization of the antiterminator by base-pairing of 4 nt (5′-UGGN-3′) within a bulged region of the antiterminator with 4 unpaired nt (5′-NCCA-3′) at the acceptor end of the unacylated tRNA (Grundy et al. 1994). Acylation of the tRNA blocks the interaction with the antiterminator, allowing the system to monitor tRNA charging to signal the requirement for expression of the regulated gene.
FIGURE 1.
Secondary structure of the 5′-UTR of the B. subtilis glyQS mRNA. The proposed secondary structure of the leader sequence (nucleotides 1–223) is shown on the left in the Antiterminator form. The Specifier Loop in Stem I contains the glycine codon sequence GGC (nucleotides 99–101) and is responsible for binding the tRNAGlyGCC anticodon GCC in the anticodon stem and loop domain (red). The Antiterminator bulge (nucleotides 151–161 [green]) contains the sequence UGGA that binds the 3′-terminal UCCA sequence (red) of the unacylated tRNA. In the absence of unacylated tRNA, the stem and bulge containing the UGGA sequence (green) take the alternate conformation of the Terminator (right). Nucleosides with asterisks (*) are conserved among T box RNAs (Grundy and Henkin 1993). The figure is not drawn to scale.
tRNAGly-dependent antitermination of the Bacillus subtilis glyQS gene has been reproduced in vitro with a purified transcription system, demonstrating that no additional cellular factors are required for interaction of the tRNA with the nascent RNA transcript (Grundy et al. 2002). Specific binding of tRNAGly to glyQS RNA generated by T7 RNAP transcription has also been demonstrated, and structural alterations in both RNA partners have been detected in the complex (Yousef et al. 2005). Base-pairing between the Specifier Sequence and the tRNA anticodon, and between the antiterminator bulge and the acceptor end of the tRNA, are required for a functional interaction between the two RNAs (Grundy et al. 2002; Yousef et al. 2005), and the tRNA must retain its overall tertiary structural features (Yousef et al. 2003). However, detailed information concerning the three-dimensional interactions between the two RNAs is lacking.
A model RNA representing the antiterminator element and its interaction with both tRNA and structural analogs of the tRNA acceptor end has been studied in detail (Gerdeman et al. 2003). Clear experimental evidence was provided for presentation of residues within the antiterminator bulge for base-pairing with the corresponding positions in the tRNA acceptor end (Fig. 1). The goal of the current study was to further investigate the structural arrangement of the Specifier Loop domain of T box RNAs and the presentation of the Specifier Sequence residues for interaction with the tRNA anticodon domain. The Specifier Loop is responsible for the accurate display of the codon sequence for pairing with the anticodon of the cognate tRNA and discrimination against noncognate tRNAs. The tRNA anticodon/Specifier Sequence interaction may have physical characteristics and chemical properties similar to those of tRNA binding to mRNA on the ribosome (Agris 2004; Murphy et al. 2004), but codon–anticodon pairing in T box RNAs occurs in the absence of proteins or ribosomal subunits. The Specifier Loop is formed in the context of the complex Stem I element of the 5′-UTR, formation of which requires long-range folding interactions within the RNA (Fig. 1). In most T box family RNAs, the Specifier Loop includes an RNA structural motif similar to the Loop E motif found in 5S rRNAs (Leontis and Westhof 1998; Yousef et al. 2005). Little information is currently available concerning the structural arrangement of Stem I and the Specifier Loop region, although preliminary structural mapping studies support the secondary structural model derived from phylogenetic analyses (Luo et al. 1998; Yousef et al. 2005), and protection of the Specifier Sequence by binding of the tRNA has been demonstrated. Here, we report the design and analysis of a bipartite model RNA that mimics the Specifier Loop region of the B. subtilis glyQS 5′-UTR, and characterize its interaction with the anticodon stem and loop domain (ASL) of the appropriate tRNA, ASLGlyGCC.
RESULTS
Design of a bimolecular system to mimic the T box Specifier Loop domain
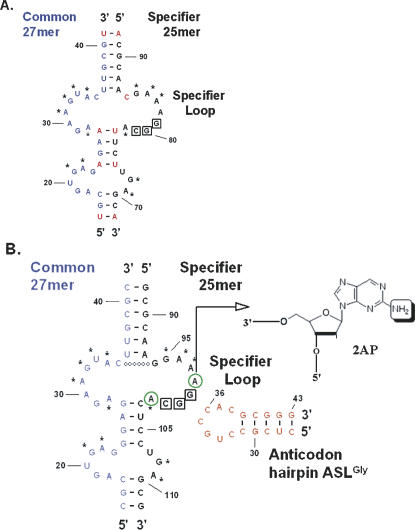
The structural arrangement of the bottom half of the B. subtilis glyQS 5′-UTR Stem I domain was generated from RNAs corresponding to the 5′ (“Common,” 27 nt) and 3′ (“Specifier,” 25 nt) regions. The Common half of this bimolecular complex corresponds to glyQS residues 15–41, while the Specifier half corresponds to residues 88–112 (Fig. 2A; Grundy et al. 2002). Residues conserved in T box family sequences were maintained in the model sequences, while certain A–U pairings within helical domains were replaced with G–C pairings at positions where G–C pairings are found in other T box sequences (T.M. Henkin, unpubl.). Specifier half molecule constructs were synthesized with or without a fluorescence reporter in the form of a single 2-aminopurine ribonucleoside (2AP) substitution at A98 or A102, on either side of the GGC glycine codon (Fig. 2B). Neither A98 nor A102 are invariant residues, although the position corresponding to A102 is conserved as a purine and pairs with the conserved U33 residue of tRNA (Yousef et al. 2005). A 17-nt RNA hairpin structure, comprising residues 27–43 of tRNAGlyGCC, was synthesized and used as a mimic of the anticodon stem and loop domain (Fig. 2B).
FIGURE 2.
Specifier Loop region of the glyQS 5′-UTR. (A) Native sequences for the 5′- and 3′-sides of the bottom portion of Stem I. Nucleosides with asterisks (*) are conserved among T box RNAs. Native sequence A•U pairs in red were substituted with G•C base pairs. (B) The biomolecular model RNAs. The Specifier half molecule contains the Specifier Loop with the glycine codon GGC. The sequences were altered to increase the stability of the complex between the Common and Specifier half molecules, including the addition of a predicted base pair between C35 and G94 denoted with open circles (○). The Specifier half molecule was synthesized with 2-aminopurine (2AP) substituting for A either 5′ or 3′ to the codon, at position 98 or position 102 (green circled “A”). The 2AP (inset) fluorescence was used to monitor interactions. The 17-nt ASLGlyGCC is shown in the secondary structure determined from NMR analyses (F. Vendeix and P.F. Agris, unpubl.).
Monitoring the reconstitution of the Stem I complex by PAGE mobility shift
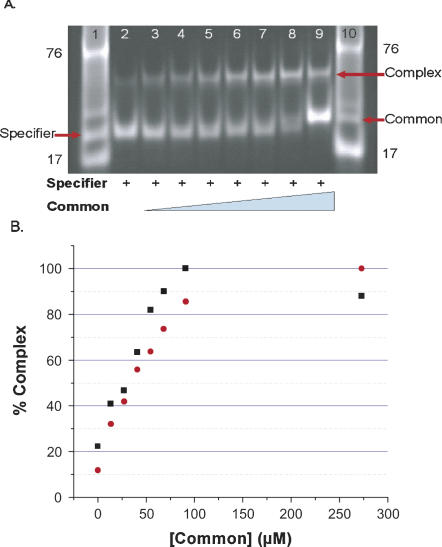
Formation of a complex between the Common and Specifier half molecules was assessed initially by gel mobility shift assays. Addition of increasing concentrations of the Common RNA to a constant amount of the Specifier 2AP98-RNA resulted in appearance of a band with slower migration, the intensity of which increased as higher concentrations of the Common half molecule were added (Fig. 3). The migration of the Specifier/Common complex was consistent with that of a unimolecular RNA of equal size (52 mer), as determined from the 17 mer and 76 mer standards, and individual Specifier (25 mer) and Common (27 mer) species in the gel. The individual Specifier and Common half molecules did not exhibit slower moving bands even at excessive (300 μM) concentrations (data not shown). Quantification of the ethidium bromide stained bands yielded binding curves for the interactions of the Common half molecule with Specifier 2AP98 and 2AP102 half molecules (Fig. 3B). Dissociation constants (K ds) and standard free energies of binding at equilibrium (ΔG°s) were obtained from the binding curves. The reconstitution of Stem I through the interaction of the Specifier half molecule with the Common half molecule was found to occur with K ds of 27.6 ± 1.0 μM and 32.3 ± 2.3 μM for the Specifier 2AP98 and 2AP102 molecules, respectively. The ΔG°s of complex formation from the gel mobility shift assay were 6.2 ± 0.1 Kcal/mol for Specifier 2AP98 and −6.1 ± 0.1 Kcal/mol for 2AP102.
FIGURE 3.
Reconstitution of Stem I from interaction of the Specifier and Common half molecules. (A) Formation of the Specifier/Common complex was observed by polyacrylamide gel electrophoresis (PAGE) mobility shift analyses. Specifier 2AP98 half molecule (183 μM) was titrated with increasing concentrations of the Common half molecule (lane 2, 0.0; 3, 23.9; 4, 47.9; 5, 71.8; 6, 95.8; 7, 120; 8, 144; 9, 287 μM). RNA standards, a 17-mer anticodon stem and loop and unfractionated tRNA ∼76 mer were run in lanes 1 and 10. In addition, lane 1 contained Specifier half molecule (25 mer), and lane 10 contained Common half molecule (27 mer). (B) Complex formation between the Common half molecule and the two Specifier half molecules, 2AP98 (black square) and 2AP102 (red circle). Ethidium bromide stained RNAs of the PAGE mobility shift analyses were quantified with ImageQuant; the results were normalized and plotted as percent Complex (bound) vs. concentration of the Common half molecule. Binding curves were analyzed with a nonlinear regresssion (Origin) to extrapolate dissociation constants. The error for multiple determinations of the K ds was determined to be ±1.0–±2.3 μM.
Monitoring of Specifier/Common complex formation by spectrofluorimetry
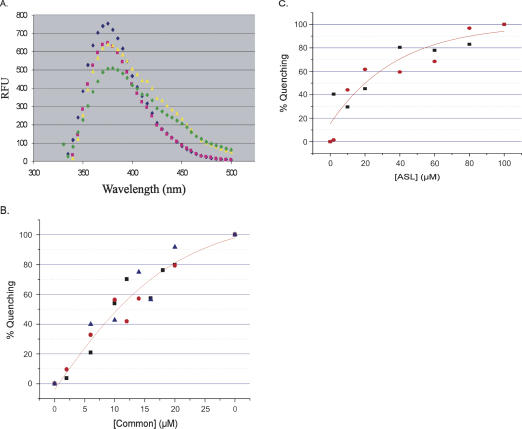
Fluorescence of the 2AP98 and 2AP102 Specifier half molecules was monitored after incubation with the Common half molecule. Formation of the complex with the Common half molecule quenched the fluorescence of both Specifier RNAs but did not alter the fluorescence spectral profile (Fig. 4A). The Specifier 2AP98 half molecule was a more sensitive reporter of complex formation than was the Specifier 2AP102 (data not shown). In addition, the pairing of A102 with the invariant U33 of the tRNA was predicted to contribute to the strength of the codon/anticodon interaction (Yousef et al. 2005) that we wanted to analyze within our experimental model. Therefore, Specifier 2AP98 was used to confirm the binding constant observed by the PAGE mobility shift assay. Titration of Specifier 2AP98 with the Common half molecule resulted in maximal quenching of the fluorescence at 20 μM Common. The affinity of Specifier 2AP98 for the Common half molecule was derived from the binding curves (Fig. 4B) to be 10.5 ± 0.7 μM with a ΔG° of −6.8 ± 0.1 Kcal/mol, almost threefold stronger than that determined by PAGE mobility shift analyses (27.6 ± 1.0 μM; −6.2 ± 0.1 Kcal/mol). Thermodynamic and free energy calculations to predict the folding interactions of the Specifier and Common half molecules in forming Stem I resulted in two closely related and almost equally stable structures. Both structures displayed the codon within the Specifier loop. However, the second most stable structure with a free energy (ΔG° at 298°K) of −19.2 Kcal/mol most closely resembled the conformation supported by genetic analysis in vivo and chemical and enzymatic probing experiments in vitro (Rollins et al. 1997; Winkler et al. 2001; Putzer et al. 2002; Yousef et al. 2005). The other structure was only modestly more stable with a ΔG° of −21.0 Kcal/mol.
FIGURE 4.
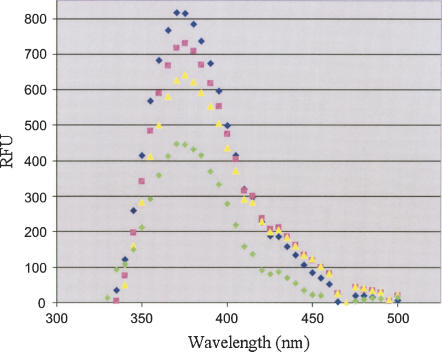
Fluorescence spectroscopy of binding of the Specifier 2AP98 to Common and to ASLGlyGCC. (A) Fluorescence emission spectra were recorded for Specifier 2AP98 alone (blue diamond), reconstitution of the 5′-UTR Stem I complex between Specifier 2AP98 and Common (pink square), the binding of ASLGlyGCC to the Specifier 2AP98 and Common complex (green diamond), and the nonbinding control of ASLPheGAA (yellow triangle). Spectra in the presence of the ASLs have unexplained shoulders in the 400–450 nm range. (B) Binding of the Specifier 2AP98 half molecule to the Common half molecule. With the Specifier 2AP98 concentration constant (2.0 μM), the concentration of the Common half molecule was increased. Data were normalized and the percent fluorescence quenching (bound) was plotted vs. Common concentration. The binding curves were analyzed with a nonlinear regression (Origin). The error (±0.7 μM) was derived from the average of three experiments (black square, red circle, blue triangle). (C) Binding of ASLGlyGCC to the Specifier 2AP98/Common complex. The concentration of the Specifier 2AP98/Common complex was held constant (2.0 μM) as ASLGlyGCC was titrated into solution. Data were normalized and the percent fluorescence quenching (bound) was plotted vs. ASLGlyGCC concentration. The binding curves were analyzed with a nonlinear regression (Origin). The dissociation constant was derived from the average of two runs of the experiment (black square, red circle).
Interaction of the Specifier Sequence with the ASL anticodon monitored by spectrofluorimetry
The addition of ASLGlyGCC to the complex formed from the Specifier 2AP98 and Common half molecules led to further quenching of the fluorescence reporter and alteration of the fluorescence emission spectrum profile (Fig. 4A). Fluorescence shoulders in the spectrum appeared at 420 and 450 nm (Fig. 4A). The Specifier 2AP98/Common complex was titrated with ASLGlyGCC in order to determine the affinity for the ASL anticodon. Fluorescence of the Specifier 2AP98 (2 μM) was not affected by increasing concentrations of ASLGlyGCC >100 μM (Fig. 4C). The affinity of the Specifier 2AP98/Commmon complex for the ASLGlyGCC was 20.2 ± 1.4 μM and yielded a ΔG° of −6.4 ± 0.1 Kcal/mol. In order to assess the specificity of the interaction, we synthesized an anticodon stem and loop corresponding to that of yeast tRNAPhe with the codon GAA (ASLPheGAA. In contrast to ASLGlyGCC addition of the negative control ASLPheGAA to the Specifier 2AP98/Common complex did not quench the fluorescence (Fig. 4A), even at a concentration (100 μM) at which maximum quenching was achieved by ASLGlyGCC. However, the same two shoulders in the spectrum were observed. Thus, we conclude that the fluorescence shoulders at 420 and 450 nm were not the result of specific interactions between the Specifier Sequence and the ASL anticodon.
Altering the codon in the Specifier loop should change the specificity for tRNA. A Specifier loop was constructed with the GUA codon for valine tRNA and 2AP at position 98 as a reporter of the interaction with the anticodon. When Stem I was reconstituted from the Val Specifier and Common half molecules, fluorescence quenching of 2AP98 was comparable to that with the Gly Specifier (Figs. 4, 5). The reconstituted Stem I was bound by ASLValUAC as evidenced by the significant quenching of the 2AP98 fluorescence. The degree of fluorescence quenching with ASLVal at 100 μM was comparable to quenching at the maximum binding achieved with 100 μM ASLGly to the Gly Specifier (Fig. 5). However, the fluorescence of the Val Specifier 2AP98 was hardly altered with the addition of a negative control, ASLArgCCG at 100 μM (Fig. 5).
FIGURE 5.
Fluorescence spectroscopy of binding of the Specifier 2AP98 with the valine codon to ASLGlyGCC. Fluorescence emission spectra were recorded for the Val Specifier 2AP98 alone (blue diamond), reconstitution of the 5′-UTR Stem I complex between Val Specifier 2AP98 and Common (pink square), the binding of ASLValUACto the Val Specifier 2AP98 and Common complex (green diamond), and the nonbinding control of ASLArgCCG (yellow triangle).
DISCUSSION
Our model system was designed for analyzing the physicochemical properties required in the functional folding of the 5′-UTR of GlyRS mRNA and in its binding to tRNAGly. A model of the 5′-UTR was designed and synthesized as two RNAs, Specifier and Common, to form the bottom portion of the Stem I element of the mRNA leader sequence (Fig. 1), which interacts with the tRNA anticodon region. Formation of the complex was assessed by PAGE and was observable and measurable by staining with ethidium bromide. The ΔG° of complex formation was determined to be −6.2 ± 0.1 Kcal/mol. A gel mobility shift assay monitored by 32P-end labeling of the Specifier half molecule also detected complex formation and produced a similar free energy of folding, −6.9 Kcal/mol. Mung bean ribonuclease digestion of the RNA indicated that the codon was accessible in the complex (data not shown). Fluorescence spectroscopy was utilized as a second approach to observing complex formation. The fluorescence of 2AP substituted for adenosine at either side of the Specifier codon was quenched upon interaction of the Specifier half molecule with the Common half molecule. The free energy of complex formation from the fluorescence derived binding constant (−6.8 ± 0.1 Kcal/mol) was comparable to that obtained by gel mobility shift assays. Thus, the empirically derived ΔGs were comparable to each other and considerably higher than the −19.2 Kcal/mol of the theoretical folding derived from empirical studies often conducted in the presence of 1 M NaCl (Mathews et al. 1999, 2004). This difference is reasonable considering that the dynamics of the system in solution assessed by gel mobility shift and by fluorescence spectroscopy and the lower counter ion concentration probably contribute to the higher free energy than that predicted by a static interaction calculation.
Neither the gel mobility shift nor the fluorescence analyses indicate if the reconstituted Stem I structure was providing a stably open Specifier Loop with the codon presented for anticodon binding, properties that are crucial to the biological relevance of the model. To investigate the interaction of the Specifier glycine codon, GGC, with the tRNAGlyGCC anticodon, we designed and synthesized the 17-nt anticodon stem and loop domain ASLGlyGCC. Observation of a specific interaction between the Specifier Sequence and the ASLGlyGCC would contribute indirect proof that the codon was accessible for binding. Addition of ASLGlyGCC to the reconstituted Stem I complex formed from Specifer 2AP98 and Common half molecules quenched the 2AP fluorescence indicating an interaction. The absence of quenching of the 2AP98 fluorescence with the negative control anticodon stem and loop of yeast tRNAPhe (ASLPheGAA) at 100 μM indicated that the interaction with ASLGlyGCC was specific. The Specifier 2AP98/Common complex bound the ASLGlyGCC RNA with a K d of 20.2 ± 0.1 μM, ΔG° = −6.4 ± 0.1 Kcal/mol. These results indicate that Specifier Loop sequence of the Stem I model RNA was physically and chemically accessible for anticodon binding and bound the ASLGlyGCC specifically. The K d of 20.2 ± 0.1 μM was surprisingly strong considering that the Common/Specifier interaction resulted in a K d of 10.5 ± 0.7 μM. The unassisted interaction of a codon with an anticodon is expected to occur with a very low affinity. The observed interaction is some five orders of magnitude stronger than that calculated for duplex formation between two trinucleotides, yet only one-tenth the affinity observed for the interaction of the anticodon of native Escherichia coli tRNAGlu with the complementary anticodon of yeast tRNAPhe (Grosjean et al. 1976).
A recent report (Yousef et al. 2005) demonstrated that the Specifier Loop glycine codon (nucleotides 99–101) was protected from Mg2+-induced cleavage in vitro when tRNAGly GCC was present. In contrast to A98, the conserved purine A102, 3′ to the codon, was protected from Mg2+ cleavage in the presence of tRNA (Yousef et al. 2005). This suggests that pairing of the position 102 purine of the 5′-UTR with the invariant U33 of the tRNA may facilitate the Specifier Loop–tRNA interaction. Thus, in our experimental model of the GlyRS mRNA 5′-UTR, A102 may bind to the ASLGlyGCC U33 forming the tetranucleotide duplex:
Specifier Loop: 2AP98 GGCA102
•••○
Anticodon Loop: CCGU33
In binding ASLGlyGCC to the 5′-UTR, we chose to monitor the fluorescence of 2AP98 to avoid interference with the U33–A102 interaction. The postulated tetranucleotide duplex may be a contributing factor to the significant binding affinity observed in our flurorescence assays.
The interaction of the glyQS 5′-UTR with tRNAGly was evident in experiments conducted with as little as 5 mM Mg2+ (Yousef et al. 2005). With our experimental model RNAs in which codon/anticodon interaction was monitored with fluorescence spectroscopy, Mg2+ was not required. Gel mobility shift and fluorescence studies indicated that the Specifier/Common complex reconstituting the 5′-UTR had a significant binding affinity (K ds = 10.5 μM). The interaction of ASLGlyGCC with the Specifier/Common complex did not generate a supershifted complex in gel electrophoresis. The failure to observe a supershift could be due to lower stability of the ASL-Stem I interaction (K d of 20.2 μM), kinetic on/off rates, or may be a consequence of the electrophoresis method.
In order to confirm the specificity of codon–anticodon interaction within the experimental model, the Specifier codon was changed to that of valine. The new Stem I construct now bound ASLVal, but not ASLArg, substantiating the availability of the codon and the selectivity of the system. The only published mutations in Stem I were studies of the the RNA structural motif called a K-turn (Winkler et al. 2001). The K-turn is composed of the dinucleotide GA on both sides of a loop, such as the two highly conserved GA sequences at the bottom of the 5′-UTR Stem I (Figs. 1, 2). A single site mutation in the GA sequence resulted in a significant decrease in expression of the B. subtilis tyrS gene in vivo. However, ASL was still able to bind the Specifier when the same single nucleoside substitution was made in vitro. Since the effect of the K-turn motif has not been tested in the context of the glyQS gene, subsequent experiments are required both in vivo and in vitro to determine the full capacity of the experimental model to mimic the in vivo results.
RNA:RNA biophysical and chemical interactions are composed of the four chemistries of adenosine, uridine, guanosine, and cytidine, contrasting with, and probably predating, the 20 chemistries of a protein's amino acids. The unique interaction of the 5′-UTR of aaRS mRNAs with unacylated tRNA in Gram-positive organisms, without a requirement for ribosomes or protein factors, may represent an early mechanism in the regulation of transcription (Henkin 1996), similar to the binding of small molecules by riboswitch RNAs (Grundy and Henkin 2004). Our experimental model supports the proposition that regulation occurs in the absence of protein and is dependent on tRNA's anticodon interaction with codon in the 5′-UTR of nascent mRNA. The Specifier Sequence/tRNA anticodon interaction is very similar to the decoding of mRNA that occurs on the ribosome between the mRNA and tRNA with the exception of the possible binding of conserved A102 to tRNA's invariant U33. Participation of U33 in ribosomal codon binding would result in a translational frame shift. However, a tetranucleotide duplex formation between the Specifier Loop and the tRNA's anticodon loop is similar to the tetranucleotide duplex formed between the UGGN sequence in the antiterminator bulge and the tRNA's universal 3′-terminal CCA sequence plus the 5′-adjacent discriminator base in the tRNA (Grundy et al. 1994; Putzer et al. 2002; Yousef et al. 2005).
Some modifications in the anticodon region of tRNAs are essential for the tRNA to form the correct tertiary structure and to function in productively binding to the codon in the ribosomal A-site (Murphy et al. 2004). The modifications essential for translation may also be critical for transcription regulation in Gram-positive organisms. The GlyRS/tRNAGly system has no requirement for tRNA modification (Grundy et al. 2002), but other systems may. The model system that we designed has many of the properties of the T box transcription regulation system studied both in vivo and in vitro. The simplified character of the model will permit investigation of the chemically and physically important aspects of 5′-UTR folding and its interactions with tRNA. In addition, a physicochemical analysis of this unique mechanism of gene regulation will enhance understanding of metabolic control in important Gram-positive bacterial pathogens.
MATERIALS AND METHODS
RNA synthesis
RNAs corresponding to the 5′ (“Common”) and 3′ (“Specifier”) sequences of the region at the base of the Stem I element of the B. subtilis glyQS 5′-UTR (Fig. 2A) were chemically synthesized. Two Common half molecules were synthesized. The wild-type sequence was comprised of the most highly conserved nucleosides and sequences (Fig. 2B). A mutant Common sequence was synthesized with an altered K-turn motif in which A22 was substituted with G22. The Specifier Loop contained either the glycine codon GGC or the valine codon GUA (Gly Specifier and Val Specifier, respectively). To increase the stability of helical elements, particular U–A or A–U base pairs were selected and replaced with C–G or G–C pairs (Fig. 2B). In addition, C94 was replaced with a G to potentially add an extra base pair in the helix above the Specifier Loop (Fig. 2B). These changes represent natural variations found in other T Box RNA sequences (F.J. Grundy and T.M. Henkin, unpubl.). The Specifier half molecule (Fig. 2B) was synthesized with and without 2-aminopurine ribonucleoside (2AP) at positions A98 or A102, flanking the codon sequence. A 17-nt RNA (ASLGlyGCC) was designed and chemically synthesized with the anticodon GCC to mimic the anticodon stem and loop domain of B. subtilis tRNAGlyGCC (Fig. 2B). An ASLValUAC was synthesized to test binding to the Val Specifier. Two negative control ASLs also were chemically synthesized, an ASLPheGAA with the GAA phenylalanine anticodon and an ASLArgCCG with the arginine anticodon CCG. RNAs were synthesized by “ACE” chemistries (Dharmacon RNA Technologies). The RNAs were deprotected as suggested by the manufacturer, lyophilized, and dissolved in H2O. The purity of the RNA was analyzed by ion exchange HPLC and denaturing (7 M urea) polyacrylamide gel electrophoresis (PAGE).
RNA folding predictions
The most thermodynamically stable structures for the Common and Specifier half molecules (individually and in combination) and the ASL RNAs were predicted by using RNAStructure 3.0 (Mathews et al. 1999, 2004).
Polyacrylamide gel mobility shift analysis
Binding of the Common and Specifier half molecules was assessed using a polyacrylamide gel electrophoresis (PAGE) mobility shift assay (Nobles et al. 2001). The gel composition was 15% polyacrylamide in TB buffer (89 mM Tris base, 89 mM boric acid at pH 8.3). PAGE was conducted at 4 ± 0.5°C using a temperature controlled gel electrophoresis apparatus (Novex Mini-cell Thermoflow, Invitrogen). Concentrations of the Specifier half molecule were kept constant and titrated with increasing amounts of the Common half molecule. The reaction mixtures were heated at 90°C for 30 min and then allowed to slow-cool to room temperature for 45 min before electrophoresis. Prior to loading, the RNA samples were diluted fourfold (v/v) with a loading buffer (TB buffer that was 50% glycerol, 0.25% [w/v] Bromophenol Blue and 0.25% [w/v] Xylene Cyanol FF). After electrophoresis, gels were stained with ethidium bromide (0.5 μg/mL, 15 min, at room temperature) and digitally photographed (BioRad). The RNA bands were quantified using ImageQuant software (Molecular Dynamics, Amersham Biosciences). Complex formation was normalized to 100% for the lane in which the Specifier RNA had been titrated completely into the complex with the Common RNA. Dissociation constants (K d) were determined using a sigmoidal model (Origin, MicroCal LLC).
Fluorescence spectroscopy
The fluorescence of 2AP in the Specifier half molecule was observed in order to monitor interactions with the Common half molecule and with the ASL. Spectra and end point fluorescence data were collected with a microplate reader with two monochrometers (SprectraMax Gemini XS, Molecular Devices). Excitation and emission wavelengths of 2AP in the Specifier half molecule were 310 nm and 375 nm, respectively. Optimum sensitivity and response for the instrument was found to occur at a concentration of 2 μM for Specifier 2AP98 and at 3 μM for Specifier 2AP102. Specifier half molecules were titrated with increasing concentrations of the Common half molecule from 2 to 30 μM. The mixture of Specifier and Common half molecules was denatured at 90°C for 30 min and then the two half molecules were allowed to anneal at room temperature for 45 min prior to measurement of fluorescence. ASL RNAs were incubated with the already formed complex of Specifier 2AP98/Common at 25°C for 5 min. The interactions of ASLGlyGCC and ASLPheGAA with the complex were monitored by collecting fluorescence emission spectra. All of the obtained spectral data were then analyzed in Microsoft Excel and in Origin to compare the change in fluorescence among the different complexes. The data were normalized to 100% for the sample exhibiting the most quenching of the fluorescent reporter, indicating the most complex formation. The molar standard Gibbs free energy change (ΔG°) of the Specifier/Common folding interaction and that of the binding of ASL to the Specifier 2AP98/Common complex were derived from ΔG° = −RTlnKeq, where T = 298°K, and
 |
ACKNOWLEDGMENTS
We thank William “Darnell” Graham for purification of the RNA samples, Helen Rho for demonstrating the feasibility of the gel mobility shift analyses, and Keith Gagnon for assisting with the 32P labeling experiments (Department of Molecular and Structural Biochemistry, NCSU). We wish to thank Dr. Jennifer Hines (Ohio University) for suggesting the use of 2-aminopurine for fluorescence monitoring and for reviewing the manuscript. The research was supported in part by grants from the NSF and the NIH (MCB-9986011 and 2RO1-GM23037 to P.F.A. and R01-GM47823 to T.M.H.). A.R.N. was the recipient of an NSF REU award.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.29906.
Abbreviations: aaRS, aminoacyl-tRNA synthetase; aatRNA, aminoacyl-tRNA; 2AP, 2-aminopurine; 2AP98, 2AP at position 98 of the 5′-UTR; 2AP102, 2AP at position 102 of the 5′-UTR; Common, the 5′-half molecule of the 5′-UTR; nt, nucleotide; RNAP, RNA polymerase; Specifier, the 3′-half molecule of the 5′-UTR containing the Specifier Sequence; Specifier Sequence, triplet codon to tRNA's anticodon; 5′-UTR, 5′-untranslated region of mRNA.
REFERENCES
- Agris P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdeman M.S., Henkin T.M., Hines J.V. Solution structure of the Bacillus subtilis T-box antiterminator RNA: Seven nucleotide bulge characterized by stacking and flexibility. J. Mol. Biol. 2003;326:189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Soll D.G., Crothers D.M. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J. Mol. Biol. 1976;103:499–519. doi: 10.1016/0022-2836(76)90214-x. [DOI] [PubMed] [Google Scholar]
- Grundy F.J., Henkin T.M. tRNA as a positive regulator of transcription antitermination in B. subtilis . Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- Grundy F.J., Henkin T.M. The T box and S box transcription termination control systems. Front. Biosci. 2003;8:d20–d31. doi: 10.2741/908. [DOI] [PubMed] [Google Scholar]
- Grundy F.J., Henkin T.M. Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 2004;7:126–131. doi: 10.1016/j.mib.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Grundy F.J., Rollins S.M., Henkin T.M. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: A new role for the discriminator base. J. Bacteriol. 1994;76:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F.J., Winkler W.C., Henkin T.M. tRNA-mediated transcription anti-termination in vitro: Codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin T.M. Control of transcription termination in prokaryotes. Annu. Rev. Genet. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- Leontis N.B., Westhof E. The 5S rRNA loop E: Chemical probing and phylogenetic data versus crystal structure. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Condon C., Grunberg-Manago M., Putzer H. In vitro and in vivo secondary structure probing of the thrS leader in Bacillus subtilis . Nucleic Acids Res. 1998;26:5379–5387. doi: 10.1093/nar/26.23.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy F.V., IV, Ramakrishnan V., Malkiewicz A., Agris P.F. The role of modifications in codon recognition and discrimination: tRNALys UUU . Nat. Struct. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- Nobles K.N., Yarian C.S., Guenther R.H., Agris P.F. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2001;30:4751–4760. doi: 10.1093/nar/gkf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer H., Condon C., Brechemier-Baey D., Brito R., Grunberg-Manago M. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 2002;30:3026–3033. doi: 10.1093/nar/gkf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins S.M., Grundy F.J., Henkin T.M. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 1997;25:411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- Winkler W.C., Grundy F.J., Murphy B.A., Henkin T.M. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M.R., Grundy F.J., Henkin T.M. tRNA requirements for glyQSantitermination: A new twist on tRNA. RNA. 2003;9:1148–1156. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M.R., Grundy F.J., Henkin T.M. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T Box leader RNA. J. Mol. Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]