Abstract
As previously established in yeast, two sequences within mRNAs are responsible for their specific localization to the mitochondrial surface—the region coding for the mitochondrial targeting sequence and the 3′UTR. This phenomenon is conserved in human cells. Therefore, we decided to use mRNA localization as a tool to address to mitochondria, a protein that is not normally imported. For this purpose, we associated a nuclear recoded ATP6 gene with the mitochondrial targeting sequence and the 3′UTR of the nuclear SOD2 gene, which mRNA exclusively localizes to the mitochondrial surface in HeLa cells. The ATP6 gene is naturally located into the organelle and encodes a highly hydrophobic protein of the respiratory chain complex V. In this study, we demonstrated that hybrid ATP6 mRNAs, as the endogenous SOD2 mRNA, localize to the mitochondrial surface in human cells. Remarkably, fusion proteins localize to mitochondria in vivo. Indeed, ATP6 precursors synthesized in the cytoplasm were imported into mitochondria in a highly efficient way, especially when both the MTS and the 3′UTR of the SOD2 gene were associated with the re-engineered ATP6 gene. Hence, these data indicate that mRNA targeting to the mitochondrial surface represents an attractive strategy for allowing the mitochondrial import of proteins originally encoded by the mitochondrial genome without any amino acid change in the protein that could interfere with its biologic activity.
Keywords: mRNA sorting to the mitochondrial surface, mitochondrial cotranslational import, nuclear encoded ATP6 gene
INTRODUCTION
A key feature of eukaryotic cells is their organization into separate subcellular compartments, each containing distinct sets of proteins. The sorting of several proteins destined to organelles involves mRNA localization. This specific localization might be preferable to protein localization; indeed, one mRNA molecule can serve as a template for multiple rounds of translation. Thus, localizing an mRNA rather than the protein to its site of action offers obvious advantages (Jansen 2001; Tekotte and Davis 2002).
Mitochondria occupy a central position in the overall metabolism of eukaryotic cells; hence, the oxidative phosphorylation (OXPHOS), the Krebs's cycle, the urea cycle, the heme biosynthesis, and the fatty acid oxidation take place within the organelle. Mitochondrial biogenesis is a complex process that requires the concerted expression of both nuclear and mitochondrial genomes. More than 99% of mitochondrial proteins are encoded by the nucleus and synthesized in the cytoplasm. Mitochondrial sorting of mRNAs encoding mitochondrial proteins might likely represent a key step to ensuring the functionality of the corresponding polypeptides inside the organelle. In this case, a cotranslational phase might assist the import of the precursors (Corral-Debrinski et al. 1999, 2000; Fünfschilling and Rospert 1999; George et al. 2002). We have demonstrated that in yeast, 47% of mRNAs encoding mitochondrial proteins are transported to the organelle surface (Sylvestre et al. 2003b). Among them, ATP2 mRNA, encoding the β-subunit of ATP synthase, exclusively localizes to the mitochondrial surface. Two sequences within the ATP2 transcript are involved in its specific subcellular localization—the region coding for the mitochondrial targeting sequence (MTS) and the 3′ untranslated region (3′UTR). Remarkably, the in vivo impairment of ATP2 mRNA sorting, due to the absence of its 3′UTR, is associated with a severe respiratory deficiency. This deficiency is the consequence of an inefficient import of the precursor, indicating that mRNA targeting to the surface of mitochondria is essential to ensure this process (Margeot et al. 2002). Interestingly, we demonstrated that several mRNAs encoding mitochondrial proteins preferentially localized to the mitochondrial surface in HeLa cells (Sylvestre et al. 2003a). We have addressed the question of whether a protein that is normally encoded by mitochondrial DNA (mtDNA) can be efficiently translocated inside the mitochondria by the use of signals that will address its mRNA to the surface of the organelle. The rationale behind this is to allow a cotranslational import mechanism. Thus, the precursor will be maintained in an import-competent conformation, impeding its aggregation before or during translocation through the TOM (translocase of the outer membrane) and TIM (translocase of the inner membrane) import complexes. The strategy of expressing mtDNA-encoded genes in the nuclear-cytoplasmic compartment is called allotopic expression. Up until today, when tried in mammalian cells, allotopic expression did not lead to an efficient mitochondrial translocation of precursors examined, probably due to the high hydrophobic nature of polypeptides encoded by mtDNA (Oca-Cossio et al. 2003; Smith et al. 2004).
We associated the MTS and the 3′UTR of the nuclear gene SOD2 to a re-engineered nucleus-localized ATP6 gene. SOD2 codes for an intramitochondrial free radical scavenging enzyme, which is the first line of defense against superoxide produced as a byproduct of OXPHOS (Li et al. 1995). We have previously shown that SOD2 mRNA localizes to the mitochondrial surface in human cells (Sylvestre et al. 2003a). Furthermore, a recent report described that in HeLa cells, SOD2 mRNA is associated to the mitochondrial surface, via its 3′UTR linked to the K homology domain of A-kinase anchor protein 121, AKAP121 (Ginsberg et al. 2003). HeLa cells were stably transfected with two different plasmids. One contains the MTS representing the first 30 amino acids of the protein in frame with the AUG codon of the recoded ATP6 gene, the other combines both the MTS and 3′UTR. We were able to demonstrate that (1) both hybrid ATP6 mRNAs localize to the mitochondrial surface, especially the one possessing the two targeting signals; (2) fusion protein synthesized from each construction localizes to mitochondria in vivo; (3) precursor forms of the ATP6 protein translated in the cytoplasm are successfully translocated inside the organelle. Interestingly, the presence of both mRNA-targeting signals leads to a 1.8-fold increase in the amount of fully mitochondrial imported protein compared with the presence of the MTS alone. Therefore, the strategy of directing a hybrid mRNA to the mitochondrial surface allows the mitochondrial import of the nucleus-encoded ATP6 gene in a highly efficient way.
RESULTS
Construction of the re-engineered mitochondrial ATP6 gene
To accomplish allotopic expression of the ATP6 gene originally located in the mitochondrial genome, we synthesized the full-length nuclear version of the gene-converting codons AUA to AUG and codons UGA to UGG to ensure the accurate translation of the transcript by cytoplasmic ribosomes. Eleven alterations were performed in the ATP6 ORF by in vitro direct mutagenesis. To force the localization of the recoded ATP6 mRNA to the mitochondrial surface, we associated with the ORF the cis-acting elements involved in the mitochondrial sorting of the SOD2 mRNA—the sequence coding for the MTS and the 3′UTR (as described in the Materials and Methods section). Consequently, we obtained two different constructions cloned in the pCMV-Tag 4A vector that we named, respectively, SOD2 MTS ATP6-3′UTR SV40 and SOD2 MTS ATP6-3′UTRSOD2. The SOD2 MTS ATP6-3′UTR SV40 contains the sequence encoding the first 30 amino acids of SOD2 in frame with the AUG codon of the recoded ATP6 gene. In this plasmid, the SV40 poly(A) signal functions as the 3′UTR. In the SOD2 MTS ATP6-3′UTRSOD2 vector, both the MTS and the 3′UTR of SOD2 were associated with the ATP6 ORF. The 3′UTR of 215 bp (Ginsberg et al. 2003) was inserted at the end of the ORF following the Flag epitope and replacing the SV40 poly(A) signal.
Subcellular distribution of hybrid ATP6 mRNAs in HeLa cells
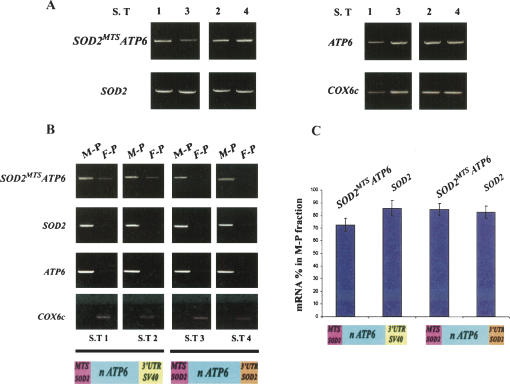
HeLa cells transfected with either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 vectors were examined to determine the steady-state levels of hybrid ATP6 mRNAs. Total RNAs were extracted from four independent populations of stably transfected cells (S.T 1 and S.T 2 for SOD2 MTS ATP6-3′UTR SV40; S.T 3 and S.T 4 for SOD2 MTS ATP6-3′UTRSOD2). Figure 1A illustrates results obtained after RT–PCR analyses. The abundance of endogenous ATP6, COX6c, and SOD2 transcripts as well as hybrid ATP6 mRNAs was examined using conditions described in Table 2 (below). No major differences in the amount of the SOD2 MTS ATP6 mRNA were observed in cell lines expressing either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 vectors.
FIGURE 1.
Subcellular distribution of hybrid ATP6 mRNAs in HeLa cells. (A) Total RNA extracted from cells expressing the SOD2 MTS ATP6-3′UTR SV40 (S.T 1 and S.T 2) or the SOD2 MTS ATP6-3′UTRSOD2 (S.T 3 and S.T 4) vectors were subjected to RT–PCR analysis to reveal amounts of hybrid ATP6 (SOD2 MTS ATP6) mRNAs and endogenous SOD2, ATP6, and COX6c mRNAs. The amount of RNAs used for the reverse transcription, PCR conditions, and specific oligonucleotides used for each gene are summarized in Table 2. (B) RNAs were purified from mitochondrion-bound polysomes (M–P) and free-cytoplasmic polysomes (F–P) of stably transfected cell lines with either SOD2 MTS ATP6-3′UTR SV40 (S.T 1 and S.T 2) or SOD2 MTS ATP6-3′UTRSOD2 (S.T 3 and S.T 4) vectors and subjected to RT–PCR analysis. The abundance of endogenous ATP6, SOD2, and COX6c mRNAs was determined in each polysomal population using the conditions shown in Table 2. (C) Densitometric analyses were performed using the Quantity One (Bio-Rad) software system. The difference between the amounts of hybrid ATP6 mRNAs in cells expressing respectively SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 constructions was significant according to the paired Student's t-test (P < 0.0034, n = 6).
TABLE 2.
RT–PCR analyses
To examine the ability of SOD2 signals associated with the recoded ATP6 gene to direct hybrid mRNAs to the mitochondrial surface, we determined their subcellular localization in the four stable cell lines obtained. For this purpose, we isolated RNAs from mitochondrion-bound polysomes (M-P) and free-cytoplasmic polysomes (F-P) and we determined by RT–PCR the steady-state levels of hybrid mRNAs in both polysomal populations (Fig. 1B). As internal controls, the subcellular distribution of endogenous mitochondrial ATP6, SOD2, and COX6c mRNAs were determined. Endogenous ATP6 mRNA exclusively localized to the mitochondrial compartment as expected. In addition, endogenous SOD2 mRNA is enriched in mitochondrion-bound polysomes (M-P), whereas COX6c mRNA is preferentially detected in free-cytoplasmic polysomes (F-P) as we have previously observed (Sylvestre et al. 2003a). The SOD2 MTS ATP6-3′UTRSOD2 vector directed the synthesis of a hybrid mRNA that was almost undetectable in free-cytoplasmic polysomes. Hybrid mRNA produced from the SOD2 MTS ATP6-3′UTR SV40 plasmid was also detected preferentially in mitochondrion-bound polysomes. However, it was also present in free-cytoplasmic polysomes (Fig. 1B). Densitometric analyses were performed to determine the amount of both endogenous SOD2 and hybrid ATP6 mRNAs in each polysomal population examined. SOD2 mRNA signal in mitochondrion-bound polysomes was 85.6% ± 6.15 in cell lines expressing the SOD2 MTS ATP6-3′UTR SV40 plasmid and 82.5% ± 4.87 in cells expressing the SOD2 MTS ATP6-3′UTRSOD2 vector. Interestingly, it was found for the ATP6 hybrid mRNA that only 72.4% ± 5.2 localized to the mitochondrial surface in cells expressing the SOD2 MTS ATP6-3′UTR SV40 vector. Instead, in cells expressing the SOD2 MTS ATP6-3′UTRSOD2 vector, 84.6% ± 4.7 of the hybrid mRNA localized to the mitochondrial surface (Fig. 1C). These values were significantly different according to the paired Student's t-test (P < 0.0034, n = 6). Thus, the combination of both the MTS and 3′UTR of SOD2 to the re-engineered ATP6 gene leads to the synthesis in the nucleus of a transcript that was almost exclusively sorted to the mitochondrial surface. Indeed, its subcellular distribution is not significantly different from the one of the endogenous SOD2 mRNA.
Detection of the fusion ATP6 proteins in vivo
We analyzed the ability of the re-engineered ATP6 product to localize to mitochondria in vivo. For this, we appended a Flag epitope in frame to the C terminus of the ATP6 ORF and examined stably transfected cells by indirect immunofluorescence (Fig. 2). HeLa cells transfected with the empty pCMV-Tag 4A vector were used as negative controls and showed a low diffused signal in cytoplasm when antibodies to Flag were used (Fig. 2, left). Stably transfected cells with either SOD2 MTS ATP6-3′UTR SV40, or SOD2 MTS ATP6-3′UTRSOD2 vectors were visualized by indirect immunofluorescence using antibodies to Flag (Fig. 2, left) and to ATP synthase subunit α (Fig. 2, middle). For each cell type visualized, a merged image in association with DAPI staining is shown at right. A typical punctuate mitochondrial pattern was observed in cells expressing the recoded ATP6 polypeptides when the Flag antibody was used. This indicates that fusion ATP6 proteins localized to mitochondria. Indeed, this typical punctuate mitochondrial pattern was also observed using specific antibodies to anti-ATP synthase subunit α. Interestingly, the merged images showed a significant colocalization between the two antibodies around the nuclear envelope and the cytoplasm as punctuate fluorescent specks. Thereby, this result confirms that both recoded ATP6 and endogenous ATP synthase α proteins present in vivo a very similar subcellular distribution. Furthermore, the localization patterns of the recoded ATP6 proteins in cells transfected with either one of the pCMV-Tag 4A vectors examined were comparable, demonstrating that the MTS alone or in combination with the 3′UTR successfully allowed re-engineered ATP6 proteins to localize to the surface of mitochondria in vivo.
FIGURE 2.
Subcellular localization of the recoded ATP6 protein in vivo. Stably transfected cells with the empty pCMV-tag4A vector, SOD2 MTS ATP6-3′UTR SV40, or SOD2 MTS ATP6-3′UTRSOD2 plasmids were visualized by indirect immunofluorescence using antibodies to Flag and ATP synthase subunit α. For each cell type visualized, a merged image in association with DAPI staining is shown at right. Indicative of the mitochondrial localization of recoded ATP6 proteins, cells transfected with either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 plasmids showed a significant colocalization of both Flag and ATP synthase α signals. In contrast, cells transfected with the empty vector exhibited a low diffuse cytoplasmic staining.
Steady-state levels of the fusion ATP6 proteins in HeLa cells
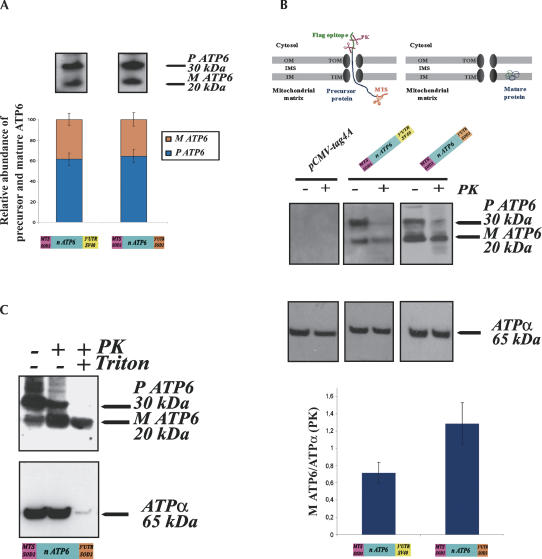
HeLa cells transfected with either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 vectors were examined to determine the steady-state levels of the fusion ATP6 proteins. Mitochondria isolated from stably transfected cells were subjected to immunoblot analyses (Fig. 3). We revealed two forms with the anti-Flag antibody of ∼30 and 20 kDa, representing the precursor and mature forms of the recoded ATP6 protein. The predicted molecular weights of both proteins are, respectively, 34 and 30 kDa larger than the ones implied by the molecular weight markers. This discrepancy has often been observed when extremely hydrophobic proteins were migrated in SDS-PAGE (Manfredi et al. 2002). In general, the electrophoretic mobility on SDS-PAGE of proteins encoded by mtDNA is higher than the one expected for their theoretical molecular weights (Mariottini et al. 1986). To determine the amounts of the recoded ATP6 polypeptides produced in HeLa cells expressing either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 vectors, we compared six independent mitochondrial extractions (Fig. 3A). Both precursor and mature polypeptides were equally abundant in mitochondria from each cell line; indeed, the expression of the SOD2 MTS ATP6-3′UTR SV40 vector leads to an accumulation of 61.4% ± 6 of the precursor form. Instead, SOD2 MTS ATP6-3′UTRSOD2 vector directed the synthesis of 64.4% ± 6.5 of the precursor. These values were not significantly different according to the paired Student's t-test. Similar results were obtained when total extracts from each cell line were examined by Western blotting (data not shown). These data are in agreement with the overall amounts of hybrid ATP6 mRNAs, detected when total RNAs from cell lines expressing either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 vectors were subjected to RT–PCR analyses (Fig. 1A). Therefore, the steady-state levels of the ATP6 precursor and its ability to recognize the TOM complex in the outer mitochondrial membrane do not depend on the presence of the SOD2 3′UTR. Notably, the relative proportions of ATP6 precursor and mature forms were analogous to the ones shown in cells for highly hydrophobic proteins en route to the mitochondria (Claros et al. 1995; Corral-Debrinski et al. 1999).
FIGURE 3.
Recoded ATP6 proteins are efficiently imported into mitochondria in vivo. (A) Six independent mitochondria purifications were performed with cells stably transfected with either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 plasmids and subjected to Western blot analysis. Signals for the ATP6 precursors and mature forms were scanned and quantified by the Quantity One System (Bio-Rad). No significant differences between the amounts of the precursor and the mature form of the recoded ATP6 proteins were observed in each cell line examined. (B) (Top) Schematic representation of mitochondrial import intermediates. The hydrophobic passenger protein can be trapped en route to the matrix. In this step, the protein can be blocked or represented an intermediate of translocation. This does not prevent the cleavage of the MTS by a mitochondrial processing peptidase; the rest of the protein remains accessible to PK digestion, and therefore, if digested, it becomes undetectable in the Western blot assay. The fraction of the protein completely translocated turns into a mature protein insensitive to PK located in the inner mitochondrial membrane. (MM) Mitochondrial matrix, (OM) outer membrane, (MIS) mitochondrial intermembrane space, (TOM) translocase of the outer membrane, (TIM) translocase of the inner membrane. (Middle) Mitochondria extracted from transfected cells with the empty pCMV-Tag 4A vector, SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 plasmids were subjected to Western blot essays. A total of 20 μg of proteins were treated with 150 μg/mL of PK at 0°C for 30 min and subjected to immunoblotting analysis using anti-ATP synthase subunit α and anti-Flag M2 antibodies. Densitometric analyses of experiments performed with six independent mitochondrial purifications are represented at the bottom. We normalized values measured for the signal of the mature form of ATP6 resistant to PK with ATPα signal revealed after PK digestion. We then compared the value obtained for cells expressing either the SOD2 MTS ATP6-3′UTR SV40 or the SOD2 MTS ATP6-3′UTRSOD2 plasmids. Signals from Western blots were scanned and quantified by the Quantity One System (Bio-Rad). The difference between the amounts of fully mitochondrial translocated ATP6 protein in cells expressing, respectively, SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 constructions was significant according to the paired Student's t-test (P < 0.0022, n = 6). (C) A total of 20 μg of mitochondria isolated from cells stably transfected with SOD2 MTS ATP6- 3′UTRSOD2 vector treated with 150 μg/mL PK and 1% Triton X100 at 0°C for 30 min and subsequently subjected to Western analysis.
Mitochondrial translocation of the fusion ATP6 protein
To determine the fraction of the C-terminally tagged ATP6 protein that has been completely translocated inside the organelle, mitochondria extractions were treated with 150 μg/mL of proteinase K (PK). Figure 3B (top) shows a schematic representation of the theoretically expected ATP6 import intermediate. The hydrophobic passenger ATP6 precursor can be trapped en route to the matrix and a mitochondrial processing peptidase can cleave the MTS. Nevertheless, the rest of the protein remained accessible to PK action and therefore became undetectable on Western blotting. Instead, the fraction of the ATP6 protein, which can be completely translocated, is insensitive to PK-induced proteolysis and can therefore be integrated into the inner mitochondrial membrane, hence remaining detectable on immunoblotting.
Figure 3B shows that precursor forms of the fusion proteins were sensitive to proteolysis in both cell lines examined. Nearly all of the ATP6 precursor signals disappeared after PK digestion, so precursors that were engaged in the process of translocation or loosely attached to the outer mitochondrial membrane, but not fully translocated, were entirely digested (Fig. 3B, middle). In contrast, a significant amount of the mature form of ATP6 is resistant to PK digestion, indicating its location inside the organelle. To examine the levels of another complex V protein in these cells, immunoblots were performed using anti-ATP synthase α antibody. This naturally imported mitochondrial protein was present at similar extents in all cells tested. Only one band of ∼65 kDa was visualized, suggesting that either we were unable to discriminate the precursor and mature forms of this protein under the electrophoretic conditions used or precursor polypeptides were very rapidly and efficiently translocated. Additionally, no major differences of the ATP synthase α signals were detected after PK treatment, confirming the integrity of the mitochondrial isolations (Fig. 3B, middle). To compare the import efficiency of the recoded ATP6 proteins in cells transfected with either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 vectors, we measured the amount of the mature form of ATP6 insensitive to PK digestion in each cell line after normalization with the amount of ATP synthase α resistant to PK proteolysis. Results for six independent mitochondrial extractions subjected to immunoblotting analyses were shown in Figure 3B, in the lower panel. Overall results show that both SOD2 signals lead to a highly efficient import of the recoded ATP6 precursor. Remarkably, the level of the mature form insensitive to PK proteolysis in cells transfected with SOD2 MTS ATP6-3′UTRSOD2 (1.28 ± 0.24) was 1.8-fold higher than in cells expressing the SOD2 MTS ATP6-3′UTR SV40 (0.71 ± 0.12). The difference measured was significant according to the paired Student's t-test (P < 0.0022, n = 6). This observation could be related to the higher enrichment in the mitochondrion-bound polysomes of the corresponding mRNA (Fig. 1B).
The question arises whether imported re-engineered ATP6 proteins were assembled into the ATP synthase complex. The complex is organized in F0–F1 domains, F1 sector is a water-soluble unit located in the matrix and has the ability to hydrolyze ATP. The F0 domain is embedded in the inner membrane and is composed by hydrophobic subunits forming a proton pathway. ATP6 is an intrinsic protein of F0, composed of five putative transmembranous α-helices. In contrast, ATP synthase α is a located in the matrix F1 domain. Studies performed with bovine heart mitochondria demonstrated that ATP6 was degraded at a very low rate when F0 subunits were subjected to trypsin treatment (Joshi and Burrows 1990). Therefore, we treated mitochondria with both PK and Triton X-100 (1%). The detergent disrupts both mitochondrial membranes and theoretically leads to the entire proteolysis of mitochondrial proteins, demonstrating their localization somewhere inside the organelle in a protease-sensitive form. Figure 3C shows that indeed ATP synthase α was fully digested by PK; however, a significant amount of ATP6 remained insensitive to PK proteolysis. This result suggests that the recoded ATP6 was assembled into complex V.
DISCUSSION
In this study, we described the construction of a nuclear version of the mtDNA-encoded ATP6 gene associated with cis-acting elements of SOD2 mRNA that will allow the sorting of the corresponding mRNA to the mitochondrial surface. This specific mRNA sorting leads to a highly efficient mitochondrial import of the ATP6 protein.
The localization of transcripts represents an extremely efficient way to target gene products to individual subcellular compartments or to a specific region of a cell or an embryo. Therefore, mRNA localization is now known to be a widespread phenomenon that occurs in unicellular organisms in plant and animal tissues as well as in developing embryos from a variety of animal species (Kloc et al. 2002). Nearly half of nuclearly transcribed mRNAs encoding mitochondrial proteins are sorted to the surface of mitochondria in the yeast Saccharomyces cerevisiae (Sylvestre et al. 2003b). This phenomenon is essential to ensure the efficient mitochondrial import of a subset of polypeptides both in vitro (Verner 1993) and in vivo (Margeot et al. 2002). The localization process is determined by at least two cis-acting elements within the transcripts—the region coding for the MTS at the 5′ extremity of the molecules and the 3′UTR (Corral-Debrinski et al. 2000; Margeot et al. 2002). Interestingly, we have previously reported the segregation of several mRNAs encoding mitochondrial proteins to the organelle surface in human cells (Sylvestre et al. 2003a). Recently, a motif within the 3′UTRs of mRNA coding for mitochondrial proteins was found highly conserved in yeasts and mammalian 3′UTRs. This motif is overrepresented within the set of genes that mRNAs localize to the surface of mitochondria, indicating its functional involvement in the specific sorting of these transcripts (Sylvestre et al. 2003b; Shalgi et al. 2005). We envision that mRNA targeting to the mitochondrial surface probably leads to a tight coupling between both translation and translocation processes, which may be required for some mitochondrially destined polypeptides, especially hydrophobic proteins (Corral-Debrinski et al. 1999; Fünfschilling and Rospert 1999; George et al. 2002; Karniely and Pines 2005).
Pathogenic point mutations of genes encoded by the mitochondrial genome have been described as the cause of many mitochondrial disorders (Smeitink et al. 2001). No effective therapy is available for these disorders (Zeviani and Carelli 2003; DiMauro 2004). A possible therapeutic approach is the allotopic expression of mtDNA genes. The basic concept is to introduce a wild-type copy of the mutated mitochondrial gene into the nucleus and to import normal copies of the protein into mitochondria from cytosol. Several reports in yeast described that some nonmitochondrial polypeptides can be relocated to the mitochondrial matrix simply by conjugating a MTS to their N terminus (Claros et al. 1995; Nagley et al. 1995; Corral-Debrinski et al. 1999). However, when this approach has been tried in human cells using different MTS and genes encoded by mtDNA, precursor polypeptides were not imported efficiently into mitochondria. Thus, up until today the spectrum of mtDNA-encoded polypeptides that can be successfully expressed and integrated into mitochondrial respiratory chain complexes is very limited (Smith et al. 2004). This limitation is thought to be the consequence of the high hydrophobic nature of mtDNA-encoded proteins, which possess transmembrane domains refractive to mitochondrial import. The precursor synthesized in the cytosol could lack the import-competent structure required for an efficient mitochondrial translocation. The concept of mesohydrophobicity is likely to be an important factor for mitochondrial import competency. Mesohydrophobicity describes the average hydrophobicity in a window of 60–80 amino acids together with the calculation of the most hydrophobic 17–amino acid segment. This calculation could predict importability of hydrophobic peptides (Claros and Vincens 1996). Using their algorithm, we analyzed this correlation to assess the mitochondrial importability of the SOD2MTSATP6 gene product and compared it with ATP6, COX8, and SOD2 polypeptides as well as the previously tested fusion protein COX8MTSATP6 (Manfredi et al. 2002). Figure 4 shows the graph obtained—as the wild-type ATP6, both fusion proteins examined cannot be translocated into mitochondria, mainly due to the high hydrophobicity of ATP6. Hence, a possibility that can allow the import of a recoded ATP6 protein into the organelle is that the precursor is engaged in a cotranslational pathway of import. Thereby, the precursor would be maintained in a loosely folded, nonaggregated conformation required for translocation through the mitochondrial import apparatus. To accomplish allotopic expression, the localization of an mRNA to the mitochondrial surface has never been tried before. In the allotopic approaches reported, even though different MTS were appended to recoded mitochondrial genes, all of the constructs examined contained at their 3′extremities the SV40 poly(A) signal that does not lead to any specific subcellular localization of the transcript (Owen et al. 2000; Guy et al. 2002; Manfredi et al. 2002; Oca-Cossio et al. 2003). Our data clearly demonstrate that the association to a recoded ATP6 gene of both the MTS and 3′UTR signals of the SOD2 gene leads to a highly efficient delivery of the hybrid mRNA to the mitochondrial surface. This improves unambiguously the feasibility of the allotopic approach for this mitochondrial gene. Indeed, not only were we able to visualize the fusion ATP6 protein in the mitochondria by indirect immunofluorescence, but definitely the amount of the processed ATP6 polypeptide inside the organelle was quite similar to the naturally imported ATP synthase subunit α, a Complex V component, as is ATP6. These data strongly indicate that recoded ATP6 precursors were successfully imported. The improvement we obtained compared with a recent report (Manfredi et al. 2002), which measured 18.5% of the precursor translocated, is certainly due to the localization of hybrid mRNAs to the mitochondrial surface. This specific localization obviously enhances the coupling between translation and import processes, therefore diminishing the block of the precursor during its translocation through the TOM and TIM complexes. It is worth mentioning that we obtained a gradual improvement on mitochondrial import of the ATP6 precursor. When both the MTS and the 3′UTR of SOD2 were combined, the amount of fully translocated ATP6 protein was 1.8-fold higher than when just the MTS was present. This is likely related to the improvement of mRNA sorting to the mitochondrial surface when both cis-acting elements of SOD2 were associated with the recoded ATP6 gene. Remarkably, proteolysis insensitivy of the translocated ATP6 protein in the presence of both PK and Triton X-100 suggested that the protein could be correctly assembled in the F0 domain of the respiratory chain Complex V.
FIGURE 4.
Mitochondrial import ability of ATP6 proteins based on the mesohydrophobicity index. A plot developed by Claros and Vincens (1996) was used to measure mitochondrial import ability of fusion ATP6 proteins. By this approach, the fusion SOD2 MTSATP6 protein would not be importable. Mesohydrophobicity, which is the average regional hydrophobicity over a 69 amino acid region, was calculated using Mito-ProtII (Claros and Vincens 1996). Values obtained are as follows: ATP6, 1.41; SOD2 MTSATP6, 1.41; COX8 MTSATP6, 1.41; SOD2, −1.26; COX8, −1.63.
Notably, by combining the cis-acting elements of the COX10 gene to the recoded ATP6 gene, we obtained a very efficient mitochondrial import ability of the fusion protein (C. Bonnet, V. Kaltimbacher, S. Ellouze, V. Forster, J-A. Sahel, and M. Corral-Debrinski, in prep.). Indeed, COX10 mRNA codes for a highly hydrophobic protein involved in Complex IV biogenesis (Valnot et al. 2000) and as SOD2, mRNA is enriched at the mitochondrial surface in human cells (Sylvestre et al. 2003a).
In conclusion, we undeniably optimized the allotopic expression approach for ATP6 by the use of mRNA-targeting signals without any amino acid change in the protein that could affect its biologic activity. This approach becomes henceforth available to rescue mitochondrial deficiencies caused by mutations in mtDNA genes.
MATERIALS AND METHODS
Plasmid construction
The full-length ATP6 mitochondrial gene was re-engineered after the synthesis of the 677-bp product by RT–PCR (Superscript III one step RT–PCR Platinium Taq HiFi, Invitrogen) using total RNAs from HeLa cells. The PCR product obtained was cloned in the PCR 2.1-Topo vector (Invitrogen). In this vector, we recoded 11 nonuniversal codons in the ATP6 gene by six rounds of in vitro–directed mutagenesis (QuikChange Multi site-directed mutagenesis kit, Stratagene). Six oligonucleotide primers were designed to change AUA codons to AUG and UGA to UGG (Table 1). To this recoded ATP6, we appended in frame the MTS of SOD2, obtained by RT–PCR. Oligonucleotide primers used are shown in Table 1. The final fragment was inserted in the pCMV-Tag 4A vector (Stratagene). This plasmid will direct the synthesis of the recoded ATP6 protein via the CMV promoter and protein detection by the presence of a Flag epitope appended to the recoded ATP6 C-terminal region. To obtain hybrid mRNAs that will also contain the 3′UTR of SOD2, we replaced the SV40 poly(A) signal of the pCMV-Tag 4A vector by 215 bp of the full-length SOD2 3′UTR, using PvuI and MluI enzymes. This sequence was obtained by RT–PCR using RNAs purified from HeLa cells and specific oligonucleotide primers containing PvuI and MluI restriction sites at each of their extremities (Table 1). The two final constructs were entirely sequenced for accuracy to verify the full-length sequences of either the fusion ATP6 genes or the 3′UTR appended to them. They were named, respectively, SOD2 MTS ATP6-3′UTR SV40 and SOD2 MTS ATP6-3′UTRSOD2.
TABLE 1.
Oligonucleotides used
Cell culture and transfection
We cultured HeLa cells with RPMI medium (GIBCO, Invitrogen) complemented with 10% of fetal bovine serum (GIBCO, Invitrogen), gentamicin (0.01%), 100 u/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine. They were transfected with FuGENE 6 transfection reagent as recommended by the manufacturer (Roche Biochemicals). Briefly, monolayer HeLa cells at ∼80% confluency were transfected with 2 μg of different plasmids purified with QIAGEN plasmid midi kit (Qiagen); 48 h later, cells were selected for G418 resistance (selectable marker present in the pCMV-Tag 4A vector) at a final concentration of 1 mg/mL (Roche Biochemicals). Stable clones were expanded for several weeks.
Immunocytochemistry
In a 24-well plate on coverslips, stably HeLa transfected cells were seeded at ∼50% confluency; 24-h later, cells were fixed with 4% paraformaldehyde in PBS for 15 min and processed for indirect immunofluorescence. After permeabilization of the cells for 5 min with Triton 1% in PBS, cells were incubated for 1 h in PBS with 1% BSA before the addition of mouse monoclonal anti-Flag M2 at a final concentration of 2.8 μg/mL (Sigma) or anti-ATP synthase subunit α at a final concentration of 0.4 μg/mL (Molecular Probes, Invitrogen) antibodies. The incubation was performed for 2 h at room temperature. After three washes in PBS, cells were incubated for 1 h with secondary antibodies: labeled goat-anti-mouse IgG1 Alexa Fluor 488 for the anti-Flag M2 antibody and labeled goat-antimouse IgG 2b Alexa Fluor 594 for the anti-ATP synthase subunit α antibody (Molecular Probes, Invitrogen) at a final concentration of 3.3 μg/mL. The cells were subsequently washed once in PBS for 5 min. For DNA staining, a second wash was done with 0.3 μg/mL of DAPI (Sigma) for 10 min. A final 10-min wash was performed in PBS, and the coverslips were mounted using Biomeda Gel/Mount. Immunofluorescence was visualized with a Leica DM 5000 B Digital Microscope. Digital images were acquired and processed with the MetaVue imaging system software.
Mitochondria isolation and Western blot analysis
A total of 100 million of stably transfected cells were treated with trypsin (GIBCO, Invitrogen) for 5 min and spun down. One wash in PBS was performed. The pellets were resuspended in a homogenization buffer (HB): 0.6 M Mannitol, 30 mM Tris-HCl (pH 7.6), 5 mM MgAc, 100 mM KCl, 0.1% fatty acid-free BSA, 5 mM β-mercaptoethanol, and 1 mM PMFS. After addition of 0.01% digitonin, cells were incubated on ice for 4 min. The homogenization was performed with 15 strokes in a Dounce glass homogenizer with a manually driven glass pestle type B. Homogenates were centrifuged for 8 min at 1000g at 4°C to pellet unbroken cells and nuclei. Since many mitochondria remain trapped in this pellet, it was resuspended and rehomogenized again with 25 additional strokes. After centrifugation under the same conditions, both supernatants were assembled and centrifuged three times to discard nuclear or cell contaminants. The supernatant obtained was centrifuged at 12,000g at 4°C for 30 min to pellet mitochondria. Four washes in HB were realized to eliminate membranes, reticulum endoplasmic, and proteases. The last two washes were performed in HB devoid of BSA and PMFS to allow the subsequent analysis with proteinase K (PK). Protein concentrations were measured using the Bradford assay. To determine whether ATP6 was translocated into the organelle, 20 μg of mitochondrial proteins were treated with 150 μg/mL PK at 0°C for 30 min. The reaction was stopped by addition of 1 mM PMFS. After denaturation at 70°C, samples were resolved in 4%–12% SDS-PAGE and transferred to a nitrocellulose membrane. Filters were probed with mouse monoclonal antibodies anti-Flag M2 (3.5 μg/mL) and anti-ATP synthase subunit α (0.5 μg/mL). Immunoreactive bands were visualized with anti-mouse coupled to horseradish peroxidase (0.1 μg/mL) followed by ECL Plus detection (Amersham International). Five independent mitochondria purifications from cells stably transfected with either SOD2 MTS ATP6-3′UTR SV40 or SOD2 MTS ATP6-3′UTRSOD2 vectors were performed. The amount of precursor and mature forms of ATP6 in mitochondria, as well as the quantities of both the mature form of ATP6 and ATP α resistant to PK proteolysis, were compared by densitometric analyses (Quantity One, Bio-Rad software system). The significance of the differences observed was validated with a paired Student's t-test.
RNA extraction and RT–PCR analyses
Mitochondria extractions were performed as described in the preceding section, with the following modifications: 400 million cells were treated with 250 μg/mL cycloheximide for 20 min at 37°C. To HB was added 200 μg/mL cycloheximide, 500 μg/mL heparine, and 1/1000 RNase inhibitor (rRNasin, Promega) as previously described (Sylvestre et al. 2003a). The last pellet of crude mitochondria associated with polysomes (M-P) was stored at −80°C until RNA extraction. Free-cytoplasmic polysomes (F-P) were obtained from the post-mitochondrial supernatant fraction by sedimentation through a step gradient of 2 M and 0.5 M sucrose as described (Sylvestre et al. 2003a). RNAs from these two fractions, as well as total RNAs from each stably transfected cell line, were obtained using RNeasy Protect Mini kit (QIAGEN). RT–PCR analyses were performed with the Superscript III one step RT–PCR Platinium Taq kit (Invitrogen) and specific oligonucleotides inside each ORF. Tables 1 and 2 show primer sequences, the expected sizes of the PCR products, the quantity of RNA used for reverse-transcription, and the number of PCR cycles performed. Densitometric analyses (Quantity One, Bio-Rad software) were performed the amount of both hybrid ATP6 and SOD2 transcripts in either mitochondrion-bound polysomes or free-cytoplasmic polysomes. Three independent RNA preparations from M-P and F-P fractions were subjected three times to RT–PCR analyses.
ACKNOWLEDGMENTS
We are grateful to Dr. Yu Chun Lone for useful discussions and comments on the manuscript and to Dr. Gonzalo M. Claros for the mitochondrial import ability predictions and useful discussions. This work was supported by funds from the INSERM (U 592), the French associations AFM (grant MNM 2004 and 2005), FRM, and Retina. V.K. is the recipient of a French government fellowship, and C.B., of a Fédération des Aveugles et Handicapés Visuels de France's award.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.18206.
REFERENCES
- Claros M.G., Vincens P. Computational methods to predict mitochondrially imported proteins and their transit peptides. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Claros M.G., Perea J., Shu Y., Samatey F.A., Popot J.-L., Jacq C. Limitations to in vivo import of hydrophobic proteins into yeast mitochondria: The case of a cytoplasmic synthesized apocytochrome b. Eur. J. Biochem. 1995;228:762–771. [PubMed] [Google Scholar]
- Corral-Debrinski M., Belgareh N., Blugeon C., Claros M.G., Doye V., Jacq C. Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo . Mol. Microbiol. 1999;31:1499–1511. doi: 10.1046/j.1365-2958.1999.01295.x. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M., Blugeon C., Jacq C. In yeast, the 3′ Untranslated Region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 2000;20:7881–7892. doi: 10.1128/mcb.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S. Mitochondrial medicine. Biochim. Biophys. Acta. 2004;1659:107–114. doi: 10.1016/j.bbabio.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Fünfschilling U., Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R., Walsh P., Beddoe T., Lithgow T. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosome with the mitochondrial surface in vivo . FEBS Lett. 2002;516:213–216. doi: 10.1016/s0014-5793(02)02528-0. [DOI] [PubMed] [Google Scholar]
- Ginsberg M.D., Feliciello A., Jones J.K., Avvedimento E.V., Gottesman M.E. PKA-dependent binding of mRNA to the mitochondrial AKAP121 protein. J. Mol. Biol. 2003;327:885–897. doi: 10.1016/s0022-2836(03)00173-6. [DOI] [PubMed] [Google Scholar]
- Guy J., Qi X., Pallotti F., Schon E.A., Manfredi G., Carelli V., Martinuzzi A., Hauswirth W.W., Lewin A.S. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann. Neurol. 2002;52:534–542. doi: 10.1002/ana.10354. [DOI] [PubMed] [Google Scholar]
- Jansen R.-P. mRNA localization: Message on the move. Nat. Rev. Mol. Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- Joshi S., Burrows R. ATP synthase complex from bovine heart mitochondria. J. Biol. Chem. 1990;265:14518–14525. [PubMed] [Google Scholar]
- Karniely S., Pines O. Single translation–dual destination: Mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005;6:420–425. doi: 10.1038/sj.embor.7400394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M., Zearfoss N.R., Etkin L.D. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- Li Y.H., Huang T.-T., Carlson E.J., Melov S., Ursell P.C., Olson J.L., Noble L.J., Yoshimura M.P., Berger C., Chan P.H., et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- Manfredi G., Fu J., Ojaimi J., Sadlock J.E., Kwong J.Q., Guy J., Schon E.A. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene to the nucleus. Nat. Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- Margeot A., Blugeon C., Sylvestre J., Jacq C., Corral-Debrinski M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21:6893–6904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariottini P.C.A., Riley M., Cottrell B., Doolittle R.F., Attardi G. Identification of the polypeptides encoded in the unassigned reading frames 2, 4, 4L, and 5 of human mitochondrial DNA. Proc. Natl. Acad. Sci. 1986;88:1563–1567. doi: 10.1073/pnas.83.6.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagley P., Farrell L.B., Gearing D.P., Nero D., Meltzer S., Devenish R.J. Assembly of functional proton-translocating ATPase complex in yeast mitochondria with cytoplasmically synthesized subunit 8, a polypeptide normally encoded within the organelle. Proc. Natl. Acad. Sci. 1995;85:2091–2095. doi: 10.1073/pnas.85.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oca-Cossio J., Kenyon L., Hao H., Moraes C.T. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics. 2003;165:707–720. doi: 10.1093/genetics/165.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R., Lewin A.P., Peel A., Wang J., Guy J., Hauswirth W.W., Stacpoole P.W., Flotte T.R. Recombinant Adeno-associated virus vector-based gene transfer for defects in oxidative metabolism. Hum. Gene Ther. 2000;11:2067–2078. doi: 10.1089/104303400750001381. [DOI] [PubMed] [Google Scholar]
- Shalgi R., Lapidot M., Shamir R., Pilpel Y. A catalog of stability-associated sequence elements in 3′UTRs of yeast mRNAs. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink J., van den Heuvel L., DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat. Genet. 2001;342:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- Smith P.M., Ross G.F., Taylor R.W., Turnbull D.M., Lightowlers R.N. Strategies for treating disorders of the mitochondrial genome. Biochim. Biophys. Acta. 2004;1659:232–239. doi: 10.1016/j.bbabio.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Sylvestre J., Margeot A., Jacq C., Dujardin G., Corral-Debrinski M. The role of the 3′ untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol. Biol. Cell. 2003a;14:3848–3856. doi: 10.1091/mbc.E03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre J., Vialette S., Corral-Debrinski M., Jacq C. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin localize to the vicinity of mitochondria. Genome Biol. 2003b;4 doi: 10.1186/gb-2003-4-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekotte H., Davis I. Intracellular mRNA localization: Motors move messages. Trends Genet. 2002;18:636–642. doi: 10.1016/s0168-9525(02)02819-6. [DOI] [PubMed] [Google Scholar]
- Valnot I., von Kleist-Retzow J.C., Barrientos A., Gorbatyuk M., Taanman J.W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., Rotig A. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:1245–1249. doi: 10.1093/hmg/9.8.1245. [DOI] [PubMed] [Google Scholar]
- Verner K. Co-translational protein import into mitochondria: An alternative view. Trends Biochem. Sci. 1993;18:364–371. doi: 10.1016/0968-0004(93)90090-a. [DOI] [PubMed] [Google Scholar]
- Zeviani M., Carelli V. Mitochondrial disorders. Curr. Opin. Neurol. 2003;16:585–594. doi: 10.1097/01.wco.0000093101.34793.c8. [DOI] [PubMed] [Google Scholar]