Abstract
In the ribozyme from the hepatitis delta virus (HDV) genomic strand RNA, a cytosine side chain is proposed to facilitate proton transfer in the transition state of the reaction and, thus, act as a general acid–base catalyst. Mutation of this active-site cytosine (C75) reduced RNA cleavage rates by as much as one million-fold, but addition of exogenous cytosine and certain nucleobase or imidazole analogs can partially rescue activity in these mutants. However, pH-rate profiles for the rescued reactions were bell shaped, and only one leg of the pH-rate curve could be attributed to ionization of the exogenous nucleobase or buffer. When a second potential ionizable nucleobase (C41) was removed, one leg of the bell-shaped curve was eliminated in the chemical-rescue reaction. With this construct, the apparent pKa determined from the pH-rate profile correlated with the solution pKa of the buffer, and the contribution of the buffer to the rate enhancement could be directly evaluated in a free-energy or Brønsted plot. The free-energy relationship between the acid dissociation constant of the buffer and the rate constant for cleavage (Brønsted value, β, = ∼0.5) was consistent with a mechanism in which the buffer acted as a general acid–base catalyst. These data support the hypothesis that cytosine 75, in the intact ribozyme, acts as a general acid–base catalyst.
Keywords: hepatitis delta virus, catalytic RNA, base rescue, Brønsted analysis, general acid–base catalysis
INTRODUCTION
The possibility that RNA side chains, or nucleobases, can function directly as catalytic groups in ribozymes has been suggested and supported by structural studies. Crystal structures of small self-cleaving ribozymes have revealed one or more nucleobases positioned in the active sites such that their direct involvement in stabilizing the transition state through H-bond formation, charge stabilization, or even proton transfer is feasible. The positioning of cytosine 75 (C75) in the hepatitis delta virus (HDV) genomic ribozyme was one such case (Ferré-D'Amaré et al. 1998; Ferré-D'Amaré and Doudna 2000; Ke et al. 2004). Also, in the hairpin ribozyme, both guanine 8 and adenine 38 are well placed in the active site to act as chemical catalysts (Rupert and Ferré-D'Amaré 2001; Ferré-D'Amaré and Rupert 2002). However, even in these ribozymes, defining the role of the nucleobase in catalysis can benefit from additional tests of their proposed functions (Perrotta et al. 1999; Nakano et al. 2000; Shih and Been 2001; Kuzmin et al. 2004, 2005; Das and Piccirilli 2005).
Ribozymes from the HDV genomic and antigenomic RNAs catalyze an internal phosphoester transfer reaction that cleaves the backbone of the RNA to generate products with a 2′,3′-cyclic phosphate group and a 5′-hydroxyl group (Kuo et al. 1988; Sharmeen et al. 1988; Wu et al. 1989). Each ribozyme contains a cytosine in the active-site pocket that has been proposed to act as a general acid–base catalyst for this reaction (Ferré-D'Amaré et al. 1998; Perrotta et al. 1999; Ferré-D'Amaré and Doudna 2000; Nakano et al. 2000; Shih and Been 2001; Ke et al. 2004; Das and Piccirilli 2005). A deletion of that cytosine in the antigenomic ribozyme (C76) or genomic ribozyme (C75), or a mutation that changes it to a G or a U, reduces cleavage activity to nearly the noncatalyzed rate (Tanner et al. 1994; Perrotta and Been 1996). However, catalyzed cleavage rates are partially restored with the addition of certain nucleobases such as cytosine and isocytosine or buffers such as imidazole and imidazole analogs with different pK a values (Perrotta et al. 1999; Shih and Been 2001). For chemical rescue of the HDV antigenomic ribozyme with the C76 deletion, the pH-rate profiles with each of the buffers followed a rate-law definition of general-base catalysis, and the apparent pKa of the reaction correlated with the pKa of the buffer in the reaction (Shih and Been 2001). Those results are consistent with several potential mechanisms in which the rate of the reaction is limited by the extent of ionization of the buffer or exogenous nucleobase. In the antigenomic ribozyme, it was shown that the rate of the reaction increased with the pKa of the buffer, suggesting that the acidity constant, and not just the ionization state, was a factor in the reaction rate. Moreover, the slope of the line generated from a free-energy plot of the rate constant as a function of the buffer's acidity constant was consistent with values expected if the buffer was acting as a general acid–base catalyst (Shih and Been 2001). Initially, the biochemical evidence linking that function of the buffer or exogenous base to the catalytic role of C76 in the intact ribozyme was the finding that a C76A mutation altered the apparent pKa of the self-cleavage reaction in a predictable fashion (Perrotta et al. 1999). More recently, Das and Piccirilli (2005) have greatly strengthened the support for C76 as a general acid–base catalyst and extended the analysis in the HDV antigenomic ribozyme by substituting the nucleobase of C76 with analogs of varying pKa. In addition, they also provided powerful evidence that the proton transfer occurs between the cytosine base at position 76 and the 5′ bridging oxygen of the scissile phosphate group as proposed by Nakano et al. (2000) for the HDV genomic ribozyme.
Cleavage activity of the HDV genomic ribozyme with C75 deleted or changed to a U was also rescued with imidazole (Nakano et al. 2000; Wadkins 2000; Ke et al. 2004) and by cytosine and several imidazole analogs (Wadkins 2000). We report here characterization of the chemical rescue results for the HDV genomic ribozyme and a Brønsted analysis with those kinetic data. A similar study was done with the HDV antigenomic ribozyme; however, there were reasons to characterize the genomic form of the ribozyme as well. First, crystal structures are available for both the precursor and product forms of the genomic ribozyme (Ferré-D'Amaré et al. 1998; Ferré-D'Amaré and Doudna 2000; Ke et al. 2004), but there are none for the antigenomic form. Second, the hypothesis of general acid–base catalysis by RNA will be further tested with results from a Brønsted analysis using a different ribozyme, even if it is similar. Third, while results from kinetic studies on either ribozyme might be expected to apply to both, there are several differences in sequence, secondary structures, and details of the cleavage reactions that distinguish the two ribozymes (Wadkins and Been 1997; Wadkins et al. 2001). As an example of this last point we report here that, for chemical-rescue reactions of the genomic ribozymes with a mutation at only position C75, the pH-rate profiles were bell shaped. A bell-shaped pH-rate profile complicated the structure-reactivity correlation of the chemical-rescue reactions and could also call into question some fundamental features of the proposed catalytic mechanisms. However, we found that the second leg of the pH-rate curve was lost upon modifying the HDV genomic ribozyme to eliminate a base-quadruple hypothesized to favor protonation of one of its nucleobases (C41) (Ferré-D'Amaré et al. 1998; Wadkins et al. 2001). The purported ionizable base, C41, is proposed to not participate directly in catalysis, due to its distance from the active site, but may enhance cleavage rates through structural effects. In rescue reactions with ribozymes lacking C41 (and C75), the pH-rate profile followed a rate-law definition of general-base catalysis that was consistent with one of the ionized forms of the buffer participating in catalysis, and the Brønsted analysis of the kinetic data supported a role for the buffer as a general acid–base catalyst.
RESULTS
Activity of the genomic ribozyme is reduced by mutations at C75
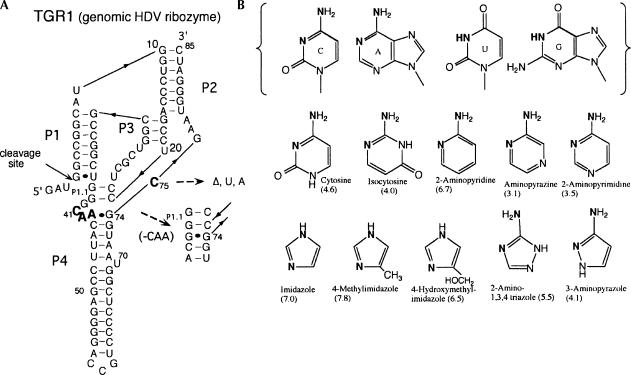
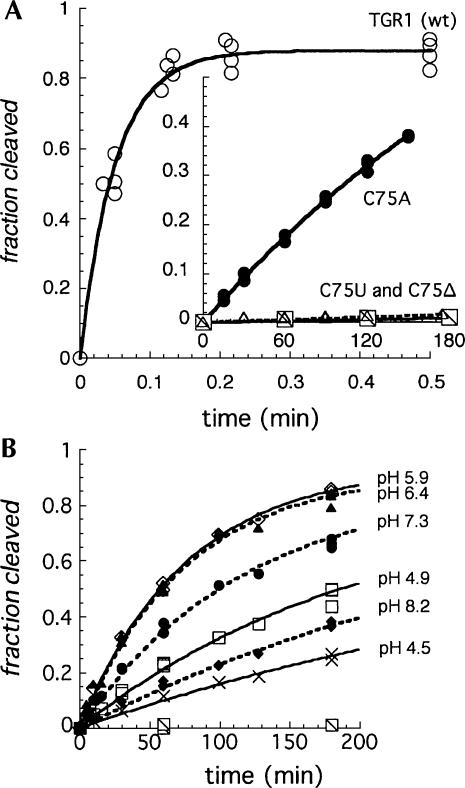
The wild-type HDV genomic ribozyme used in these studies (TGR1, Fig. 1A) (Wadkins and Been 1997) cleaved to completion (∼80% in these reactions) in <15 sec in 10 mM MgCl2 at 37°C (Fig. 2A). For time points taken manually, this reaction was more than half complete with the first time point (3–4 sec), but the 19 min−1 value obtained for the rate constant from those data provided a reasonable estimate for comparison of activity with the much slower C75 mutants. Changing the cytosine at position 75 (Fig. 1A) to a uracil (C75U) or deleting the nucleotide (C75Δ) reduced activity to near the background rates of RNA degradation (0.5 × 10−5 min−1 to 5 × 10−5 min−1) (Fig. 2A, inset). The C75A mutant cleaved 100–1000 times faster (3 × 10−3 min−1) than the deletion or U substitution, but it was still significantly slower than the wild-type construct. We and others have suggested that adenine is able to provide catalytic function similar to that of cytosine in the HDV ribozymes (Perrotta and Been 1996; Perrotta et al. 1999; Nakano et al. 2000; Shih and Been 2002). However, for this study, the large decrease in the rate constant for cleavage of the C75U and C75Δ ribozymes provided a sensitive probe for chemical (exogenous base) rescue studies in this ribozyme.
FIGURE 1.
(A) Sequence and secondary structure of the HDV genomic ribozyme and variants used in this study. The complete sequence of the wild-type ribozyme (TGR1) is shown. Changes at position 75 included deleting the nucleotide (C75Δ) or substituting with U or A (C75U, C75A). The sequence C41A42A43 was deleted from the C75Δ variant to generate the C75ΔC41Δ construct. (B) Structures of the four natural nucleobases and the exogenous bases used in the rescue reactions. The solution pKa values for the bases are given in parentheses.
FIGURE 2.
Kinetics of cleavage of TGR1 and the C75 variants. (A) Cleavage of TGR1 and C75 mutants in the absence of exogenous base in 10 mM MgCl2, 50 mM Tris-HCl (pH 7.5) at 37°C. The curves are fit to an exponential with rate constants of 19 min−1 for the wild-type ribozyme (open circles) and 3.2 × 10−3 min−1 for the C75A variant (closed circles). For the C75U and C75Δ variants, nonspecific degradation interfered with quantifying specific cleavage; limits for the rate constants were estimated at about 5 × 10−6 and 5 × 10−5 min−1, respectively, from the linear fit. (B) Cleavage of the C75Δ variant rescued with exogenous cytosine at the pH given in the figure. Rescue reactions contained 25 mM MgCl2, 37 mM cytosine, and 40 mM AMT/MTA buffers (see Materials and Methods).
Cytosine rescues activity of C75 mutants
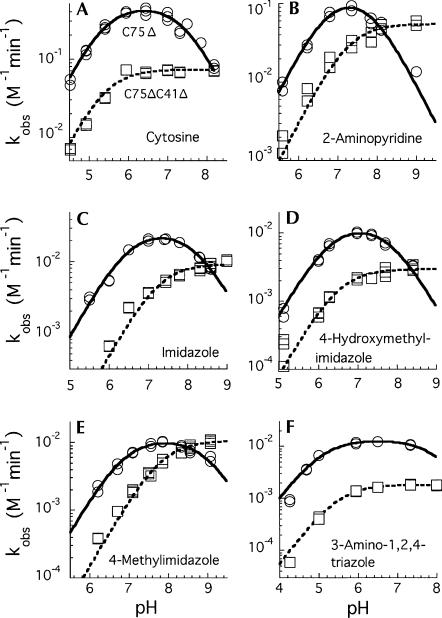
With the C75Δ mutant, addition of free cytosine (37 mM final concentration) to the reaction increased the cleavage rate as much as 1000-fold (1.3 × 10−2 min−1 at pH ∼6) (Fig. 2B). The reaction rates with cytosine increased with increasing pH from pH 4.5 to about pH 6.4, but then decreased as the pH increased further (Figs. 2B, 3A). Several cytosine analogs (Fig. 1B) were tested for rescue of cleavage activity, and, of those tested, 2-aminopyridine was particularly effective. With this analog, rates were comparable to the cytosine rescue, but the pH optimum for the reactions were slightly higher (Fig. 3B). Isocytosine also rescued activity, but the reaction with the C75Δ mutant was not further characterized (data for isocytosine rescue of another ribozyme variant is presented below).
FIGURE 3.
pH-rate curves comparing the rescued cleavage rates for C75Δ (circles) and C75ΔC41Δ (squares) in the presence of exogenous base. The concentration of cytosine (A) was 37 mM and the reactions also contained 40 mM AMT or MTA buffer. The concentration of the bases in the other reactions (B–F) was 0.4 M. Rate constants were normalized to 1 M total base. Curves were fit to the data using the equations k obs = k max/(1 + 10(pKa1-pH) + 10(pH-pKa2) + 10(pKa1-pKa2)) for C75Δ, and k obs = k max/(1 + 10(pKa-pH)) for C75ΔC41Δ. Apparent pKa values are given in Table 1.
Several other nucleobase analogs were less effective in the rescue reaction. After 24 h at 37°C, there was ∼15% conversion of precursor to product with 0.4 M aminopyrazine and <5% with 0.4 M 2-aminopyrimidine. Little or no cleavage was detected with pyridine, 3-aminopyridine, or 2-, 3- or 4-hydroxypyridine (data not shown). Given the effectiveness of both 2-aminopyridine (pKa = 6.7) and cytosine (pKa = 4.6) in these reactions, the lack of rescue by either pyridine (pKa = 5.2) or 3-aminopyridine (pKa = 5.8) was somewhat surprising. However, that result may indicate that the presence and position of the exocyclic amino group relative to the ring nitrogen is important for rescue by the cytosine analogs (Fig. 1B). While it was important to demonstrate rescue of cleavage with free cytosine, the available cytosine analogs did not provide a useful collection of related compounds for a Brønsted analysis.
Imidazole rescue of cleavage activity
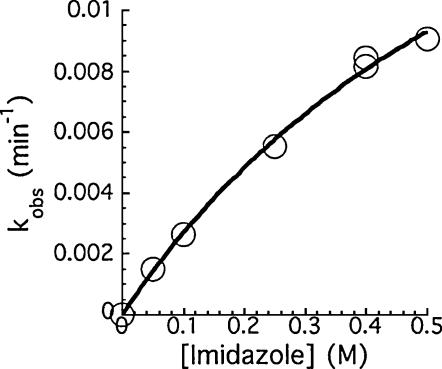
Consistent with earlier reports for the HDV ribozymes, addition of imidazole to the reaction rescued cleavage activity of both the C75Δ mutant (Fig. 4) and the C75U mutant (data not shown). For the wild-type ribozyme (TGR1), addition of 200 mM imidazole did not increase the rate of cleavage (data not shown). With the slower C75A mutant, addition of imidazole (200 mM) resulted in only a small (twofold) increase in the cleavage rate (data not shown). With the inactive C75Δ mutant, imidazole restored cleavage activity, and the rate of cleavage increased with increasing imidazole concentration (Fig. 4). The reaction did not reach saturation at 0.5 M imidazole, suggesting that binding is likely to be very weak. A concentration of 0.4 M base was used for most of the subsequent studies in which several water-soluble imidazole analogs were screened for their ability to rescue cleavage activity. Rescue was detected with 4-methylimidazole, 4-hydroxylmethylimidazole, 2-amino-1,3,4-triazole, and 3-aminopyrazole (Table 1). Similar results were seen for the C75U variant (data not shown) but only rescue of the C75Δ variants was further characterized.
FIGURE 4.
Imidazole rescue of the C75Δ variant. The dependence of the cleavage rate on imidazole concentrations (pH 7.4).
TABLE 1.
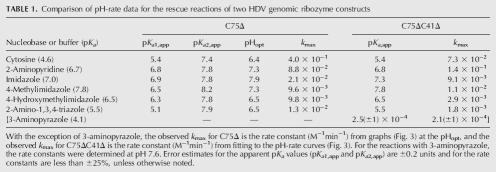
Comparison of pH-rate data for the rescue reactions of two HDV genomic ribozyme constructs
The pH-rate curves are bell shaped and shift with the pKa of the base
As noted above, cytosine was particularly effective in the rescue reactions with the C75Δ mutant, but the pH-rate curve for the cleavage in 37 mM cytosine was bell shaped (Fig. 3A). The pH-rate curve in 0.4 M 2-aminopyridine was also bell shaped, but it was both narrower and shifted to higher pH relative to the cytosine data (Fig. 2B; Table 1). Likewise, bell-shaped pH-rate curves were characteristic of imidazole and imidazole-analog rescued cleavage reactions of the C75Δ ribozyme (Fig. 3C–F; Table 1). It was evident that the curves varied in both their width and pH optimum for the different bases. The bell-shaped curves contrasted with the shape of the pH-rate curves generated in the rescue reactions with the C76Δ mutant form of the HDV antigenomic ribozyme (Perrotta et al. 1999; Shih and Been 2001). In those rescue reactions, the rate of cleavage of the antigenomic C76Δ ribozyme increased with pH, at low pH, and leveled off at pH values greater than the pKa of the base. Thus, the pH-rate profiles are described as following a rate-law definition of general-base catalysis (Shih and Been 2001).
The bell-shaped pH-rate curves seen with the genomic C75Δ ribozyme complicated interpretation of the rescue data, but indicated that the rate of cleavage is limited by the ionization state of two, or at least two, groups with pKa values in the experimental pH range. For some, but not all, of the rescue reactions, one of the two apparent pKa values is close to the solution pKa of the base (Table 1). However, apparent pKa values cannot be well defined in some of these reactions because of uncertainty in the curve fitting when the inflection points are too close together. Nevertheless, the observed rate constants reflected the ionization state of two groups with somewhat similar pKa values.
A modification of the HDV genomic ribozyme simplified the pH-rate profiles
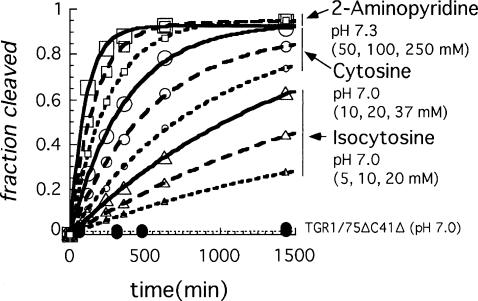
The genomic HDV ribozyme contains a “pedestal” for the active-site pocket (Ferré-D'Amaré et al. 1998). Protonation of the N3 position of C41 could stabilize a base quadruple (A43•C41•G73•C44) in the base of this pedestal structure by allowing the N3 of C41 to form an additional H-bond to O6 of G73. The antigenomic ribozyme lacks the 3-nt sequence equivalent to C41–A42–A43 (CAA) in the genomic ribozyme and, as a result, would not be able to form a similar arrangement to favor an ionized base at this position. This extra CAA segment in the genomic ribozyme is not critical for cleavage activity under some cleavage conditions, and, for the reaction in 10 mM MgCl2, deletion of the CAA sequence resulted in slightly faster cleavage (Wadkins and Been 1997). However, under conditions less favorable for cleavage activity (1M NaCl and no MgCl2), a pH-dependent contribution to cleavage rates correlated with the presence of the CAA sequence, and this effect was detected in both the genomic and antigenomic ribozymes (Wadkins and Been 1997). To test if ionization of a group associated with this feature was the source of the second apparent pKa observed in the chemical-rescue reactions, the genomic C75Δ ribozyme was further modified by again deleting the CAA segment so that the quadruple involving C41 could not form. This construct, referred to as C75ΔC41Δ, was examined for chemical rescue cleavage activity. As expected, cleavage activity of the C75ΔC41Δ variant of TGR1 was very slow (∼1 × 10−5 and 2 × 10−4 min−1 at pH 7.0 and 8.7, respectively) (Fig. 5; data not shown) but could be rescued with addition of cytosine, isocytosine, or 2-aminopyridine to the reaction (Fig. 5). While the rate constants for chemical-rescue cleavage in the C75ΔC41Δ construct were lower than those for the C75Δ construct over most of the pH range tested (Fig. 3; data not shown), the pH-rate profiles now followed a rate-law definition of general-base catalysis as was observed previously with rescue of the HDV antigenomic ribozyme. At higher pH values (>∼8), the rates for the two HDV genomic ribozyme variants converged, and for some buffers the C75ΔC41Δ variant was observed to cleave faster than the C75Δ variant. Thus, the deletion of the CAA sequence substantially altered the pH-rate profile for the chemical-rescue reactions, and the apparent pKa for chemical-rescue cleavage corresponded well to the solution pKa of the buffers (Table 1).
FIGURE 5.
Cleavage of the C75ΔC41Δ variant of TGR1 is rescued with increasing concentrations of cytosine and cytosine analogs. In the absence of exogenous base, the rate of cleavage at pH 7.0 (40 mM AMT buffer) was < 6 × 10−6 min−1 (filled circles). Data is shown for isocytosine at 5, 10, and 20 mM (triangles), cytosine at 10, 20, and 37 mM (circles) (AMT buffer at pH 7.0), and 2-aminopyridine at 50, 100, and 250 mM (pH 7.3, no additional buffering) (squares). Pseudo first-order rate constants ranged from ∼3 × 10−4 to 1 × 10−2 min−1 for the rescue reactions shown.
The change in the pH-rate curve suggested that the ionization state of a group (presumably C41) in the base-multiple or pedestal structure contributed to the higher cleavage rates seen with the C75Δ ribozyme in the rescue reactions at lower pH values. At higher pH, this advantage can be lost. The data just presented are consistent with the previous idea that low pH favored protonation of C41, which then, by stabilizing the pedestal for the active site, contributes positively to the overall reaction rate (Ferré-D'Amaré et al. 1998; Wadkins et al. 2001).
Brønsted analysis
Elimination of the second leg in the pH-rate curve simplified interpretation of the pH-rate data for the HDV genomic ribozyme. With the C75ΔC41Δ construct, maximal cleavage rates in the chemical-rescue reactions occurred in the higher pH range for each nucleobase or buffer tested. Among the imidazole analog buffers, there was a consistent correlation between increasing rate constants at high pH (k max) and increasing pKa of the base used in the reaction (Table 1). Such a relationship would be expected if the nucleobase analogs were acting as general acid–base catalysts in the reaction but not if catalysis depended only on the ionization state of the nucleobase (Jencks 1969; Fersht 1985). The sensitivity of the reaction to the pKa of the base is described by the Brønsted value (β), which is the slope of the line from the equation
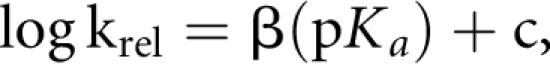 |
where krel is a normalized second-order rate constant (here, we used k max at 1 M base from the pH-rate curves) for each base and the slope of the line is β. The Brønsted plot of the rescue data (Fig. 6) revealed a linear correlation between the log of the strength of the base and the log of the rate of the reaction, and a slope or Brønsted value (β) of ∼0.5 (0.47 ± 0.03).
FIGURE 6.
Brønsted plot of the rescue data for the C75ΔC41Δ variant. For each reaction, the log of the normalized rate constants was plotted against the pKa of the base used in the reaction. The slope of the line fit to these data is 0.47 (R = 0.97). For 2-amino-1,2,4-triazole, 4-hydroxylmethylimidazole, imidazole, and 4-methylimidazole, k max values from independent pH-rate curves of the rescue reactions were used. For 3-aminopyrazole, the rate constant used was the k obs obtained from the initial slope of 24–48-h reactions at pH 7.6 (∼15% cleavage in 24 h).
DISCUSSION
The hypothesis that the cytosine nucleobase at position 75 in the HDV genomic ribozyme acts as a general acid–base catalyst was examined by quantifying the contribution of buffer as an exogenous-nucleobase analog to cleavage rates in a C75 deletion mutant. To test this idea, a Brønsted analysis was applied to the chemical-rescue kinetic data. This approach to study an enzyme's catalytic mechanism was first used by Toney and Kirsch (1989) to evaluate general acid–base catalysis in aspartate aminotransferase. Chemical (base) rescue has also been used in structure–function studies of the hammerhead ribozyme containing abasic residues in the conserved core region to probe and identify transition state interactions (Peracchi et al. 1996, 1998). More recently, chemical rescue has been used with the hairpin ribozyme in defining the catalytic roles of the G8 and A38 side chains (Lebruska et al. 2002; Kuzmin et al. 2004, 2005). For the hairpin ribozyme, the data were interpreted as supporting a model of electrostatic catalysis in which an exogenous nucleobase could rescue activity by countering the negative charge that develops on the cleavage-site phosphate in the transition state (Kuzmin et al. 2004, 2005). For the HDV ribozymes, a structural or electrostatic catalytic role for buffer or exogenous nucleobases is also possible for rescue of the C75 and C76 mutants. However, if either role were the sole catalytic functions, the rate of the reaction might be expected to show a dependence on the ionization state of the buffer but not on its pKa. The Brønsted analysis quantifies the contribution of the acidity constant to the rate enhancement.
This study initially was complicated by the finding that, in addition to the exogenous nucleobase or buffer, there was a second ionizable group that contributed to the shape of the pH-rate profiles in the rescue of the C75 mutants. There could be several candidates for a second ionizable group. One possibility is that there were, originally, two titratable nucleobases acting as catalytic groups in the active site; for example, a proton donor and an acceptor acting in concert. However, that prospect appears unlikely unless the crystal structures of the HDV ribozyme shows an arrangement of the nucleotides far removed from its active forms (Ferré-D'Amaré et al. 1998; Ferré-D'Amaré and Doudna 2000; Ke et al. 2004). (A metal ion-bound water in the active site may participate in catalysis, but its pKa would likely be high relative to the experimental pH range and, thus, would not be the source of the second leg of the bell-shaped pH-rate curve in these studies; Nakano et al. 2001; Ke et al. 2004). A more probable explanation is that the observed second titratable group is a nucleobase previously identified as part of a structural feature near, but not directly part of, the active site. In the structures of the HDV genomic ribozyme, the Doudna group had identified a base quadruple (A43•C41•G73•C44) within a larger multibase interaction located between the ends of P4 and P1.1 that could help stabilize a pedestal for the active-site pocket of the ribozyme (Ferré-D'Amaré et al. 1998).
Protonation of the N3 position of cytosine 41 was predicted to stabilize the base quadruple (Ferré-D'Amaré et al. 1998; Lupták et al. 2001). There is additional independent support of that proposal; an identical nucleobase-quadruple interaction in two frame-shifting pseudoknots studied by NMR provides strong evidence that ionization of the cytosine is important for stability in that context as well (Nixon and Giedroc 2000; Nixon et al. 2002). Importantly, the wild-type HDV antigenomic ribozyme cannot form this quadruple. Here, we provided additional evidence that there is ionization of a group in the pedestal structure of the genomic ribozyme, and the results suggest that the bell shape of the pH-rate profile reflects ionization states of both C41 (or an associated group) and the exogenous nucleobase or buffer (substituting for C75). Removal of C41–A43 resulted in a pH-rate curve for the rescue reaction that followed a rate-law definition of general-base catalysis. The results were also consistent with the idea that the ionized form of C41 functions to enhance the rate of cleavage, although evidence for inhibition of cleavage rates by the nonionized form was also seen at higher pH. The distance from C41 to the scissile phosphate group would appear to be quite far (about 15–17 Å) for that protonated cytosine to participate directly in catalysis, for example, in the electrostatic stabilization of the anionic transition state. Moreover, the space between C41 and the scissile phosphate is largely occupied by part of the short P1.1 duplex. Given this location of C41, it appears that its contribution to catalysis is indirect, although additional study would be necessary to reveal how this might work.
For the rescue of the C75ΔC41Δ variant, there was a noticeable positive correlation between the reaction rate and the pKa of the buffer. A Brønsted plot (log k obs vs. pKa) revealed a linear free-energy relationship between the rate constant for the reaction and the equilibrium constant for the buffer, with a characteristic slope or Brønsted coefficient (β) of about 0.5. In this case, the value can be taken to reflect the extent of positive charge transfer between donor and acceptor atoms in the transition state (Jencks 1969; Fersht 1985), and it supports a mechanism of general acid–base catalysis by the buffer or exogenous nucleobase in these reactions. It is also picturesquely described as evidence for a proton “in flight” in the transition state. A value of or close to zero would suggest a mechanism in which the buffer was not involved in transferring a proton in the transition state, and, thus, not acting as a general acid–base catalyst. Examples might be the ionized buffer molecule acting as an electrostatic catalyst by shielding developing charge in the transition state or the un-ionized base stabilizing the transition state by filling a space left by the deletion mutation. A value close to one would suggest a role involving extensive new bond formation in the transition state. An example would be the buffer acting as the nucleophile in the reaction. Values of 0.4–0.8 were experimentally determined in rescue reactions with enzymes proposed to use a histidine or lysine for general acid–base catalysis (Toney and Kirsch 1989; Newmyer and de Montellano 1996; Huang and Tu 1997). A Brønsted value (∼0.5) was found previously for chemical rescue with the HDV antigenomic ribozyme (Shih and Been 2001).
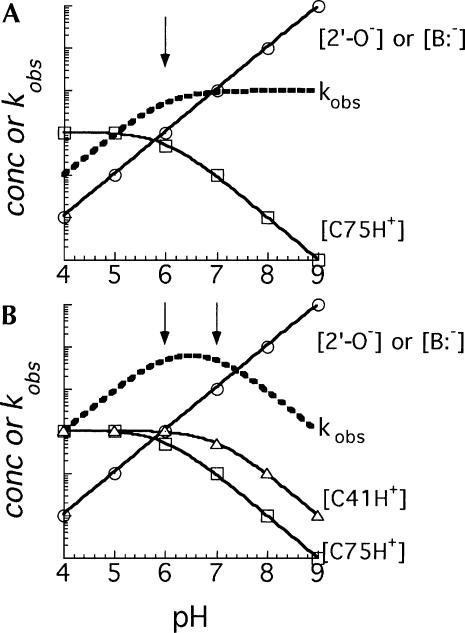
A recent study of the catalytic mechanism of the HDV antigenomic ribozyme supports a model for catalysis in which the active-site cytosine (C76) acts as a general acid–base catalyst at the 5′ bridging oxygen of the scissile phosphate (Das and Piccirilli 2005). We assume that the mechanisms of the two HDV ribozymes are similar, and, therefore, C75 and the exogenous nucleobases or buffer acts at this same position in the genomic ribozyme. Thus, a model for catalysis of the reaction in the direction of cleavage involves transfer of a proton from the protonated nucleobase to the 5′ bridging oxygen, making the oxygen a better leaving group in the rate-determining step for cleavage. The nucleobase can then be viewed as functioning as a general-acid catalyst for the forward (cleavage) reaction and as a general-base catalyst in the reverse (ligation) reaction. It is important to emphasize that the observed pH-rate profile for the chemical-rescue reactions with the C75ΔC41Δ ribozyme, which is described as a general-base pH-rate profile, is not in conflict with a mechanistic role for the nucleobase as a general-acid catalyst in the cleavage direction of the reaction. Resolution of what may at first seem a contradiction is nicely addressed by Jencks (1969), who illustrates why catalysis by a specific-base plus general-acid mechanism follows a pH-rate profile described as general base. His description is restated here for the reactions of this study. As long as the pH of the reaction is below the pKa of the buffer or exogenous nucleobase, the buffer (nucleobase) catalyst would exist mostly in the protonated form and that concentration would not vary appreciably (<2-fold, until the pH exceeds the pKa) (Fig. 7A). Thus, with increasing pH, but at pH still below the pKa of the buffer (nucleobase), the rate of the reaction increases because the concentration of “activated substrate” increases log-linearly with deprotonation of the 2′-OH group. This increase in the concentration of the activated form of the ribozyme in the reaction continues as the pH increases, and the increase in the rate of cleavage reflects this activation. However, at pH values above the pKa of the buffer (nucleobase), the concentration of the acid form of the general acid–base catalyst will decrease, also log-linearly. Thus, as the pH increases in this higher pH range, the concentration of the active form of the general-acid catalyst decreases by loss of a proton at the same rate as the 2′-OH group is activated by loss of a proton, and the sum of those effects on the overall rate of the reaction exactly cancel each other. As a result, the pH-rate profile, which reflects the sum of the effects of pH on the two positions, follows the rate-law definition of general-base catalysis (Jencks 1969; Nakano et al. 2000). For this description, the ionization of the 2′-OH group is assumed to be rapid and occurring prior to the rate-determining step in the cleavage reaction, and, thus, that step appears kinetically as specific-base catalysis. However, the same explanation for the pH-rate profiles can apply to the reaction catalyzed by a concerted general-acid general-base mechanism in which the cytosine (or buffer) is still acting as the general-acid catalyst, but a hydrated metal ion, functioning as a general-base catalyst, is “activated” with increasing pH, and it then accepts the proton from the 2′-OH group (Nakano et al. 2000, 2001, 2003; Nakano and Bevilacqua 2001). The data we present here do not distinguish between these mechanisms for activation of the 2′-OH group. However, in either mechanism, the interpretation of the role of the cytosine as a general acid–base catalyst and its effect on the pH-rate curves would be the same.
FIGURE 7.
Idealized examples of how the shape of a pH-rate curve would relate to the extent of protonation of ionizable groups. (A) A general-base pH-rate curve (dotted line) resulting from specific-base plus general-acid catalysis. An increase in the concentration of the activated substrate (activation of the 2′-OH group by base) is represented by the line through circles, while the decrease in concentration of the conjugate-acid form of the nucleobase catalyst with a pK a of 6 (arrow) is represented by the line through squares. (B) A bell-shaped pH-rate curve (dotted line) resulting from specific-base plus general-acid catalysis and a second ionizable group that, when protonated, increases activity of the ribozyme. Lines through circles and squares as in A but in addition there is a third ionizable group (e.g., C41) with a pKa of 7 (triangles).
Although we have presented the results in the context of a mechanism consistent with the findings of Das and Piccirilli (2005) that demonstrate that the cytosine acts at the 5′ bridging oxygen, it is important to note that a recent structure of a precursor form of the HDV genomic ribozyme (Ke et al. 2004) appears to position the nucleobase at position 75 for action at the 2′-OH group. As such, the cytosine could activate the 2′-OH group by accepting its proton. If transfer of this proton from the 2′ oxygen to C75 was rate determining for the reaction, C75 would be acting as a general-base catalyst in the cleavage reaction. The rescue studies we present do not distinguish between the two models for the site of catalysis because, again, both the forward and reverse reactions would be predicted to follow the rate-law definition of general-base catalysis (Jencks 1969).
There is yet another related issue of ambiguity involving use of the data from the rescue studies in the interpretation of the results of the Brønsted analysis. Were the buffers to act as “true” general-base catalysts in the cleavage reaction, it is obvious how the catalytic mechanism could generate the kinetic data and the subsequent Brønsted plot; that is, the rate constants would increase with the pKa, or increasing basicity. Likewise, for “true” general-acid catalysis the slope of the line for a similar Brønsted plot would be negative because the rate constants would decrease with decreasing acidity constant (increasing pKa) of the buffer. What can be less obvious, initially at least, is why the plot should show an increase in rate with increasing pKa of the buffer in a specific-base plus general-acid catalyzed reaction because it is, after all, the conjugate acid of the buffer that would be the proton donor. Simply noting that this is the case because “true” general-base catalysis and specific-base plus general-acid catalysis are kinetically equivalent may not provide an intuitively satisfying explanation for the Brønsted plot. However, because it is easier to see the relationship between increasing rate constants and increasing basicity in “true” general-base catalysis, it is useful to view the result in terms of the kinetically equivalent reverse reaction. For the Brønsted analysis it would not matter in which reaction direction the kinetics are measured because the buffer, in acting as a catalyst, lowers the activation-energy barrier for the reaction regardless of the direction of the reaction. That is, if increasing the pKa of the buffer increases the rate of cleavage, it would also increase the rate of ligation (assuming ligation is a true reversal of the cleavage reaction). For the reverse of the cleavage reaction, the buffer catalyzes the same rate-determining step as in the forward reaction, yet now that step, deprotonation of the 5′ OH group, would be described as “true” general-base catalysis. Thus, the Brønsted plot immediately appears consistent with catalysis by the buffer for reverse reaction, and, for the reasons just described, a Brønsted analysis of the data from the reaction measured in either direction would give the same results. The bottom line is that the ambiguity inherent in the kinetic data is not eliminated when those data are used in the Brønsted analysis; nevertheless, the Brønsted value should correctly describe the transition state.
From data presented here and elsewhere for the HDV genomic and antigenomic ribozymes (Ferré-D'Amaré et al. 1998; Perrotta et al. 1999; Nakano et al. 2000; Shih and Been 2001; Wadkins et al. 2001; Das and Piccirilli 2005), we suggest that the bell-shaped pH-rate curves for rescue of cleavage activity in the C75Δ mutants of the genomic ribozyme can be best explained by the effect of pH on three ionizable groups (Fig. 7B). Those three groups are: (1) the 2′-OH group nucleophile (or a general-base catalyst with a relatively high pKa that acts at the 2′-OH group; Nakano et al. 2000, 2001); (2) the buffer that substitutes for C75 and acts as a proton donor in the cleavage reaction; and (3) a nucleobase (C41) associated with the pedestal structure. The ascending leg of the bell-shaped curve, at low pH, reflects activation of the 2′-OH group (or a hydrated metal ion general acid–base catalyst, as noted above). The leveling off and descending leg of the curve, as the pH increases further, is then attributed to a combination of the loss of a proton from the conjugate base of the buffer acting as the general-acid at the 5′ bridging oxygen and to a presumably indirect effect of removing a proton from the ionized form of C41.
MATERIALS AND METHODS
Enzymes and reagents
T7 RNA polymerase was prepared by M. Puttaraju from an overexpressing clone provided by W. Studier, Brookhaven National Laboratory, Upton, NY (Davanloo et al. 1984). All other enzymes and chemicals were purchased from commercial suppliers.
Bases and nucleobases used in the rescue reactions were purchased from Aldrich. Stock solutions were prepared and the pH adjusted with HCl or NaOH. For reactions with cytosine and isocytosine, additional buffering was provided with a three-buffer system; 25 mM acetic acid/25 mM MES (2-(N-morpholino)ethanesulfonic acid)/50 mM Tris (AMT buffer, pH 4.0–8.0) or 50 mM MES/25 mM Tris/25 mM AMP (2-amino-2-methyl-1-propanol) (MTA buffer, pH 7.0–10.0) (Ellis and Morrison 1982).
Ribozymes
The construction of the plasmid containing the ribozyme sequences used in this study (TGR1) was described previously (Wadkins and Been 1997). Site-directed mutagenesis with single-stranded uracil-containing DNA and synthetic oligonucleotides (Kunkel et al. 1987; Vieira and Messing 1987) was used to generate the C75 variants and to delete the C41–A43 sequence, as previously described (Wadkins and Been 1997; Wadkins et al. 2001). Plasmid DNA was linearized with BamHI and transcribed in vitro with T7 RNA polymerase in the presence of [α-32P]-ATP or [α-32P]-CTP to prepare labeled run-off transcripts. Products of the transcription reaction were fractionated on denaturing polyacrylamide gels, the band containing precursor RNA was excised, and the RNA was eluted and stored frozen in 0.1 mM EDTA. To block self-cleavage of the TGR1 ribozyme during transcription, the reaction included a second plasmid that was cotranscribed to yield a short RNA that base-paired to part of the ribozyme (Wadkins and Been 1997). This blocking RNA was removed upon gel purification.
Ribozyme rescue reactions
To prepare the RNA, it was heated for 1 min at 95°C in 0.1 mM EDTA, allowed to cool to room temperature (10 min), and then incubated at 37°C for 10 min in 5 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.1 M NaCl. The renatured RNA was then diluted 1:25 into a reaction buffer containing 25 mM MgCl2, 1 mM EDTA, 0.5 mM spermidine, and the base (typically 400 mM) at various pH values. All reactions were at 37°C. To avoid evaporation/condensation in the tubes for the longer time points, the reactions were incubated in a 37°C incubator rather than a water bath. The final pH was measured from a mock reaction lacking only the RNA. For reactions in cytosine and isocytosine, 50 mM AMT or MTA buffer (see above) were included in the reaction. Aliquots were removed at various times and quenched with an equal volume of formamide containing 50 mM EDTA and tracking dyes, and these were frozen until all samples were collected. Precursor and product RNAs were separated by gel electrophoresis, and the extent of cleavage was quantified using a PhosphorImager (Molecular Dynamics). First-order rate constants were determined from nonlinear curve fitting to the exponential form of the equation using KaleidaGraph (Synergy Software). Reaction conditions and time courses for TGR1 and the C75A, C75U, and C75Δ variants without exogenous base were as previously described (Wadkins and Been 1997; Wadkins et al. 1999, 2001), and differed mainly in that the RNAs were preincubated in the absence of divalent metal ions and the reactions initiated with addition of MgCl2 to 11 mM.
ACKNOWLEDGMENTS
We thank A. Brown, S. Wilkinson, S. Crary, S. Chamberlin, and J. Rudolph for helpful comments on various versions of this manuscript. This work was supported by a grant from the NIH (GM047233).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.14106.
REFERENCES
- Das S.R., Piccirilli J.A. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A.H., Dunn J.J., Studier F.W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K.J., Morrison J.F. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Doudna J.A. Crystallization and structure determination of a hepatitis delta virus ribozyme: Use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Rupert P.B. The hairpin ribozyme: from crystal structure to function. Biochem. Soc. Trans. 2002;30:1105–1109. doi: 10.1042/bst0301105. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Fersht A. Enzyme structure and mechanism. W.H. Freeman; New York: 1985. [Google Scholar]
- Huang S., Tu S.C. Identification and characterization of a catalytic base in bacterial luciferase by chemical rescue of a dark mutant. Biochemistry. 1997;36:14609–14615. doi: 10.1021/bi9722554. [DOI] [PubMed] [Google Scholar]
- Jencks W.P. Catalysis in chemistry and enzymology. McGraw-Hill; New York: 1969. [Google Scholar]
- Ke A., Zhou K., Ding F., Cate J.H., Doudna J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A., Roberts J.D., Zakour R.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kuo M.Y.-P., Sharmeen L., Dinter-Gottleib G., Taylor J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 1988;62:4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Fedor M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Kuzmin Y.I., Da Costa C.P., Cottrell J.W., Fedor M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lebruska L.L., Kuzmine I.I., Fedor M.J. Rescue of an abasic hairpin ribozyme by cationic nucleobases: Evidence for a novel mechanism of RNA catalysis. Chem. Biol. 2002;9:465–473. doi: 10.1016/s1074-5521(02)00130-8. [DOI] [PubMed] [Google Scholar]
- Lupták A., Ferré-D'Amaré A.R., Zhou K., Zilm K.W., Doudna J.A. Direct pKa measurement of the active-site cytosine in a genomic hepatitis delta virus ribozyme. J. Am. Chem. Soc. 2001;123:8447–8452. doi: 10.1021/ja016091x. [DOI] [PubMed] [Google Scholar]
- Nakano S., Bevilacqua P.C. Proton inventory of the genomic HDV ribozyme in Mg(2+)-containing solutions. J. Am. Chem. Soc. 2001;123:11333–11334. doi: 10.1021/ja0166850. [DOI] [PubMed] [Google Scholar]
- Nakano S.-I., Chadalavada D.M., Bevilacqua P.C. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- Nakano S.-I., Proctor D.J., Bevilacqua P.C. Mechanistic characterization of the HDV genomic ribozyme: Assessing the catalytic and structural contributions of divalent metal ions within a multi-channel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- Nakano S., Cerrone A.L., Bevilacqua P.C. Mechanistic characterization of the HDV genomic ribozyme: Classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- Newmyer S.L., de Montellano P.R.O. Rescue of the catalytic activity of an H42A mutant of horseradish peroxidase by exogenous imidazoles. J. Biol. Chem. 1996;271:14891–14896. doi: 10.1074/jbc.271.25.14891. [DOI] [PubMed] [Google Scholar]
- Nixon P.L., Giedroc D.P. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- Nixon P.L., Rangan A., Kim Y.G., Rich A., Hoffman D.W., Hennig M., Giedroc D.P. Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot. J. Mol. Biol. 2002;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- Peracchi A., Beigelman L., Usman N., Herschlag D. Rescue of abasic hammerhead ribozymes by exogenous addition of specific bases. Proc. Natl. Acad. Sci. 1996;93:11522–11527. doi: 10.1073/pnas.93.21.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi A., Matulic-Adamic J., Wang S., Beigelman L., Herschlag D. Structure-function relationships in the hammerhead ribozyme probed by base rescue. RNA. 1998;4:1332–1346. doi: 10.1017/s1355838298980979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A.T., Been M.D. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A.T., Shih I.-h., Been M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- Rupert P.B., Ferré-D'Amaré A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Sharmeen L., Kuo M.Y.-P., Dinter-Gottlieb G., Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 1988;62:2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih I.-h., Been M.D. Involvement of a cytosine side chain in proton transfer in the rate-determining step of ribozyme self-cleavage. Proc. Natl. Acad. Sci. 2001;98:1489–1494. doi: 10.1073/pnas.98.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih I.-h., Been M.D. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- Tanner N.K., Schaff S., Thill G., Petit-Koskas E., Crain-Denoyelle A.-M., Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr. Biol. 1994;4:488–497. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Toney M.D., Kirsch J.F. Direct Brønsted analysis of the restoration of activity to a mutant enzyme by exogenous amines. Science. 1989;243:1485–1488. doi: 10.1126/science.2538921. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wadkins T.S. “Features unique to the two forms of the Hepatitis Delta Virus ribozymes: effects on self-cleavage activity.” Ph.D. thesis. Department of Biochemistry and Program in Cell and Molecular Biology; Duke University, Durham, NC: 2000. [Google Scholar]
- Wadkins T.S., Been M.D. Core-associated non-duplex sequences distinguishing the genomic and antigenomic self-cleaving RNAs of hepatitis delta virus. Nucleic Acids Res. 1997;25:4085–4092. doi: 10.1093/nar/25.20.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadkins T.S., Perrotta A.T., Ferré-D'Amaré A.R., Doudna J.A., Been M.D. A nested double-pseudoknot is required for self-cleavage activity of both the genomic and antigenomic HDV ribozymes. RNA. 1999;5:720–727. doi: 10.1017/s1355838299990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadkins T.S., Shih I., Perrotta A.T., Been M.D. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- Wu H.N., Lin Y.J., Lin F.P., Makino S., Chang M.F., Lai M.M. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc. Natl. Acad. Sci. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]