Abstract
The human 25S U4/U6.U5 tri-snRNP is a major building block of the U2-type spliceosome and contains, in addition to the U4, U6, and U5 snRNAs, at least 30 distinct proteins. To learn more about the molecular architecture of the tri-snRNP, we have investigated interactions between tri-snRNP proteins using the yeast two-hybrid assay and in vitro binding assays, and, in addition, have identified distinct protein domains that are critical for the connectivity of this protein network in the human tri-snRNP. These studies revealed multiple interactions between distinct domains of the U5 proteins hPrp8, hBrr2 (a DExH/D-box helicase), and hSnu114 (a putative GTPase), which are key players in the catalytic activation of the spliceosome, during which the U4/U6 base-pairing interaction is disrupted and U4 is released from the spliceosome. Both the U5-specific, TPR/HAT-repeat-containing hPrp6 protein and the tri-snRNP-specific hSnu66 protein interact with several U5- and U4/U6-associated proteins, including hBrr2 and hPrp3, which contacts the U6 snRNA. Thus, both proteins are located at the interface between U5 and U4/U6 in the tri-snRNP complex, and likely play an important role in transmitting the activity of hBrr2 and hSnu114 in the U5 snRNP to the U4/U6 duplex during spliceosome activation. A more detailed analysis of these protein interactions revealed that different HAT repeats mediate interactions with specific hPrp6 partners. Taken together, data presented here provide a detailed picture of the network of protein interactions within the human tri-snRNP.
Keywords: pre-mRNA splicing, U4/U6.U5 tri-snRNP, protein–protein interactions
INTRODUCTION
Introns of nuclear pre-mRNAs are recognized and precisely removed by the spliceosome. This molecular machine consists of the U1, U2, U4/U6, and U5 snRNPs and numerous non-snRNP splicing factors (Will and Lührmann 2006). The spliceosomal machine is a particularly protein-rich RNP, held together by an intricate and highly dynamic network of protein–protein, protein–RNA, and RNA–RNA interactions. Recently, mass spectrometry identified >200 spliceosomal proteins, of which at least 60 are associated with specific snRNPs (Hartmuth et al. 2002; Jurica et al. 2002; Makarov et al. 2002; Rappsilber et al. 2002; Zhou et al. 2002). The U1 and U2 snRNPs initially bind to the pre-mRNA substrate and generate the pre-spliceosome, or “complex A.” This is followed by the stable integration of the pre-formed 25S U4/U6.U5 tri-snRNP, in which the U4 and U6 snRNAs are base paired, to form the pre-catalytic spliceosome, or “complex B.” A major structural change, in which the U1 and U4 snRNPs are released, transforms complex B into the catalytically competent spliceosome, also termed complex B*. After the first step of splicing, the spliceosome is converted into complex C. The second step generates the mature mRNA product and is followed by the release of the remaining snRNPs from the spliced-out intron.
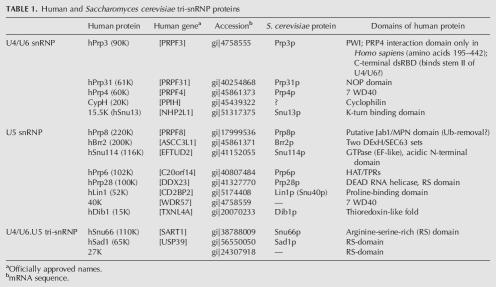
One of the major building blocks of the human spliceosome is the 25S U4/U6.U5 tri-snRNP. The purified tri-snRNP from HeLa cells contains, in addition to the U4, U6, and U5 snRNAs, ∼30 distinct proteins (for review, see Will and Lührmann 2006). The tri-snRNP is thus a particularly protein-rich particle. The nomenclature and the domain structure of the known human tri-snRNP proteins are summarized in Table 1. One set of seven Sm proteins is associated with both the U4 and U5 snRNAs, while one set of the LSm 2–8 proteins is bound to the U6 snRNA. Five proteins (in addition to the Sm or Lsm proteins) were found to bind stably to the U4/U6 snRNAs, namely 15.5K (alias hSnu13), CypH (alias 20K), hPrp4 (60K), hPrp31 (61K), and hPrp3 (90K). U5 snRNA is associated with the U5-specific proteins hPrp8 (220K), hBrr2 (200K), hSnu114 (116K), hPrp6 (102K), hPrp28 (100K), hLin1 (52K), 40K, and hDib1 (15K); of these only hLin1 is not found in the U4/U6.U5 tri-snRNP (Laggerbauer et al. 2005). Three proteins termed hSnu66 (110K), hSad1 (65K), and 27K associate more stably with the tri-snRNP than with individual U4/U6 or U5 particles, and are thus designated “tri-snRNP-specific.” All proteins of the 25S tri-snRNP have been highly conserved during evolution, and all have orthologs in the yeast Saccharomyces cerevisiae except the 27K and 40K proteins, and possibly the CypH protein (Gottschalk et al. 1999; Stevens and Abelson 1999).
TABLE 1.
Human and Saccharomyces cerevisiae tri-snRNP proteins
Once the tri-snRNP binds to the nascent pre-spliceosome, it undergoes dramatic structural changes during the life cycle of the spliceosome. Within the U4/U6 snRNP, the U4 and U6 snRNAs form two intermolecular RNA helices (stems I and II), both of which are disrupted during the activation of the spliceosome, with subsequent release of U4. U6 snRNA then interacts with U2 snRNA and the 5′ end of the intron to form part of the catalytic center. Concomitantly, U1 snRNA dissociates from the 5′ splice site. In addition, the major loop of the U5 snRNA is involved in aligning the two exons for ligation (Nilsen 1998; Tycowski et al. 2006).
The U4/U6.U5 tri-snRNP contains several proteins that facilitate RNA/RNP rearrangements during splicing. Studies in yeast indicate an involvement of Prp28p, a DExH-box RNA helicase, in the dissociation of the U1 snRNP from the 5′ splice site (Strauss and Guthrie 1994; Staley and Guthrie 1999). There is evidence that the DExH-box RNA helicase hBrr2 (the ortholog of Brr2p in yeast) and the GTPase hSnu114 (Snu114p in yeast) are driving forces behind the disruption of the U4/U6 snRNA helices prior to spliceosome activation (Xu et al. 1996; Fabrizio et al. 1997; Laggerbauer et al. 1998; Raghunathan and Guthrie 1998; Bartels et al. 2002, 2003). hPrp8 and its yeast counterpart Prp8 have been shown to contact all elements of the pre-mRNA involved in splicing, i.e., the 5′ and 3′ splice sites, as well as the branch site (Teigelkamp et al.1995a; Siatecka et al. 1999; Query and Konarska 2004; Grainger and Beggs 2005), and Prp8 is thought to play an important role in modulating the activity of those U5 proteins involved in these rearrangements (Kuhn et al. 2002 and references therein). However, the actual mechanism by which these U5-specific “motor” proteins disrupt the U4/U6 RNA helices is still not understood. It may involve either direct contact between these proteins and the snRNAs or indirect contact through a network of protein–protein interactions, capable of relaying a “power stroke” from a helicase and/or GTPase to the RNA.
In yeast, a study by van Nues and Beggs (2001) revealed interactions between some of the yeast tri-snRNP proteins. In contrast, relatively little is known about the protein–protein and protein–RNA network within the human tri-snRNP. The human U4/U6-specific proteins CypH, hPrp4, and hPrp3 form a stable RNA-free trimeric subcomplex, and hPrp4 and hPrp3 have been shown to interact directly with each other (Horowitz et al. 1997; Lauber et al. 1997; Teigelkamp et al. 1998; Gonzalez-Santos et al. 2002). An additional stable RNA-free subcomplex is formed by the U5 proteins hSnu114, hPrp8, hBrr2, and 40K (Achsel et al. 1998). Three human U4/U6-specific proteins have been shown to contact the U4 and/or U6 snRNAs. These are the 15.5K and hPrp31 proteins, which can be cross-linked to the U4 snRNA, and the hPrp3 protein, which can be cross-linked to the U6 snRNA (Nottrott et al. 2002).
Compared with the information currently available about protein–protein and protein–RNA interactions within the U4/U6 and U5 snRNPs, even less is known about the interactions that bridge the two snRNPs. To date, only an interaction between the U5-specific protein hPrp6 and U4/U6-specific hPrp31 protein has been reported (Makarova et al. 2002; Schaffert et al. 2004). There is no indication of RNA–RNA interactions between the two particles, and only one inter-particle RNA–protein cross-link has been observed, namely between the yeast protein Prp8 (corresponding to human 220K) and the U6 snRNA (Vidal et al. 1999). Thus, protein–protein interactions likely dominate in the communication between the U4/U6 and the U5 snRNPs. The role played by these protein–protein interactions in determining the conformational and configurational changes in the spliceosomal snRNAs during the splicing cycle remains to be elucidated.
To obtain a comprehensive picture of protein interactions in the human tri-snRNP, and in particular of the protein interactions bridging the U4/U6 and U5 snRNP, we performed yeast two-hybrid (Y2H) and in vitro binding assays. Our data reveal the basic design of the human tri-snRNP protein network and, in addition, identify distinct protein domains that are critical for the connectivity of this network in human tri-snRNPs.
RESULTS
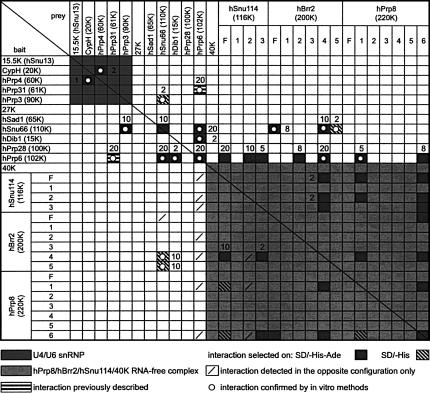
To detect protein–protein interactions between components of the human U4/U6.U5 tri-snRNP, the yeast two-hybrid (Y2H) assay was used. Full-length cDNAs encoding each of the known human tri-snRNP proteins, except for Sm and LSm proteins, were subcloned into the pGBKT7 and pGADT7 vectors to generate “bait” and “prey” fusions. Vectors were then co-transformed into the yeast strain AH109, and protein–protein interactions were identified by growth on minimal medium lacking tryptophan, leucine, and histidine (SD/−His), or, for higher stringency, lacking both histidine and adenine (SD/−His−Ade). Additionally, positives were re-examined in the presence of 3-amino-1,2,4-triazole (3-AT). This competitive inhibitor of the His3 reporter helps to discriminate strong and weak interactors. A full-length hBrr2 cDNA, as well as those encoding hPrp4 and hPrp8, were obtained by PCR from a Marathon-Ready cDNA library (see Materials and Methods). All other cDNAs employed in this study have been previously described, and their accession numbers are given in Table 1. False negatives and false positives can arise from various conditions during Y2H screens. In an independent approach several protein–protein interactions were therefore also investigated by in vitro binding studies (i.e., glutathione-S-transferase [GST]-tagged protein pull-down and co-immunoprecipitation [co-IP]) using proteins translated in vitro and/or recombinant proteins expressed in Escherichia coli.
Analysis of interactions between U4/U6 proteins
We first focused on interactions between the five tri-snRNP proteins associated with the U4/U6 snRNA. Consistent with our previous co-immunoprecipitation results (Teigelkamp et al. 1998), an interaction between the CypH and hPrp4 protein was also observed by Y2H analysis with each protein serving in turn as bait or prey (Fig. 1). This interaction was also detected in vitro by GST pull-down assays (not shown). Remarkably, the CypH–hPrp4 interaction was the only one that could be detected among the five U4/U6 proteins by the two-hybrid method (see area highlighted dark gray in Fig. 1). Notably, no interaction was found between the 15.5K protein and any of the other proteins. As binding between hPrp4 and the central domain of hPrp3 had previously been demonstrated using co-immunoprecipitation and isothermal titration calorimetry (Gonzalez-Santos et al. 2002), we also performed in vitro binding assays with His-tagged hPrp3 protein. Significantly, in vitro translated HA-tagged hPrp4 was co-precipitated specifically and efficiently by His-tagged hPrp3, confirming this interaction (not shown).
FIGURE 1.
Two-hybrid interactions within the human tri-snRNP. All 29 full-length (F) proteins and protein fragments served as bait and prey in 29 × 29 Y2H combinations. Fragments are numbered as shown in Figure 2. Numbers adjacent to symbols indicate the highest tested 3-AT concentration (in mM) still supporting the interaction.
Interactions between U5 proteins in the hPrp8/hBrr2/hSnu114/40K tetramer
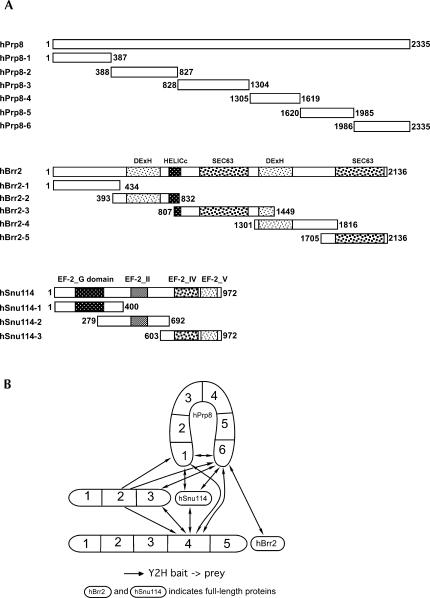
The U5-specific proteins hPrp8, hBrr2, hSnu114, and 40K form an RNA-free protein complex that remains stable even in 0.2 M NaSCN, with the interaction between the hPrp8 and hSnu114 proteins resistant to dissociation in up to 0.4 M NaSCN (Achsel et al. 1998). Here, we have used Y2H assays to analyze the interactions between these proteins in more detail. Clear protein–protein interactions were not observed when full-length hPrp8, hBrr2, hSnu114, and 40K were used as bait or prey (Fig. 1, lower right quadrant). Failure to detect interactions between full-length proteins, in particular very large ones such as hPrp8 (274 kDa) and hBrr2 (244.5 kDa), is not uncommon, and has been observed previously (e.g., van Nues and Beggs 2001). In order to overcome this problem, we cloned several deletion mutants of the three large proteins into the bait and prey vectors. hBrr2 was divided into five overlapping fragments, on the basis of the structural domains deducible from its amino acid sequence, which includes two helicase domains followed by a SEC63 domain; the function of the latter domain is currently unknown but possibly anchors proteins to RNP complexes (Ponting 2000; Fig. 2A). The second SEC63 domain of Brr2p is involved in interactions with spliceosomal proteins Prp16p and Snp1p (van Nues and Beggs 2001). Thus, hBrr2 fragments 2 and 4 consist of its two helicase domains, while fragments 3 and 5 harbor its two SEC63 domains (Fig. 2A). Similarly, the hSnu114 protein was divided up taking into account the boundaries of the conserved GTPase structural features known from the ribosomal translocase EF-2 (Fig. 2A). Thus, fragment 1 contains the EF-2 GTP-binding domain, while fragments 2 and 3 harbor domain II and domains IV+V (according to the EF-2 nomenclature), respectively. In contrast to these two proteins, the sequence of hPrp8, although highly conserved in all eukaryotes, for a long time did not offer significant clues as to its structural organization. Only recently, protein motives such as the MPN domain (Mpr-1, Pad-1, N-terminal) and RRM (RNA Recognition Motif) have been discovered in Prp8p (Grainger and Beggs 2005; Bellare et al. 2006; Boon et al. 2006). We divided hPrp8 into six arbitrary fragments of roughly equal size (315–477 amino acids; Fig. 2A).
FIGURE 2.
(A) Schematic representation of the hPrp8, hBrr2, and hSnu114 proteins and their fragments used in two-hybrid assays. (DExH) DExH box helicase domain; (HELICc) helicase superfamily C-terminal domain; (SEC63) domain of unknown function in Sec63p and other proteins; (EF-2_G) eukaryotic elongation factor 2 GTP-binding domain; (EF-2_II [IV, V]) eukaryotic elongation factor 2 domains II, IV, and V. (B) Observed interactions between the hPrp8, hBrr2, and hSnu114 proteins.
When bait and prey constructs with full-length and truncated proteins were again tested in the Y2H assay, interactions between the hPrp8, hBrr2, and hSnu114 proteins were clearly observed (1, 2B). The N- and C-terminal fragments of hPrp8 (hPrp8–1 and hPrp8–6) appear to be the major protein–protein interaction sites of this protein. Both interact not only with fragment 2 of hSnu114, but also with fragment 4 of hBrr2. This fragment contains the C-terminal helicase domain of hBrr2 and in turn interacts with fragments 2 and 3 of the hSnu114 protein. Interestingly, the terminal fragments of hPrp8 also interacted with each other, raising the possibility that hPrp8 forms an intramolecular bridge. The C-terminal fragment of hPrp8, in addition, interacts with the C-terminal fragment of hSnu114 (hSnu114–3). These interactions are shown schematically in Figure 2B. Full-length hSnu114 was found to interact with terminal fragments of hPrp8 (Fig. 1), indicating that hSnu114 can be stably expressed and properly folded in yeast. The Y2H interactions observed between hPrp8 and hSnu114 are consistent with our previous findings that hSnu114 interacts with hPrp8, as shown by Far Western blots and by the fact that a heterodimeric complex of both proteins, that is stable even in the presence of 0.4 M of the chaotropic salt NaSCN, can be isolated from U5 snRNPs (Achsel et al. 1998). In yeast, the association of Snu114p with a region in the N-terminal half of Prp8p was detected during Y2H and pull-down experiments (Grainger and Beggs 2005; Boon et al. 2006).
Previously, we detected an interaction between hPrp8 and both the 40K protein and hSnu114 in Far Western blotting assays (Achsel et al. 1998). Binding partners of the 40K protein, however, could not be identified by Y2H analysis using either full-length or truncated proteins (Fig. 1). This result was not due to the absence of 40K in yeast; the production of stable 40K was confirmed by Western blotting (not shown). Except for four interactions involving hBrr2–4 and fragments of hSnu114, the generally weak interactions within the hPrp8/hBrr2/hSnu114/40K tetramer are all suppressed by 2 mM 3-AT (Fig. 1). Arbitrarily dividing a protein into fragments may possibly result in weakly or non-interacting fragments. However, the fact that these fragments are involved in stronger interactions with other proteins of the tri-snRNP demonstrates the general validity of this approach.
Reassessment of Y2H positives in the presence of 3-AT
To further assess the strength of Y2H interactions, positives were grown in the presence of different concentrations of 3-AT, which increases the stringency of the assay. Yeast colonies transformed with hPrp6 and hDib1, hSnu114–2, hSnu114–3, hBrr2–2, hPrp8–1, or hPrp8–6 were able to grow in the presence of 2–10 mM 3-AT, while those transformed with hPrp6 and protein hPrp31, hSnu66, hPrp6, hSnu114, or hBrr2–4 grew in the presence of 20 mM 3-AT. Yeast colonies transformed with hSnu66 and hPrp3, hSnu66, hBrr2, hBrr2–4, or hBrr2–5 grew in the presence of 2–10 mM 3-AT (Fig. 1). Except where indicated, interactions within the hPrp8/hBrr2/hSnu114/40K complex were all suppressed in the presence of 2 mM 3-AT. Nevertheless, fragments originating from these proteins showed stable interactions with other proteins. The use of 3-AT, however, cannot exclude that an observed “strong” interaction simply reflects a high level of expression of stable protein.
hPrp6 bridges the U5 and U4/U6 snRNPs in the tri-snRNP
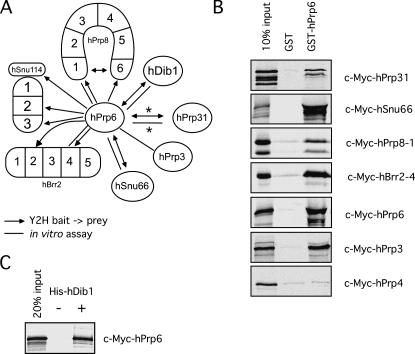
Mutational analyses of the yeast Prp6 protein, and previous biochemical analyses and RNAi-mediated knockdown of hPrp6 in our laboratory, all point to a crucial role for Prp6 in establishing a connection between the U5 and U4/U6 snRNPs (Galisson and Legrain 1993; Makarov et al. 2000; Makarova et al. 2002; Schaffert et al. 2004). Indeed, hPrp6 has been shown to interact with the U4/U6-associated hPrp31 protein (Makarova et al. 2002). However, little is currently known about the interactions of the hPrp6 protein within the U5 snRNP. To obtain a better resolved picture of the bridging function of hPrp6, we investigated its interactions with various U5 and U4/U6 proteins using the two-hybrid system. We used full-length hPrp6 as bait and prey to identify interaction partners within the set of all full-length tri-snRNP proteins, and hBrr2, hSnu114, and hPrp8 protein fragments. hPrp6 unidirectionally interacted with distinct domains of each of the three large U5-specific proteins—i.e., full-length hSnu114 as well as fragments 2 and 3, hBrr2 fragments 2 and 4, and hPrp8 fragments 1 and 6 (Fig. 1). Additional interactions were observed with the U5-specific protein hDib1, the tri-snRNP-specific protein hSnu66, and the U4/U6-specific protein hPrp31 (Fig. 1). The resultant network of interactions is shown schematically in Figure 3A. hPrp6 also interacted with itself, suggesting that it potentially dimerizes or forms an intramolecular bridge.
FIGURE 3.
Interactions of the hPrp6 protein. (A) Summary of yeast two-hybrid and in vitro interactions involving hPrp6. (*) These interactions were identified previously (Makarova et al. 2002). (B) GST pull-downs performed with GST alone or GST-hPrp6 (as indicated) and in vitro translated c-Myc-tagged tagged hPrp3, hPrp4, hPrp31, hPrp6, hSnu66, fragment hPrp8–1, and fragment hBrr2–4 (indicated on the right). The input lane contains 10% of the total amount of in vitro translated protein added to each reaction. The input and bound proteins were analyzed by SDS-PAGE and visualized by autoradiography. (C) Co-IP experiments with purified His-tagged hDib1 and in vitro translated [35S]-methionine-labeled c-Myc-hPrp6 protein. His-tagged hDib1 was precipitated with anti-His antibodies, and co-precipitating proteins were again visualized by autoradiography following SDS-PAGE. A mock precipitation without His-tagged hDib1 is included as a negative control. The input lane represents 20% of the c-Myc-hPrp6 added to the reaction.
Next we studied interactions between hPrp6 and U5 and U4/U6 proteins in vitro, employing GST pull-down assays with GST-hPrp6. As shown in Figure 3B, GST-hPrp6, but not GST alone, bound [35S]-labeled, in vitro translated hPrp31 (which served as a positive control; Makarova et al. 2002), as well as c-Myc-hSnu66, c-Myc-hBrr2–4, and c-Myc-hPrp8–1, consistent with the results obtained by Y2H assays for these protein pairs (see Fig. 1). Moreover, GST-hPrp6 also co-precipitated in vitro translated hPrp6, supporting the idea that this protein may interact with itself. We note that GST-hPrp6 did not co-precipitate in vitro translated hDib1, hSnu114, or hSnu114–2 and hSnu114–3 (data not shown). This could indicate either that the strength of the interaction between the tested protein parts is too weak to withstand the in vitro conditions employed or that the N-terminal GST tag might hinder complex formation between GST-hPrp6 and hDib1 or hSnu114. Indeed, an interaction between hPrp6 and hDib1 was observed in a co-precipitation experiment with His-tagged hDib1 expressed in E. coli. As shown in Figure 3C, [35S]-labeled c-Myc-hPrp6 was efficiently co-precipitated with His-tagged hDib1. An equivalent co-precipitation experiment with tagged hSnu114 protein could not be carried out because of solubility problems with recombinantly expressed hSnu114.
Our Y2H assays revealed a strong interaction of hPrp6 with the U4/U6 snRNP-specific hPrp31 protein but not with hPrp3 and hPrp4 (Makarova et al. 2002; Fig. 1). As an interaction between hPrp3 and hPrp4 was only detected by co-precipitation experiments, but not by Y2H assays, we investigated whether hPrp6 interacts with the latter two proteins by performing GST pull-down experiments. As shown in Figure 3B, GST-hPrp6, but not GST alone, efficiently bound [35S]-labeled, in vitro translated hPrp3; no interaction with hPrp4 was observed. Thus, hPrp6 interacts with at least two U4/U6-specific proteins, hPrp31 and hPrp3.
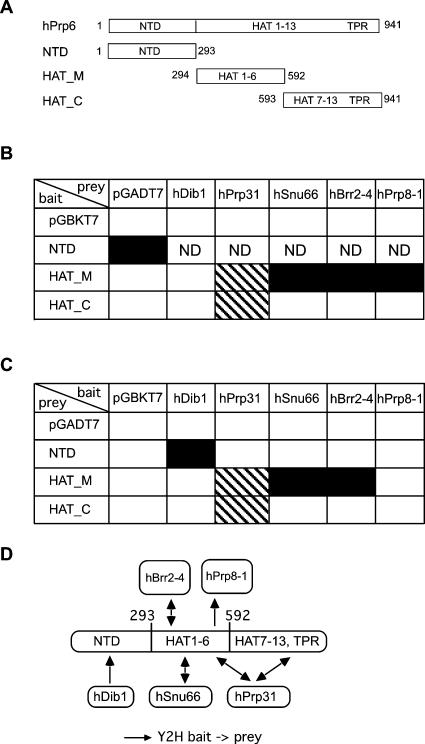
The hPrp6 protein contains an N-terminal domain followed by 13 repeats of the HAT (Half-A-TPR) motif and one TPR (tetratricopeptide) motif, as shown in Figure 4A. The HAT motif is found in several RNA-binding proteins (Lamb et al. 1995; Preker and Keller 1998), and its structure and sequence are similar to those of TPR repeats, which mediate protein–protein interactions (Lamb et al. 1995; Blatch and Lassle 1999; D'Andrea and Regan 2003). To investigate which regions of hPrp6 interact with the various proteins, hPrp6 was divided into three fragments (Fig. 4A): an N-terminal domain lacking HAT-repeats (NTD), a second domain comprising HAT-repeats 1–6 (HAT_M), and a third domain comprising HAT-repeats 7–13 and the TPR-repeat. All hPrp6 protein–protein interactions identified by both Y2H and GST pull-down or co-IP experiments were further examined with the hPrp6 fragments used either as bait (Fig. 4B) or as prey (Fig. 4C). The N-terminal domain interacted with the hDib1 protein, while the central HAT_M fragment bound to hSnu66, fragment 4 of hBrr2, and fragment 1 of hPrp8 (see Fig. 4D for summary of interactions). The hPrp31 protein interacted with both the HAT_M and the HAT_C fragment. In summary, it appears that distinct regions of hPrp6 are dedicated to interactions with different proteins.
FIGURE 4.
hPrp6 domains and protein–protein interactions. (A) Schematic representation of the protein fragments derived from full-length hPrp6 used in two-hybrid assays. (B) The yeast strain AH109 was transformed with pGBKT7 carrying hPrp6 fragments and pGADT7 carrying hDib1, hPrp31, hSnu66, and fragments hBrr2–4 and hPrp8–1. Empty vectors were included as a negative control. Combinations labeled ND were not analyzed due to self-activation of the bait. (Black squares) Interactions selected on SD/−His−Ade, (hatched squares) those selected on SD/−His. (C) Interactions of reciprocal bait–prey pairs. (D) Schematic drawing of the observed interactions involving hPrp6.
The tri-snRNP-specific hSnu66 protein interacts with both U5-specific and U4/U6-specific proteins
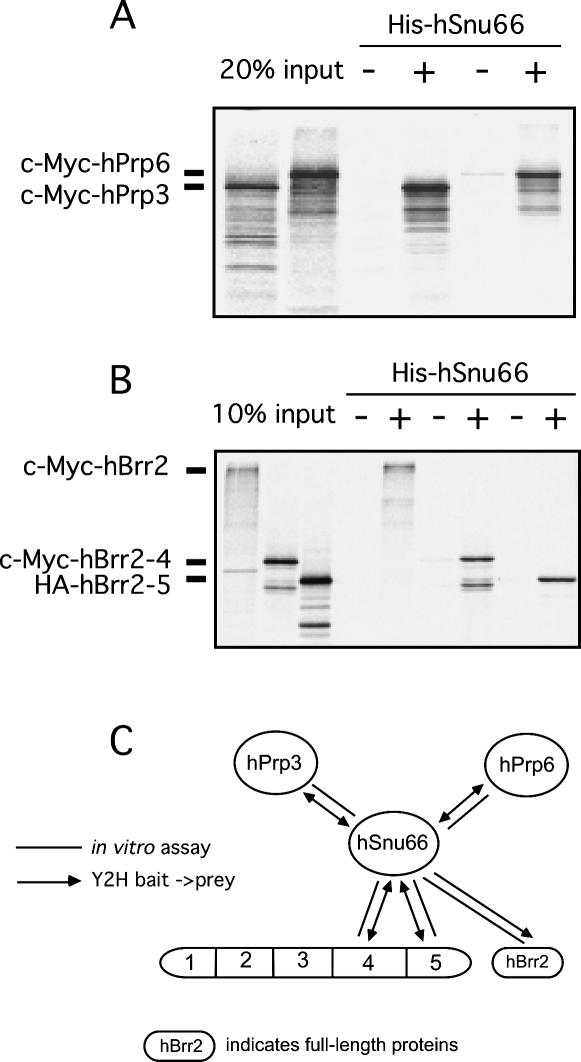
Three proteins, hSnu66, hSad1, and 27K, associate with the tri-snRNP, but less so—or not at all—with its individual snRNP components (Fetzer et al. 1997; Makarova et al. 2001). Y2H assays were also employed to identify the binding partners of these three proteins. Full-length hSnu66 was found to interact with the U4/U6-specific protein hPrp3 and also with the full-length U5-specific proteins hPrp6 and hBrr2 (Fig. 1). In the case of hBrr2, the interaction with hSnu66 appears to involve the C-terminal region encompassed by fragments 4 and 5. Binding interactions were additionally investigated by co-immunoprecipitation assays. Consistent with the Y2H results, [35S]-labeled full-length c-Myc-hPrp3, c-Myc-hPrp6, and c-Myc-hBrr2 protein, as well as c-Myc-hBrr2–4 and HA-hBrr2–5 fragments, were co-precipitated together with His-tagged hSnu66 by anti-hSnu66 antibodies (Fig. 5A, B). Y2H assays did not reveal any interactions involving the 27K or the hSad1 protein (Fig. 1).
FIGURE 5.
Analysis of interactions between the hSnu66 protein and in vitro-translated fusion proteins of (A) hPrp3 and hPrp6, and (B) full-length hBrr2, as well as fragments hBrr2–4, hBrr2–5. Purified His-tagged hSnu66 was incubated with anti-hSnu66 antibodies bound to protein A-Sepharose and [35S]methionine-labeled proteins produced by in vitro translation. The precipitated proteins were fractionated by SDS-PAGE and visualized by autoradiography. Mock precipitations without His-tagged hSnu66 protein were included as a negative control. (C) Summary of hSnu66 interactions.
Interactions of the hPrp3 protein
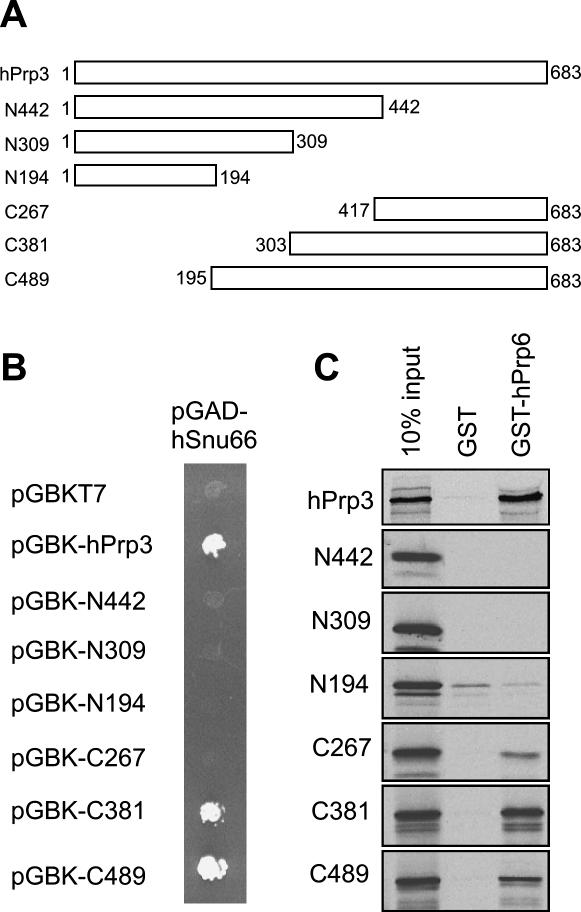
Y2H assays revealed binding between full-length hPrp3 and hSnu66 (Fig. 1). To delineate the boundaries of the hSnu66-interacting domain, hPrp3 was divided into four fragments according to (Gonzalez-Santos et al. 2002) and two additional fragments comprising the N- and C-terminal halves of the protein (Fig. 6A). All six fragments were subcloned into pGBKT7 as bait fusions and tested with hSnu66, hPrp4, or hPrp6 prey fusions in the Y2H system. Only the C-terminal fragments (C489 and C381) and full-length hPrp3 (as positive control) interacted with full-length hSnu66 protein (Fig. 6B). Interactions with hPrp4 or hPrp6 were not observed (not shown).
FIGURE 6.
Protein–protein interactions of the hPrp3 protein. (A) Schematic drawing of the protein fragments derived from the full-length hPrp3 protein. (B) Yeast two-hybrid analysis of interactions between hPrp3 bait constructs (labeled according to panel A) and hSnu66 prey. (C) In vitro pull-downs of hPrp3-derived constructs with GST-hPrp6 (performed as described in Fig. 3).
As described above, an interaction between the U4/U6-specific hPrp3 and hPrp6 proteins was detected by GST pull-down assays. To delineate the region of hPrp3 that interacts with hPrp6, [35S]-labeled c-Myc-tagged hPrp3 deletion mutants were translated in vitro and used in GST pull-down experiments with GST-hPrp6. The C-terminal hPrp3 fragments (C267, C381, and C489) bound to GST-hPrp6 (but not GST alone), while binding of the N-terminal fragments was not above background levels (Fig. 6C). It therefore appears that the C-terminal region of hPrp3, comprising amino acids 417–683, suffices for binding to hPrp6, while an extended C terminus, comprising amino acids 303–683, is required for interaction with hSnu66.
DISCUSSION
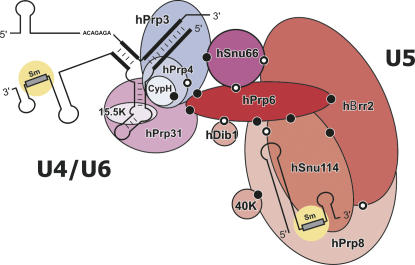
In this study, we have investigated protein–protein interactions in the human tri-snRNP, using the yeast two-hybrid technique. Y2H experiments were performed with 38 polypeptides (full-length proteins and fragments thereof) derived from 15 protein components of the human tri-snRNP. In total, 12 pairs of interacting proteins were detected by Y2H analysis. For eight of them, an interaction was corroborated by in vitro binding experiments, again using either full-length proteins or protein fragments. Three additional interactions were observed in vitro only. Our Y2H results are shown in tabular form in Figure 1, and the network of interactions is illustrated schematically in Figure 7. No interactions were detected for the hPrp28, hSad1, and 27K proteins. Thus, these proteins are either not expressed in yeast (due to the presence of RS domains) or require complex binding sites comprised of multiple proteins. In yeast, PRP28 and SAD1 have been shown to be synthetically lethal or to synthetically enhance SNU114 mutations (Brenner and Guthrie 2005), suggesting that they may also physically interact with Snu114p. Taken together, our data provide a detailed picture of the protein network within the human tri-snRNP and thus expand our understanding of the molecular architecture of the spliceosome's largest subunit.
FIGURE 7.
Schematic depiction of protein–protein interactions within the human tri-snRNP. All dots indicate protein–protein interactions demonstrated in this study. Those in black and white indicate interactions also observed between the respective orthologous proteins in yeast. The Snu114p-Prp8p interaction has been reported by Dix et al. (1998), Prp6p–Dib1p by Uetz et al. (2000), Prp3p–Prp4p by Ito et al. (2001), and Prp8p–Brr2p, Brr2p–Snu66p, Snu66p–Prp6p by van Nues and Beggs (2001).
U4/U6 protein interactions
Interactions among U4/U6 snRNP proteins were detected between hPrp4 and hPrp3 (in vitro binding assay only) and between the CypH protein and hPrp4 (Y2H and in vitro) (Figs. 1, 7). The failure to detect an interaction between hPrp4 and hPrp3 via Y2H could be due to folding problems or lack of a necessary post-translational modification. The observed interactions underlie the biochemically stable CypH•hPrp4•hPrp3 heterotrimeric complex (Horowitz et al. 1997; Teigelkamp et al. 1998). Interactions of this group of proteins with 15.5K and hPrp31 could not be detected. This does not, however, rule out that these proteins engage in protein–protein interactions within the native U4/U6 snRNP. It is particularly important to consider the hierarchical nature of interactions within this particle. It has been shown that the human 15.5K protein binds directly to a specific sequence element of the U4 snRNA through a novel RNA-binding domain (Vidovic et al. 2000; Nottrott et al. 2002). The CypH•hPrp4•hPrp3 complex and hPrp31 bind only after this initiating step (Nottrott et al. 2002). We have experimental evidence that within U4/U6 snRNPs distinct surface areas of the 15.5K protein are involved in interactions with hPrp31 and the CypH•hPrp4•hPrp3 heterotrimer, respectively (A. Schultz, S. Nottrott, N. Watkins, and R. Lührmann, unpubl.). In some instances these interactions appear to be dependent on the presence of the RNA. The task of the 15.5K protein appears to be to bring the snRNA into the right conformation for subsequent binding of the other U4/U6 proteins, rather than to act merely as a bridge between these proteins and the U4 and U6 snRNA. In support of this idea, within the U4/U6 snRNP, hPrp31 and hPrp3 can be cross-linked to the U4 and U6 snRNAs, respectively (Nottrott et al. 2002).
Evolutionary conservation of interactions in the U5 heterotetrameric complex
Four U5-specific proteins, namely hPrp8, hBrr2, hSnu114, and 40K, are known to form a stable RNA-free heterotetramer (Achsel et al. 1998). Of this quartet, hPrp8, hBrr2, and hSnu114 bind directly to one another, and, furthermore, each of them binds to hPrp6 (summarized in Fig. 7). In contrast to the other three proteins, 40K appears to contact only one protein (hPrp8), with the caveat that weak interactions may not have been detected by the methods used in this work (due, for example, to subtle differences in the conformation of the expressed fusion proteins). One of the pairwise interactions—that between hPrp8 and hSnu114—had previously been observed via Far Western blotting (Achsel et al. 1998). The multiple interactions of hBrr2 and of hSnu114 with hPrp8 have interesting implications. Both of these proteins belong to families that often require other proteins as coactivators. There is thus the interesting possibility that the hPrp8 protein may be an activating factor for hBrr2 and/or for the hSnu114 EF-2 homolog. Moreover, considering that the hPrp8 protein (like its yeast counterpart) also contacts in a sequential manner the 5′ and 3′ splice sites (Teigelkamp et al.1995a, b; Reyes et al. 1996), its potential activation of the hBrr2 RNA-unwindase or the hSnu114 protein might be triggered by certain functional states of the spliceosome.
A more detailed analysis of the protein interactions involving hPrp8 and hBrr2 suggests that both proteins, despite their considerable size, make contacts with other proteins only through distinct limited regions. In the case of hPrp8, the immediate N or C termini (fragments hPrp8–1 and hPrp8–6) interact preferably with hBrr2, hSnu114, and hPrp6 within the U5 snRNP (Figs. 2B, 3A). Interestingly, the terminal fragments of hPrp8 also interact with each other, raising the possibility that they form an intramolecular bridge. We have also dissected the C-terminal region in more detail and again analyzed its interactions with the aforementioned binding partners. Interestingly, three out of four Y2H interactions, namely those with hPrp8–1, hSnu114–3, and hBrr2–4, were confirmed using just the very C-terminal stretch of ∼100 amino acids of hPrp8 as prey (data not shown). Thus, this limited stretch of amino acids represents a very important contact region. This finding is of interest considering the fact that certain patients suffering from Retinitis pigmentosa possess mutations just in this region of hPrp8 (Grainger and Beggs 2005). The observed disease phenotype could therefore originate from altered protein–protein interactions in the tri-snRNP. It will be interesting to determine in future studies how these mutations affect splicing. Our analyses indicate that hPrp8 is the central component of the heterotetrameric hPrp8•hBrr2•hSnu114•40K complex of the U5 snRNP, with confirmed contacts to all complex components. This tetramer appears to remain intact throughout the entire spliceosomal cycle and also during the final release of the post-spliceosomal 35S complex (Makarov et al. 2002).
Our two-hybrid experiments reveal a critical role of the C terminus of hBrr2 in protein–protein interactions. This includes interactions within the RNA-free heterotetramer (through interactions with hPrp8 and hSnu114) and with hSnu66 and hPrp6. hBrr2 displays the same domain structure as its yeast ortholog (Fig. 2A). Of the two helicase domains, the C-terminal one (fragment hBrr2–4) exhibits only poor conservation of the helicase consensus motifs in sub-domains V and VI (following the nomenclature of Tanner and Linder [2001]). In yeast, only the N-terminal helicase domain of Brr2p is required for unwinding the U4/U6 snRNA duplex (Raghunathan and Guthrie 1998; Kim and Rossi 1999). The poorly conserved second helicase domain, therefore, possibly serves exclusively as a protein–protein interaction domain. Our data also revealed that the hBrr2–5 fragment interacts with hSnu66 in Y2H and in vitro binding assays. Fragment hBrr2–5 comprises a SEC63 domain, which is suspected to mediate protein–protein interactions, particularly in the context of RNA–protein complexes (Ponting 2000).
In a previous Y2H study, a network of interactions involving a set of yeast spliceosomal proteins, including the U5-specific proteins Prp8p and Brr2p and the tri-snRNP-specific protein Snu66p, were observed (van Nues and Beggs 2001). These data revealed that Brr2p is involved in numerous protein–protein interactions, and was therefore assigned the role as a central organizer not only of the tri-snRNP, but also of spliceosomal protein dynamics in general. The C-terminal region of yeast Brr2p, comprising the second helicase-like domain (corresponding to our fragment hBrr2–4), appears to be involved in most Brr2p interactions with tri-snRNP proteins, and the extreme C terminus (our fragment hBrr2–5) is involved in the recruitment of first- and second-step factors, and contacts proteins of the U1 snRNP. Our Y2H experiments confirm the critical function of the second helicase-like domain of hBrr2 in protein–protein interactions and thus reveal a high degree of evolutionary conservation of interactions in yeast and man involving this domain.
Filling the gap between U4/U6 and U5
The interaction of the U5 and U4/U6 snRNPs to form the U4/U6.U5 tri-snRNP appears to be solely protein-mediated, as no interaction between the U5 and U4/U6 snRNAs is known. This snRNP bridging may occur via protein–protein and/or protein–RNA interactions. Based on our Y2H data, hPrp6 and hSnu66 are key players in establishing contacts between U5 and U4/U6 proteins. That is, hSnu66 interacts with hPrp3, hPrp6, and hBrr2 (Fig. 5C), and thus it may also contribute to bridging the snRNP components of the tri-snRNP. Unlike hPrp6, however, hSnu66 is not required for tri-snRNP stability (Makarova et al. 2001); thus, its primary function remains to be elucidated. hPrp6 instead emerges as the most important bridge between the human U5 snRNP and U4/U6 snRNP. This is supported by our previous biochemical analyses of hPrp6 (Galisson and Legrain 1993; Makarov et al. 2000; Makarova et al. 2002; Schaffert et al. 2004). In addition to numerous interactions within the U5 snRNP, full-length hPrp6 also interacts with the tri-snRNP-specific hSnu66 and U4/U6-specific hPrp3 and hPrp31 proteins (summarized in Fig. 3A). Mutational analyses of yeast Prp6p support the idea that Prp6p assumes an equally important bridging role in yeast (Galisson and Legrain 1993). hPrp6 contains as many as 13 HAT and one TPR repeats, motifs found in several other spliceosomal proteins (Gottschalk et al. 1998). Domains of this kind make a protein particularly well suited to serve as a binding partner in multiple, simultaneous protein–protein interactions during the assembly of macromolecular machines (Scheufler et al. 2000; D'Andrea and Regan 2003). The targets for binding are apparently small peptide stretches, with a specificity that is still not understood. hPrp6 interacts with seven tri-snRNP proteins (Figs. 3A, 7). Our protein–protein interaction data for fragments of hPrp6 show that different HAT/TPR repeats mediate interactions with specific hPrp6 partners (Fig. 4). All repeats participate in binding hPrp31, whereas only the repeats of the central HAT_M fragment bind hSnu66, fragment 4 of hBrr2, and fragment 1 of hPrp8. Our lab previously reported that antibodies raised against the HAT_C fragment efficiently precipitate free U5 snRNP, but not the U4/U6.U5 tri-snRNP, from HeLa nuclear extracts (Makarov et al. 2000). Binding of the hPrp31 protein to the C-terminal HAT/TPRs could explain this observation. A differential contribution of individual TPR domains of an extended TPR array was also reported for the yeast spliceosome assembly factor Clf1p (Vincent et al. 2003). The N-terminal fragment of hPrp6, predicted to contain a coiled-coil domain (Makarov et al. 2000), also is involved in protein–protein interactions and binds to hDib1 (Fig. 3C).
The U5-specific proteins hBrr2 and hSnu114 are considered to be the driving forces behind the disruption of the U4/U6 snRNA helices during activation. Regardless of the precise mechanism, a structural rearrangement in the U4/U6 snRNP must take place for the U4/U6 helix to be disrupted. One possible mechanism involves the direct docking of an enzyme onto the double-stranded U4/U6 RNA substrate, followed by triphosphate consumption and catalysis. In an alternative mechanism, a helicase or GTPase is the active center in which the triphosphate is hydrolyzed, thus providing a “power stroke,” which is then passed on through components of the tri-snRNP to the U4/U6 RNA double helix, either unwinding the helix itself or displacing or reorganizing its surrounding proteins (Will and Lührmann 2001). Indeed, DExH/D helicases were recently shown to function as “RNPases” and displace proteins from RNPs (Jankowsky et al. 2000, 2001; Fairman et al. 2004). Our observation that hPrp6 simultaneously interacts with the U5 proteins hBrr2 and hSnu114, and also with the U4/U6 proteins hPrp31 and hPrp3 (Fig. 3), makes this protein an excellent candidate for transmitting such a power stroke. In this respect it is also interesting to note that hPrp31 and hPrp3 contact directly the regions of the U4 and U6 snRNAs involved in the formation of stem I of the U4/U6 interaction domain (Nottrott et al. 2002; A. Schulz, S. Nottrott, K. Hartmuth, and R. Lührmann, unpubl.). Both proteins could thus act as final effectors in this chain of protein–protein interactions. Consistent with this idea, hPrp31 and hPrp3 are significantly destabilized during the catalytic activation step and apparently released from the spliceosome (Makarov et al. 2002). However, the sequence of events is clearly even more complex, as >20 additional proteins become stably integrated into the spliceosome upon the transition from pre-catalytic complex B to activated B* (Makarov et al. 2002). Nonetheless, a direct action of motor proteins or the transmission of a power stroke might provide a useful working hypothesis for future experiments.
MATERIALS AND METHODS
cDNA cloning
The complete cDNA of the hBrr2 was cloned from a human Marathon-Ready cDNA library (Clontech) using 5′-RACE PCR techniques with published partial cDNA sequences. Specifically, three overlapping fragments were PCR-amplified from Marathon-Ready cDNA by using three pairs of oligonucleotides: HPV-29/HPV-30, HPV-31/HPV-32, and HPV-33/HPV-34 (see below), which are complementary to the partial cDNA sequence of hBrr2 (gi|20521659; Nagase et al. 1998). The three PCR products were cloned into pNoTA/T7 and subsequently cloned jointly into pBluescript SK(−), resulting in a 4.8-kb 3′ region of the hBrr2 cDNA. To obtain the 5′-end sequence, 5′-RACE PCR was performed using Marathon-Ready cDNA and the gene-specific primer HPV-52 (see below). The resulting 2.0-kb 5′ RACE product was sequenced and fused with the 3′ sequence to yield full-length pSK-hBRR2 cDNA. The full-length cDNA encodes a protein that has 2136 amino acids and a calculated molecular mass of 244.5 kDa. The protein product of a transcription/translation reaction in vitro with this cDNA co-migrates on an SDS-PAGE gel with hBrr2 isolated from native U5 snRNPs (not shown). The sequence has been deposited with GenBank (accession no. gi|45861371).
Because of inconsistencies between the various sequences of the hPrp4 protein deposited in public databases, the cDNA of hPrp4 was cloned again by PCR from the same cDNA library used for the hBrr2 protein. This sequence has been deposited with GenBank (accession no. gi|45861373). It is essentially identical to the one published by Horowitz et al. (1997). To clone hPrp4, two oligonucleotides, HPV-324 and HPV-325 (see below), corresponding respectively to the 5′ and 3′ end of the ORF (gi|2708304), were synthesized and used in PCR with human Marathon-Ready cDNA as template. The amplified cDNA was cloned into the PCR cloning vector pNoTA/T7 (5′→3′ Inc.) to generate pNoTA–hPrp4. To clone hPrp8, three overlapping fragments comprising the 5′, central, and 3′ regions of full-length hPrp8 were amplified from the Marathon-Ready cDNA library by PCR using three pairs of oligonucleotides: HPV-3/HPV-4, HPV-5/HPV-6, and HPV-7/HPV-8 (see below), designed according to the deposited sequence (gi|3661609). The PCR products were cloned into the pNoTA/T7 vector. The three fragments were then released from pNoTA/T7 and jointly cloned into pBluescript SK(−) to generate the full-length hPrp8 cDNA. The sequence of this cDNA was identical to that of the hPrp8 cDNA clone published by Luo et al. (1999).
Oligonucleotides used for cloning hPrp4, hBrr2, and hPrp8:
HPV-3: CCCTTCGAAGCATATGGCCGGAGTGTTTCCTTATC
HPV-4: CTGAGACTGGTTCAACCGAGAC
HPV-5: CCCACTAGTTATAAGCATGACACCAAGTTGCTC
HPV-6: CCCCTCGAGACATTCCACTTATAGGAGGC
HPV-7: ATATAAGATGAACTCTTCCTGTG
HPV-8: CCCCTCGAGTTCGAAAGGCTTCGGCCTCGGGAGGCTG
HPV-29: CAGAGGTCCTGTCTTAGAAGCTTT
HPV-30: GTGCACTGACAACTCCAACTTGGG
HPV-31: ATGTGTCCTCTGCGCCAGTTCCGG
HPV-32: ATTGGGGTTCTGTGTCATGCGGCG
HPV-33: GCCAAGCCTGTGTACCATGCTATC
HPV-34: CCCCTCGAGTCAATCTGAATCACTGTCTGTCTC
HPV-52: AGGGTCAACACGTAGAAAGGTGGC
HPV-324: CCCGGATCCATATGGCTTCCTCGCGAGCCTCTTCC
HPV-325: CCCCTCGAGCTATTCAGCCATCCACAGCTTGAA
Yeast two-hybrid analysis
To construct plasmids for two-hybrid analysis, the corresponding ORFs or fragments thereof were amplified by PCR and introduced into pGADT7 (a GAL4 activation domain “prey” vector) or pGBKT7 (a GAL4 DNA-binding domain “bait” vector). The accuracy of each construct was confirmed by sequencing. Two-hybrid analyses were performed with the Matchmaker Two-Hybrid System 3 (Clontech). This employs the ADE2 and HIS3 reporter genes in the yeast strain AH109 to control the stringency of selection: The ADE2 gene, under the control of the GAL2 promoter, reduces the number of false positives, while the HIS3 gene, under the control of the GAL1 promoter, provides sensitive growth selection that helps to identify weak positive signals. Two-hybrid assays were carried out according to procedures described by the manufacturer. To screen for protein–protein interactions, the bait and prey plasmids were co-transformed into the yeast strain AH109 by using the lithium acetate method. Co-transformation reaction mixtures were plated onto minimal synthetic dropout (SD) medium lacking leucine and tryptophan and incubated for 3–5 d at 30°C. Co-transformants were then replicated on two different selective media: SD/−His for activation of the GAL1-HIS3 reporter gene, and SD/−His−Ade for the activation of both the GAL1-HIS3 and the GAL2-ADE2 reporter gene. Y2H positives in Figure 1 were re-examined on SD/−Leu−Trp−His medium in the presence of various concentrations of 3-AT. In each assay, empty pGADT7 and pGBKT7 vectors were used as negative controls, and pGADT7-T/pGBKT7–53 as a positive control. At least two independent co-transformants were tested for each assay.
Transcription and translation in vitro
The T7 promoter of pGADT7 and pGBKT7 is positioned immediately upstream of the epitope tag (HA or c-Myc, respectively) and the insertion site of the cloned cDNA. All constructs could therefore be used as transcription templates in transcription/translation reactions in vitro (rabbit reticulocyte TNT-System, Promega) to produce [35S]-labeled HA- or c-Myc-tagged proteins, using [35S]-methionine (1000 Ci/mmol, Amersham Pharmacia Biotech).
GST pull-down assays
The full-length hPrp6 protein was subcloned into pGEX-6P-1 (Amersham Pharmacia Biotech). Recombinant GST-hPRP6 protein was expressed in E. coli BL21 and affinity-purified on glutathione–Sepharose 4B following the manufacturer's instructions. For GST pull-down assays, lysates containing the GST-hPrp6 fusion protein were bound to glutathione–Sepharose 4B beads for 30 min at room temperature and then washed 3× with PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3) and 4× with binding buffer (20 mM HEPES pH 7.9, 100 mM KCl, 2.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 1 mM DTT, and protease inhibitor cocktail). For each reaction, 20 μL of beads (containing ∼10 μg GST-hPrp6 protein) and 10 μL of [35S]-labeled protein, prepared as described above, were incubated in 300 μL of binding buffer for 2 h at 4°C. Beads were washed 5× with binding buffer, and bound proteins were eluted in SDS sample buffer and analyzed by SDS-PAGE followed by autoradiography.
Co-immunoprecipitations
For the co-IPs shown in Figure 3C, 10 μg of His-tagged hDib1 protein (Reuter et al. 1999), together with 1.6 μg anti-pentahistidine antibodies and 25 μL protein A–Sepharose, were incubated in 400 μL IP buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Triton X-100) for 2 h at 4°C. Beads were pelleted by centrifugation and resuspended in 300 μL IP buffer. 10 μL of c-Myc-hPrp6 protein prepared by translation in vitro was added to the sample, and the incubation was continued for 2 h. The beads were subsequently washed 5× with 0.8 mL of IP buffer. Precipitated proteins were eluted in SDS sample buffer and analyzed by SDS-PAGE followed by autoradiography.
The cloning, expression, and purification of the hSnu66 protein were described previously (Makarova et al. 2001). Purified His-tagged hSnu66 protein was dialyzed against buffer containing 20 mM Tris-HCl pH 8.0, 300 mM NaCl, and 0.05% NP-40. [35S]-labeled full-length protein and fragments thereof were prepared by translation in vitro as described above. 100 μL of protein A–Sepharose beads were pre-incubated for 3 h at 4°C with 100 μL of affinity-purified anti-hSnu66 antibody (Makarova et al. 2001) in a total volume of 1 mL of phosphate-buffered saline (PBS), pH 8.0, containing 0.5 mg/mL BSA and 50 μg/mL total yeast tRNA. Beads were then washed 4× with 1 mL PBS and resuspended in 100 μL IP buffer, resulting in a final volume of 200 μL of anti-hSnu66 protein A–Sepharose beads. For each reaction, 30 μL of these beads was incubated for 2 h at 4°C in 250 μL IP buffer together with 10 μL of [35S]-labeled proteins in the presence or absence of His-tagged hSnu66 protein. Beads were then washed 4× with IP buffer, and bound proteins were analyzed as described above.
ACKNOWLEDGMENTS
We are grateful to Cindy Will for critical comments. We thank Irene Öchsner for technical assistance. This work was supported by grants from the Fonds der Chemischen Industrie, the BMBF (grant 031U215B to R.L.), and the Ernst Jung Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.55406.
REFERENCES
- Achsel T., Ahrens K., Brahms H., Teigelkamp S., Lührmann R. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 1998;18:6756–6766. doi: 10.1128/mcb.18.11.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Klatt C., Lührmann R., Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C., Urlaub H., Lührmann R., Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 2003;278:28324–28334. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- Bellare P., Kutach A.K., Rines A.K., Guthrie C., Sontheimer E.J. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch G.L., Lassle M. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Boon K.L., Norman C.M., Grainger R.J., Newman A.J., Beggs J.D. Prp8p dissection reveals domain structure and protein interaction sites. RNA. 2006;12:198–205. doi: 10.1261/rna.2281306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner T.J., Guthrie C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea L.D., Regan L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Dix I., Russell C.S., O'Keefe R.T., Newman A.J., Beggs J.D. Protein–RNA interactions in the U5 snRNP of Saccharomyces cerevisiae . RNA. 1998;4:1675–1686. doi: 10.1017/s1355838298412998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Laggerbauer B., Lauber J., Lane W.S., Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman M.E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Fetzer S., Lauber J., Will C.L., Lührmann R. The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA. 1997;3:344–355. [PMC free article] [PubMed] [Google Scholar]
- Galisson F., Legrain P. The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res. 1993;21:1555–1562. doi: 10.1093/nar/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Santos J.M., Wang A., Jones J., Ushida C., Liu J., Hu J. Central region of the human splicing factor Hprp3p interacts with Hprp4p. J. Biol. Chem. 2002;277:23764–23772. doi: 10.1074/jbc.M111461200. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Tang J., Puig O., Salgado J., Neubauer G., Colot H.V., Mann M., Séraphin B., Rosbash M., Lührmann R., et al. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Neubauer G., Banroques J., Mann M., Lührmann R., Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger R.J., Beggs J.D. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K., Urlaub H., Vornlocher H.P., Will C.L., Gentzel M., Wilm M., Lührmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D.S., Kobayashi R., Krainer A.R. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA. 1997;3:1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Gross C.H., Shuman S., Pyle A.M. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- Jankowsky E., Gross C.H., Shuman S., Pyle A.M. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Licklider L.J., Gygi S.R., Grigorieff N., Moore M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Rossi J.J. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A.N., Reichl E.M., Brow D.A. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. 2002;99:9145–9149. doi: 10.1073/pnas.102304299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B., Achsel T., Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B., Liu S., Makarov E., Vornlocher H.P., Makarova O., Ingelfinger D., Achsel T., Lührmann R. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA. 2005;11:598–608. doi: 10.1261/rna.2300805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.R., Tugendreich S., Hieter P. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem. Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Lauber J., Plessel G., Prehn S., Will C.L., Fabrizio P., Groning K., Lane W.S., Lührmann R. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA. 1997;3:926–941. [PMC free article] [PubMed] [Google Scholar]
- Luo H.R., Moreau G.A., Levin N., Moore M.J. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA. 1999;5:893–908. doi: 10.1017/s1355838299990520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov E.M., Makarova O.V., Achsel T., Lührmann R. The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J. Mol. Biol. 2000;298:567–575. doi: 10.1006/jmbi.2000.3685. [DOI] [PubMed] [Google Scholar]
- Makarov E.M., Makarova O.V., Urlaub H., Gentzel M., Will C.L., Wilm M., Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Makarova O.V., Makarov E.M., Lührmann R. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 2001;20:2553–2563. doi: 10.1093/emboj/20.10.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O.V., Makarov E.M., Liu S., Vornlocher H.P., Lührmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6.U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–1157. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T., Ishikawa K., Suyama M., Kikuno R., Miyajima N., Tanaka A., Kotani H., Nomura N., Ohara O. Prediction of the coding sequences of unidentified human genes. XI. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998;5:277–286. doi: 10.1093/dnares/5.5.277. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. RNA–RNA interactions in nuclear pre-mRNA splicing. In: Simons R.W., Grunberg-Manago M., editors. RNA structure and function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. pp. 279–307. [Google Scholar]
- Nottrott S., Urlaub H., Lührmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: A biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P. Proteins of the endoplasmic-reticulum-associated degradation pathway: Domain detection and function prediction. Biochem. J. 2000;351:527–535. [PMC free article] [PubMed] [Google Scholar]
- Preker P.J., Keller W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 1998;23:15–16. doi: 10.1016/s0968-0004(97)01156-0. [DOI] [PubMed] [Google Scholar]
- Query C.C., Konarska M.M. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol. Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- Raghunathan P.L., Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A.I., Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter K., Nottrott S., Fabrizio P., Lührmann R., Ficner R. Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein. J. Mol. Biol. 1999;294:515–525. doi: 10.1006/jmbi.1999.3258. [DOI] [PubMed] [Google Scholar]
- Reyes J.L., Kois P., Konforti B.B., Konarska M.M. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA. 1996;2:213–225. [PMC free article] [PubMed] [Google Scholar]
- Schaffert N., Hossbach M., Heintzmann R., Achsel T., Lührmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Siatecka M., Reyes J.L., Konarska M.M. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Stevens S.W., Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E.J., Guthrie C. PRP28, a “DEAD-box” protein, is required for the first step of mRNA splicing in vitro. Nucleic Acids Res. 1994;22:3187–3193. doi: 10.1093/nar/22.15.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N.K., Linder P. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S., Newman A.J., Beggs J.D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995a;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., Whittaker E., Beggs J.D. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res. 1995b;23:320–326. doi: 10.1093/nar/23.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., Achsel T., Mundt C., Gothel S.F., Cronshagen U., Lane W.S., Marahiel M., Lührmann R. The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA. 1998;4:127–141. [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Kolev N.G., Conrad N.K., Fok V., Steitz J.A. The ever-growing world of small nuclear ribonucleoproteins. In: Gesteland R.F., Cech T.R., Atkins J.F., et al., editors. The RNA world. 3d ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 327–368. [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Nues R.W., Beggs J.D. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae . Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal V.P., Verdone L., Mayes A.E., Beggs J.D. Characterization of U6 snRNA–protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I., Nottrott S., Hartmuth K., Lührmann R., Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- Vincent K., Wang Q., Jay S., Hobbs K., Rymond B.C. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics. 2003;164:895–907. doi: 10.1093/genetics/164.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. RNP remodeling with DExH/D boxes. Science. 2001;291:1916–1917. doi: 10.1126/science.1059682. [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. Spliceosome structure and function. In: Gesteland R.F., et al., editors. The RNA world. 3d ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Xu D., Nouraini S., Field D., Tang S.J., Friesen J.D. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]