Abstract
Aberrant forms of proteins ubiquitin B and β-amyloid precusor protein, UBB+1 and APP+1, are implicated in human neurodegenerative diseases. They have their carboxyl-terminal regions derived from an alternative reading frame. Transcription slippage has been invoked to explain the production of these proteins from abnormal mRNA. However, ribosomal frameshifting on wild-type mRNA may account for the great majority of the aberrant protein. Ribosomal frameshifting may also be involved in the progression of triplet expansion diseases such as Huntington's and spinocerebellar ataxias. In a particular spinocerebellar ataxia, SCA3, Toulouse and colleagues recently discovered −1 frameshifting in a transcript containing an expanded CAG-repeat. Antibiotics that affect mammalian ribosomes may have complex effects on frameshifting and disease progression.
Keywords: ribosomal frameshifting, Alzheimer's disease, molecular misreading, UBB+1, APP+1, polyglutamine disease
A broad spectrum of human neurodegenerative diseases shares strikingly similar histopathological features that may underlie their molecular pathogenesis. A common finding is the presence of insoluble proteinaceous deposits, such as the neurofibrillary tangles and neuritic plaques of Alzheimer's disease and the intranuclear inclusions of Huntington's disease. The inclusions are associated with ubiquitin and components of the proteasome, leading to the hypothesis that inhibited proteolysis is involved in disease progression (van Leeuwen et al. 2006). Alzheimer's disease is multifactorial involving a combination of genetic and nongenetic effects (Fischer et al. 2003). It has been proposed that the aberrant forms of certain proteins inhibit the function of the proteasome, leading to their accumulation. In Alzheimer's disease, aberrant forms of β-amyloid precursor protein and ubiquitin, APP+1 and UBB+1, respectively, are detected. In the literature, these proteins are called “+1 proteins” because they have carboxyl-terminal amino acids encoded by an alternate reading frame of the mRNA (e.g., Fischer et al. 2003). This terminology is unfortunate because the carboxyl-terminal amino acids derive, in fact, from the −1 reading frame as illustrated for ubiquitin B (UBB) in Figure 1.
FIGURE 1.
The C-terminal region of UBB+1 is derived from the −1 reading frame. The peptide sequence used to raise antibodies to UBB+1 is shaded. Asterisks denote termination codons in the indicated reading frame.
These “+1” proteins are routinely stated to arise from standard translation of a proportion of mRNAs that have a dinucleotide deletion due to a process termed “molecular misreading.” This process, as defined by van Leeuwen et al. (2000), appears to operate cotranscriptionally via transcriptional slippage rather than post-transcriptionally, through editing of RNA transcripts. Standard translation of the mRNA lacking 2 nucleotides (nt) present in the wild-type RNA would produce the trans-frame protein. However, the proportion of deleted mRNAs is very small (<1 in 100,000) and is no different between Alzheimer's patients and nondemented controls (Gerez et al. 2005). Although not considered previously, a more attractive possibility for production of these aberrant proteins is frameshifting by a small proportion of ribosomes translating wild-type mRNA.
RIBOSOMAL FRAMESHIFTING
Though the translational reading frame is normally maintained at high accuracy, frameshifting does occur and the frequency depends on the nature of the sequence (Atkins et al. 1972). For instance, at the sequence A_AAA_AAC (codons shown in the zero frame), −1 frameshifting occurs at ∼2% in a mammalian in vitro system (Brierley et al. 1992). (However, frameshifting measurements on various slippery sequences in vivo generally show reduced levels compared with in vitro [Parkin et al. 1992; Grentzmann et al. 1998; Kim et al. 1999]). This involves dissociation of pairing of the anticodons of both the P- and A-site tRNAs from the zero frame codons AAA and AAC, respectively, and re-pairing to mRNA at AAA and AAA in the −1 reading frame. Frameshifting involving only a single tRNA re-pairing in the −1 frame is well known (Jacks et al. 1988; Weiss et al. 1987, 1990; Licznar et al. 2003). The efficiency is generally lower than for tandem shifts and is sequence-specific. However, from extrapolation of analyses in bacteria and a mammalian example, frameshifting is expected to be substantially higher than the frequency of transcriptional slippage observed (Gerez et al. 2005) in UBB and APP.
The efficiency of frameshifting can be greatly enhanced by certain mRNA structural elements located 3′ of the shifty sequence, e.g., the stem–loop structure that stimulates −1 frameshifting at the gag-pol junction in HIV (Dulude et al. 2002; Gaudin et al. 2005; Staple and Butcher 2005). The stimulatory effects on frameshifting by other 3′ RNA structural elements are well established (Brierley and Pennell 2001; Atkins et al. 2006). RNA pseudoknots have been recently demonstrated to promote −1 frameshifting in decoding the mammalian genes edr (Manktelow et al. 2005) and paraneoplastic antigen Ma3 (Wills et al. 2006). Frameshift stimulatory RNA structures induce the ribosome to pause, but pausing alone does not account for their effect on frameshifting (Lopinski et al. 2000; Kontos et al. 2001).
If the translational model for UBB+1 is correct, frameshifting must occur between nt 175 and 227, the region between the stop codon in the −1 frame and the zero frame proteolytic cleavage site used to generate ubiquitin monomers from polyubiquitin precursor (Fig. 2A). Though classic shift-prone sequences do not occur in this region, sequences that allow for at least two base pairs between the codon and anticodon in the new frame are potential “shifty” sites. There are two such sequences within the region, G_AAG (−1 re-pairing from AAG to GAA) or AGA_GG (+2 re-pairing from AGA to AGG) (Fig.2C). In the first case, an additional 12 amino acids would differ from UBB+1 predicted by translation of the aberrant mRNA (Fig. 2B). In the second case, a single amino acid difference would result, tryptophan from +2 shifting, instead of glycine, from the aberrant mRNA (Fig. 2B,C). A similar scenario could be responsible for the production of APP+1 (Fig. 2D).
FIGURE 2.
Regions of UBB and APP mRNAs. (A) Wild-type (WT) sequence of UBB with the corresponding amino acid sequence shown above. The proteolytic cleavage site used to generate ubiquitin monomers from polyubiquitin precursor is indicated by the arrow. The GU dinucleotide that is deleted in a proportion of UBB mRNAs is boxed. Nucleotide positions (numbering from the A of the initiating AUG) are shown. A UGA stop codon in the −1 frame is shown in the black box. (B) UBB ΔGU mRNA generated by the dinucleotide deletion from wild type. The mRNA is translated in a standard fashion but yields amino acids, shaded, derived from the −1 reading frame of wild-type mRNA. The total number of amino acids encoded by the −1 frame is 20 (18 not shown). (C) Potential frameshift sites in the WT UBB mRNA are shaded. The codon pairings in the zero frame are underlined and the pairings in the −1 frame are overlined. The 5′ barrier to shifting to the −1 frame is shown in the black box. The amino acids encoded by the −1 reading frame due to either potential frameshift event are shaded. (D) Wild-type APP mRNA. Potential frameshift sites in the WT APP mRNA are shaded. The codon pairings in the zero frame are underlined and the pairings in the −1 frame are overlined. The 5′ barrier to shifting to the −1 frame is shown in the black box. The amino acids encoded by the −1 reading frame due to either potential frameshift event are shaded.
TRIPLET REPEAT EXPANSION DISEASES
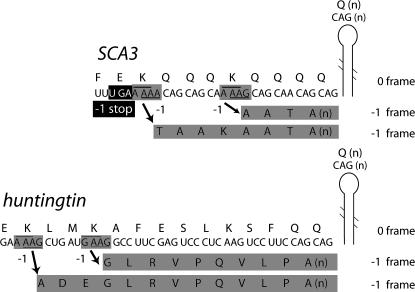
A subset of the unstable repeat expansion diseases is the polyglutamine (polyQ) group, exemplified by Huntington's disease. The polyglutamine-containing protein is found in aggregates associated with the disease, and the length of the tract is correlated with age of onset and disease severity (Gatchel and Zoghbi 2005). In another polyglutamine disease, spinocerebellar axtaxia 3 (SCA3) (also known as Machado-Joseph disease [MJD]), it has been shown that ribosomal shifting to the −1 frame occurs on transcripts containing at least 59 CAG (encoding glutamine) repeats. Frameshifting results in decoding of GCA repeats in the −1 frame yielding a trans-frame polyalanine-containing protein. The amount of trans-frame product is dependent on the length of the CAG repeat (Toulouse et al. 2005). tRNAGln with the anticodon 3′GUC5′ decodes CAG (another tRNAGln contains the anticodon 3′GUU*5′ [where U* is an unknown modification] and can decode CAA or CAG). To effect the −1 (or +2) frameshift on the run of CAGs, the 3′GUC5′ anticodon would have to realign to GCA where no Watson-Crick base pairs would be possible (re-pairing of the anticodon 3′GUU*5′ to mRNA at GCA would allow for one, third position Watson-Crick pair). Therefore, the possibilities for −1 frameshifting include peptidyl-tRNA realignment with no or only one Watson-Crick base pair in the new frame (Herr et al. 2004), noncognate frameshifting (Atkins et al. 1979), or frameshifting on a sequence 5′ of the run of CAGs. In a mutant SCA3 transcript containing an expansion to 65 CAG repeats, the CAGs form an elongated stem–loop structure (Michlewski and Krzyzosiak 2004). Since certain RNA structures are known to promote frameshifting, it is not unlikely that −1 frameshifting occurs on a shift-prone sequence(s) 5′ of the CAG repeats. Indeed, there are two potentially “shifty” sites, A_AAG 6 nt 5′ and A_AAA 18 nt 5′, of the CAG repeats in SCA3 (Fig. 3A). Similar shift-prone sequences are also found in huntingtin mRNA (Fig. 3B). (In spinocerebellar ataxia 6, there is a potential tandem shift site, G_GGG_CCC, 3 nt 5′ of the CAG repeats in the α1A-voltage-dependent calcium channel subunit [CACNA1A] mRNA [Zhuchenko et al. 1997]). Frameshifting to the −1 frame in mutant SCA3 transcripts has been shown to be dependent on the number of CAG repeats (Toulouse et al. 2005) consistent with the prediction that an elongated stem–loop formed by 59 or more CAG repeats acts as an efficient frameshift stimulator.
FIGURE 3.
Regions of the SCA3 and huntingtin CAG-expanded mRNAs. Potential frameshift sites in SCA3 and huntingtin are shaded. The pairing in the zero frame and −1 frame are underlined and overlined, repectively. The 5′ barrier to shifting to the −1 frame in SCA3 is boxed in black. The amino acids encoded by the −1 reading frame due to potential frameshift events are shaded. The RNA structure is positioned as shown to contain only uninterrupted CAG repeats that form an elongated stem–loop structure (Michlewski and Krzyzosiak 2004).
Translational frameshifting to the −1 frame (via either a −1 or a +2 shift) before the run of CAGs would produce a shortened protein containing polyalanine (due to a termination codon in the new frame). Polyalanine tracts have been implicated in nine human diseases (Brown and Brown 2004). In one of these diseases, oculopharyngeal muscular dystrophy (OPMD), a short expansion to a total of 12–17 alanines in poly(A) binding protein nuclear-1 causes inclusion formation and neurodegeneration in humans and mice (Dion et al. 2005). These findings together with the observation of −1 frameshifting in poly-CAG containing mRNAs suggest that polyalanine proteins may contribute to progression of polyglutamine diseases (Toulouse et al. 2005).
Additional or alternative roles for translational pausing due to the CAG repeat RNA structure(s) could be to promote mRNA cleavage (Doma and Parker 2006) and/or peptidyl-tRNA drop-off. N-terminal fragments of huntingtin are found in high molecular weight soluble complexes whereas N-terminal fragments of glutamine-expanded huntingtin are found as soluble monomers (Cong et al. 2005). It is certainly possible that these mutant N-terminal fragments arise from ribosomal frameshifting, mRNA cleavage and/or drop-off as opposed to, or in conjunction with, proteolytic processing. These soluble fragments of mutant huntingtin may be an important contributor to molecular pathogenesis (Schaffar et al. 2004).
CONCERNS ABOUT ANTIBIOTIC EFFECTS
The subtle effects on mammalian ribosomes of certain antibiotics that enhance frameshifting by targeting various regions of the ribosome merit further exploration. Minor effects on mammalian protein synthesis, in general, can be of little consequence compared with the large rewards from antibiotics that block protein synthesis of pathogenic bacteria, since a large battery of proteases would rapidly destroy most proteins arising from frameshifting. However, an increase in the amounts of a small minority of trans-frame proteins, whether aberrant or normally utilized, may be far from inconsequential.
The ribosomal P-site plays a key role in mediating frameshifting. The antibiotics, sparsomycin and anisomycin, that affect the P-site also influence frameshifting (Dinman et al. 1997). Anisomycin, an inhibitor of the accommodation of the frameshifted tRNA to the codon in the −1 frame, reduced frameshifting on SCA3 mRNA while sparsomycin, that slows peptidyl transfer allowing time for tRNA realignment, increased frameshifting (Toulouse et al. 2005). However, aminoglycosides, such as clinically utilized gentamycin, that affect tRNA acceptance in the ribosomal A-site, have complex effects that may be relevant to trans-frame disease progression. By reducing the fidelity of acceptance of an incoming tRNA, aminoglycosides can reduce the length of time taken for the ribosomal A-site to accept a tRNA (Pape et al. 2000). This would mean there is less time for dissociation of the pairing of the anticodon of peptidyl-tRNA and realigning with, and re-pairing to, mRNA. Hence, a reduction in frameshifting would occur at certain sites. However, the other consequence of increased acceptance of near-cognate tRNA is that, when such a tRNA enters the ribosomal P-site, it will be more prone to dissociate—a prelude to realignment and re-pairing to mRNA in a new frame (Farabaugh and Björk 1999; Baranov et al. 2004). Hence, increased frameshifting would occur at a subset of sites. At other sites, these contrasting effects may balance each other out and there would be no net effect on frameshifting levels. One illustration of an aminoglycoside causing increased frameshifting at a specific site comes from a study of Clostridium perfringens containing a frameshift mutation in its beta2 toxin gene. Infected horses treated with gentamycin had an increased amount of the toxin gene product synthesized by the bacteria, worsening the symptoms rather than alleviating them (Vilei et al. 2005).
In the translational model proposed, compounds that reduce frameshifting could be beneficial. However, there have been very few studies of antibiotic-like compounds (Irvine et al. 1998) that parallel certain ribosomal rRNA mutations that decrease frameshifting (Weiss-Brummer et al. 1995).
If the frameshifting model proposed is correct, then treatment with certain antibiotics could either exacerbate or delay onset of a variety of neurodegenerative diseases.
PERSPECTIVE
Earlier work showed that translational frameshifting and transcriptional slippage are alternatives used in decoding dnaX polymerase subunit genes in Escherichia coli and Thermus thermophilus, respectively (Larsen et al. 2000). In both cases the product of nonstandard expression is in a 1:1 ratio with the product of standard expression, and both have distinct functions as polymerase subunits. In contrast, for the synthesis of UBB+1 and APP+1, both processes are likely involved. Transcriptional slippage was demonstrated previously, but we propose that translational frameshifting is likely the major contributor to the synthesis of these trans-frame proteins. In addition, consideration of the potential of frameshift sites 5′ of expanded CAG repeats and the potential for clinically utilized antibiotics to affect consequential frameshifting in humans are topics that merit further investigation.
ACKNOWLEDGMENTS
We thank Raymond Gesteland, Pavel Baranov, Michael Howard, and Tara Atkins for discussions. This work was supported by NIH grant GM71853 (to R.F. Gesteland). J.F.A. was supported by an award from Science Foundation Ireland.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.84406.
REFERENCES
- Atkins J.F., Elseviers D., Gorini L. Low activity of β-galactosidase in frameshift mutants of Escherichia coli . Proc. Natl. Acad. Sci. 1972;69:1192–1195. doi: 10.1073/pnas.69.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J.F., Gesteland R.F., Reid B.R., Anderson C.W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979;18:1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Atkins J.F., Gesteland R.F., Jackson R.J., Wills N.M. The shapely mRNA; Knotting ventured, knotting gained. In: Gesteland R.F., et al., editors. The RNA world. 3d ed. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2006. pp. 437–467. [Google Scholar]
- Baranov P.V., Gesteland R.F., Atkins J.F. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L.Y., Brown S.A. Alanine tracts: The expanding story of human illness and trinucleotide repeats. Trends Genet. 2004;20:51–58. doi: 10.1016/j.tig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Cong S.Y., Pepers B.A., Roos R.A.C., van Ommen G.J.B., Dorsman J.C. Small N-terminal mutant huntingtin fragments, but not wild type, are mainly present in monomeric form: Implications for pathogenesis. Exp. Neurol. 2005 doi: 10.1016/j.expneurol.2005.11.008. (doi:10.1016/j.expneurol.2005.11.008) [DOI] [PubMed] [Google Scholar]
- Dinman J.D., Ruiz-Echevarria M.J., Czaplinski K., Peltz S.W. Peptidyl-transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: Development of model systems. Proc. Natl. Acad. Sci. 1997;94:6606–6611. doi: 10.1073/pnas.94.13.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion P., Shanmugam V., Gaspar C., Messaed C., Meijer I., Toulouse A., Laganiere J., Roussel J., Rochefort D., Laganiere S., et al. Transgenic expression of an expanded (GCG)13 repeat PABPN1 leads to weakness and coordination defects in mice. Neurobiol. Dis. 2005;18:528–536. doi: 10.1016/j.nbd.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Doma M.K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulude D., Baril M., Brakier-Gingras L. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 2002;30:5094–5102. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J., Björk G.R. How translational accuracy influences reading frame maintenance. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D.F., de Vos R.A.I., van Dijk R., de Vrij F.M.S., Proper E.A., Sonnemans M.A.F., Verhage M.C., Sluijs J.A., Hobo B., Zouambia M., et al. Disease-specific accumulation of mutant ubiquitin as a marker for proteasomal dysfunction in the brain. FASEB J. 2003;17:2014–2024. doi: 10.1096/fj.03-0205com. [DOI] [PubMed] [Google Scholar]
- Gatchel J.R., Zoghbi H.Y. Diseases of unstable repeat expansion: mechanisms and common practices. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- Gaudin C., Mazauric M.H., Traikia M., Guittet E., Yoshizawa S., Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- Gerez L., de Haan A., Hol E.M., Fischer D.F., van Leeuwen F.W., van Steeg H., Benne R. Molecular misreading: The frequency of dinucleotide deletions in neuronal mRNAs for β-amyloid precursor protein and ubiquitin B. Neurobiol. Aging. 2005;26:145–155. doi: 10.1016/j.neurobiolaging.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Herr A.J., Wills N.M., Nelson C.C., Gesteland R.F., Atkins J.F. Factors that influence selection of coding resumption sites in translational bypassing: minimal conventional peptidyl-tRNA:mRNA pairing can suffice. J. Biol. Chem. 2004;279:11081–11087. doi: 10.1074/jbc.M311491200. [DOI] [PubMed] [Google Scholar]
- Irvine J.H., Horsfield J.A., McKinney C.Z., Tate W.P. A novel strategy to interfere with human immunodeficiency virus type 1 propagation. N.Z. Med. J. 1998;111:222–224. [PubMed] [Google Scholar]
- Jacks T., Power M.D., Masiarz F.R., Luciw P.A., Barr P.J., Varmus H.E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Kim Y.G., Su L., Maas S., O'Neill A., Rich A. Specific mutations in a viral RNA pseudoknot drastisically change ribosomal frameshifting efficiency. Proc. Natl. Acad. Sci. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos H., Napthine S., Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Wills N.M., Nelson C., Atkins J.F., Gesteland R.F. Nonlinearity in genetic decoding: Homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl. Acad. Sci. 2000;97:1683–1688. doi: 10.1073/pnas.97.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznar P., Mejlhede N., Prère M.F., Wills N.M., Gesteland R.F., Atkins J.F., Fayet O. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 2003;22:4770–4778. doi: 10.1093/emboj/cdg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopinski J.D., Dinman J.D., Bruenn J.A. Kinetics of ribosomal pausing during programmed −1 translational frameshifting. Mol. Cell. Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manktelow E., Shigemoto K., Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 2005;33:1553–1563. doi: 10.1093/nar/gki299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G., Krzyzosiak W.J. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Pape T., Wintermeyer W., Rodnina M.V. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- Parkin N.T., Chamorro M., Varmus H.E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: Demonstration by expression in vivo . J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F.U. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol. Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Staple D.W., Butcher S.E. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Toulouse A., Au-Yeung F., Gaspar C., Roussel J., Dion P., Rouleau G.A. Ribosomal frameshifting on MJD-1 transcripts with long CAG tracts. Hum. Mol. Genet. 2005;14:2649–2660. doi: 10.1093/hmg/ddi299. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F.W., Fischer D.F., Benne R., Hol E.M. Molecular misreading: a new type of transcript mutation in gerontology. Ann. N.Y. Acad. Sci. 2000;908:267–281. doi: 10.1111/j.1749-6632.2000.tb06654.x. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F.W., van Tijn P., Sonnemans M.A.F., Hobo B., Mann D.M.A., Van Broeckhoven C., Kumar-Singh S., Cras P., Leuba G., Savioz A., et al. Frameshift proteins in autosomal dominant forms of Alzheimer disease and other tauopathies. Neurology. 2006;66:S86–S92. doi: 10.1212/01.wnl.0000193882.46003.6d. [DOI] [PubMed] [Google Scholar]
- Vilei E.M., Schlatter Y., Perreten V., Straub R., Popoff M.R., Gibert M., Grone A., Frey J. Antibiotic-induced expression of a cryptic cpb2 gene in equine beta2-toxigenic Clostridium perfringens . Mol. Microbiol. 2005;57:1570–1581. doi: 10.1111/j.1365-2958.2005.04789.x. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn D.M., Atkins J.F., Gesteland R.F. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn D.M., Atkins J.F., Gesteland R.F. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Zollner A., Haid A., Thompson S. Mutation of a highly conserved base in the yeast mitochondrial 21S rRNA restricts ribosomal frameshifting. Mol. Gen. Genet. 1995;248:207–216. doi: 10.1007/BF02190802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills N.M., Moore B., Hammer A., Gesteland R.F., Atkins J.F. A functional −1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J. Biol. Chem. 2006;281:7082–7088. doi: 10.1074/jbc.M511629200. [DOI] [PubMed] [Google Scholar]
- Zhuchenko O., Bailey J., Bonnen P., Ashizawa T., Stockton D.W., Amos C., Dobyns W.B., Subramony S.H., Zoghbi H.Y., Lee C.C. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the α 1A-voltage-dependent calcium channel. Nat. Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]