Abstract
In eukaryotic cells, mRNA localization can provide local protein synthesis. Metallothionein-1 (MT-1) mRNA is associated with the perinuclear cytoskeleton, and this is essential for subsequent nuclear import of the protein. The present study defines the cis-acting localization signal and a trans-acting binding protein. Gel retardation and UV cross-linking assays using MT-1 3′UTR transcripts and CHO cell extracts revealed formation of a complex containing a ∼50-kDa protein. Only localization-positive mutant transcripts competed for binding of this protein. Using an RNA affinity technique, Western blotting, mass spectrometry, and a supershift assay, the protein was identified as Elongation factor 1α (eEF1α). Mutation and deletion analysis showed that two regions, nucleotides 21–36 and 66–76, were required for both binding and localization. RNA-folding prediction combined with chemical and enzymatic probing experiments suggest that these regions are in juxtaposition within a stem/internal loop structure. Mutations that are predicted to alter this structure abrogate protein binding. Our hypothesis is that the cis-acting signal in MT-1 3′UTR is formed by this stem/internal loop, that it binds eEF1α, and that eEF1α–cytoskeleton interactions play a role in perinuclear mRNA localization.
Keywords: mRNA localization, metallothionein-1, 3′untranslated region, elongation factor 1α, RNA binding protein
INTRODUCTION
In eukaryotic cells the tightly regulated distribution of specific mRNA species to different regions of the cytoplasm provides a mechanism for localized synthesis of proteins close to where they are required (Hesketh 1994; Oleynikov and Singer 1998; Jansen 2001; Carson and Barbarese 2005). For example, in neurons and oligodendroglia, some mRNAs are transported over considerable subcellular distances (Shan et al. 2003), and in fibroblasts, transcripts encoding β-actin and creatine kinase isoform M are transported to the cell periphery (Kislauskis et al. 1993; Wilson et al. 1995), whereas, in contrast, c-myc, c-fos, Metallothionein-1 (MT-1), and vimentin mRNAs remain localized to the perinuclear cytoplasm (Hesketh et al. 1994; Mahon et al. 1997; Zehner et al. 1997; Dalgleish et al. 2001). The mRNAs localized around the nucleus, including MT-1 mRNA, are associated with cytoskeletal-bound polysomes or the cytoskeleton itself (Hesketh et al. 1991; Mahon et al. 1997; Dalgleish et al. 2001). In the case of MT-1, perinuclear localization of the mRNA has been shown to be required for subsequent import of the protein into the nucleus during the G1/S transition and for the ability of MT-1 to protect cells from DNA damage (Levadoux et al. 1999; Levadoux-Martin et al. 2001).
In the majority of cases, localization of mRNAs is due to cis-acting signals within the 3′untranslated regions (3′UTR) (Oleynikov and Singer 1998; Kwon et al. 1999; Chabanon et al. 2004). It is envisaged that in addition to such a cis-acting signal a trans-acting factor(s) binding to these localization elements is also required for localization (Oleynikov and Singer 1998; Jansen 2001; Chabanon et al. 2004); together, the cis-acting RNA localization element and the trans-acting binding protein would form the basis of a complex that determines localization. In the cases of mRNAs that demonstrate a perinuclear localization, the precise nature of the cis-acting signal within the 3′UTR has not been defined. However, in respect to MT-1 mRNA, it has been established by in situ hybridization of cells expressing a series of deletion constructs that the region of the 3′UTR between nucleotides 45 and 76 is required for localization, and that deletion of nt 66–76 abrogates localization, whereas in contrast, deletion of nt 76–86 has little effect (Fig. 1; Nury et al. 2005).
The trans-acting factor that binds to the MT-1 mRNA localization element has not been previously identified. Since MT-1 mRNA is associated with cytoskeletal-bound polysomes (Mahon et al. 1997), this factor would be required, either directly or indirectly, to be associated with the cytoskeleton so that it could anchor the MT-1 mRNA in the perinuclear cytoplasm. The aims of the present work were to investigate the protein-binding properties of transcripts corresponding to the MT-1 3′UTR, to identify the ∼50-kDa protein that demonstrated specific binding, and to describe the region of the 3′UTR required for binding as well as its relationship with the cis-acting RNA localization signal.
RESULTS
Cellular proteins form complexes with the MT-1 3′UTR
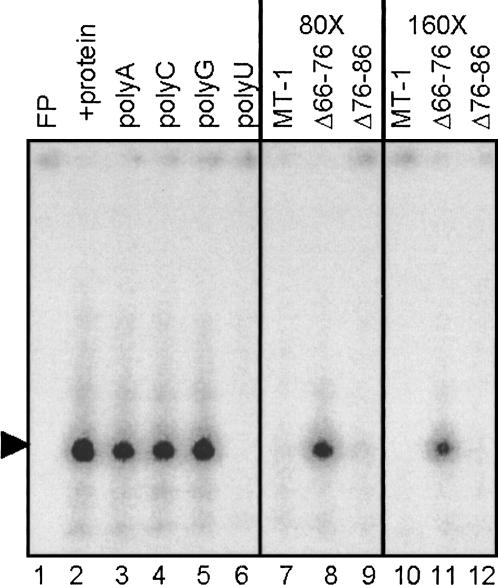
Perinuclear localization of the rat MT-1 mRNA requires a signal within the 3′UTR (Mahon et al. 1997), but the whole 3′UTR is not required, and the first 86 nt are sufficient for localization (Nury et al. 2005). To establish whether cellular proteins bind to the 3′UTR of MT-1 mRNA, electrophoretic mobility shift assays (EMSA) were performed using S100 extracts from CHO cells and a radiolabeled probe corresponding to nt 1–111 of the 3′UTR that excluded the polyadenylation signal. Following RNase T1 digestion, ribonucleoprotein complexes were separated by native PAGE, and this revealed the presence of one major complex (Fig. 2, lane 2). MT-1 3′UTR transcripts with specific deletions were utilized as competitors to determine the specificity of complex formation and whether it is linked to perinuclear localization. Both wild-type transcripts and transcripts with nt 76–86 deleted (a deletion that does not alter the localization) (Nury et al. 2005) competed effectively for complex formation at 80- and 160-fold molar excesses (Fig. 2, lanes 7,9,10,12). In contrast, transcripts with nt 66–76 deleted, which are not localized (Nury et al. 2005), were unable to compete for binding of the complexes (Fig. 2, lanes 8,11). When homoribopolymers were added to the binding reaction as competitors, only polyU competed with the probe for binding of these complexes (Fig. 2, lanes 3–6). This competition of polyU for the complex in the EMSA is somewhat surprising because the MT-1 3′UTR is not especially U-rich (27%). It may be that the protein or proteins bind to U residues in this sequence or that polyU disrupts the structure of the 3′UTR when added to the binding reaction.
FIGURE 2.
RNA–protein complex formation monitored by gel retardation assay. Complex formation was studied using [α-32P]CTP-labeled MT-1 3′UTR RNA (12 fmol) and 2 μg S100 extract protein from CHO cells and prepared as described in Materials and Methods. RNase T1 digestion was performed after the binding reaction and products separated by native PAGE. Lane 1 contains MT-1 3′UTR RNA alone (free probe; FP) and lane 2 shows formation of retardation complexes in the presence of cell extract. The effects of competition with a 100-fold mass excess of homoribopolymers polyA, C, G, and U are shown in lanes 3–6, respectively. Poly U dramatically reduces the major gel retardation complex. In lanes 7–12, either an 80- or 160-fold molar excess of MT-1, Δ66–76, and Δ76–86 unlabeled RNAs was used to compete with MT-1 RNA for protein binding. Note that MT-1 and Δ76–86 compete more effectively than Δ66–76.
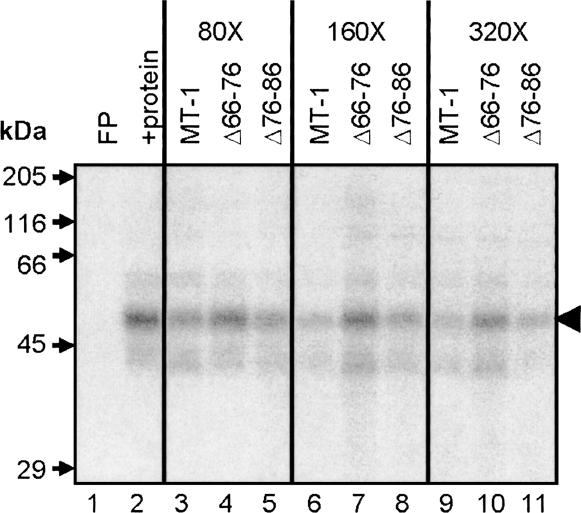
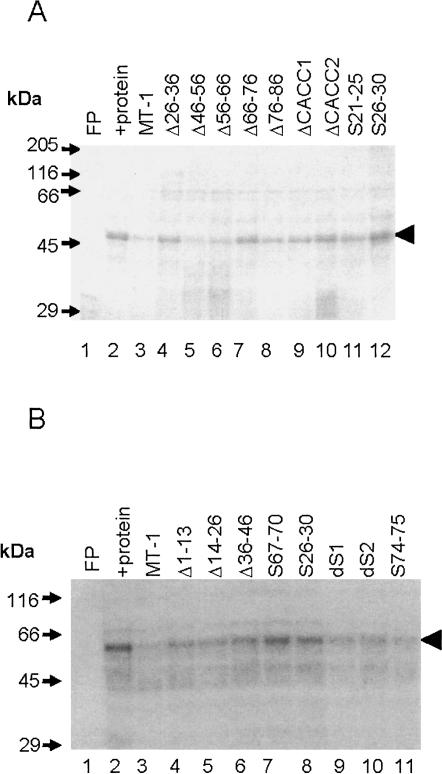
To investigate the nature of the protein(s) forming the ribonucleoprotein complexes, UV cross-linking assays were performed. Separation of labeled proteins by SDS-PAGE revealed a major band of ∼50 kDa bound to the MT-1 3′UTR (Fig. 3, lane 2). Competitive UV cross-linking showed that wild-type MT-1 transcripts and transcripts with nt 76–86 deleted competed very effectively for binding of the ∼50-kDa protein, but that transcripts with nt 66–76 deleted only competed to a very small extent. These data implicate the ∼50-kDa protein as forming a specific complex with the MT-1 3′UTR. Furthermore, the correlation of the effects of these deletions on binding, as assessed by both EMSA and UV cross-linking with effects on localization (Nury et al. 2005), suggests that the binding of the ∼50-kDa protein is specifically involved in perinuclear localization.
FIGURE 3.
UV cross-linking analysis of proteins binding to MT-1 3′UTR. An [α-32P] CTP-labeled MT-1 3′UTR RNA probe (50 fmol) was incubated with 35 μg protein (CHO cell S100 extract), prepared as described in Materials and Methods. Complexes were fixed by UV irradiation and the probe was removed by RNase A digestion. Labeled proteins were separated by SDS-PAGE and detected by autoradiography. Lane 1 contains MT-1 3′UTR RNA alone (free probe; FP) and lane 2 shows proteins labeled by the MT-1 3′UTR probe after incubation with S100 extract. Labeled RNA binds to a major band of ∼50 kDa and weaker bands of ∼35–40 and 60–65 kDa. Binding to the protein indicated by the black arrowhead (∼50 kDa) is competed for effectively by a 80–320 molar excess of unlabeled competitor MT-1 3′UTR and Δ76–86 RNAs, but markedly less so by Δ66–76 (lanes 3–11).
Identification of the ∼50-kDa protein binding to MT-1 3′UTR
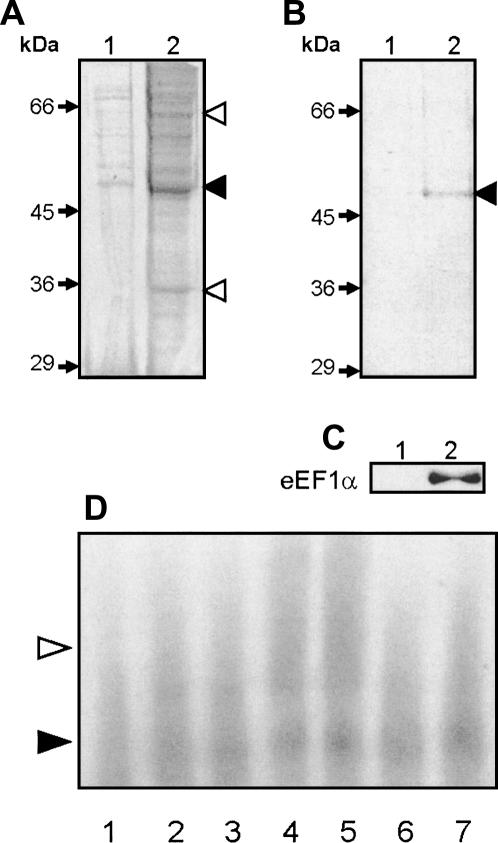
Streptavidin-coated paramagnetic beads linked to MT-1 3′UTR transcripts with incorporated biotin-16-UTP were used to pull out proteins binding to the MT-1 3′UTR. Beads were incubated with CHO cell S100 extract and, after removing unbound proteins by extensive washing, proteins were eluted from the beads by boiling in dissociation buffer and subjected to SDS-PAGE. Two negative controls using beads with either no RNA attached or with beads linked to RNA corresponding to the β-globin sequences were carried out in parallel. As shown in Figure 4A, a number of proteins bound to the beads linked to RNA corresponding to the MT-1 3′UTR (lane 2), but when compared with the negative control with no RNA (lane 1), the MT-1 3′UTR bound significantly more of a ∼50-kDa protein, as well as proteins of ∼35–40 kDa and ∼60–65 kDa. Using binding buffer containing 100 μM calcium chloride and including a postincubation step with 10 mg/mL heparin to reduce nonspecific binding of proteins to RNA, the number of proteins recovered in the eluate from the beads linked to MT-1 3′UTR (Fig. 4B) was drastically reduced: essentially, only one protein was bound to the MT-1 3′UTR under these conditions (lane 2), a protein of ∼50 kDa. This protein was absent in the eluate from control beads (lane 1) linked to RNA corresponding to the β-globin sequences known not to contain a perinuclear localization signal (Hesketh et al. 1994; Mahon et al. 1997; Dalgleish et al. 2001).
FIGURE 4.
Identification of the ∼50-kDa protein binding to MT-1 3′UTR. (A–C) Data from an RNA-affinity experiment in which proteins from a CHO S100 extract (1 mg) were incubated with biotinylated MT-1 3′UTR RNA anchored to SA-PMP (see Materials and Methods) and eluted proteins separated by SDS-PAGE. (A) A gel stained with Coomassie Brilliant Blue in which lane 1 shows the eluate from no-RNA negative control beads, and lane 2 contains proteins eluted from beads conjugated to MT-1 3′UTR RNA. The black arrowhead points to the major ∼50-kDa band and white arrowheads indicate bands of 35–40 and 60–65 kDa in the MT-1 3′UTR eluate that are reduced or absent in the negative eluate. In B, 100 μM CaCl2 was added to the protein binding reaction, and a 15-min postincubation in which heparin was added to a final concentration of 10 mg/mL was performed. Lane 2 shows the eluate from beads conjugated to MT-1 3′UTR, compared to that from β-globin 3′UTR RNA (lane 1). The black arrowhead indicates the ∼50-kDa band present in the MT-1 3′UTR eluate. In C, eluates from beads conjugated to either MT-1 3′UTR RNA (lane 2) or β-globin 3′UTR RNA (lane 1) were subjected to Western blotting using 0.5 μg/mL anti-eEF1α antibody and chemiluminescence detection. In D, binding reactions were carried out using [α-32P]CTP-labeled MT-1 3′UTR RNA (12 fmol) and 2 μg S100 protein (from CHO cells, prepared as described in Materials and Methods). For supershift reactions protein extract was preincubated with 8 μg IgG. Complexes were separated by native PAGE. Lane 1 contains labeled MT-1 3′UTR transcripts alone (free probe); lanes 2–7 transcripts are incubated with S100 extract either in the absence of added antibodies (lanes 2,3), or in the presence of 0.5 μg anti-eEF1α antibodies (lanes 4,5) or anti-biotin antibodies (lanes 6,7). Formation of an RNP complex with S100 extract is indicated by the black arrowhead (lanes 2,3), and the formation of a larger complex (supershift) with anti-eEF1α antibodies (lanes 4,5) is indicated by the white arrowhead. There was no apparent supershift by antibiotin antibodies (lanes 6,7).
To identify the ∼50-kDa protein, the Coomassie-stained band was extracted from the gel, digested with trypsin, and subjected to MALDI-TOF mass spectrometry. Comparison of six peptides, constituting 21% of the total amino acids in the resultant tryptic peptide mass fingerprint, with known rodent proteins in the Mascot database (excluding those from mice and rats) identified the protein as Chinese hamster eukaryote elongation factor 1α (eEF1α) with a Mowse score of 72 (P = 0.00037). eEF1α has a molecular mass of 50.08 kDa. Western blotting with an anti-eEF1α antibody confirmed both that the single protein bound by MT-1 3′UTR under these conditions was eEF1α (Fig. 4C, lane 2) and that there was no evidence of eEF1α binding to a control β-globin sequence (lane 1). Gel retardation experiments with cell extracts incubated with anti-eEF1α antibodies showed that such antibodies caused increased retardation of the complex or “supershift” (Fig. 4D), but that this did not occur with a comparable concentration of a control IgG. This “supershift” with anti-eEF1α antibodies provides further evidence of specificity of complex formation between eEF1α and the MT-1 3′UTR.
Structural analysis of MT-1 3′UTR
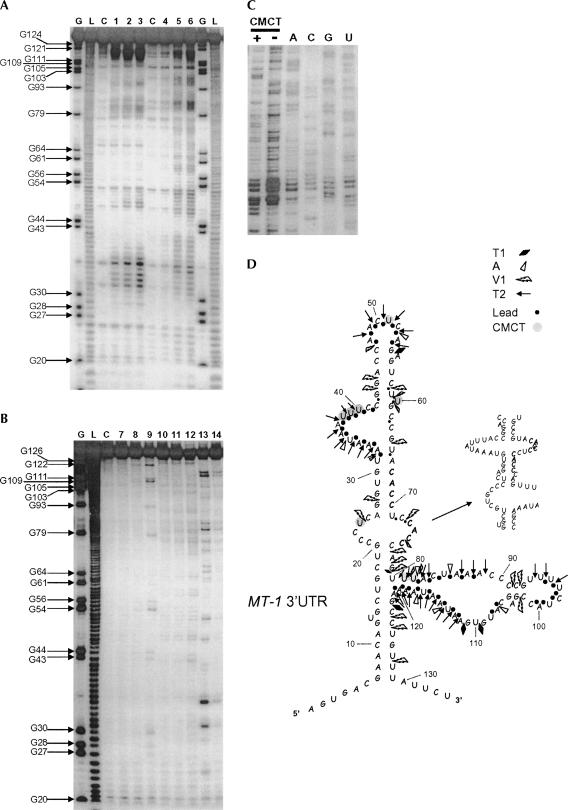
Nucleotides 66–76 in the MT-1 3′UTR are required for binding of eEF1α (Fig. 2) and localization (Nury et al. 2005). The sequence of this region is ACACCUCCACC, containing a CACC repeat. A recent bioinformatic analysis (Betley et al. 2002) has suggested that CAC repeats may be a common feature of localization signals, particularly in mRNAs localized in Xenopus oocytes but also including that of β-actin mRNA. The ACACCUCCACC sequence is sufficient to drive localization of reporter transcripts to the perinuclear cytoplasm but not as effective as the full MT-1 3′UTR (Nury et al. 2005), suggesting that the structural context of this region is important. In order to determine how the MT-1 3′UTR folds in solution, enzymatic and chemical probing of MT-1 3′UTR was carried out. Representative gels are shown in Figure 5A,B, and C. Compilation of the cleavage data obtained from this analysis together with the predicted folding of this region by the Mfold program produced the proposed secondary structure for MT-1 3′UTR shown in Figure 5D.
FIGURE 5.
(Legend on next page)
In this model, several clear single-stranded regions were revealed by cleavage patterns obtained with either RNase T2 or lead (Fig. 5A,D): they correspond to a bulge formed by the residues A33 to C41, an apical loop formed by residues A48 to A53, and most of a large region encompassing the nt U81–A121. Major cuts obtained with the RNases T1 (specific cleavage 3′ to Gs in single-strand regions) and A (specific for Us and Cs in single-strand regions) also occurred inside or close to these regions (Fig. 5B,D). In contrast, with the exceptions of C91, C92, and C102, most of the residues sensitive to the double-stranded-specific RNase V1 appeared outside the regions delineated by the single-stranded region-specific probes (other ribonucleases and lead). Such cuts (C123, C124, and U127; G43, G44, and C58; Fig. 5B) define basal and apical stem regions within the structure (Fig. 5D). Both stems on each side of the central internal loop containing the second CACC (nt 73–76) motif also present cleavage points for the RNase V1. Cleavage with V1 also suggests some flexibility in the structure of this internal stem–loop region such that either the first CACC of the CACC repeat within nt 66–76 is in a stem and the second is in an internal loop or vice versa. Chemical probing with CMCT (reacts mainly at N3-U and to a weaker extent at N1-G) showed a lack of reactivity of U29, U57, U59, U65, and U71, consistent with an upper stem containing the first CACC motif. Overall, the probing analysis suggests that the MT-1 3′UTR folds into a complex structure with various stem and loop regions, and that within this structure the regions 20–30 and 66–76 are in close juxtaposition forming a stem and internal loop.
Mutational analysis of the 3′UTR region required for protein binding
The region of the 3′UTR required for binding of eEF1α was investigated by screening a range of mutants (see Fig. 1) for binding activity using competitive UV cross-linking assays. As shown in Figure 6, deletion of either nt 26–36 or either of the CACC motifs present within the 66–76 region, but particularly the second CACC comprising nt 73–76, produced transcripts that competed very poorly in UV cross-linking assays, i.e., did not bind to eEF1α. Deletion of nt 1–13, 14–26, or 36–46 produced transcripts that retained some binding activity, and deletion of nt 46–56, 56–66, or 76–86 produced transcripts that maintained the ability to compete with wild-type MT-1 3′UTR transcripts for complex formation, indicating that these deletions had little effect on protein binding.
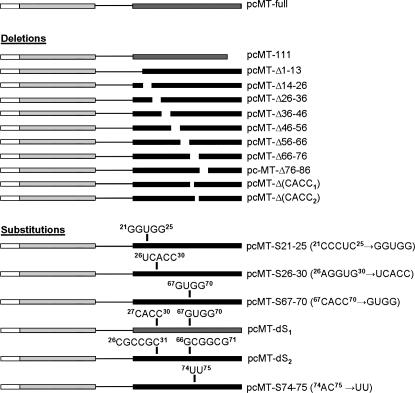
FIGURE 1.
Details of gene constructs. pcMT-full contained the rat MT-1 coding region and 3′UTR (Nury et al. 2005). pcMT-111 was made by PCR cloning (see Materials and Methods). Site-directed mutagenesis was used to delete successive 11–13-nucleotide regions of the MT-1 3′UTR to create pcMT-Δ1–13, -Δ14–26, -Δ26–36, -Δ36–46, -Δ46–56, -Δ56–66, -Δ66–76 (Nury et al. 2005), and -Δ76–86 (Nury et al. 2005), as well as nt 67–70 (first CACC motif) and 73–76 (second CACC motif) to produce pcMT-Δ(CACC1) and pcMT-Δ(CACC2), respectively. Site-directed mutagenesis was also used for substitutions of nt 21–25 (Nury et al. 2005), 26–30 (Nury et al. 2005), 67–70, and 74–75 of MT-1 3′UTR so as to create pcMT-S21–25, -S26–30, -S67–70, and -S74–75, respectively. Two double substitutions were made in which either both nt 27–30 and 67–70 or nt 26–31 and 66–71 were mutated. Details of all the sequence substitutions are shown in the figure.
FIGURE 6.
UV cross-linking analysis of the effects of deletions and base substitutions on protein binding to MT-1 3′UTR. An [α-32P] CTP-labeled MT-1 3′UTR RNA probe (50 fmol) was incubated with 35 μg protein (CHO cell S100 extract) prepared as described in Materials and Methods and 160× molar excess of unlabeled competitor transcripts. Complexes were fixed by UV irradiation, and the probe was removed by RNase A digestion. Labeled proteins were separated by SDS-PAGE and detected by autoradiography (A,B). In the absence of protein there was no binding detected (free probe; FP). Note the major labeled band of ∼50 kDa (arrowhead) after incubation of the probe with protein extract (+protein). Binding to this protein is effectively competed by a 160× molar excess of unlabeled competitor MT-1 3′UTR (A, B, lane 3) but markedly less so by Δ66–76, Δ26–36, ΔCACC1, ΔCACC2, S21–25, S26–30 (A, lanes 4,7,9–12), S67–70, and S26–30 (B, lanes 7,8) (A, B; transcript abbreviations as shown in Fig. 1). Transcripts Δ46–56, Δ56–66, Δ76–86 (A, lanes 5,6,8) dS1, dS2, and S74–75 (B, lanes 9–11) competed effectively for protein binding.
Substitutions that were predicted to destroy the stem formed between nt 26–30 and 66–70 (S26–30, S67–70) caused a drastic reduction in binding to eEF1α, as judged by competitive UV cross-linking assays (Fig. 6B). Two double substitutions were made in which either both nt 26–31 and 66–71 or nt 27–30 and 67–70 were mutated so that a stem structure was reformed. These transcripts (dS1 and dS2) competed considerably more effectively for binding than S26–30 or S67–70 (Fig. 6), and this was the case whether the CACC sequence had been altered (dS2) or not (dS1). Substitution of the CCCUC of nt 21–25 by GGUGG (S21–25) is predicted to replace the internal loop with a stem. Transcripts with this mutation were also found to reduce binding, while transcripts with a substitution of nt 74–75, predicted to alter the second CACC sequence but maintain the internal loop (S74–75), showed strong binding. Overall, the data from these deletion and substitution mutants suggest that nt 21–30 and 66–76 are critical for binding, and that substitutions which are predicted to alter base-pairing between these regions (Fig. 5) modulate protein binding.
Loss of binding is associated with loss of localization
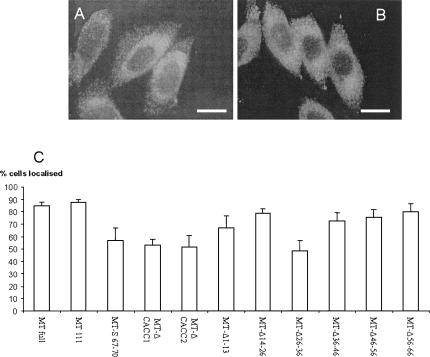
The effects of a range of deletions and substitutions on transcript localization was determined in CHO cells transfected with gene constructs in which the MT-1 coding region was linked to the full 3′UTR containing appropriate mutations. These cells, which express undetectable levels of endogenous MT-1 (Levadoux et al. 1999), were subjected to immunocytochemistry to assess MT-1 protein distribution. Since MT-1 protein localization has been shown to be dependent upon mRNA localization (Levadoux et al. 1999; Levadoux-Martin et al. 2001; Nury et al. 2005), protein distribution, as assessed by immunocytochemistry, is a reliable indirect measure of mRNA localization. As shown in Figure 7, quantification of the MT-1 protein distribution showed that in cells transfected with constructs containing either the full MT-1 3′UTR or nt 1–111, >80% of the cells exhibited the characteristic ring of perinuclear MT-1. Deletion of nt 1–13, 14–26, 36–46, 46–56, or 56–66 had little or no effect on localization. In contrast, deletion of nt 26–36, deletion of either of the CACC motifs within nt 66–76, or the substitution S67–70 caused a marked loss of localization so that only ∼50% of the cells showed a perinuclear ring of MT-1. A comparable approach to quantification of mRNA localization has previously found localization in a limited proportion of cells during deletion and mutation analysis of the β-actin 3′UTR (Kislauskis et al. 1994). Such effects are probably due to a partly effective or weak signal directing mRNA localization in some cells but not others, depending on the cells' precise physiological state. Therefore, the present data suggest that deletion of nt 26–36, deletion of either of the CACC motifs within nt 66–76, or substitution S67–70 all alter critical features of the perinuclear localization signal present in the MT-1 3′UTR such that it is considerably less efficient.
FIGURE 7.
Effects of deletions and substitutions in the 3′UTR on localization of MT-1. Results show immunostaining of cells after incubation with anti-XPress antibody and FITC-conjugated goat anti-mouse secondary IgG. Examples are shown of (A) cells exhibiting a distinct perinuclear localization of MT-1 (CHO cells expressing pcMT-111), and (B) CHO cells showing a diffuse, nonlocalized MT-1 distribution (cells expressing pcMT-Δ26–36). Bars represent 10 μm. (C) The staining pattern was quantified in cells transfected with a range of constructs by examining at least 100 cells for perinuclear localization of MT-1 and scored as showing localization or not. Mean % of cells exhibiting localization ± SEM are shown from at least three separate experiments.
DISCUSSION
The data from EMSA and UV cross-linking experiments both show that proteins in a CHO cell extract form a complex with RNA transcripts corresponding to the MT-1 3′UTR. Furthermore, a protein of ∼50 kDa is a significant component of this complex. Competitive binding studies with transcripts containing specific deletions and substitutions showed that mutations which lower binding (S21–25, S26–30; and Δ26–36, S67–70, ΔCACC1, and ΔCACC2; Nury et al. 2005; Fig. 7) had the most effect on localization of MT-1 in transfected CHO cells, suggesting that this protein binding functions in perinuclear localization.
The ∼50-kDa protein present in complexes with MT-1 transcripts was identified as eEF1α by demonstration of a supershift with anti-eEF1α antibodies in EMSA assays and by elution from MT-1 3′UTR-bound beads followed by Western blotting and MALDI-TOF mass spectrometry. Although originally characterized as the translation factor catalyzing the GTP-dependent aminoacyl-tRNA binding to ribosomes during elongation, eEF1α has been reported to bind specifically both to a stem–loop structure within the 3′UTR of the West Nile RNA virus (Blackwell and Brinton 1997) and to the localized β-actin mRNA (Liu et al. 2002). Eukaryotic EF1α is highly abundant in the cell (∼1% of total protein in mammalian cells), and is therefore in excess of other components of the cell's translation machinery (Condeelis 1995). This suggests that it may have other roles apart from in translation. In addition, eEF1α is known to be associated with the actin and microtubule cytoskeletons (Condeelis 1995), implying that it may act as an anchor for certain mRNAs such as β-actin in the cell periphery (Liu et al. 2002) and MT-1 around the nucleus, preventing their diffusion throughout the cytoplasm. Further studies are being carried out to determine if eEF1α binds to the 3′UTR of other perinuclear RNAs.
Mutational analysis of the MT-1 3′UTR using both UV cross-linking assays and immunocytochemistry of transfected cells showed that the regions of the 3′UTR corresponding to nt 66–76 and 21–36 were critical for both binding and localization. In contrast, nt 1–20, 36–66 or 86–134 had little or no influence on either localization or binding Figs. (6, 7) (Nury et al. 2005). This extends earlier observations that nt 66–76 were necessary for MT-1 mRNA localization (Nury et al. 2005) in showing that an additional part of the 3′UTR, namely nt 21–36, is required for localization, and in demonstrating that both of these regions are also required for protein binding. A combination of enzymatic and chemical cleavage data and computer folding prediction (Fig. 5D) indicates that the MT-1 3′UTR forms a series of loops, internal loops, bulges, and stem regions such that the regions implicated in localization are in close juxtaposition forming a stem–internal loop structure.
Substitutions that were predicted to destroy the stem formed between 26–30 and 66–70 caused a considerable reduction in binding of eEF1α, whereas double substitutions that reform the stem showed strong binding. Notably, when the stem was reformed, binding occurred whether the CACC sequence was altered or not. In addition, a substitution predicted to replace the internal loop with a stem was also found to reduce binding, while transcripts with altered sequence but with the internal loop maintained strong binding. Together, the binding and structural data suggest that the internal stem–loop formed between the CACC repeat and the sequence within nt 21–30 plays a role in eEF1α binding to the MT-1 3′UTR. Interestingly, eEF1α has been reported to bind to a double-stranded region of the West Nile RNA virus that contains a CAC motif close to a single-stranded bulge (Blackwell and Brinton 1997).
In conclusion, the present data indicate that in solution the MT-1 3′UTR folds such that nt 20–36 and 66–76 are in juxtaposition within an internal loop and stem, and that the two regions are both critical for binding of eEF1α. Taken together with localization data and previous observations (Liu et al. 2002; Nury et al. 2005), these findings form the basis for our hypothesis that (1) this stem/internal loop structure is part of the perinuclear localization signal in the MT-1 3′UTR, and (2) that eEF1α binds to this signal and anchors the MT-1 mRNA to the cytoskeleton in the perinuclear cytoplasm thus allowing synthesis of the protein near to its site of function. It has been reported that eEF1α binds to the localized actin mRNA (Liu et al. 2002), that eEF1γ binds to the localization element of vimentin mRNA (Al-Maghrebi et al. 2002), and that the translation initiation factor eIF4AIII binds to the exon junction complex through the Magoh-Y14 dimer, an interaction that is required for the localization of oskar mRNA to the posterior pole of Drosophila oocytes (Palacios et al. 2004). The present data provide further evidence for the view that certain translation factors are multifunctional, and that they may have key roles in mRNA localization as well as in translation.
MATERIALS AND METHODS
Gene constructs
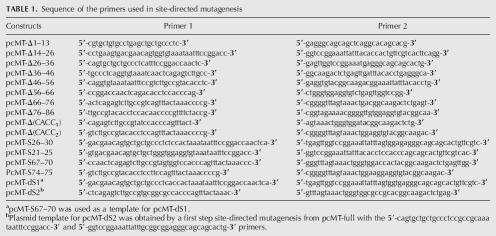
Mutant constructs (deletions or substitutions) of MT-1 3′UTR were made by site-directed mutagenesis using the plasmid pcMT-full as a template and following the manufacturer's instructions (Stratagene, Quikchange). pcMT-full contained the rat MT-1 coding region and full 3′UTR in pcDNA4/Hismax-TOPO (Nury et al. 2005). The primers used for the mutagenesis are listed in Table 1, and the details of the different constructs are presented in Figure 1. A further construct, namely pcMT-111 (nt 112–134 of MT-1 3′UTR deleted), was generated by PCR from pcMT-full with the forward 5′-AGTACTTTTCCCGACTTCAGCAG-3′ and reverse 5′-CACATGCTCGGTAGAAAACGG-3′ primers. This PCR product was directly cloned into pcDNA4/Hismax-TOPO following the manufacturer's instructions (Invitrogen). All constructs were verified by sequencing using the T7 forward and BGH reverse primers.
TABLE 1.
Sequence of the primers used in site-directed mutagenesis
In vitro transcription of labeled and unlabeled RNAs
Templates for transcription of rat MT-1 3′UTR nt 1–111 and the various mutants were generated by PCR from pcMT-full, or the appropriate mutant construct, with forward 5′-TAATACGACTCACTATAGGAGTGACGAACAGTGCTGCTG-3′ containing the T7 promoter sequence (underlined) and reverse 5′-CACATGCTCGGTAGAAAACGG-3′ primers. PCR products were purified using QIAquick columns (Qiagen). For electrophoretic mobility-shift assays and UV cross-linking experiments, transcripts were labeled with [α-32P]CTP (800 Ci/mmol) using the MAXIscript kit (Ambion). Unlabeled RNA or transcripts incorporating biotin-16UTP (Roche) were produced using the MEGAshortscript kit (Ambion). RNA was isolated by phenol-chloroform extraction and ethanol precipitation. Incorporation of radionucleotide into RNA was assessed by scintillation counting, and unlabeled RNA was quantified spectrophotometrically. Integrity was verified by denaturing gel electrophoresis with comparison against known standards. For structural studies, the template for transcription of the whole MT-1 3′UTR was produced from pcMT-full using the above forward primer and the reverse primer 5′-AGAATAAACAGGCTTTTATTATTCACATGCTCGGTAGAAAACGG-3′.
S100 cell extracts
Chinese hamster ovary (CHO) cells were grown in 175 cm2 flasks (Greiner Bio-one) in Ham's F-12 medium supplemented with 10% fetal calf serum and in an atmosphere of 5% CO2 at 37°C. Cells were grown to ∼90% confluence, and cytoplasmic protein extracts were prepared following the method of Behar et al. (1995) with modifications. Cells were resuspended in lysis buffer (130 mM NaCl, 5 mM MgCl2, 30 mM Tris-HCl pH 7.6 containing 2 mM DTT, 0.5% [v/v] NP-40 and EDTA-free protease inhibitor cocktail [Roche]) and lysed by passing them through a 21-gauge needle seven times. Large debris were removed by centrifugation at 5000g for 10 min, and the supernatant fluid was diluted with 3 volumes of 40 mM NaCl lysis buffer and centrifuged at 100,000g for 1 h.
Electrophoretic mobility-shift assays (EMSA) and UV cross-linking
EMSA reactions were carried out with 2 μg protein extract and 12 fmol of 32P-labeled RNA (heated to 70°C and allowed to cool and refold slowly) in 40 mM NaCl lysis buffer (see above) in a total volume of 8 μL at 22°C for 15 min. Following the binding reaction, 40 units (U) of RNase T1 were added and incubation continued for 5 min. Two microliters of 20% (w/v) Ficoll were then added, and complexes were separated by electrophoresis at 4°C for 2 h at 20 V/cm through 5% (w/v) nondenaturing polyacrylamide gels. For competition experiments, unlabeled RNA was added just prior to the labeled probe. For supershift assays, 8 μg mouse anti-eEF1α (Upstate) IgG or a control IgG (anti-biotin) were incubated with cell extract for 2.5 h at 22°C prior to binding reactions (as above), RNase T1 digestion and addition of Ficoll.
UV cross-linking reactions (13 μL total volume) were carried out at 22°C using 35 μg protein and 50 fmol of 32P-labeled RNA in 40mM NaCl lysis buffer (see above): binding reactions and subsequent digestion with RNase T1 were performed as for the EMSA, before addition of 10 mg/mL heparin and incubation for a further 5 min. The reactions were placed on ice and irradiated with UV light for 12 min to cross-link protein–RNA complexes. Unprotected RNA was then removed by digestion with 10 μg RNase A for 30 min at 37°C. The reactions were adjusted to 1× dissociation buffer (45 mM Tris-HCl pH 6.8, 10% [w/v] glycerol, 1% [w/v] SDS, 1% [v/v] β-mercaptoethanol, 0.01% [w/v] bromophenol blue) and then denatured at 95°C for 5 min before being subjected to SDS-PAGE. Gels were dried and analyzed by autoradiography.
Protein isolation using paramagnetic beads
Prewashed MagneSphere streptavidin-coated paramagnetic particles (SA-PMP, Promega, 0.6 mL) were resuspended in 0.1 mL 0.5× SSC containing 100 μg BSA and 100 μg yeast tRNA and incubated at room temperature for 1 h with shaking. After washing twice with 0.3 mL 0.5× SSC, the SA-PMP were incubated with 20 μg biotinylated RNA (preheated to 70°C and cooled slowly) in 0.3 mL 0.5× SSC for 10 min at room temperature. Unbound RNA was removed by washing twice with 0.3 mL 0.5× SSC. SA-PMP with RNA bound were then incubated at 4°C for 1 h with 1 mg cell extract in 40 mM NaCl lysis buffer (see above) with 0.5 mg/mL yeast tRNA, 0.2 mg/mL BSA, and 800 units/mL RNasin (Promega) in a total volume of 0.5 mL. The supernatant fluid was removed, and after 5×1 mL washes with 40 mM NaCl lysis buffer proteins bound to the RNA were eluted by boiling in 1× dissociation buffer (see above).
Proteins were separated by SDS-PAGE and either stained with Coomassie Brilliant Blue or transferred to PVDF membrane for Western blotting. Bands from Coomassie-stained gels were excised for in-gel trypsin digestion followed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (carried out by J. Gray, Institute for Cell and Molecular Biosciences, University of Newcastle). PVDF membranes were incubated with monoclonal antibodies to anti-eEF1α (Upstate, 0.5 μg/mL), followed by anti-mouse HRP-conjugated antibody (Sigma) and detection using a POD chemiluminescence kit (Roche) and Kodak XOmat A2–5 film.
Immunocytochemistry
Studies of MT-1 protein distribution were carried out in cells grown in multiwell chamber slides so that the different cell lines could be studied under identical conditions and the quantification of staining would be directly comparable. Immunocytochemistry was carried out using an anti-Xpress antibody (Invitrogen) directed against the Xpress peptide epitope tag from the pcDNA4/HisMax-TOPO plasmid. Cells were washed three times with PBS and fixed with 1% (w/v) paraformaldehyde in PBS for 20 min, before being permeabilized with 1% paraformaldehyde/0.05% Triton X-100 in PBS for 10 min at 4°C. The cells were washed in PBS and incubated with 10% (v/v) normal sheep serum in PBS for 30 min to allow saturation of nonspecific antibody binding sites. After a brief wash in PBS containing 1% BSA, cells were incubated with anti-Xpress antibody diluted 1/400 in PBS/1% BSA for 45 min at room temperature. After four washes in PBS/1% BSA, cells were incubated with FITC-conjugated goat anti-mouse IgG (Sigma) at a dilution of 1/150 in PBS/1% BSA. Finally, cells were rinsed four times in PBS/1% BSA and mounted in Citifluor. Two controls were carried out routinely: untransfected CHO cells taken through the whole procedure described above and transfected cells that were not incubated with the primary antibody but with PBS/1% BSA. Standard microscopy was performed using an Olympus BX51 microscope, and digital images of the cells were captured under the ×100 oil immersion lens using an Olympus DP50 digital camera and Analysis Viewfinder Lite SIS Software. Staining was quantified by examining fields of view at random and assigning localization characteristics (presence or absence of a distinct ring of perinuclear staining) of ∼100 cells in each of at least three separate experiments. Cells were classified as exhibiting perinuclear localization of protein or no localization.
RNA secondary structure analysis
RNA corresponding to MT-1 3′UTR was separated on an 8% polyacrylamide gel in 7 M urea, the band excised, and the RNA recovered by soaking overnight in elution buffer (0.5 M ammonium acetate, 1 M EDTA, 0.1% SDS) and precipitation with ethanol. RNA was labeled at the 5′-end with [γ-32P]ATP after 5′-phosphate removal using the KinaseMax kit (Ambion) according to the manufacturer's instructions. Following labeling, RNA was purified by spin-column chromatography (chromaspin-30 columns; BD Bioscience). 32P-labeled RNA (∼105 cpm) was incubated with RNase at room temperature in 20 mM Tris-HCl pH 7.5/10 mM MgCl2/100 mM KCl in the presence of 2–5 μg of yeast tRNA: either 0.02, 0.05, and 0.1 U of RNase T1 (1 U/μL; Ambion) for 5 min, 0.005, 0.01, and 0.02 U of RNase V1 (0.1 U/μL; Ambion) for 5 min, 0.02 and 0.05 μg of RNase A (1 μg/μL; Ambion), or 0.1, 0.2, and 0.4 U of RNase T2 (30 U/μL; Sigma) for 3 min. Enzymatic cleavage reactions were stopped on ice by addition of 20 μL of inactivation/precipitation solution (Ambion). RNA was ethanol precipitated in the presence of 1 μg of Glycoblue (Ambion), air dried, and resuspended in loading buffer. RNA cleavage products were separated on a 10% polyacrylamide gel (19:1; 7 M urea and 1× TBE). Gels were analyzed by autoradiography after drying. For lead cleavage experiments, RNA was supplemented with 4 μg of yeast tRNA and incubated for 5 min at room temperature with 2, 5, or 10 mM Pb2+ in 20 mM HEPES-NaOH pH 7.5/7 mM Magnesium acetate/50 mM potassium acetate. Lead hydrolysis was stopped by chilling the samples on ice and addition of 2 μL of 100 mM EDTA. Subsequent steps of the procedure were as described above. Alkaline and G-specific RNase T1 ladders were generated in parallel to identify the cleavage positions. Limited alkaline hydrolysis of 32P-labeled RNA was performed by incubation in sodium carbonate (pH 9) at 90°C for 4 min. Ladders of G were produced by preincubation of the labeled RNA at 55°C for 10 min in 20 mM sodium citrate pH 5/1 mM EDTA/7 M urea, and a further 10 min incubation at 55°C in the presence of 0.5 U of RNase T1. Chemical reactions with carbodiimide (CMCT; 10 mg/mL) were performed using the standard procedure described by Huntzinger et al. (2005). Detection of the modified nucleotides was accomplished by hybridization and extension of the 5′-end labeled primer 5′-GGATCCTAGGTACAGGCCC-3′ (anneals on the RNA to a plasmid sequence located downstream of MT-1 3′UTR). Positions of the modifications were identified by running in parallel a dideoxysequencing reaction. The reverse transcripts were separated on a 10% sequencing gel.
ACKNOWLEDGMENTS
This work was supported by the BBSRC (C13737 and C14236). Z.C.-L. thanks the Wellcome Trust for continued support. We thank Edward Burton for help with making some of the deletion constructs.
Footnotes
Abbreviations: 3′UTR, 3′untranslated region; CHO, Chinese hamster ovary; CMCT, 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate; eEF1α, eukaryote elongation factor 1α; eEF1γ, eukaryote elongation factor 1γ; eIF4AIII, eukaryote initiation factor 4AIII; EMSA, electrophoretic mobility-shift assay; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; MT-1, metallothionein-1; RNase, ribonuclease; SA-PMP, streptavidin-coated paramagnetic particles; SDS-PAGE, sodium dodecyl sulphate polyacrylamide gel electrophoresis.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2730106.
REFERENCES
- Al-Maghrebi M., Brule H., Padkina M., Allen C., Holmes W.M., Zehner Z. The 3′untranslated regin of human vimentin mRNA interacts with protein complexes containing eEF1 and HAX1. Nucleic Acids Res. 2002;320:5017–5028. doi: 10.1093/nar/gkf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar L., Marx R., Sadot E., Barg J., Ginzburg I. cis-Acting signals and trans-acting proteins are involved in tau mRNA targeting in neurites of differentiating neuronal cells. Int. J. Dev. Neurosci. 1995;13:113–127. doi: 10.1016/0736-5748(95)00001-w. [DOI] [PubMed] [Google Scholar]
- Betley J.N., Frith M.C., Graber J.H., Choo S., Deshler J.O. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- Blackwell J.L., Brinton M.A. Translation elongation factor-1 α interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson J.H., Barbarese E. Systems analysis of RNA trafficking in neural cells. Biol. Cell. 2005;97:51–62. doi: 10.1042/BC20040083. [DOI] [PubMed] [Google Scholar]
- Chabanon H., Mickleburgh I., Hesketh J.E. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief. Funct. Genomic. Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- Condeelis J. Elongation-factor 1-α, translation and the cytoskeleton. Trends Biochem. Sci. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- Dalgleish G., Veyrune J.L., Blanchard J.M., Hesketh J.E. mRNA localisation by a 145-nucleotide region of the c-fos 3′untranslated region. J. Biol Chem. 2001;276:13593–13599. doi: 10.1074/jbc.M001141200. [DOI] [PubMed] [Google Scholar]
- Hesketh J.E. Translation and the cytoskeleton: A mechanism for targeted protein synthesis. Mol. Biol. Rep. 1994;19:233–243. doi: 10.1007/BF00986965. [DOI] [PubMed] [Google Scholar]
- Hesketh J.E., Campbell G.P., Whitelaw P.F. c-myc mRNA in cytoskeletal-bound polysomes in fibroblasts. Biochem. J. 1991;274:607–609. doi: 10.1042/bj2740607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh J.E., Campbell G., Piechaczyk M., Blanchard J.M. Targeting of c-myc and β-globin coding sequences to cytoskeletal-bound polysomes by c-myc 3′ untranslated region. Biochem. J. 1994;298:143–148. doi: 10.1042/bj2980143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E., Possedko M., Winter F., Moine H., Ehresmann C., Romby P. Probing RNA structures with enzymes and chemicals in vitro and in vivo . In: Hartmann, et al., editors. Handbook of RNA biochemistry. Wiley–VCH; Weinheim, Germany: 2005. pp. 151–171. [Google Scholar]
- Jansen R.-P. Message on the move. Nat. Rev. Mol. Cell Biol. 2001 mRNA localization;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- Kislauskis E.H., Li Z., Taneja K.L., Singer R.H. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis E.H., Zhu X.C., Singer R.H. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S., Barbarese E., Carson J.H. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J. Cell Biol. 1999;147:247–256. doi: 10.1083/jcb.147.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levadoux M., Mahon P.C., Beattie J.H., Wallace H.M., Hesketh J.E. Nuclear import of metallothionein requires its mRNA to be associated with the perinuclear cytoskeleton. J. Biol. Chem. 1999;274:34961–34966. doi: 10.1074/jbc.274.49.34961. [DOI] [PubMed] [Google Scholar]
- Levadoux-Martin M., Hesketh J.E., Beattie J.H., Wallace H.M. Influence of metallothionein-1 localization on its function. Biochem. J. 2001;355:473–479. doi: 10.1042/0264-6021:3550473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Grant W.M., Persky D., Latham V.M., Singer R.H., Condeelis J. Interactions of elongation factor 1 α with F-actin and β-actin mRNA: Implications for anchoring mRNA in cell protrusions. Mol. Biol. Cell. 2002;13:579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon P.C., Partridge K., Beattie J.H., Glover L.A., Hesketh J.E. The 3′untranslated region plays a role in the targeting of metallothionein-1 mRNA to the perinuclear cytoplasm and cytoskeletal-bound polysomes. Biochim. Biophys. Acta. 1997;1358:153–162. doi: 10.1016/s0167-4889(97)00058-x. [DOI] [PubMed] [Google Scholar]
- Nury D., Chabanon H., Levadoux-Martin M., Hesketh J.E. A 11-nucleotide section of the 3′untranslated region is required for perinuclear localisation of rat metallothionein-1 mRNA. Biochem. J. 2005;387:419–428. doi: 10.1042/BJ20040630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y., Singer R.H. RNA localization: Different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- Palacios I.M., Gatfield D., St Johnston D., Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Shan J., Munro T.P., Barbarese E., Carson J.H., Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J. Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.A., Brindle K.M., Fulton A.M. Differential localisation of the mRNA of the M and B forms of creatine kinase in myoblasts. Biochem. J. 1995;308:599–605. doi: 10.1042/bj3080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehner Z.E., Shepherd R.K., Gabryszuk J., Fu T.F., Al-Ali M., Holmes W.M. RNA–protein interactions within the 3′ untranslated region of vimentin mRNA. Nucleic Acids Res. 1997;25:3362–3370. doi: 10.1093/nar/25.16.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]