Abstract
RNase MRP is a eukaryotic endoribonuclease involved in nucleolar and mitochondrial RNA processing events. RNase MRP is a ribonucleoprotein particle, which is structurally related to RNase P, an endoribonuclease involved in pre-tRNA processing. Most of the protein components of RNase MRP have been reported to be associated with RNase P as well. In this study we determined the association of these protein subunits with the human RNase MRP and RNase P particles by glycerol gradient sedimentation and coimmunoprecipitation. In agreement with previous studies, RNase MRP sedimented at 12S and 60–80S. In contrast, only a single major peak was observed for RNase P at 12S. The analysis of individual protein subunits revealed that hPop4 (also known as Rpp29), Rpp21, Rpp20, and Rpp25 only sedimented in 12S fractions, whereas hPop1, Rpp40, Rpp38, and Rpp30 were also found in 60–80S fractions. In agreement with their cosedimentation with RNase P RNA in the 12S peak, coimmunoprecipitation with VSV-epitope-tagged protein subunits revealed that hPop4, Rpp21, and in addition Rpp14 preferentially associate with RNase P. These data show that hPop4, Rpp21, and Rpp14 may not be associated with RNase MRP. Furthermore, Rpp20 and Rpp25 appear to be associated with only a subset of RNase MRP particles, in contrast to hPop1, Rpp40, Rpp38, and Rpp30 (and possibly also hPop5), which are probably associated with all RNase MRP complexes. Our data are consistent with a transient association of Rpp20 and Rpp25 with RNase MRP, which may be inversely correlated to its involvement in pre-rRNA processing.
Keywords: RNase MRP, RNase P, snoRNP, RNA processing, ribonuclease, ribonucleoprotein
INTRODUCTION
Eukaryotic ribosome synthesis starts with transcription of the rDNA genes by RNA polymerase I. The human 18S, 5.8S, and 28S rRNAs are transcribed as a long polycistronic RNA, designated 47S pre-rRNA. In order to yield the mature 18S, 5.8S, and 28S rRNAs, the 47S pre-rRNA undergoes a series of endo- and exonucleolytic processing steps, and in addition many nucleotides are chemically modified. These post-transcriptional modifications include pseudouridinylation and 2′-O-ribose methylation. Most of these processing and maturation steps are catalyzed by small nucleolar ribonucleoprotein complexes (snoRNPs). These protein–RNA complexes reside in the nucleolus and are typically composed of multiple protein subunits associated with a single snoRNA species that serves as a guide in substrate recognition. Based upon common characteristics in their RNA moiety, snoRNPs are categorized into three major classes (Tollervey and Kiss 1997; Terns and Terns 2002). The first class is the box H/ACA snoRNPs, which are involved in the pseudouridinylation of rRNA (Ganot et al. 1997). The second class is the box C/D snoRNPs, which guide the modification of rRNA by methylation of riboses (Kiss-Laszlo et al. 1996). Some box C/D snoRNPs, for example, the U3 snoRNP, are involved in endonucleolytic cleavage steps of the pre-rRNA (Hughes and Ares 1991). Two snoRNPs cannot be classified as box H/ACA or box C/D snoRNPs (Tollervey and Kiss 1997). These snoRNPs are RNase MRP (ribonuclease for mitochondrial RNA processing) and RNase P. RNase P is a ubiquitous enzyme that functions in the maturation of tRNA by the removal of the 5′ leader sequence of pre-tRNAs (Altman 2000; Jarrous 2002). In the yeast Saccharomyces cerevisiae, RNase MRP has been reported to cleave site A3 within internal transcribed spacer 1 (ITS1) of pre-rRNA, thereby contributing to the maturation of the 5′-end of the 5.8S rRNA (Lygerou et al. 1996a). In addition to this nucleolar function, RNase MRP has been reported to be involved in the generation of RNA primers for mitochondrial DNA replication by cleavage of an RNA transcript that originates from the mitochondrial origin of replication (Chang and Clayton 1987). Recently, a third function for S. cerevisiae RNase MRP, the cleavage of Clb2 mRNA, has been described (Gill et al. 2004). In spite of the reported mitochondrial and mRNA cleavage functions, the vast majority of RNase MRP is found in the nucleolus, the site of pre-rRNA processing (Reimer et al. 1988).
RNase MRP from human, Arabidopsis thaliana, and Nicotiana tabacum has been reported to be associated with different complexes sedimenting at ∼12S and 60–80S in glycerol gradients (Kiss et al. 1992; Lee et al. 1996). For the U3 snoRNP, 12S (low molecular weight) complexes were identified as core particles consisting of the U3 snoRNA and the most stably associated proteins, whereas also in this case high-molecular-weight (60–80S) complexes were observed, which most likely represent pre-ribosomes (Tyc and Steitz 1989; Lukowiak et al. 2000; Granneman et al. 2003). Interestingly, Granneman and coworkers found that a subset of the human U3 snoRNP-associated proteins only cosediments with the 60–80S complexes and not with the 12S complexes. Therefore, these proteins were proposed to be required for the function of the U3 snoRNP in ribosome synthesis. The sedimentation of the human RNase MRP RNA in similarly sized complexes suggested that the 12S and 60–80S complexes correspond to a core particle and the pre-ribosomal complexes, respectively. Today, 10 proteins have been identified as RNase MRP components in several organisms, and it is generally believed that most of these proteins are stably associated with both RNase MRP and RNase P (Lygerou et al. 1996b; Eder et al. 1997; Chamberlain et al. 1998; Jarrous et al. 1998, 1999, 2001; van Eenennaam et al. 1999, 2001; Guerrier-Takada et al. 2002). To obtain experimental evidence on the protein composition of these particles in humans, their sedimentation patterns in glycerol gradients were determined. The results of these experiments not only revealed striking differences between the sedimentation behavior of RNase MRP and RNase P, but also demonstrated extensive heterogeneity in the association of the protein subunits with the different complexes. Taken together, our data demonstrate for the first time that differences in protein subunit composition between the human RNase MRP and RNase P complexes exist and in addition show that the protein composition of RNase MRP in part depends on its association with 60–80S pre-ribosomal complexes.
RESULTS
Glycerol gradient sedimentation analysis of RNase MRP and RNase P
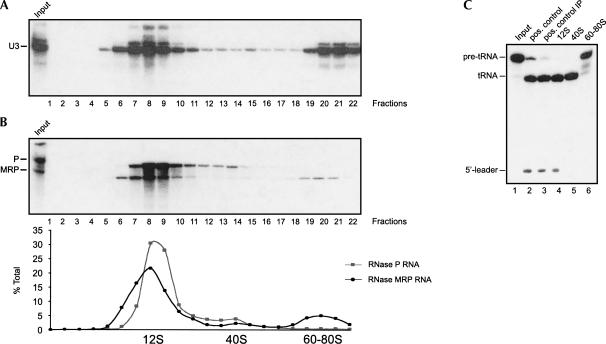
To investigate the heterogeneity of the human RNase MRP and RNase P ribonucleoprotein complexes, we fractionated total cell extracts from HEp-2 cells in 5%–40% glycerol gradients. The sedimentation behavior of these complexes was first visualized by Northern blot hybridization using probes specific for the RNase MRP and RNase P RNA components. In parallel the fractionation of rRNAs was monitored by agarose gel electrophoresis and ethidium bromide staining; the positions of the 18S and 28S rRNAs were used as markers for 40S and 60S complexes. A U3 snoRNA probe was used as an additional marker for the positions of 12S and 60–80S complexes in the gradient (Fig. 1A; Granneman et al. 2003). As shown in Figure 1B, the majority of RNase MRP RNA and RNase P RNA was found in 12S fractions (fractions 6–10). Quantification of the hybridization signals by PhosphorImaging revealed that ∼65% of the total pool of RNase MRP RNA and ∼75% of the RNase P RNA sedimented in these fractions. In addition, both RNase MRP RNA and RNase P RNA showed minor peaks sedimenting at ∼40S (fraction 14, RNase P RNA; fraction 15, RNase MRP RNA). A third peak (containing ∼17% of the total RNase MRP RNA pool) was observed for RNase MRP RNA in 60–80S fractions (18–22). The glycerol gradient sedimentation analysis was carried out several times, and the results appeared to be very reproducible. The sedimentation pattern of RNase P RNA in 60–80S fractions was confirmed by analyzing RNase P activity in the 12S, 40S, and 60–80S fractions. Rpp38-containing complexes were immunoprecipitated from pooled 12S, 40S, and 60–80S fractions (fractions 7–9, 13–15, and 19–21, respectively), and the presence of RNase P activity in the immunoprecipitated material was analyzed by an in vitro RNase P activity assay using pre-tRNA as a substrate. The results in Figure 1C show that Rpp38-containing complexes from 12S and 40S fractions were able to cleave the pre-tRNA into the 5′ leader sequence and the mature tRNA (lanes 4,5). The absence of the 5′ leader sequence in the reaction products of the precipitate from the 40S fractions is most likely due to the coprecipitation of an activity that is responsible for the degradation of the released 5′ leader sequence. In contrast, anti-Rpp38 immunoprecipitates from 60–80S fractions did not display RNase P activity (Fig. 1C, lane 6). Note that the lack of RNase P activity in the latter fractions was not due to the absence of Rpp38, because this protein was found in all of these fractions (see below).
FIGURE 1.
Analysis of the human RNase MRP and RNase P complexes by glycerol gradient sedimentation. HEp-2 extracts were fractionated in 5%–40% (v/v) glycerol gradients. Total RNA was isolated from the fractions and analyzed by Northern blot hybridization. (A) Northern blots were hybridized with a U3 snoRNA-specific riboprobe. The signals were visualized by autoradiography. The position of the U3 snoRNA is indicative for 12S fractions. (B) Northern blots were hybridized successively with RNase MRP RNA- and RNase P RNA-specific probes. Signals were visualized by autoradiography and quantified by PhosphorImaging. The relative distributions of RNase MRP RNA and RNase P RNA were calculated. The sedimentation of 40S and 60S particles was determined by the presence of 18S and 28S rRNAs, respectively. (C) Pooled fractions 7–9, 13–15, and 19–21 (representing the 12S, 40S, and 60–80S peaks, respectively) were subjected to anti-Rpp38 immunoprecipitation. Subsequently, the immunoprecipitated material was analyzed for the presence of coprecipitated RNase P activity (lanes 4–6). In lane 1, purified pre-tRNA substrate (input) was loaded. As a positive control, pre-tRNA substrate cleaved by partially purified HeLa RNase P is shown in lane 2. As a positive control for anti-Rpp38 immunoprecipitation, the partially purified RNase P was immunoprecipitated, and the pre-tRNA processing assay was performed on the beads (lane 3). The positions of pre-tRNA, tRNA, and the 5′ leader sequence are indicated on the left.
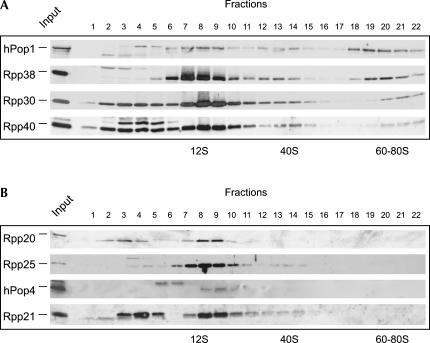
The differences observed in the sedimentation behavior of RNase MRP RNA and RNase P RNA prompted us to investigate the RNase MRP and RNase P protein subunit distribution. To analyze the sedimentation behavior of these protein subunits, glycerol gradient fractions were separated by SDS-PAGE, and the proteins were visualized by immunoblotting using polyclonal rabbit antisera. Surprisingly, striking differences were observed between the sedimentation patterns of individual protein subunits. As shown in Figure 2A, substantial amounts of all RNase MRP and RNase P protein subunits sedimented at 12S (at which the RNase MRP and RNase P RNAs also showed major peaks). However, in contrast to hPop1, Rpp38, Rpp30, and Rpp40, which showed an RNase MRP RNA-like sedimentation pattern with a peak in the 60–80S region, Rpp20, Rpp25, hPop4 (also known as Rpp29), and Rpp21 did not detectably sediment in high-molecular-weight fractions (Fig. 2B). In addition, more subtle differences between the sedimentation behavior of individual protein subunits were observed. Quantification of the data by PhosphorImaging demonstrated that the Rpp38 distribution was virtually indistinguishable from that of the RNase MRP RNA, with a major peak at 12S and a minor peak at 60–80S. The intensity of the hPop1 peak at 60–80S, on the other hand, was about equal to that of the 12S peak. Rpp30 and Rpp40 both showed the three peaks observed also for the RNase MRP RNA and Rpp38, but in addition were present in fractions containing complexes/proteins sedimenting slower than 12S. These fractions may contain the free proteins. In addition to the sedimentation of Rpp20, Rpp25, hPop4, and Rpp21 in 12S fractions, we also detected weak signals for these proteins in the 40S fractions. The signals observed in the low-molecular-weight fractions (2–6) with antibodies against Rpp20, hPop4, and Rpp21 do not migrate at the positions of the respective proteins and therefore, most likely, are due to cross-reactivity. Although antisera raised against Rpp14 and hPop5 were available, these sera appeared to be unable to detect the respective proteins in the gradient fractions. Because of the small difference in peak sedimentation of 12S RNase MRP and RNase P RNA (Fig. 1B, lanes 8 and 8–9, respectively), we tried to obtain a higher resolution in this region of the gradients by running 12%–18% glycerol gradients. Interestingly, the results of these analyses strongly suggested that hPop4 and Rpp21 completely cosedimented with the RNase P RNA and not with the RNase MRP RNA (data not shown). In summary, the RNase MRP/RNase P protein subunits can be divided into two major groups: proteins that cosediment with 12S, 40S, and 60–80S complexes and proteins that solely cosediment with 12S and 40S complexes.
FIGURE 2.
Distribution of RNase MRP/RNase P protein subunits in glycerol gradients. The protein distribution in 5%–40% glycerol gradients was analyzed by immunoblotting using polyclonal rabbit sera against the individual subunits. (A) Proteins that sedimented at 12S, 40S, and 60–80S. (B) Proteins that sedimented at 12S and 40S only. Note that the available polyclonal rabbit sera against hPop5 and Rpp14 failed to detect the corresponding proteins.
Differential association of protein subunits in transfected cells
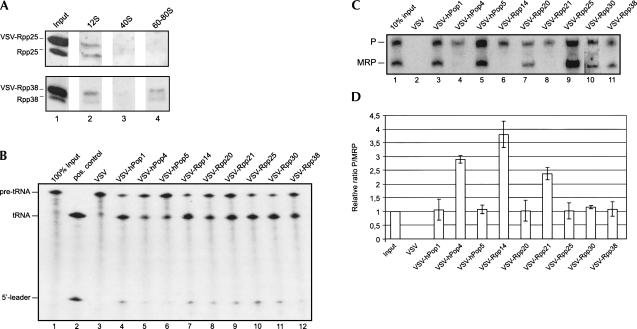
To further investigate the presumptive differential association of the protein subunits with 12S RNase MRP and RNase P, cDNAs encoding all RNase MRP/RNase P protein subunits were cloned in the pCI-Neo vector, in frame with an N-terminal VSV-tag encoding sequence as described before (Welting et al. 2004). HEp-2 cells were transfected with the individual constructs, and after culturing for 24 h, the incorporation of the VSV-tagged proteins into RNase MRP/RNase P-sized complexes was checked by analyzing the glycerol gradient sedimentation behavior of transiently expressed VSV-Rpp25 or VSV-Rpp38. Extracts from cells expressing VSV-Rpp25 or VSV-Rpp38 were separated on 5%–40% glycerol gradients, and the fractionation of the VSV-tagged as well as the endogenous proteins was analyzed by Western blotting using polyclonal rabbit sera against the respective proteins. The results showed that, like the endogenous proteins, VSV-Rpp25 associated with complexes sedimenting at 12S, and VSV-Rpp38 associated with complexes sedimenting at 12S and 60–80S (Fig. 3A). These results indicated that the tags did not interfere with the incorporation of these proteins into RNase MRP and RNase P particles. In agreement with this, the RNAs associated with these particles coprecipitated with the tagged proteins when the latter were immunoprecipitated with anti-VSV-tag antibodies (see below). To further demonstrate the association of the VSV-tagged protein subunits with these ribonucleoprotein particles, extracts from transiently transfected cells were subjected to immunoprecipitation with anti-VSV-tag monoclonal antibodies and the RNase P activity in the immunoprecipitates was determined in vitro. As can be seen in Figure 3B, in contrast to the immunoprecipitate from the extract from cells transfected with the control VSV-tag vector, the immunoprecipitates from all cells transfected with the individual VSV-fusion proteins displayed RNase P activity. These data show that all VSV-tagged RNase MRP/RNase P protein subunits analyzed are capable of associating with enzymatically active RNase P particles. Note that Rpp40 was not included in these analyses, because we failed to express VSV-tagged Rpp40 to detectable levels in this system. To study the association of these VSV-tagged proteins with both RNase MRP and RNase P, the coprecipitating RNAs were isolated from anti-VSV-tag immunoprecipitates and analyzed by Northern blot hybridization using riboprobes specific for the RNA components of RNase MRP and RNase P. The results in Figure 3C show that VSV-hPop1, VSV-hPop5, VSV-Rpp20, VSV-Rpp25, VSV-Rpp30, and VSV-Rpp38 all coprecipitate both RNase MRP RNA and RNase P RNA. The relative efficiencies by which these two RNAs are coprecipitated are about equal for all of these proteins, indicating that these subunits are associated with both RNase MRP RNA and RNase P RNA. This was confirmed by quantification of the data by PhosphorImaging of the RNase MRP RNA and RNase P RNA signals for three independent experiments (Fig. 3D). Surprisingly, for VSV-Rpp21, VSV-Rpp14, and VSV-hPop4, we never detected coprecipitation of RNase MRP RNA. To exclude the possibility that VSV-Rpp21, VSV-Rpp14, and VSV-hPop4 did not coprecipitate RNase MRP RNA because of interference of the VSV-tag, the experiment was repeated, but now with the VSV-tag fused to the C terminus instead of the N terminus of these proteins. Also in these cases, no coprecipitation of RNase MRP RNA was observed (data not shown). The lack of RNase MRP RNA coimmunoprecipitation with VSV-tagged Rpp21 and hPop4 is consistent with results obtained by 12%–18% glycerol gradient sedimentation analyses, which showed the specific cosedimentation of Rpp21 and hPop4 with the 12S RNase P RNA, indicating that these proteins are preferentially associated with the RNase P holoenzyme.
FIGURE 3.
Coimmunoprecipitation of RNase MRP RNA and RNase P RNA with transiently expressed VSV-tagged protein subunits. Incorporation of VSV-tagged protein subunits in 12S and 60–80S complexes was determined by fractionation of extracts from transiently transfected HEp-2 cells expressing VSV-Rpp25 or VSV-Rpp38 on 5%–40% glycerol gradients. In parallel, these extracts were subjected to immunoprecipitation with anti-VSV-tag antibodies. (A) The presence of VSV-tagged and endogenous proteins in gradient fractions was determined by immunoblotting using polyclonal antisera against Rpp25 or Rpp38. In lane 1, the input material was loaded. In lanes 2–4, 12S, 40S, and 60–80S peak fractions were analyzed (fractions 8, 15, and 20, respectively). The positions of the VSV-tagged and endogenous proteins are indicated on the left. (B) An in vitro pre-tRNA processing assay was performed with the immunoprecipitated material. The reaction products were separated by denaturing gel electrophoresis. In lane 1, 100% of the input pre-tRNA was loaded. (Lane 2) As a positive control, pre-tRNA was incubated with partially purified RNase P. As a negative control, lane 3 shows the reaction products after incubation with an immunoprecipitate from an extract of HEp-2 cells that was transiently transfected with the control VSV-tag vector. Lanes 4–12 show the results for the immunoprecipitated material from cells expressing VSV-hPop1, VSV-hPop4, VSV-hPop5, VSV-Rpp14, VSV-Rpp20, VSV-Rpp21, VSV-Rpp25, VSV-Rpp30, VSV-Rpp38, respectively. The positions of the pre-tRNA, the mature tRNA and the released 5′ leader sequence are indicated on the left. (C) RNA coimmunoprecipitated with the anti-VSV antibodies was isolated and analyzed by Northern blot hybridization using RNase MRP RNA- and RNase P RNA-specific probes. Lane 1 contains total RNA from 10% of input material. In lanes 2–11 coimmunoprecipitated RNAs from cells transfected with the “empty” VSV vector or with constructs encoding VSV-hPop1, VSV-hPop4, VSV-hPop5, VSV-Rpp14, VSV-Rpp20, VSV-Rpp21, VSV-Rpp25, VSV-Rpp30, and VSV-Rpp38 were loaded. The positions of RNase MRP and RNase P RNA are indicated on the left. (D) Relative ratios of coprecipitated RNase P and RNase MRP RNAs quantified by PhosphorImaging of Northern blot hybridization data. The ratio of the signals of these RNAs in the input material was arbitrarily set at 1. The ratios were determined for three independent experiments, and the error bars indicate the standard deviation.
DISCUSSION
Previous studies have identified 10 human proteins that are believed to be stably associated with RNase MRP and RNase P (Lygerou et al. 1996b; Eder et al. 1997; Jarrous et al. 1998, 1999, 2001; van Eenennaam et al. 1999, 2001; Guerrier-Takada et al. 2002). For most of these proteins, coimmunoprecipitation experiments demonstrated that they are, indeed, associated with both RNase MRP RNA and RNase P RNA in human cell lines (Pluk et al. 1999; van Eenennaam et al. 2002). Moreover, all 10 proteins copurified with catalytically active RNase P, suggesting that they are part of the core structure of the holoenzyme. Recently, we and others have identified mutual interactions between protein subunits and interactions between protein subunits and the RNA components by two-hybrid analyses and in in vitro assays (Welting et al. 2004; Jiang and Altman 2001; Jiang et al. 2001). In this study, we have addressed the molecular composition of the complexes in HEp-2 cell lysates. Our data not only show a preferential association of hPop4, Rpp21, and Rpp14 with RNase P, but also provide evidence for the transient association of Rpp25 and Rpp20 with RNase MRP. The results suggest that the latter proteins dissociate from the RNase MRP complex when it binds to 60–80S complexes.
When the RNA component is taken as a molecular marker for all particles, RNase MRP cosedimented with 12S and 60–80S complexes and a minority with 40S complexes, whereas RNase P was found mainly at 12S and a small amount at 40S. The cosedimentation of RNase MRP with 60–80S complexes in glycerol gradients has also been reported for RNase MRP from HeLa cells and from plants (Kiss et al. 1992; Lee et al. 1996). This is consistent with its involvement in pre-rRNA processing, because these complexes are believed to represent ribosomal precursors associated with processing factors. The lack of RNase P RNA in 60–80S fractions does not support a role for RNase P in human pre-rRNA processing, which was previously suggested both in yeast (Chamberlain et al. 1996) and in mammals (Lee et al. 1996).
Up to now, most of the 10 protein subunits identified for the mammalian RNase MRP/RNase P complexes were reported to be associated with both ribonucleoprotein particles. The only clear reported difference between both complexes is the identity of the associated RNA (Baer et al. 1990; Topper and Clayton 1990). Interestingly, in S. cerevisiae, Snm1p and RMP1 have been identified as unique protein components of RNase MRP (Schmitt and Clayton 1994; Salinas et al. 2005) and Rpr2p as a unique protein component of RNase P (Chamberlain et al. 1998). However, the recent purification of RNase MRP from S. cerevisiae demonstrated that significant amounts of Rpr2p are present in the purified complex, suggesting that at least a subset of RNase MRP is also associated with Rpr2p (Salinas et al. 2005). In agreement with the difference in protein composition, electron microscopic analyses of affinity-purified “POP complexes” from yeast showed the existence of large and small complexes, which may reflect the size difference due to different protein compositions of both yeast complexes (Aloy et al. 2004). The present results indicate that a subset of the 10 protein subunits (hPop4, Rpp21, and Rpp14) is more efficiently associated with RNase P than with RNase MRP. hPop4 and Rpp21 not only specifically cofractionate with 12S RNase P RNA (note that Rpp14 could not be detected in glycerol gradient fractions), but also preferentially associate with RNase P RNA in transfection experiments with VSV-tagged proteins. Our results indicate that the separation of RNase MRP and RNase P in the 12S region may not only be due to the size differences between the RNAs, but may also be caused by the differential association of protein subunits. Alternatively, we cannot exclude the possibility that stoichiometric differences between RNase MRP and RNase P affect the distribution of protein components in glycerol gradient sedimentation. Most interestingly, in vitro RNase P reconstitution experiments have demonstrated that a particle composed of human RNase P RNA and recombinant hPop4 and Rpp21 generated a functionally active complex (Mann et al. 2003). Similarly, the archaeal hPop4 and Rpp21 orthologs of Pyrococcus horikoshii (Ph1771 and Ph1601, respectively) have been shown to be essential components of an active, reconstituted RNase P particle (Kouzuma et al. 2003). In combination with the reported binding activities of Rpp14 to precursor tRNA (Jarrous et al. 2001) and hPop4 and Rpp21 to both mature and precursor tRNA (Jarrous et al. 2001; Sharin et al. 2005), these data suggest that the 12S RNase P particle represents the human holoenzyme. Indeed, the 12S fractions catalyzed the cleavage of a pre-tRNA substrate in vitro.
In agreement with their preferential association with RNase P particles, the hPop4 and Rpp21 proteins were undetectable in the 60–80S fractions. This suggests that these proteins are not required for the role of RNase MRP in pre-rRNA processing in human cells. In previous studies, hPop4 has been shown to interact with both the RNase P and the RNase MRP particle (van Eenennaam et al. 1999) and to display direct binding to the RNase P and RNase MRP RNAs (Welting et al. 2004). Moreover, the yeast Pop4p protein has been described to be required for pre-rRNA processing (Chu et al. 1997), and, recently, a TAP-tagged version of the Pop4p protein was used to purify the yeast RNase MRP complex (Salinas et al. 2005). In part, these conflicting results may be explained by a physical or functional link between subsets of the RNase MRP and RNase P complexes. In 60–80S complexes, a direct interaction between RNase MRP and RNase P was previously proposed by Lee et al. (1996). Since we were not able to detect RNase P in 60–80S complexes, the interaction between subsets of the RNase MRP and RNase P may be relatively unstable or transient. Alternatively, a small amount of complexes containing both RNase MRP and RNase P may also sediment in regions of the gradients that were not analyzed in detail in this study. Indeed, the sedimentation of both RNase MRP and RNase P at 40S indicates that additional complexes do exist. Finally, our data do not completely rule out the possibility that a small subset of RNase MRP particles does contain hPop4 (and Rpp21 and Rpp14). Such an interaction may be mediated by a direct interaction of hPop4 with the RNase MRP RNA, which we previously observed by in vitro RNA-binding experiments (Welting et al. 2004). It should also be noted that significant differences in RNase MRP and RNase P protein composition exist between the mammalian and yeast complexes (Walker and Engelke 2006). Therefore, the association of Pop4p with the yeast RNase MRP complex does not necessarily mean a similar association in mammalian cells as well. Protein subunits that are associated with both holoenzymes in yeast may be differentially associated with these enzymes in mammalian cells.
Four of the other proteins that were found in 12S fractions were also present in the presumptive pre-ribosomal complexes at 60–80S. Whereas the distribution over 12S and 60–80S complexes for most of these proteins was similar to that of RNase MRP RNA, the relative abundance of hPop1 in 60–80S material seemed to be higher than that for the other proteins. Currently it is not known whether this is due to the association of hPop1 with other complexes sedimenting at 60–80S or to substoichiometric amounts of this protein in 12S complexes.
Surprisingly, Rpp25 and Rpp20 were not detected at all in 60–80S fractions, even though they have been demonstrated to be stably associated with RNase MRP (van Eenennaam et al. 2002). Rpp25 and Rpp20 are evolutionarily related proteins, both belonging to the Alba superfamily (Aravind et al. 2003), which directly bind to each other (Welting et al. 2004) and most likely bind as a heterodimer to the P3 domain of RNase MRP RNA (Pluk et al. 1999; van Eenennaam et al. 2002). In RNase MRP RNA, this region is in close proximity to nucleotides 68–71 and the P4 stem, which are believed to be part of the catalytic center of the enzyme. Therefore, it is tempting to speculate that the Rpp25–Rpp20 heterodimer has to dissociate from the complex before the enzyme can associate with and cleave substrate RNAs, such as the pre-rRNA in the 60–80S complexes. Previously, Rpp20 has been reported to contain ATPase activity (Li and Altman 2001). It would be interesting to investigate whether this activity is required for the dissociation of the Rpp25–Rpp20 heterodimer from the RNase MRP complex during pre-rRNA processing.
Because previous studies showed that both Rpp25 and Rpp20 are associated with catalytically active RNase P, it is an interesting question whether the cleavage of substrate RNAs by RNase MRP is blocked by the association of the Rpp25–Rpp20 heterodimer. Currently, it is unclear what the function of these proteins may be. They may be involved in stabilization of the “free” RNase MRP (and RNase P) enzymes, they may play a role in their subcellular localization, they may be involved in the recruitment of RNase MRP/RNase P to substrate RNAs, or they may regulate the storage and/or activity of these enzymes. An overview of the differences in human RNase MRP composition is given in Figure 4. Because of the lack of glycerol sedimentation data on hPop5, we could not specifically assign this protein to either the 12S or the 60–80S RNase MRP populations.
FIGURE 4.
Differential association of protein subunits with the human RNase P and RNase MRP particles. The models used in this figure are adapted from and based upon a previously described model for the human RNase MRP complex (Welting et al. 2004). Protein subunits that are associated with 12S RNase P and both 12S and 60–80S RNase MRP are shown in light gray (1, hPop1; 5, hPop5; 30, Rpp30; 38, Rpp38; and 40, Rpp40). The proteins associated with both 12S RNase P and 12S RNase MRP are depicted in black (25, Rpp25; 20, Rpp20). The subunits preferentially associated with 12S RNase P are shown in dark gray (4, hPop4; 14, Rpp14; and 21, Rpp21). Only the major complex compositions based upon the current data are shown. A minority of RNase MRP may also contain hPop4, Rpp21 and Rpp14. (A) Schematic representation of the 12S human RNase P complex. (B) Composition of the human RNase MRP complexes. The differential association of the Rpp25–Rpp20 heterodimer is based upon 5%–40% glycerol gradient sedimentation data (Fig. 2). Note that because of the lack of glycerol gradient sedimentation data on hPop5, we could not assign this protein to either the 12S or the 60–80S RNase MRP species.
In addition to the complexes discussed above, a small subset of both RNase MRP and RNase P was reproducibly sedimenting at 40S. Both the RNAs and most of the protein components were detected in these fractions. Currently it is unknown with which other molecules or complexes the enzymes are associated in these 40S complexes. With regard to the 40S sedimentation of RNase MRP and RNase P, it is interesting to note that the B. subtilis RNase P particle has been reported to interact with the 30S small ribosomal subunit of this prokaryote (Barrera and Pan 2004).
A schematic representation of the differences observed in the composition of RNase MRP and RNase P complexes is presented in Figure 4. We conclude that hPop4, Rpp21, and Rpp14 are preferentially associated with the human RNase P and that cells probably contain heterogeneous pools of RNase MRP complexes, which differ in their protein composition depending on either their functional activity or their association with other complexes. Taken together, our data provide the first evidence for differences in protein composition of the human RNase MRP and RNase P complexes and indicate that Rpp20 and Rpp25 transiently associate with the RNase MRP complex. Further experiments will be required to shed light on the functional implication of these differences in particle composition.
MATERIALS AND METHODS
Glycerol density gradient centrifugation
HEp-2 monolayer cells were grown in T75 flasks to 70% confluence in DMEM (GIBCO BRL) supplemented with 10% fetal calf serum. After harvesting, 107 cells were resuspended in 750 μL of gradient buffer (20 mM HEPES/KOH at pH 7.6, 150 mM NaCl, 0.5 mM DTE) and disrupted by sonication (Branson microtip). Subsequently, Triton X-100 was added to a final concentration of 0.2% (v/v) and insoluble material was removed by centrifugation at 15,000g in a microcentrifuge. The 5%–40% (v/v) glycerol gradients were prepared using the Biocomp Gradient Master 107 in gradient buffer supplemented with 0.2% Triton X-100. The cleared cell lysate (750 μL) was layered on a 12-mL gradient. The gradients were centrifuged in a Sorvall TH641 rotor for 17 h at 100,000g at 4°C. After centrifugation, 500-μL fractions were collected manually and stored at −20°C. The 12%–18% (v/v) glycerol gradients were prepared as described above. However, on the latter gradients, lysate from only 106 HEp-2 cells was loaded.
RNA isolation and Northern blotting
To isolate total RNA from the glycerol gradient fractions, 35 μL per fraction was subjected to Trizol (Invitrogen) extraction. The aqueous phase was collected and the RNA was precipitated by the addition of one volume of isopropanol in the presence of 100 μg of glycogen. After centrifugation, the pellet was washed with 80% ethanol. Subsequently, the pellet was dissolved in 30 μL of formamide loading buffer. The RNAs were separated by denaturing urea-PAGE and subsequently transferred to Hybond-N membranes (Pharmacia) by electroblotting in 18.4 mM Na2HPO4, 6.5 mM NaH2PO4. After blotting, the RNAs were covalently bound to the membranes in a UV cross-linker and stored at 4°C.
SDS-PAGE and immunoblotting
To determine the distribution of the RNase MRP/RNase P protein subunits in the glycerol gradients, samples from gradient fractions were separated by SDS-PAGE and transferred to nitrocellulose membranes by electroblotting. Immunodetection of hPop1, hPop4, hPop5, Rpp14, Rpp20, Rpp21, Rpp25, Rpp30, Rpp38, and Rpp40 was performed using polyclonal rabbit antisera against these proteins (Lygerou et al. 1996b; Eder et al. 1997; Jarrous et al. 1998, 1999, 2001; van Eenennaam et al. 1999, 2001; Guerrier-Takada et al. 2002) and a monoclonal mouse antibody against Rpp20 (ModiQuest). Horseradish peroxidase-conjugated polyclonal swine anti-rabbit immunoglobulin antibodies were purchased from Dako (Glostrup) and applied as secondary antibody. Bound antibodies were visualized by enhanced chemiluminescence detection procedures.
In vitro transcription and antisense probing of RNAs
Antisense radiolabeled riboprobes were generated from plasmids carrying the sequences of the human RNase MRP RNA, RNase P RNA, and U3 snoRNA (Verheijen et al. 1994; Jacobson et al. 1997). The plasmids were linearized with the appropriate restriction endonucleases and transcribed in vitro as described before (Welting et al. 2004). After purification of the transcripts using a G-50 spin column, the antisense RNAs were stored at −20°C or directly used in hybridization experiments. For probing with the antisense riboprobes, Northern blots were blocked at 65°C with hybridization buffer (6× SSC, 0.1 mg/mL sheared herring sperm DNA, 0.2% SDS, 10× Denhardt's). After blocking for 1 h, the riboprobe was added and hybridization was performed overnight at 65°C. The blots were subsequently washed twice with 1× SSC, 0.2% SDS and twice with 0.1× SSC. The RNAs were visualized by autoradiography, and the signals were quantified using the Bio-Rad GS-363 PhosphorImager.
Transfection constructs, transient transfection, and immunoprecipitation
RNase MRP/RNase P protein cDNAs were cloned into a pCI-neo mammalian expression vector (Promega) containing the vesicular stomatitis virus (VSV) G-epitope sequence as described before (Welting et al. 2004). HEp-2 monolayer cells were cultured in a T75 flask and harvested at 70% confluence. For transfection, 20 μg of purified plasmid DNA was added individually to 5 × 106 HEp-2 cells in 800 μL of culture medium. The cells were electroporated at 260 V at a capacity of 950 μF using a Bio-Rad Genepulser II. After transfection, the cells were cultured in a T75 flask and harvested 24 h post-transfection. To check incorporation of the transiently expressed proteins into ribonucleoprotein particles, extracts were either loaded on 5%–40% (v/v) glycerol gradients (as described above) or subjected to immunoprecipitation. For immunoprecipitation, the cells were washed with PBS and cell pellets were resuspended in 700 μL of lysis buffer containing 25 mM Tris-HCl (pH 7.6), 100 mM KCl, 1 mM DTE, 2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, and 0.05% (v/v) NP-40. Subsequently, the cells were disrupted by sonication (Branson microtip), and insoluble material was removed by centrifugation (15,000g, 10 min). Simultaneously, anti-VSV mouse monoclonal antibodies or rabbit anti-Rpp38 antibodies were coupled to protein-A–agarose beads in 500 μL of IPP500 (500 mM KCl, 10 mM Tris-HCl at pH 8.0, 0.05% [v/v] NP-40). Prior to immunoprecipitation, the beads were washed with IPP150 (150 mM KCl, 10 mM Tris-HCl at pH 8.0, 0.05% [v/v] NP-40). For immunoprecipitation, the beads were mixed with 500 μL of total cell extract, and IPP150 was added to a total volume of 750 μL. After 2 h of incubation by end-over-end rotation at 4°C, the beads were washed three times with 500 μL of IPP150 and subsequently analyzed for pre-tRNA processing activity or subjected to RNA isolation and Northern blot hybridization as described above.
pre-tRNA processing assay
RNase P activity in immunoprecipitated material was monitored by the ability to process a pre-tRNA substrate from Schizosaccharomyces pombe tRNASer (Krupp et al. 1986) to mature tRNA and the released 5′ leader sequence. The substrate was transcribed in vitro from a linearized template using SP6 RNA polymerase in the presence of [α-32P]UTP. The RNA was purified from the reaction mixture by denaturing urea-PAGE, excision from the gel, and elution (overnight) in a buffer containing 100 mM NaCl and 0.1% SDS. After elution, the RNA was precipitated and dried pellets were stored at −20°C. For monitoring pre-tRNA processing activity in immunoprecipitated material, the beads containing the precipitates were resuspended in a buffer containing the purified pre-tRNA substrate, 20 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 1 mM DTE, and 50 mM KCl. The cleavage reaction was carried out for 20 min at 37°C under continuous agitation. The reaction was stopped, and the reaction products were resolved by urea-PAGE. The RNAs were visualized by autoradiography.
ACKNOWLEDGMENTS
We thank Sidney Altman and Cecilia Guerrier-Takada (Yale University, New Haven, Connecticut) and Nayef Jarrous (Hebrew University-Hadassah Medical School, Jerusalem, Israel) for their kind gifts of cDNAs encoding RNase P protein subunits and for antisera against RNase MRP/RNase P protein subunits. We are grateful to Reinout Raijmakers and Erwin van Dijk (Radboud University Nijmegen, Nijmegen, The Netherlands) for critical reading of the manuscript and for helpful comments. This work was supported in part by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2293906.
REFERENCES
- Aloy P., Bottcher B., Ceulemans H., Leutwein C., Mellwig C., Fischer S., Gavin A.C., Bork P., Superti-Furga G., Serrano L., et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- Altman S. The road to RNase P. Nat. Struct. Biol. 2000;7:827–828. doi: 10.1038/79566. [DOI] [PubMed] [Google Scholar]
- Aravind L., Iyer L.M., Anantharaman V. The two faces of Alba: The evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer M., Nilsen T.W., Costigan C., Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera A., Pan T. Interaction of the Bacillus subtilis RNase P with the 30S ribosomal subunit. RNA. 2004;10:482–492. doi: 10.1261/rna.5163104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain J.R., Pagan R., Kindelberger D.W., Engelke D.R. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain J.R., Lee Y., Lane W.S., Engelke D.R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes & Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.D., Clayton D.A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chu S., Zengel J.M., Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- Eder P.S., Kekuda R., Stolc V., Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc. Natl. Acad. Sci. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P., Bortolin M.L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Gill T., Cai T., Aulds J., Wierzbicki S., Schmitt M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S., Gallagher J.E., Vogelzangs J., Horstman W., van Venrooij W.J., Baserga S.J., Pruijn G.J. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60–80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Eder P.S., Gopalan V., Altman S. Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA. 2002;8:290–295. doi: 10.1017/s1355838202027954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.M., Ares M., Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M.R., Cao L.G., Taneja K., Singer R.H., Wang Y.L., Pederson T. Nuclear domains of the RNA subunit of RNase P. J. Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jarrous N. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N., Eder P.S., Guerrier-Takada C., Hoog C., Altman S. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- Jarrous N., Eder P.S., Wesolowski D., Altman S. Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA. 1999;5:153–157. doi: 10.1017/s135583829800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N., Reiner R., Wesolowski D., Mann H., Guerrier-Takada C., Altman S. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA. 2001;7:1153–1164. doi: 10.1017/s1355838201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Altman S. Protein–protein interactions with subunits of human nuclear RNase P. Proc. Natl. Acad. Sci. 2001;98:920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Guerrier-Takada C., Altman S. Protein–RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., Filipowicz W. 7–2/MRP RNAs in plant and mammalian cells: Association with higher order structures in the nucleolus. EMBO J. 1992;11:3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Kouzuma Y., Mizoguchi M., Takagi H., Fukuhara H., Tsukamoto M., Numata T., Kimura M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003;306:666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- Krupp G., Cherayil B., Frendewey D., Nishikawa S., Soll D. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe . EMBO J. 1986;5:1697–1703. doi: 10.1002/j.1460-2075.1986.tb04413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Matera A.G., Ward D.C., Craft J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: A possible coordinate role in ribosome biogenesis. Proc. Natl. Acad. Sci. 1996;93:11471–11476. doi: 10.1073/pnas.93.21.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Altman S. A subunit of human nuclear RNase P has ATPase activity. Proc. Natl. Acad. Sci. 2001;98:441–444. doi: 10.1073/pnas.021555498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowiak A.A., Granneman S., Mattox S.A., Speckmann W.A., Jones K., Pluk H., Venrooij W.J., Terns R.M., Terns M.P. Interaction of the U3-55K protein with U3 snoRNA is mediated by the box B/C motif of U3 and the WD repeats of U3-55K. Nucleic Acids Res. 2000;28:3462–3471. doi: 10.1093/nar/28.18.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Allmang C., Tollervey D., Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996a;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Pluk H., van Venrooij W.J., Seraphin B. hPop1: An autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996b;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Mann H., Ben Asouli Y., Schein A., Moussa S., Jarrous N. Eukaryotic RNase P: Role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol. Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Pluk H., van Eenennaam H., Rutjes S.A., Pruijn G.J., van Venrooij W.J. RNA–protein interactions in the human RNase MRP ribonucleoprotein complex. RNA. 1999;5:512–524. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G., Raska I., Scheer U., Tan E.M. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp. Cell Res. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Salinas K., Wierzbicki S., Zhou L., Schmitt M.E. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- Schmitt M.E., Clayton D.A. Characterization of a unique protein component of yeast RNase MRP: An RNA-binding protein with a zinc-cluster domain. Genes & Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- Sharin E., Schein A., Mann H., Ben Asouli Y., Jarrous N. RNase P: Role of distinct protein cofactors in tRNA substrate recognition and RNA-based catalysis. Nucleic Acids Res. 2005;33:5120–5132. doi: 10.1093/nar/gki828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M.P., Terns R.M. Small nucleolar RNAs: Versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Kiss T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Topper J.N., Clayton D.A. Characterization of human MRP/Th RNA and its nuclear gene: Full length MRP/Th RNA is an active endoribonuclease when assembled as an RNP. Nucleic Acids Res. 1990;18:793–799. doi: 10.1093/nar/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K., Steitz J.A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eenennaam H., Pruijn G.J., van Venrooij W.J. hPop4: A new protein subunit of the human RNase MRP and RNase P ribonucleoprotein complexes. Nucleic Acids Res. 1999;27:2465–2472. doi: 10.1093/nar/27.12.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eenennaam H., Lugtenberg D., Vogelzangs J.H., van Venrooij W.J., Pruijn G.J. hPop5, a protein subunit of the human RNase MRP and RNase P endoribonucleases. J. Biol. Chem. 2001;276:31635–31641. doi: 10.1074/jbc.M103399200. [DOI] [PubMed] [Google Scholar]
- van Eenennaam H., Vogelzangs J.H., Lugtenberg D., Van Den Hoogen F.H., van Venrooij W.J., Pruijn G.J. Identity of the RNase MRP- and RNase P-associated Th/To autoantigen. Arthritis Rheum. 2002;46:3266–3272. doi: 10.1002/art.10673. [DOI] [PubMed] [Google Scholar]
- Verheijen R., Wiik A., De Jong B.A., Hoier-Madsen M., Ullman S., Halberg P., van Venrooij W.J. Screening for autoantibodies to the nucleolar U3- and Th(7-2) ribonucleoproteins in patients’ sera using antisense riboprobes. J. Immunol. Methods. 1994;169:173–182. doi: 10.1016/0022-1759(94)90261-5. [DOI] [PubMed] [Google Scholar]
- Walker S.C., Engelke D.R. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welting T.J., van Venrooij W.J., Pruijn G.J. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]