Abstract
It has been proposed that the 7-methylguanosine cap and poly(A) tail of mRNAs have important functions in translation and transcript stability. To directly test these roles of the cap and poly(A) tail, we have constructed plasmids with a ribozyme within the coding region or 3′ UTR of reporter genes. We show that the unadenylated 5′ cleavage product is translated and is rapidly degraded by the cytoplasmic exosome. This exosome-mediated decay is independent of the nonstop mRNA decay pathway, and, thus, reveals an additional substrate for exosome-mediated decay that may have physiological equivalents. The rapid decay of this transcript in the cytoplasm indicates that this unadenylated cleavage product is rapidly exported from the nucleus. We also show that this cleavage product is not subject to rapid decapping; thus, the lack of a poly(A) tail does not always trigger rapid decapping of the transcript. We show that the 3′ cleavage product is rapidly degraded by Xrn1p in the cytoplasm. We cannot detect any protein from this 3′ cleavage product, which supports previous data concluding that the 5′ cap is required for translation. The reporter genes we have utilized in these studies should be generally useful tools in studying the importance of the poly(A) tail and 5′ cap of a transcript for export, translation, mRNA decay, and other aspects of mRNA metabolism in vivo
Keywords: uncapped, unadenylated, exosome
INTRODUCTION
Eukaryotic mRNAs have a modified nucleotide (the cap) at their 5′ end and a poly(A) tail at their 3′ end. Both the cap and poly(A) tail have been proposed to have important roles in a wide variety of mRNA processing reactions including transcription elongation, splicing, nuclear export, translation, and mRNA degradation (for review, see Mangus et al. 2003; Coller and Parker 2004; Aguilera 2005). For example, it is widely believed that in order for a transcript to leave the nucleus, it must be properly capped and polyadenylated, that efficient translation requires both a cap and a poly(A) tail on the mRNA, and that removal of the cap structure is an important step in mRNA degradation that is triggered by the removal of the poly(A) tail. These conclusions are mainly based on four types of experiments.
First, some of the evidence for the roles of the cap and poly(A) tail in translation and transcript stability is based on analysis of yeast mutants defective in capping or polyadenylation enzymes, or in the poly(A) binding protein that mediates most poly(A) effects. Surprisingly, unadenylated mRNAs in a pap1 mutant strain cofractionate with polyribosomes, suggesting that the poly(A) tail is not required for translation under these conditions (Patel and Butler 1992; Proweller and Butler 1994, 1997). Similarly, genetic approaches suggest that sequential removal of the poly(A) tail and cap are rate-limiting steps in mRNA decay. Specifically, removal of the poly(A) tail precedes degradation of the mRNA, and a ccr4 pan2 double mutant that prevents removal of the poly(A) tail also stabilizes the mRNA (Decker and Parker 1993; Tucker et al. 2001). Similarly, a dcp1 mutation that prevents removal of the cap structure stabilizes the mRNA (Beelman et al. 1996). While this experimental approach has the advantage of studying the roles of the 5′ cap and poly(A) tail in vivo, it has the disadvantage that the capping enzymes, poly(A) polymerase (Pap1p), and poly(A) binding protein (Pab1p) are encoded by essential genes, while the dcp1Δ and ccr4Δ, pan2Δ strains grow slowly (Sachs et al. 1987; Shibagaki et al. 1992; Mao et al. 1995; Beelman et al. 1996; Tsukamoto et al. 1997; Tucker et al. 2001). Consequently, many of these studies are carried out under conditions where the cells are dead or dying. In addition, with any genetic analysis it is difficult to distinguish direct effects from indirect effects.
A second approach to study the role of the mRNA cap and poly(A) tail is to use in vitro systems utilizing cell extracts. Various studies using rabbit reticulocyte, human HeLa cell extracts, or yeast extracts have shown that uncapped and unadenylated transcripts can be translated at very low levels. Translation is elevated upon addition of a cap or poly(A) tail, but optimal translation occurs when both a cap and poly(A) tail are present (Doel and Carey 1976; Munroe and Jacobson 1990; Iizuka et al. 1994; Michel et al. 2000). Cell extracts have also been utilized to study the roles of the 5′ cap and poly(A) tail on transcript stability (Boeck et al. 1996; Korner and Wahle 1997; Chen et al. 2001; Gao et al. 2001; Wang and Kiledjian 2001). However, it is often unclear whether the enzymes identified in vitro are the major enzymes in vivo (e.g., Tucker et al. 2001; Yamashita et al. 2005; Stoecklin et al. 2006). In addition, each step in gene expression is highly regulated and individual steps in gene expression are often interconnected; in vitro systems are unlikely to fully reflect this in vivo situation.
A third line of experiments to address the role of the cap and poly(A) tail is to introduce mRNA by electroporation or microinjection into cells. Studies using a wide variety of cells have shown that addition of a 5′ cap greatly increases translation of an mRNA, and this increase is amplified upon addition of a poly(A) tail (Nudel et al. 1976; Huez et al. 1981; Drummond et al. 1985; Galili et al. 1988; Gallie 1991; Masison et al. 1995). However, it is not clear to what extent this reflects a direct role in translation, since, at least in yeast, mRNA stability seems to be a major contributor to the effect of the poly(A) tail in this experimental approach (Brown and Johnson 2001). Furthermore, while these experiments technically occur in vivo, the mRNP proteins that normally become associated with mRNA during its biogenesis in the nucleus are unlikely to associate with electroporated RNA.
A fourth approach that has been especially useful to resolve the role of deadenylation and decapping in mRNA decay is a transcriptional pulse chase, where the state of the mRNA can be followed over time. These experiments have clearly shown that deadenylation and decapping normally precede degradation of the mRNA (Decker and Parker 1993; Muhlrad et al. 1994, 1995). With this approach it is hard to distinguish between causal relationships and purely chronological relationships. However, as pointed out above, mutants that are defective in deadenylation or decapping have more stable mRNAs, suggesting a causal relationship in this case (Decker and Parker 1993b; Beelman et al. 1996; Tucker et al. 2001).
We have generated reporter constructs that allow for the study of the roles of the cap and poly(A) tail in RNA processing reactions in vivo in wild-type yeast cells. This was achieved by inserting a ribozyme into the reporter gene of interest. Cleavage by such a ribozyme leads to a capped, but not adenylated 5′ fragment, which allows for characterization of the role of the poly(A) tail and an uncapped, but polyadenylated 3′ fragment that allows for characterization of the role of the cap. Importantly, this approach does not rely on the use of conditional mutants in essential genes. Using this system, we confirm some of the roles of the cap and poly(A) tail, but also find some unanticipated results. Importantly, we show that the translation product of an uncapped mRNA does not accumulate to detectable levels, but that an unadenylated mRNA is translated surprisingly well. We show that an uncapped mRNA is very rapidly degraded by Xrn1p, but that the simple absence of a poly(A) tail is not sufficient to trigger rapid decapping. We also show that unadenylated mRNAs are rapidly degraded by the cytoplasmic exosome. These studies clarify the normal role of the cap and poly(A) tail in a wild-type yeast cell. Furthermore, they suggest that similar approaches can be used to study the role of the cap and poly(A) tail in a variety of mRNA processing reactions, even in organisms where mutants in capping and polyadenylation are not available.
RESULTS
Experimental approach
To investigate the role of the cap and poly(A) tail and the lack of a stop codon on mRNA metabolism, we generated various plasmid constructs that were designed to produce mRNAs lacking a cap, poly(A) tail, and/or stop codon. We chose to do this by having the endogenous RNA polymerase II transcribe an initial transcript that was both capped and polyadenylated, but that was cleaved into an unadenylated 5′ fragment and an uncapped 3′ fragment. This experimental approach has the advantage that these mRNAs are transcribed in vivo in a wild-type yeast strain background, where the bulk of the mRNA has both a cap and poly(A) tail.
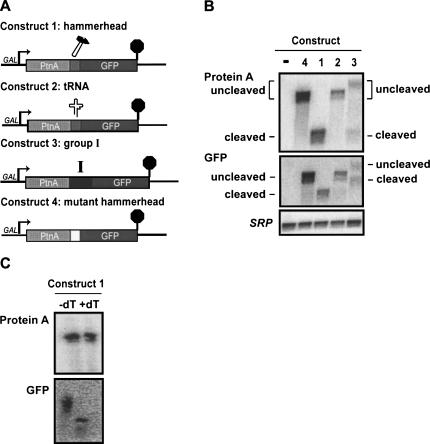
We used three different strategies to trigger post-transcriptional cleavage of the mRNA. In the first strategy, a hammerhead ribozyme was introduced into the mRNA (Fig. 1A, Construct 1). This hammerhead ribozyme was identical to that used by Duvel et al. (2002) and is known to be active in yeast. In a second approach, we inserted a tRNA into a reporter gene (Fig. 1A, Construct 2). tRNAs are normally processed at their 5′ and 3′ ends by the endoribonucleases P and Z, respectively (Xiao et al. 2001; Schiffer et al. 2002). In a third approach, we inserted a mutant Group I intron into a reporter gene (Fig. 1A, Construct 3). Selection of the 3′ splice site of the Tetrahymena Group I intron depends on a guanosine residue just 5′ of the 3′ splice site and on stem P9.0, which precedes this guanosine residue. Thus, Group I introns that lack the last few nucleotides are capable of cleaving at the 5′ splice site, but cannot complete splicing (Price and Cech 1988; Michel et al. 1989, 1990; Burke et al. 1990). Based on this, we used a Group I intron derivative lacking the last five nucleotides of the intron. In each of these three strategies, we initially inserted the cleavage sequence between the coding sequence for the ZZ domain of Protein A and GFP.
FIGURE 1.
Transcripts are efficiently cleaved by hammerhead ribozyme. (A) Construct 1: A hammerhead ribozyme sequence was inserted at the junction of Protein A (lacking a stop codon) and GFP. Construct 2: A Gly-tRNA sequence was inserted at the junction of Protein A and GFP. Construct 3: A mutated Group I intron, allowing for cleavage, but preventing ligation, was inserted at the junction of Protein A and GFP. Construct 4: The ACC sequence immediately following the ribozyme cleavage site in Construct 1 was changed to a CAA sequence, preventing cleavage and allowing for readthrough to the stop codon of GFP. (B) Total RNA from wild-type cells containing either empty vector (−), or the indicated construct was isolated for Northern hybridization analysis. Blots were hybridized to detect Protein A mRNA (oAV72), GFP mRNA (oAV223), or for the 7S RNA subunit of the Signal Recognition Particle (SRP) (oAV224) as a loading control. (C) Total RNA from wild-type cells containing Construct 1 was isolated and subjected to RNase H treatment either in the presence or absence of oligo(dT). Subsequent blots were hybridized with probes for Protein A (oAV72) or GFP (oAV224).
Hammerhead and Group I ribozymes cleave efficiently in vivo
To test whether any of our strategies are effective in generating cleaved mRNAs, we probed Northern blots of wild-type yeast containing the constructs described above with Protein A and GFP probes. This analysis indicated that the hammerhead and Group I ribozyme constructs behave as designed, but that the tRNA construct was not cleaved.
In yeast containing the hammerhead ribozyme construct, one single mRNA species is detected with a probe specific for Protein A mRNA (Fig. 1A,B, Construct 1). Three lines of evidence suggest that this is indeed the expected 5′ product resulting from hammerhead ribozyme cleavage. First, hybridization with a probe specific for Protein A mRNA yields a band of about the expected size for the 5′ cleavage product (485 nt), but too small for uncleaved mRNA. Second, rehybridizing the same blot with GFP does not detect this species, but instead detects a separate species that is the expected size for the 3′ fragment resulting from hammerhead cleavage (892 nt, not including the length of the poly[A] tail). Third, as an important control, a construct with a defective hammerhead ribozyme was included. This yields an mRNA that is much larger and hybridizes with both the Protein A and GFP probes (Fig. 1A,B, Construct 4; expected size is 1377 nt, not including the poly[A] tail). Although the hammerhead ribozyme has been used previously to study the metabolism of the unadenylated 5′ cleavage product (Duvel et al. 2002; Dower et al. 2004), the 3′ cleavage product has not, to our knowledge, been detected or studied previously. The detection of the 3′ cleavage product here provided an opportunity to study the fate of uncapped mRNAs in vivo (see below).
Although the results above are consistent with the conclusion that the hammerhead and Group I ribozymes cleave in vivo, this experiment alone cannot exclude that cleavage occurs during RNA extraction from the cells. In the remainder of this manuscript, we provide further evidence that is consistent with cleavage occurring in vivo and that appears inconsistent with the possibility of cleavage occurring during or post-RNA extraction. Specifically, our data show that the 5′ cleaved fragment, but not the full-length precursor is translated, and that mutations in the mRNA decay machinery differently affect the degradation of either the 5′ or 3′ cleavage fragment (see below).
The construct with the Group I-derived ribozyme also yielded the expected cleaved mRNA species, although cleavage was not as efficient, and uncleaved mRNA was also detectable (Fig. 1A,B, Construct 3). As expected, the 5′ cleavage product detected with the Protein A probe was similar in size to that from the hammerhead ribozyme construct. Rehybridization of this blot with a probe specific for GFP detected the 3′ cleavage product and the uncleaved mRNA. It is important to note that the Group I-derived ribozyme (420 nt) is much larger than the hammerhead ribozyme (52 nt). Since cleavage occurs between the Protein A and Group I ribozyme sequences, the 3′ cleavage product of the Group I ribozyme construct is significantly larger than the 3′ cleavage product from the hammerhead ribozyme construct. Similarly, the uncleaved fraction of the Group I ribozyme (Construct 3) is significantly larger than the uncleavable hammerhead control of Construct 4. Thus, the Group I-derived ribozyme is also effective at RNA cleavage, but in this context, is less efficient than the hammerhead ribozyme. From these results we conclude that the Group I ribozyme is also capable of cleaving itself, but does so less efficiently than the hammerhead ribozyme.
Overall, the results presented so far indicate that the hammerhead ribozyme construct behaves as expected and is cleaved more efficiently than the Group I ribozyme construct. Therefore, we used the hammerhead ribozyme for the majority of the experiments described below.
The 5′ cleavage product of a hammerhead ribozyme is not polyadenylated
If the hammerhead and Group I ribozymes cleave a longer initial transcript, the 5′ cleavage fragment should lack a poly(A) tail, while the 3′ fragment may contain a poly(A) tail added downstream of the GFP coding region. We confirmed this prediction with an RNase H/oligo(dT) assay for the presence of poly(A) tails on RNA. RNase H cleaves RNA only in an RNA/DNA hybrid and, thus, the combination of RNaseH and oligo(dT) specifically cleaves poly(A) tails. Figure 1C (top) shows that the mobility of the 5′ fragment of the hammerhead ribozyme construct does not change upon RNase H/dT treatment, indicating that this RNA does not have a poly(A) tail. This result essentially confirms previously published data (Duvel et al. 2002; Dower et al. 2004). As an important control, the blot was rehybridized with a probe specific for GFP mRNA, which shows a clear shift in mobility upon treatment with RNase H and oligo(dT) (Fig. 1C, bottom). This indicates that the RNase H assay worked as designed, and that the 3′ cleavage product of this hammerhead construct is polyadenylated. This polyadenylation of the 3′ cleavage fragment was expected, since all of our constructs include signals for cleavage and polyadenylation downstream of the GFP coding region.
Rapid degradation of the 5′ cleavage product of a hammerhead ribozyme depends on cytoplasmic exosome function
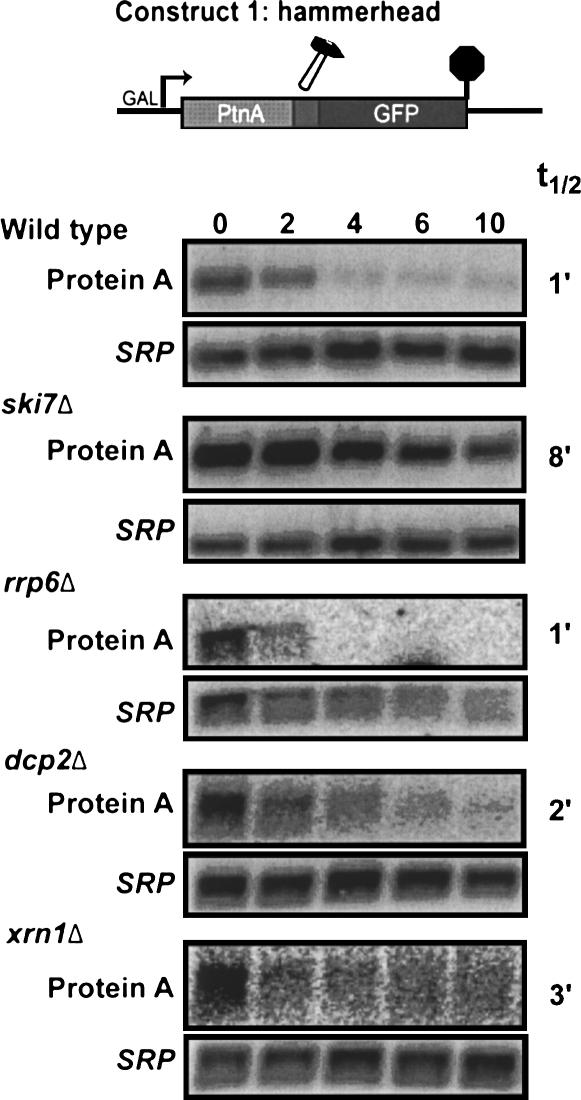
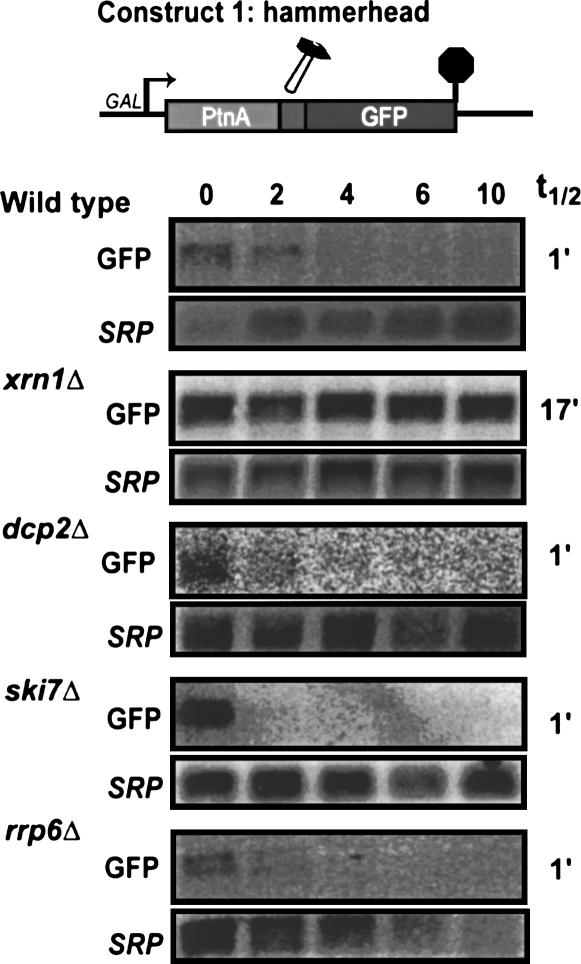
Current models for mRNA degradation state that the removal of the poly(A) tail triggers rapid decapping by Dcp2p, followed by Xrn1p-mediated degradation (Coller and Parker 2004). These models therefore predict that the 5′ cleavage product of a hammerhead ribozyme, which is not polyadenylated, is rapidly decapped and degraded in a Dcp2p- and Xrn1p-dependent manner. To test this prediction, we studied the degradation of the 5′ cleavage product of the hammerhead ribozyme (Construct 1) in wild-type yeast, as well as mutants defective in decapping (i.e., dcp2Δ), cytoplasmic 5′ to 3′ exonucleolytic decay (i.e., xrn1Δ), cytoplasmic exosome function (i.e., ski7Δ), or nuclear exosome function (i.e., rrp6Δ).
To facilitate measurements of mRNA decay rates, Construct 1 contained the GAL1 promoter. Thus, the reporter gene is transcribed when yeast is grown in the presence of galactose, but is rapidly turned off after addition of glucose. Figure 2 shows that the 5′ cleavage product of the hammerhead ribozyme is very unstable in a wild-type strain and disappears with a half life of ∼1 min after transcription is shut off. This degradation occurs at essentially the same rate in strains defective in decapping or cytoplasmic 5′ to 3′ exoribonuclease activity (dcp2Δ and xrn1Δ, respectively). The degradation rate was also unaltered in a strain defective in the nuclear exosome (i.e., rrp6Δ). In contrast, the 5′ hammerhead cleavage product is significantly more stable in a mutant defective in cytoplasmic exosome function (i.e., ski7Δ). Thus, the rapid degradation of the 5′ cleavage product does not depend on Dcp2p and Xrn1p, but instead depends on cytoplasmic exosome function.
FIGURE 2.
The 5′ cleavage product of a hammerhead ribozyme is degraded by the cytoplasmic exosome. Construct 1 was transformed into the indicated strains. Stability of the 5′ cleavage product was measured by isolating RNA at various times after inhibition of transcription of the reporter gene. Blots were hybridized with probes for Protein A mRNA (oAV72) and for the 7S RNA subunit of SRP (oAV224) to serve as a loading control.
The observation that rapid degradation of the 5′ cleavage fragment depends on cytoplasmic exosome function has three important implications. First, these data show that the 5′ cleavage product is degraded in the cytoplasm; therefore, this mRNA may be a useful tool to study the role of the poly(A) tail in translation (see below) and other aspects of cytoplasmic mRNA metabolism. Second, the observation that an unadenylated mRNA is relatively stable in a ski7Δ mutant indicates that the absence of a poly(A) tail does not trigger rapid decapping of this mRNA (see Discussion). Third, the rapid cytoplasmic exosome-dependent degradation may reflect an additional role for the exosome. As a first step toward figuring out what the physiological equivalents of the 5′ cleavage product are, we sought to determine what features of this 5′ cleavage product are responsible for its rapid degradation.
Degradation of the 5′ cleavage product of a hammerhead ribozyme is distinct from nonstop mRNA decay
Construct 1 has the hammerhead ribozyme inserted into the coding region of the Protein A-GFP fusion reporter. Therefore, the 5′ cleavage product does not contain a stop codon. We have previously shown that mRNAs that lack a stop codon are rapidly degraded by the cytoplasmic exosome (Frischmeyer et al. 2002; van Hoof et al. 2002), which raises the possibility that the 5′ cleavage product of the hammerhead ribozyme is degraded by the nonstop mRNA decay pathway. We tested this possibility with two experimental approaches.
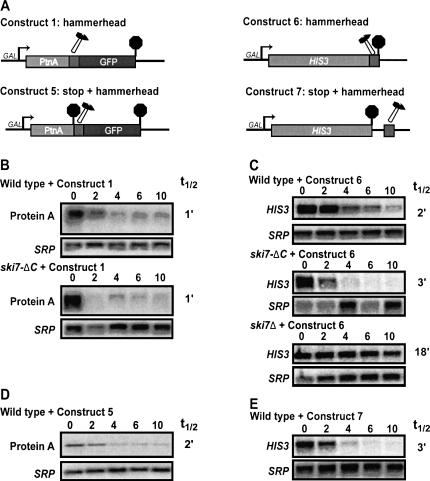
The degradation of normal mRNAs and nonstop mRNAs by the cytoplasmic exosome largely require the same trans-acting factors. The only known exception to this is that nonstop mRNA degradation requires both the N- and C-terminal domains of Ski7p, while all other cytoplasmic exosome functions require only the N-terminal domain of Ski7p (Araki et al. 2001; van Hoof et al. 2002). Therefore, we tested the effect of a ski7-ΔC mutation on the degradation of the 5′ cleavage products of our hammerhead ribozyme construct. Figure 3, A and B, shows that the ski7-ΔC has no effect on the degradation of the 5′ cleavage fragment. This experiment suggests that degradation of this 5′ cleavage product does not depend on the nonstop mRNA decay machinery.
FIGURE 3.
Degradation of the 5′ cleavage product of a hammerhead ribozyme is distinct from nonstop mRNA decay. (A) Construct 1: See Figure 1. Construct 5: A stop codon was inserted upstream of the ribozyme. Construct 6: A hammerhead ribozyme sequence was inserted immediately before the termination codon of HIS3. Construct 7: A hammerhead ribozyme sequence was inserted within the HIS3 3′ UTR, 55 bp 3′ of the stop codon. (B–E) Stability of the 5′ cleavage product was measured by isolating RNA at various times after inhibition of transcription of the reporter gene. Blots were hybridized with probes for Protein A mRNA (oAV72) and for the 7S RNA subunit of SRP (oAV224) to serve as a loading control.
It has previously been shown that mRNAs that have a stop codon inserted just 5′ of the poly(A) tail are not subject to nonstop mRNA decay (Frischmeyer et al. 2002). Therefore, in a second set of experiments, we tested the relevance of nonstop mRNA decay by introducing a stop codon upstream of the hammerhead ribozyme (Fig. 3A, Construct 5). The 5′ cleavage product from this construct now contains a stop codon and, thus, by definition, is no longer a substrate for nonstop mRNA decay. As shown in Figure 3D, this 5′ cleavage product containing a stop codon is still unstable; thus, its rapid degradation does not depend on the absence of a stop codon. The 5′ cleavage product of Construct 5 is stabilized in a ski7Δ strain, but not in a ski7-ΔC strain, indicating that it is degraded by the same pathway as the cleavage product that lacks a stop codon (data not shown). Overall, these results show that the 5′ cleavage product is still rapidly degraded if nonstop mRNA is blocked either in cis or in trans. Therefore, the cytoplasmic exosome degrades unadenylated mRNAs generated by the hammerhead ribozyme independent of nonstop mRNA decay.
Degradation of the 5′ cleavage product of a hammerhead ribozyme is independent of Protein A sequences
A second possible explanation for the rapid exosome-dependent degradation of the 5′ cleavage product of Construct 1 is that there is some specific sequence in the 5′ fragment that triggers its rapid degradation. In all of the above experiments, the 5′ cleavage product encodes the ZZ domain of Protein A from Staphylococcus aureus. To test whether this Protein A sequence contained some fortuitous instability sequence, we introduced a hammerhead ribozyme into the yeast HIS3 gene. In Construct 6 (Fig. 3A), the hammerhead sequence was introduced just 5′ of the HIS3 stop codon, thus generating a 5′ cleavage product that lacks a stop codon. In Construct 7 (Fig. 3A), the hammerhead ribozyme was inserted into the 3′ UTR, 55 nucleotides downstream of the stop codon. The fate of the 5′ cleavage product derived from these constructs paralleled that of the Protein A constructs. Specifically, the HIS3 5′ cleavage product was unstable, and stabilized by ski7Δ, but was not stabilized by ski7-ΔC or by the presence of a stop codon (Fig. 3C,E). Therefore, the specific sequence of the Protein A-derived construct is not responsible for the rapid cytoplasmic exosome-dependent mRNA degradation.
Degradation of the 5′ cleavage product of a hammerhead ribozyme is not triggered by a 2′ 3′ cyclic phosphate end
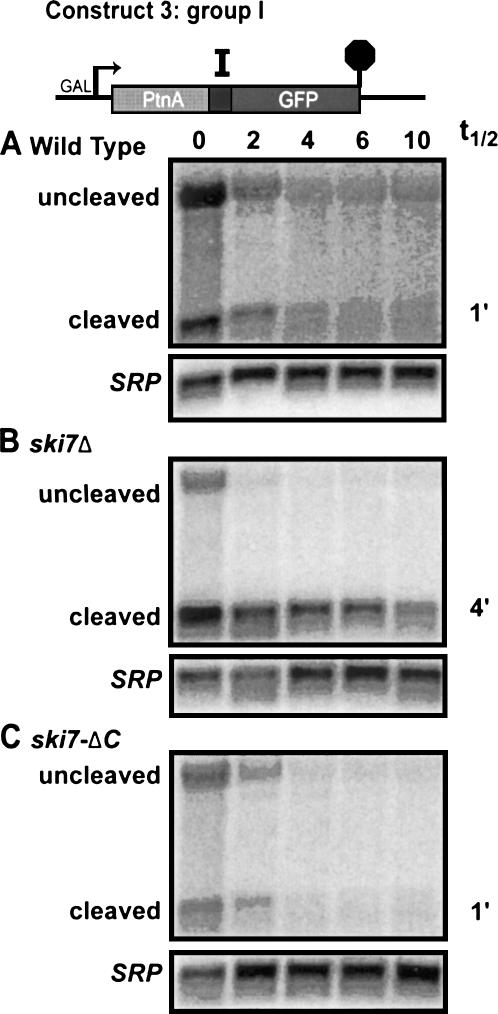
A third possible explanation for the rapid degradation of the 5′ cleavage product of the hammerhead ribozyme involves the fact that the hammerhead ribozyme generates a 5′ cleavage product ending in a 2′ 3′ cyclic phosphate (Hutchins et al. 1986). Since cellular mRNAs do not normally contain 2′ 3′ cyclic phosphates at their 3′ ends, the cell may specifically recognize this aberrant feature and target the mRNA to the cytoplasmic exosome. To determine whether this is the case, we analyzed decay rates of the 5′ cleavage product of the Group I intron-derived ribozyme. The first transesterification reaction of Group I introns is initiated when the 3′ OH of a guanine mononucleotide attacks the phosphodiester bond to be broken. This results in a 5′ cleavage product with 2′ and 3′ hydroxyls, which is identical to the natural 3′ end of mRNAs (Zaug and Cech 1982). Thus, the Group I intron-derived ribozyme of Construct 3 results in cleavage without the production of a 2′ 3′ cyclic phosphate and, thus, allows us to test whether such a cyclic phosphate is required for the rapid decay of the 5′ cleavage fragment. Figure 4A shows that the 5′ cleavage product of the Group I ribozyme is rapidly degraded in a wild-type strain. Importantly, in a ski7Δ strain, this mRNA fragment is stabilized (Fig. 4B). Thus, the 5′ cleavage products with 2′ 3′ cyclic phosphates and 2′ and 3′ hydroxyls behave the same. It should be noted that in addition to the 5′ cleavage product, the Protein A probe also detects some full-length (uncleaved) Protein A-Group I-GFP mRNA that disappears with similar kinetics from the wild-type and ski7Δ strains. We do not know whether this disappearance reflects the cleavage rate of the Group I ribozyme or the degradation rate of uncleaved mRNA. Overall, this experiment indicates that a 2′ 3′ cyclic phosphate is not responsible for triggering the exosome-dependent degradation of 5′ products of ribozyme cleavage.
FIGURE 4.
Ribozyme cleaved transcript with a 3′ hydroxyl and a 2′3′ cyclic are both degraded by the same pathway. Total RNA was isolated from designated strains containing Construct 3 and subjected to stability assays. Top bands correspond to uncleaved transcript and bottom bands correspond to 5′ cleaved product (Protein A). All blots were hybridized with probes for Protein A mRNA (oAV72) as well as for the 7S RNA subunit of SRP (oAV224).
Similar to the hammerhead ribozyme Construct 1, the Group I intron construct does not contain a stop codon in the Protein A region. In order to determine whether rapid degradation of the Group I ribozyme cleavage product requires the nonstop mRNA decay pathway, we also analyzed its stability in a ski7-ΔC strain. This experiment shows that the 5′ cleavage product is not stabilized in this strain (Fig. 4C), indicating that the 5′ cleavage product of the Group I ribozyme is also degraded by the cytoplasmic exosome independent of the nonstop mRNA decay machinery. Together, several different experiments indicate that the rapid degradation of unadenylated 5′ cleavage fragments of ribozymes requires the cytoplasmic exosome in trans, and does not depend on nonstop mRNA decay, specific sequences, or a 2′ 3′ cyclic phosphate in cis.
The unadenylated 5′ cleavage product of a hammerhead ribozyme is translated
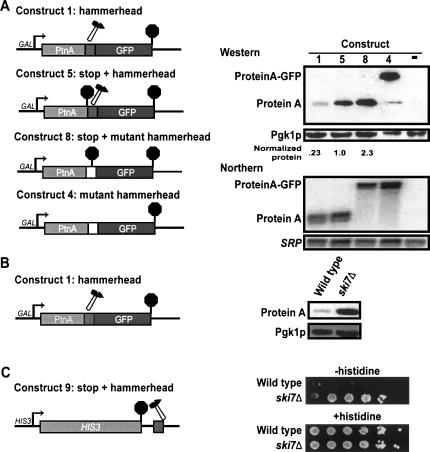
The experiments described above establish that the 5′ cleavage product of a hammerhead ribozyme is degraded in the cytoplasm and, thus, should be available for translation. Western blot analysis shows that this fragment is indeed translated. Figure 5 shows that either the 5′ cleavage product without (Construct 1) or with a stop codon (Construct 5) is translated and results in readily detectable levels of protein. The Western blot analysis included two critical controls, which carry mutant hammerhead ribozymes that are defective for cleavage. The first control (Construct 8) encodes a protein that is identical to that produced from the 5′ cleavage product of Construct 1 and serves as a size marker for the expected translation product. This control construct was generated by mutating the three nucleotides immediately following the cleavage site. This mutation both disrupts the secondary structure required to form an active ribozyme and introduces a stop codon. In addition, this control provides a measure of the expected steady-state level of Protein A produced from a polyadenylated mRNA. The second control (Construct 4) encodes a protein that is identical to that produced from mRNA that fails to be cleaved by the hammerhead ribozyme. This control construct was generated by mutating the three nucleotides immediately following the cleavage site to a different sense codon. Therefore, the mRNA is uncleavable and codes for a Protein A-GFP fusion protein. This control construct produced the expected Protein A-GFP fusion protein, although in some experiments a small amount of a Protein A decay fragment was also detectable (Fig. 5A; note that less protein was loaded to correct for higher expression levels). In contrast, cells containing a construct with an active hammerhead ribozyme construct accumulated only the Protein A product and never any detectable Protein A-GFP fusion protein. This control rules out the possibility that the Protein A from Construct 1 is a breakdown product of Protein A-GFP translated from a small amount of uncleaved mRNA. Therefore, these results indicate that the unadenylated 5′ cleavage product of a hammerhead ribozyme is translated.
FIGURE 5.
The 5′ cleavage product of a hammerhead ribozyme is translated. (A) Construct 8: A stop codon was inserted at the ribozyme cleavage site to prevent cleavage and to terminate translation of the uncleaved mRNA. Other constructs as described above. (Right) Western analysis was performed with antibodies specific for Protein A, on wild-type strains containing the indicated constructs or an empty vector. Construct 4 serves only as a size marker for the ProteinA-GFP fusion protein. Because this fusion protein is expressed at much higher levels, 10-fold less protein was loaded. Northern analysis was performed on the same cultures and probed for Protein A mRNA (oAV72) and the 7S RNA subunit of SRP (oAV224). (B) Western analysis of Construct 1 in wild-type and ski7Δ strains with antibodies specific for Protein A and for the loading control Pgk1p. (C) Construct 9: A hammerhead ribozyme was inserted 55 bp into the HIS3 3′ UTR. Wild-type (BY4741) and ski7Δ (1852) strain, each containing Construct 9, were grown 5 d on medium lacking or containing histidine.
Protein levels from a construct with an active ribozyme were roughly similar to the control construct that contained a stop codon inactivating the ribozyme (Fig. 5A, cf. Constructs 5 and 8). Upon quantification of Protein A levels, and correction for differences in the steady state level of the mRNA, we observed that the protein level produced from the unadenylated cleavage product of Construct 5 was only two- to threefold lower than that observed for the uncleavable mRNA of Construct 8. Thus, the absence of a poly(A) tail may reduce translation somewhat, but an unadenylated mRNA can clearly be translated. We also found an about fourfold difference in the protein level produced from Construct 1 and Construct 5. Both of these constructs produce an unadenylated mRNA, but Construct 5 includes a stop codon, while Construct 1 does not. The reduced protein level for Construct 1 may reflect a reduced translation rate on mRNAs that lack a stop codon or an increased protein degradation rate if the protein is not properly released from the ribosome. Overall, these results suggest that the absence of either a poly(A) tail or a stop codon reduces the steady-state protein level, probably by reducing translation rates, and that these effects are additive.
Our conclusions that the 5′ cleavage product is translated and that the 5′ cleavage product is stabilized in a ski7Δ strain predict that the translation product should be more abundant in a ski7Δ strain than in a wild-type strain. Consistent with this prediction and, thus, these two conclusions, Figure 5B shows that Protein A derived from Construct 1 is indeed up-regulated by ski7Δ. Similar results were obtained with Construct 5 (data not shown).
As a second assay for the translation of ribozyme cleavage products, we analyzed the growth phenotype of strains containing the HIS3-ribozyme reporter (Construct 9). This Construct 9 is identical to Construct 7, except that it contains the HIS3 promoter instead of the GAL1 promoter. Since the HIS3-ribozyme reporter is the only source of His3p protein, growth in the absence of added histidine is a result of the translation of the HIS3-ribozyme reporter mRNA. Consistent with the conclusion that the 5′ fragment is translated, we found that ski7Δ strains that carried the HIS3-ribozyme construct grew better in the absence of histidine than wild-type strains carrying the same construct (Fig. 5C). This observation indicates that the ski7Δ strain accumulates higher levels of His3p, and thus confirms both that this fragment is translated and that the cytoplasmic exosome is responsible for degrading this mRNA.
Rapid degradation of the 3′ cleavage product of a hammerhead ribozyme depends on the cytoplasmic 5′ to 3′ exoribonuclease Xrn1p
In the major pathway of mRNA degradation, the removal of the cap structure by Dcp2p exposes the body of the mRNA for rapid degradation by the cytoplasmic 5′ to 3′ exonuclease Xrn1p (Decker and Parker 1993; Muhlrad et al. 1994; Steiger et al. 2003). Since the 3′ cleavage product of the hammerhead ribozyme contains a 5′ hydroxyl, it would be expected to be degraded rapidly by Xrn1p, independent of Dcp2p (Hutchins et al. 1986). To test this hypothesis, we measured the decay rate of the 3′ cleavage product of the Protein A-ribozyme-GFP construct in wild-type yeast, as well as in mutants defective in decapping (i.e., dcp2Δ), cytoplasmic 5′ to 3′ exonucleolytic decay (i.e., xrn1Δ), cytoplasmic exosome function (i.e., ski7Δ), or nuclear exosome function (i.e., rrp6Δ) (Fig. 6).
FIGURE 6.
The 3′ cleavage product of a hammerhead ribozyme is degraded by Xrn1p. Construct 1 was transformed into the indicated strains. Stability of the 3′ cleavage product was measured by isolating RNA at various times after inhibition of transcription of the reporter gene. Blots were hybridized with probes for GFP mRNA (oAV223) or the 7S RNA subunit of SRP (oAV224).
In wild-type strains, this cleavage product is rapidly degraded. In a dcp2Δ strain, the stability of this product is not affected, indicating that the decapping enzyme is not required for the normal decay of this product. This product is also unstable in ski7Δ and rrp6Δ mutants, which verifies that this cleavage product is not being degraded by the nuclear or cytoplasmic exosomes. In an xrn1Δ strain, the 3′ cleavage product is stabilized, suggesting that Xrn1p normally degrades this product. These results suggest that the capacity of Xrn1p to degrade uncapped mRNAs is independent of previous steps in the deadenylation dependent decapping mRNA decay pathway.
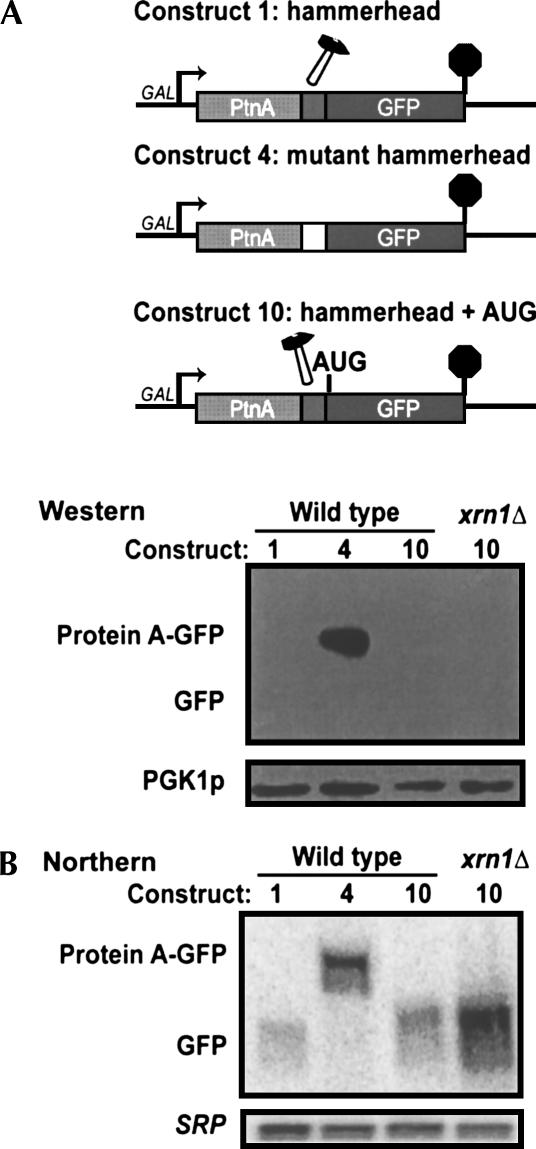
The protein encoded by the 3′ cleavage product of a hammerhead ribozyme is not detectable
Since the 3′ cleavage product of the hammerhead ribozyme lacks a 5′ cap, this product may be a useful in vivo tool to study the requirement for the 5′ cap in translation. Therefore, we set out to determine whether 3′ cleavage products resulting from cleavage by a ribozyme can be translated. The constructs described above did not include the start codon of GFP. We therefore made Construct 10, which is similar to Construct 1, except that it has an AUG start codon just 3′ of the hammerhead sequence and in frame with the GFP coding region. Figure 7 shows that no GFP protein is detectable in strains containing GFP with or without an in-frame start codon (Constructs 1 and 10, respectively). As a positive control we included Construct 4, which contains a mutant ribozyme and encodes a Protein A-GFP fusion protein. This fusion protein was easily detectable, indicating that the Western blot worked. Since the 3′ cleavage product of the hammerhead ribozyme is degraded by Xrn1p, we reasoned that a more sensitive assay for translation of this mRNA would be to look for the encoded GFP in an xrn1Δ strain. Figure 7 shows that even in an xrn1Δ strain, the GFP protein is undetectable. These results are consistent with the proposed importance of the cap for translation (see Discussion).
FIGURE 7.
The protein encoded by the 3′ cleavage product of a hammerhead ribozyme is not detectable. (A) Constructs 1 and 4 were constructed as stated previously. An AUG codon was inserted before the first codon of GFP to form Construct 10. Western analysis with antibodies specific for GFP or Pgk1p was performed on wild-type or xrn1Δ strains containing the indicated constructs. (B) Northern analysis of the same strains as in A hybridizing for GFP mRNA (oAV223) or the 7S RNA subunit of SRP (oAV224).
DISCUSSION
Ribozymes are useful tools to study mRNA metabolism in vivo
In this study, we have generated transcripts that lack a 5′ cap or poly(A) tail by inserting a hammerhead ribozyme or mutated Group I intron within a coding region or 3′ UTR. In agreement with previous studies, the hammerhead ribozyme cleaves a transcript very efficiently, leaving no detectable full-length product (Duvel et al. 2002; Dower et al. 2004). We also show that a mutated Group I intron is capable of cleaving a transcript in yeast. This suggests that Group I-derived ribozymes might be useful tools for studying eukaryotic RNA metabolism. One key difference between the hammerhead ribozyme and the Group I-derived ribozyme is that the hammerhead ribozyme produces an mRNA with a 2′ 3′ cyclic phosphate, while the Group I-derived ribozyme produces an mRNA with 2′ and 3′ hydroxyls at its 3′ end. Using the Group I-derived ribozyme may be preferable in some instances, because 2′ 3′ cyclic phosphates are not usually found in mRNA.
As an alternative to using ribozymes, we also tested whether we could target RNase P and/or RNase Z to an mRNA by inserting the sequence for a tRNA into the target mRNA. RNase P and RNase Z generate the 5′ and 3′ ends of tRNAs, respectively, and recognize the overall secondary structure of tRNA (Xiao et al. 2001; Schiffer et al. 2002). The chimeric mRNA/tRNA is apparently not efficiently processed by both RNase P and RNase Z. We do not fully understand why this would be. One possible explanation is that RNase P and RNase Z do not cleave the mRNA–tRNA hybrid because the RNA substrate and the enzymes are in different locations within the cell. A second possibility is that the tRNA in the mRNA–tRNA hybrid is misfolded, either because of the specific sequences we used, or because mRNP proteins prevent the tRNA portion from folding, thus preventing recognition by RNases P and Z.
Although ribozymes have previously been used to study the fate of the 5′ cleavage product, we extended those studies by also studying the 3′ cleavage product. A simple prediction of the torpedo model of transcription termination is that the 3′ cleavage product does not accumulate. Therefore, we were surprised that this 3′ cleavage product is readily detectable. Specifically, the torpedo model predicts that the uncapped 5′ end that results from cleavage and polyadenylation is an important trigger for transcription termination (Kim et al. 2004; Teixeira et al. 2004; West et al. 2004). This model states that the downstream uncapped 5′ end generated during the cleavage step targets the sequences downstream of the cleavage site to degradation by Rat1p, and that transcription termination results when Rat1p catches up with the transcribing polymerase. Our observation that the 3′ cleavage product is detectable and is degraded by Xrn1p suggests that the presence of a hammerhead ribozyme is not sufficient to trigger transcription termination.
Both Dower et al. (2004) and Duvel et al. (2002) have previously used hammerhead ribozymes in yeast. Based on cosedimentation with polyribosomes, Duvel et al. (2002) concluded that the ribozyme-cleaved mRNAs are efficiently exported from the nucleus. In contrast, Dower et al. (2004) reported accumulation of the 5′ cleavage product in dots in the nucleus using fluorescence in situ hybridization (FISH). Our data show that the 5′ cleavage product is rapidly degraded in the cytoplasm, and thus the mRNA must be exported rapidly. There are numerous differences in experimental details that might explain this apparent discrepancy. Importantly, while we suspect that hammerhead ribozyme cleavage is very rapid in vivo, we cannot exclude that it is the uncleaved precursor that is released from the site of transcription, exported, and subsequently cleaved. However, while this work was in preparation, Bird et al. (2005) showed that the 5′ cleavage product of a different ribozyme is also exported in Xenopus oocytes and human cells, suggesting that the export of unadenylated mRNAs is an evolutionarily conserved phenomenon. Clearly, future studies are needed to fully define the role of the poly(A) tail and the cleavage and polyadenylation process in nuclear export.
In addition to tools in basic research, ribozymes have been explored as tools to control gene expression in gene-therapy protocols. For example, Yen et al. (2004) introduced a large number of ribozymes into different positions of a mammalian reporter mRNA. Some ribozymes were effective in reducing gene expression when inserted into the 5′ UTR, but had much less effect when inserted into the 3′ UTR. Our results suggest a possible explanation for this, since we would predict that upon cleavage of the ribozyme in the 5′ UTR, the mRNA would no longer be translated, while cleavage of the ribozyme in the 3′ UTR may have less effect on the translation of the mRNA. Thus, a more detailed understanding of the fates of ribozyme-cleaved mRNAs may have important implications for such gene-therapy approaches.
The role of the cap, poly(A) tail, and stop codon in translation
The role of the cap in translation is well supported by biochemical experiments. However, in vivo analysis of the role of the cap in a wild-type yeast cell has been very limited. We therefore tested whether the uncapped 3′ hammerhead cleavage product can be translated. We failed to detect any GFP encoded by this uncapped RNA, which is consistent with the cap being very important for efficient translation.
The role of the poly(A) tail in translation, especially in a rapidly growing yeast cell, is less well established. In translation extracts, polyadenylated mRNA is more efficiently translated than unadenylated mRNAs (see Introduction). In vivo, the poly(A) tail is bound by the poly(A) binding protein Pab1p, which in turn interacts with the translation factor eIF4G, suggesting that the poly(A) tail has a role in translation (Tarun and Sachs 1996). On the other hand, while Pab1p is an essential gene, a deletion of the PAB1 gene can be suppressed by a wide variety of other mutations (Sachs and Davis 1989). Therefore, Pab1p cannot be absolutely required for translation. The poly(A) polymerase (Pap1p) is also an essential gene, but studies utilizing a temperature-sensitive allele (pap1-1) have shown that these strains accumulate unadenylated transcripts, which are translated (Patel and Butler 1992; Proweller and Butler 1994, 1997). Consistent with these latter observations, we find that the unadenylated 5′ cleavage fragment of a hammerhead ribozyme is translated surprisingly well.
Our data indicate that a cleavage product that lacks a stop codon produces lower steady-state levels of proteins than cleavage products that contain a stop codon. This is not entirely surprising, since the stop codon is thought to be important for the release of ribosomes from mRNA. Thus, in the absence of a stop codon, the ribosome may stall at the end of an mRNA and prevent completion of translation. An alternative explanation for the reduced levels of protein is that there may be differences in protein-degradation rates. Our Constructs 1 and 8 encode the exact same amino acid sequence, but the stop codon in Construct 8 would cause the covalent bond between the last amino acid and the last translating tRNA to be hydrolyzed. Thus, the protein would be released from the ribosome as a free protein. In contrast, since Construct 1 does not contain a stop codon, the protein might be released while still covalently attached to the last tRNA, which may affect protein-degradation rates. Future experiments will be needed to distinguish between these possibilities. Inada and Aiba (2005) have recently reported that mRNAs that either lack a stop codon or have a stop codon just before the poly(A) tail do not produce detectable levels of protein. We can clearly detect the protein produced from an mRNA that lacks a stop codon (Construct 1) or an mRNA with a short 3′ UTR (Construct 5), although the steady-state protein levels are reduced.
The role of the cap and poly(A) tail in mRNA decay
Current models for mRNA degradation state that mRNA degradation is initiated by a gradual removal of the poly(A) tail. In the main pathway of mRNA decay, this deadenylation then triggers the mRNA to enter P-bodies, removal of the cap structure by Dcp2p, and degradation of the mRNA by the 5′ to 3′ exoribonuclease Xrn1p (Muhlrad et al. 1994; Dunckley and Parker 1999; Sheth and Parker 2003; Steiger et al. 2003). In an alternate pathway that is two- to fivefold slower, deadenylation is followed by degradation of the body of the mRNA from the 3′ end by the exosome (Muhlrad et al. 1995; Jacobs Anderson and Parker 1998; Cao and Parker 2001).
It is currently not clear whether 5′ to 3′ decay can happen without prior targeting to P-bodies and decapping. We find that the uncapped 3′ cleavage product of a hammerhead ribozyme is rapidly degraded, and that this degradation is slower in an xrn1Δ strain, but not in a dcp2Δ strain. This indicates that the final 5′ to 3′ decay can happen independent of the decapping step. Two recent publications indicate that this is a more general phenomenon. Orban and Izaurralde (2005) showed that after RISC induced cleavage of a Drosophila mRNA, the 3′ cleavage product is degraded in an Xrn1p-dependent, but Dcp2-independent manner. Similarly, Doma and Parker (2006) showed that a stalled ribosome can trigger endonuclease digestion of an mRNA, and that the 3′ cleavage product is degraded by Xrn1p. Further experiments are needed to determine whether such Dcp2-independent 5′ to 3′ decay occurs in P-bodies or is also independent of P-bodies.
A prediction of current models of mRNA degradation is that an unadenylated mRNA will be rapidly decapped and degraded. This is not what we observed for the unadenylated 5′ cleavage products of either a hammerhead ribozyme or Group I-derived ribozyme. Specifically, in a ski7Δ strain, where degradation rates reflect decapping rates, degradation of these 5′ cleavage products was relatively slow. Consistent with our data is the observation that the unadenylated 5′ cleavage fragment resulting from RISC cleavage is also fairly stable when the exosome is inactivated, and therefore not subject to rapid decapping (Orban and Izaurralde 2005). One interpretation is that it is not the absence of the poly(A) tail per se that triggers decapping, but that the process of deadenylation or the resulting oligo(A) tail triggers decapping, possibly by altering the mRNP structure or mRNA location. For example, it is known that certain proteins (e.g., Lsm1-7) are recruited to the mRNP upon completion of deadenylation, and that the mRNA is targeted to P-bodies (Tharun and Parker 2001; Sheth and Parker 2003). Our results suggest that the absence of a poly(A) tail is not sufficient to trigger targeting to P-bodies and decapping. Instead, the process of deadenylation, or some event occurring at the same time as deadenylation, may result in Lsm protein recruitment and/or RNA localization to P-bodies.
In this study, we show that the 5′ fragment resulting from ribozyme cleavage is rapidly degraded. This is in contrast to previous studies that concluded that such cleavage products are relatively stable (Donahue and Fedor 1997; Duvel et al. 2002). These previous conclusions were based solely on steady-state mRNA levels, which do not always accurately reflect decay rates and certainly do not reveal the enzymes by which mRNAs are degraded. Although we have established that the 5′ fragment that results from ribozyme cleavage is degraded by the cytoplasmic exosome, it is not entirely clear what feature of the mRNA is important for its degradation. Specific sequences do not appear to be required, as similar results were obtained with the exogenous Protein A gene and the endogenous HIS3 gene. The nature of the very 3′ end (2′, 3′ cyclic phosphate vs. 2′ and 3′ hydroxyls) also appears irrelevant, since similar results were obtained with a hammerhead- and Group I-derived ribozyme. We were somewhat surprised that the cleavage fragment with a 2-3′ cyclic phosphate is rapidly degraded by the cytoplasmic exosome, since in vitro, the exosome is inhibited by a 3′ phosphate (Mitchell et al. 1997). Possible explanations include that a 3′ phosphate is inhibitory, but a cyclic phosphate is not, or that the cyclic phosphate generated by the hammerhead ribozyme is rapidly removed in vivo. One other possible feature that targets the 5′ cleavage product to the exosome is the absence of a poly(A) tail and/or its associated poly(A) binding protein (Pab1p). Consistent with this hypothesis, Coller et al. (1998) have previously shown that tethering Pab1p to an unadenylated mRNA will stabilize this mRNA. However, the phenotype of a pab1Δ mutant is not consistent with the absence of Pab1p triggering exosome-mediated decay. Specifically, in a pab1Δ strain, mRNA is still degraded by deadenylation and decapping (Caponigro and Parker 1995). Thus, the complete absence of Pab1p in the cell does not target normal mRNAs to the cytoplasmic exosome. Interestingly, natural substrates of the cytoplasmic exosome include the L-A and Killer virus RNAs that also lack a 3′ poly(A) tail (Ridley et al. 1984; Masison et al. 1995).
In addition to having pathways for degradation of normal transcripts, eukaryotic cells have pathways to degrade aberrant mRNAs. One of these pathways is nonstop mRNA decay, which is the very rapid degradation of mRNAs that lack a stop codon by the cytoplasmic exosome. Both mutations in cis (i.e., the introduction of a stop codon) and mutations in trans (i.e., ski7Δc) indicate that the nonstop mRNA mechanism is not required for the degradation of unadenylated mRNA fragments described here. However, this does not mean that unadenylated mRNAs are immune to nonstop decay. Importantly, nonsense-mediated mRNA decay and nonstop mRNA decay are easily studied when analyzing aberrant versions of an otherwise stable reporter gene such as PGK1. However, aberrant versions of an already unstable reporter gene such as MFA2 are much less useful in this regard. Trying to analyze whether the 5′ cleavage product is immune to nonstop mRNA decay is not unlike trying to study nonstop mRNA decay or nonsense-mediated mRNA decay by creating aberrant versions of MFA2 and claiming that it is immune because it is not significantly destabilized relative to the wild-type MFA2. Similarly, we suspect that unadenylated nonstop mRNAs are targeted to the exosome by two independent mechanisms, and that the mechanism that targets unadenylated mRNAs to the exosome might mask any effect of nonstop mRNA decay. In this view, only simultaneous blocks in both pathways will stabilize these mRNAs.
MATERIAL AND METHODS
Yeast strains
ski7Δ, xrn1Δ, and rrp6Δ in the BY4741 strain background were obtained from Open Biosystems. The ski7-ΔC strain (yav577) has been previously described (van Hoof et al. 2002). The dcp2Δ strain yRP1346 and the isogenic wild-type strain yRP840 have been described previously (Hatfield et al. 1996; Dunckley and Parker 1999).
Plasmids
All Protein A-GFP plasmids are modified from pMOV (Lydall and Weinert 1997). The ZZ domain of Protein A was PCR amplified from plasmid pEZZ18 (Amersham) with oligonucleotides oRP999 and oRP1000 (Table 1), and cloned into the XbaI and BamHI sites, and GFP and the 3′ UTR of ADH1 gene were isolated from pFA6a-GFP(S65T)-KanMX6 (Longtine et al. 1998) as a BamHI–BglII fragment and inserted into the BamHI site. The resulting plasmid (pAV214) contains the GAL promotor, ZZ domain, GFP, and ADH1 3′ UTR.
Construct 1 (pAV216) was created by inserting oligonucleotides oAV40 and oAV41 into the BamHI site of pAV214.
Construct 2 (pAV265) was created by PCR amplifying the SUF5 gene with oligonucleotides oAV152 and oAV153. The resulting PCR product was then digested with BamHI and BglII and ligated into pAV214 cut with BamHI.
Construct 3 (pAV275) was created by PCR amplifying the Group I intron from pBGST7 (Been and Cech 1986) with oligonucleotides oAV253 and oAV254. The Group I intron was then inserted as a BamHI fragment into the BamHI site of pAV214.
Construct 4 (pAV218) was created by inserting oligonucleotides oAV44 and oAV45 into the BamHI site of pAV214.
Construct 5 (pAV226) was created by inserting oligonucleotides oAV85 and oAV86 into the BamHI site of pAV214 and then inserting oAV40 and oAV41 into the recreated BamHI site.
Construct 6 (pAV263) was created by PCR amplifying the HIS3 coding region, ribozyme, and HIS3 3′ UTR using pAV241 as template and primers oAV183 and oAV203. This insert was digested with SacI and XbaI and ligated into pMOV (Lydall and Weinert 1997), then cut with the same enzymes. pAV241 was created by overlap extension PCR: Primers oAV87 and oAV155 were used to amplify the coding region of HIS3, with 236 bp upstream and a hammerhead ribozyme downstream. Primers oAV154 and oAV103 were used to amplify the hammerhead ribozyme followed by the stop codon and 3′ UTR of HIS3. These PCR products were used as templates for overlap PCR to generate the full-length insert that was digested with SacI and EcoRI. This insert was then ligated into pRS416 cut with the same enzymes.
Construct 7 (pAV279) was created by PCR amplifying nonstop HIS3 from pAV239 (see below) with primers oAV183 and oAV274. This PCR product was then digested using SacI and XbaI and ligated into pMOV (Lydall and Weinert 1997) cut with the same enzymes.
Construct 8 (pAV217) was created by inserting oligonucleotides oAV42 and oAV43 into the BamHI site of pAV214.
Construct 9 (pAV239) was created by overlap extension PCR. Primers oAV87 and oAV97 were used to amplify the entire coding region of HIS3 including 236 bp upstream and 55 bp of HIS3 3′ UTR, followed by a hammerhead ribozyme downstream. Primers oAV98 and oAV103 were used to amplify the hammerhead ribozyme followed by the remaining 3′ UTR of HIS3. These PCR products were used as templates for overlap PCR to generate the full-length insert that was digested with SacI and EcoRI. This insert was then ligated into pRS416 cut with the same enzymes (Sikorski and Hieter 1989).
Construct 10 (pAV280) was created by cloning oligonucleotide oAV232 into the XmaI site of pAV216.
RNA analysis
Yeast strains were grown in SC-URA+2% galactose to mid-log phase. The medium was replaced with SC-URA+4% glucose, RNA was extracted, blotted by standard methods, and blots were hybridized with 32P 5′ end-labeled oligonucleotides specific for Protein A (oAV72), GFP (oAV223), HIS3 (oAV111), or the RNA subunit of the signal recognition particle as a loading control (oAV224). Signals were detected and quantitated using a STORM PhosphorImager (Amersham), and corrected for loading by quantitating the RNA subunit of the signal recognition particle.
RNaseH cleavage assay
Forty micrograms of total RNA were annealed to oligo(dT) (oAV220) and then subjected to RNase H treatment for 1 h. Assays included an additional cleavage oligonucleotide specific for the middle of the GFP (oAV238) coding region so that cleavage by RNase H resulted in a smaller product that allowed for better separation on polyacrylamide gels.
Protein analysis
Western blot analysis was performed according to standard techniques with antibodies for Protein A (Sigma), GFP (gift from W. Margolin), and Pgk1p (Molecular Probes). Each band was quantitated in triplicate using Alpha EaseFC software (Alpha Innotech) and an average reading was obtained. Average numbers for each reporter were then normalized for loading by similarly quantitating Pgk1p, and then to corresponding transcript levels to give corrected protein levels.
TABLE 1.
Oligonucleotides used
ACKNOWLEDGMENTS
We thank Norm Pace and Barbara Golden for valuable suggestions for constructing cleavage products lacking a cyclic phosphate; William Margolin for supplying anti-GFP antibody; and Rick Lloyd, Ann-Bin Shyu, Miles Wilkinson, and the van Hoof lab for helpful comments on the manuscript. We also acknowledge the Pew Scholarship in the Biomedical Sciences and NIH (GM069900) for funding.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.46306.
REFERENCES
- Aguilera A. Cotranscriptional mRNP assembly: From the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Araki Y., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A., Stevens A., Caponigro G., LaGrandeur T.E., Hatfield L., Fortner D.M., Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Been M.D., Cech T.R. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986;47:207–216. doi: 10.1016/0092-8674(86)90443-5. [DOI] [PubMed] [Google Scholar]
- Bird G., Fong N., Gatlin J.C., Farabaugh S., Bentley D.L. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol. Cell. 2005;20:747–758. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Boeck R., Tarun S., Jr., Rieger M., Deardorff J.A., Muller-Auer S., Sachs A.B. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- Brown J.T., Johnson A.W. A cis-acting element known to block 3′ mRNA degradation enhances expression of polyA-minus mRNA in wild-type yeast cells and phenocopies a ski mutant. RNA. 2001;7:1566–1577. [PMC free article] [PubMed] [Google Scholar]
- Burke J.M., Esherick J.S., Burfeind W.R., King J.L. A 3′ splice site-binding sequence in the catalytic core of a group I intron. Nature. 1990;344:80–82. doi: 10.1038/344080a0. [DOI] [PubMed] [Google Scholar]
- Cao D., Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J.M., Gray N.K., Wickens M.P. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes & Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Doel M.T., Carey N.H. The translational capacity of deadenylated ovalbumin messenger RNA. Cell. 1976;8:51–58. doi: 10.1016/0092-8674(76)90184-7. [DOI] [PubMed] [Google Scholar]
- Doma M.K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue C.P., Fedor M.J. Kinetics of hairpin ribozyme cleavage in yeast. RNA. 1997;3:961–973. [PMC free article] [PubMed] [Google Scholar]
- Dower K., Kuperwasser N., Merrikh H., Rosbash M. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA. 2004;10:1888–1899. doi: 10.1261/rna.7166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.R., Armstrong J., Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T., Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K., Valerius O., Mangus D.A., Jacobson A., Braus G.H. Replacement of the yeast TRP4 3′ untranslated region by a hammerhead ribozyme results in a stable and efficiently exported mRNA that lacks a poly(A) tail. RNA. 2002;8:336–344. doi: 10.1017/s1355838202021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P.A., van Hoof A., O'Donnell K., Guerrerio A.L., Parker R., Dietz H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Galili G., Kawata E.E., Smith L.D., Larkins B.A. Role of the 3′-poly(A) sequence in translational regulation of mRNAs in Xenopus laevis oocytes. J. Biol. Chem. 1988;263:5764–5770. [PubMed] [Google Scholar]
- Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes & Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gao M., Wilusz C.J., Peltz S.W., Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L., Beelman C.A., Stevens A., Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huez G., Bruck C., Cleuter Y. Translational stability of native and deadenylylated rabbit globin mRNA injected into HeLa cells. Proc. Natl. Acad. Sci. 1981;78:908–911. doi: 10.1073/pnas.78.2.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins C.J., Rathjen P.D., Forster A.C., Symons R.H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N., Najita L., Franzusoff A., Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae . Mol. Cell. Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T., Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson J.S., Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Korner C.G., Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lydall D., Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: Further evidence for roles in DNA replication and/or repair. Mol. Gen. Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Schwer B., Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison D.C., Blanc A., Ribas J.C., Carroll K., Sonenberg N., Wickner R.B. Decoying the cap-mRNA degradation system by a double-stranded RNA virus and poly(A)-mRNA surveillance by a yeast antiviral system. Mol. Cell. Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Hanna M., Green R., Bartel D.P., Szostak J.W. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989;342:391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Netter P., Xu M.Q., Shub D.A. Mechanism of 3′ splice site selection by the catalytic core of the sunY intron of bacteriophage T4: The role of a novel base-pairing interaction in group I introns. Genes & Dev. 1990;4:777–788. doi: 10.1101/gad.4.5.777. [DOI] [PubMed] [Google Scholar]
- Michel Y.M., Poncet D., Piron M., Kean K.M., Borman A.M. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D., Jacobson A. mRNA poly(A) tail, a 3′enhancer of translational initiation. Mol. Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudel U., Soreq H., Littauer U.Z. Globin mRNA species containing poly(A) segments of different lengths. Their functional stability in Xenopus oocytes. Eur. J. Biochem. 1976;64:115–121. doi: 10.1111/j.1432-1033.1976.tb10279.x. [DOI] [PubMed] [Google Scholar]
- Orban T.I., Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Butler J.S. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.V., Cech T.R. Determinants of the 3′ splice site for self-splicing of the Tetrahymena pre-rRNA. Genes & Dev. 1988;2:1439–1447. doi: 10.1101/gad.2.11.1439. [DOI] [PubMed] [Google Scholar]
- Proweller A., Butler S. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae . Genes & Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- Proweller A., Butler J.S. Ribosome concentration contributes to discrimination against poly(A)-mRNA during translation initiation in Saccharomyces cerevisiae . J. Biol. Chem. 1997;272:6004–6010. doi: 10.1074/jbc.272.9.6004. [DOI] [PubMed] [Google Scholar]
- Ridley S.P., Sommer S.S., Wickner R.B. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B., Davis R.W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Davis R.W., Kornberg R.D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S., Rosch S., Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 2002;21:2769–2777. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki Y., Itoh N., Yamada H., Nagata S., Mizumoto K. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanylytransferase subunit from Saccharomyces cerevisiae . J. Biol. Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M., Carr-Schmid A., Schwartz D.C., Kiledjian M., Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–237. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Mayo T., Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z., Jr., Sachs A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Teixeira A., Tahiri-Alaoui A., West S., Thomas B., Ramadass A., Martianov I., Dye M., James W., Proudfoot N.J., Akoulitchev A. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- Tharun S., Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Shibagaki Y., Imajoh-Ohmi S., Murakoshi T., Suzuki M., Nakamura A., Gotoh H., Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme β subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem. Biophys. Res. Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Wang Z., Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Xiao S., Houser-Scott F., Engelke D.R. Eukaryotic ribonuclease P: Increased complexity to cope with the nuclear pre-tRNA pathway. J. Cell. Physiol. 2001;187:11–20. doi: 10.1002/1097-4652(200104)187:1<11::AID-JCP1055>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Yen L., Svendsen J., Lee J.S., Gray J.T., Magnier M., Baba T., D'Amato R.J., Mulligan R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Zaug A.J., Cech T.R. The intervening sequence excised from the ribosomal RNA precursor of Tetrahymena contains a 5-terminal guanosine residue not encoded by the DNA. Nucleic Acids Res. 1982;10:2823–2838. doi: 10.1093/nar/10.9.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]