Abstract
Sequences and structures in the mRNA can alter the accuracy of translation. In some cases, mRNA secondary structures like hairpin loops or pseudoknots can cause frequent errors of translational reading frame (programmed frameshifting) or misreading of termination codons as sense (nonsense readthrough). In other cases, the primary mRNA sequence stimulates the error probably by interacting with an element of the ribosome to interfere with error correction. One such primary mRNA sequence, the Ty3 stimulator, increases programmed +1 frameshifting 7.5 times in the yeast Saccharomyces cerevisiae. Here we show that this stimulator also increases the usage of non-AUG initiation codons in the bacterium Escherichia coli but not in S. cerevisiae. These data suggest that in E. coli, though not in yeast, an element of the ribosome's elongation accuracy mechanism ensures initiation accuracy.
Keywords: translational accuracy, programmed frameshifting, Saccharomyces cerevisiae, Escherichia coli, codon discrimination, non-AUG initiation
INTRODUCTION
Accurate protein synthesis being essential to cell survival, the translational machinery has evolved multiple mechanisms to ensure faithful translation. These mechanisms involve both kinetic proofreading and induced fit during binding of aminoacyl-tRNAs in the ribosomal A-site (for review, see Rodnina and Wintermeyer 2001; Wintermeyer et al. 2004). With the advent of high-resolution structures of bacterial ribosomes, it has become possible to correlate structures in the ribosome with mechanisms of error regulation (for review, see Ogle and Ramakrishnan 2005). Available lower resolution structures of ribosomes from the eukaryote Saccharomyces cerevisiae have emphasized the universality of the core structures of the ribosome (Spahn et al. 2004), which implies that the basic functions of error correction might also be conserved between prokaryotes and eukaryotes.
Some mRNA sequences program ribosomes to make extremely frequent translational errors, either frequent translational frameshifts or readthrough of termination codons (for review, see Farabaugh 1996; Baranov et al. 2002). Analysis of the mechanisms causing these programmed errors has provided additional insights into the mechanism of error correction. These mRNA sequences can interfere with acceptance of correct aminoacyl-tRNAs or promote the acceptance of errant ones.
In many cases, the sequence context surrounding a programmed error site strongly stimulates the translational error. One such sequence is a 14-nucleotide sequence immediately downstream from the programmed +1 frameshift site of the retrotransposon Ty3, which we term the Ty3 stimulator, that increases frameshifting up to 7.5-fold (Farabaugh et al. 1993; Li et al. 2001). Our previous analysis showed that the primary RNA sequence of the stimulator caused this effect rather than either the decoded peptide sequence in the region or the local secondary structure of the mRNA (Li et al. 2001). For a primary sequence to have such an effect, we presume it must interact with a target molecule, perhaps a region of the ribosome. The structure of the bacterial mRNA•70S ribosome complex shows that the region immediately downstream of the ribosomal decoding sites (the A-site and P-site) passes through a tunnel through the 30S ribosomal subunit consisting of elements of the 16S rRNA and ribosomal proteins rpS3, rpS4, and rpS5 (Yusupova et al. 2001). Yeast 80S ribosomes have a similar structure (Spahn et al. 2004). The three ribosomal proteins compose the ribosome's mRNA helicase function (Takyar et al. 2005). During frameshifting, about half of the Ty3 stimulator should lie within the entrance tunnel, suggesting that its target might be the rRNA and ribosomal proteins located there. Previously, we had hypothesized that the Ty3 stimulator functioned by pairing with a complementary sequence in a component of the entrance tunnel, Helix 18 (Li et al. 2001). Since Helix 18 forms part of the ribosomal accuracy center responsible for modulating A-site decoding (Ogle and Ramakrishnan 2005), our data were consistent with the Helix 18•Ty3 stimulator pairing hypothesis.
Nucleotides in rRNA Helix 18 also appear to regulate accuracy during translation initiation (O'Connor et al. 1997), and the structure contacts initiation factor 1 (IF1) bound to the ribosomal A-site (Carter et al. 2001), a factor that increases the rate of initiation codon selection in bacteria (Pon and Gualerzi 1984). Furthermore, a eukaryotic homolog of IF1, eIF1A, binds to the A-site and may play the same function (Roll-Mecak et al. 2001). In our model that the Ty3 stimulator interferes with the function of some element of the A-site, we hypothesized that it might alter initiation accuracy in the same way as it alters frame maintenance.
Here we present evidence that the Ty3 stimulator reduces codon discrimination during initiation in bacteria. The phenotype observed differs from those caused by rRNA or initiation factor mutations, suggesting that it may identify a novel mechanism governing accuracy during initiation.
RESULTS
Effect of the Ty3 stimulator on initiation accuracy in S. cerevisiae
To test if the Ty3 stimulator affects initiation in S. cerevisiae, we modified pMB38, a lacZ reporter plasmid used to assay frameshift errors (Belcourt and Farabaugh 1990), introducing oligonucleotides creating an AUG initiation codon with or without the Ty3 stimulator downstream and variants of these two plasmids replacing the AUG codon with each of nine non-AUG codons (UUG, CUG, GUG, ACG, AAG, AGG, AUU, AUC, and AUA). The presence or absence of the Ty3 stimulator sequence had no significant effect on expression from any of the constructs in S. cerevisiae cells. Expression from an AUG initiation codon was stimulated slightly (from 85 units to 140 units of β-galactosidase), but expression of constructs with each of the near-AUG codons remained barely detectable (less than one unit of activity) and showed little or no effect of the Ty3 stimulator. We conclude from this experiment that the Ty3 stimulator has no significant effect on the efficiency or the accuracy of translation initiation in S. cerevisiae.
The Ty3 stimulator weakly stimulates translational initiation efficiency in E. coli
We can interpret our results in the yeast system in two ways. First, initiation might be fundamentally insensitive to the effects of the Ty3 stimulator because its target acts only during elongation. Alternatively, the eukaryotic initiation system, by restricting initiation only to AUG, might preclude the stimulator from affecting initiation accuracy. We hypothesized that if the second explanation were true, the less strict rules for codon selection during prokaryotic initiation might allow the Ty3 stimulator to have a measurable effect. We therefore converted our reporter plasmids for use in the bacterial system, introducing a consensus SD site (AGGAGG) 6 nt upstream of the initiation codon in each of our constructs, an optimum distance for initiation (Ringquist et al. 1992). The reporter constructs also include the Ty3 stimulator starting at the second nucleotide downstream of the AUG. As a control lacking the Ty3 stimulator, we constructed a mutation replacing eight of its 14 nucleotides in two blocks of four nucleotides. When tested in a frameshift reporter system, this 8-nt mutation eliminates all of the Ty3 stimulator effect, suggesting that it completely inactivates the sequence (data not shown). We also constructed reporters in which each of the two four-nucleotide blocks were mutated together or individually. The three mutations we term b1b2, b1, and b2. Reporters were constructed with the wild type and each of the three mutant forms of the stimulator combined with an AUG initiation codon and each of the nine non-AUG codons.
A comparison of the AUG reporters revealed that the Ty3 stimulator has a slight effect on translational efficiency. A reporter with the Ty3 stimulator expresses 2.2-fold more β-galactosidase than one with the completely inactive stimulator, b1b2 (1700 ± 26 versus 790 ± 36 units). A comparison of the two single mutant constructs, b1 and b2, shows that the effect of the stimulator depends only on the 5′ half; the b1 mutant reduces expression about twofold (to 780 ± 51 units), but the b2 mutant has no significant effect (1900 ± 46 units). This modest efficiency effect does not depend on the presence of the Shine-Dalgarno interaction. We replaced the canonical SD, AGGAGG, with a sequence, AUCCCU, recognized extremely poorly by bacterial ribosomes (Lee et al. 1996) resulting in an ∼600-fold reduction in expression, yet the stimulator still induces a 2.2-fold increase in initiation (2.9 ± 0.2 units for the wild type versus 1.3 ± 0.1 units for the b1b2 mutant). These data show that independent of the SD, the Ty3 stimulator only very weakly affects initiation.
The Ty3 stimulator increases the use of non-AUG codons in E. coli
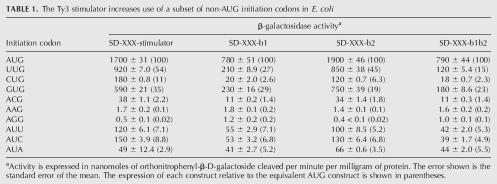
Next we tested the effect of the mutant and wild-type Ty3 stimulator sequences on the efficiency of initiation at non-AUG codons. As shown in Table 1, in the absence of the Ty3 stimulator (mutant b1b2, column 4) initiation at AUG in E. coli is substantially more efficient than at the nine non-AUG codons tested, but the efficiency of recognition of those codons varied widely. After AUG the most efficiently expressed reporters have the non-AUG initiation codons GUG (23%) and UUG (15%). Expression is less efficient with AUA, AUU, AUC, CUG, and ACG and significantly lower with AAG and AGG. These data are quite similar to those obtained by Sussman et al. (1996).
TABLE 1.
The Ty3 stimulator increases use of a subset of non-AUG initiation codons in E. coli
The presence of the Ty3 stimulator significantly altered the pattern of relative expression. The Ty3 stimulator increased expression of all of the constructs with the exception of those involving the AAG and AGG initiation codons (Table 1, column 1). Expression dependent on the GUG, ACG, AUU, AUC, and AUA initiation codons increased only slightly more than did the AUG reporter, averaging 2.9-fold. Expression of two of the non-AUG constructs (UUG and CUG) increased significantly more than did AUG. Initiation on CUG and UUG increased by 10-fold and 7.7-fold, respectively. Increased translational efficiency cannot explain all of these increases; when adjusted for the increase in expression of the AUG construct, there remains a residual increase in CUG initiation and UUG initiation of 4.5-fold and 3.5-fold, respectively.
The effect in E. coli does not reflect an alteration in downstream translation elongation
The efficiency of initiation depends on a variety of factors: strength and spacing of the upstream SD interaction, sequence of the initiation codon, identity of the first few codons downstream of the initiation codon, and presence of mRNA sequences that alter the kinetics of some step in initiation (see Stenstrom et al. 2001a and references therein). Since the Ty3 stimulator lies downstream of the initiation codon, it could exert its effect either as a coding region during the first few steps of elongation or as an RNA sequence by directly interacting with the translational machinery.
A simple test of the downstream coding region model is to shift the Ty3 stimulator into a different reading frame. The codons present in the −1 and +1 reading frames are quite different from those in the zero frame. If the rate of decoding of the codons used in the Ty3 stimulator were the basis of its effect, then we would expect that a shift of this type would eliminate its function. Of course, such a shift also changes the spacing between the decoding sites and the stimulator, which could also interfere with its function. We therefore constructed a set of reporters in which we insert from one to 15 nucleotides between the initiation codon and the stimulator. Because the stimulator has a UAA stop codon in the −1 reading frame, a +1 insertion would put the codon in frame with translation. In order to study a +1 spacing mutation, we mutated the UAA to a UAC sense codon to allow continued reading into the downstream reporter gene; a normal spacing control with the same mutation shows that the mutation does not alter the stimulator effect (data not shown). The source of the 15-nucleotide inserted sequence was codons 4–8 of the E. coli lacZ gene, which includes the region distal to two AUG codons at codons 1 and 3. A similar analysis using codons 2–6 of the S. cerevisiae HIS4 gene gave consistent results (data not shown).
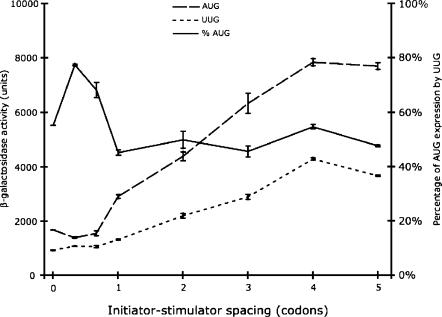
Insertions of one or two nucleotides shift the stimulator from the zero frame into the −1 or +1 reading frames, respectively, and insertion of three nucleotides restores the reading frame. Each insertion was introduced into a pair of reporters using either AUG or UUG as the initiation codon; we chose UUG because the Ty3 stimulator increased its recognition as an initiator by the largest amount, implying that interfering with stimulator function might have the maximum effect. Figure 1 shows that the level of AUG initiation varied among the spacing tester constructs. The mutations introduce changes to the second and third codons downstream of the initiator, which is known to affect translation efficiency (Stenstrom et al. 2001b; Stenstrom and Isaksson 2002); we presume that the differences reflect this effect. In general, we see no significant decrease in the Ty3 stimulator's effect on UUG initiation; the ratio of UUG to AUG expression actually increased in the one-nucleotide and two-nucleotide insertions, contrary to the prediction of the downstream coding region hypothesis. These data demonstrate that the effect of the stimulator is unrelated to its coding potential. The spacing between the initiation codon and the stimulator is even more flexible since further insertions of two, three, four, or five codons also had little or no effect on relative UUG initiation (Fig. 1). Again, we observed equivalent increases in the efficiency of initiation of AUG and UUG as the spacing increased, which may reflect the effect of early rounds of elongation on apparent initiation efficiency.
FIGURE 1.
Lack of effect of varying spacing between the Ty3 stimulator and the initiation codon. The figure shows expression of β-galactosidase from plasmids with increasing spacing between the Ty3 stimulator and the initiation codon (see Table 2 for sequences of the initiation regions). For each construct, the observed units of β-galactosidase (nanomoles of orthonitrophenyl-β-galactopyranoside cleaved per minute per OD600 of culture) are shown; constructs using an AUG (dashed line) or UUG (dotted line) initiation codon. The expression of the UUG constructs is shown as a percentage of the expression of the corresponding AUG construct (solid line).
DISCUSSION
In the yeast S. cerevisiae, the Ty3 stimulator sequence increases the frequency of translational frameshift errors. No experiment has explicitly identified the target of the stimulator, but it probably interacts directly with the ribosome since the primary sequence of the mRNA itself causes the effect (Li et al. 2001). The structure of the mRNA•ribosome complex (Yusupova et al. 2001; Takyar et al. 2005) predicts that at the time of the frameshift, the stimulator is within the entrance tunnel of the ribosome, a structure composed of two layers consisting of rRNA and residues of the ribosomal proteins rps3, rps4, and rps5. Each of these proteins plays a role in maintaining translational accuracy, as indicated by the existence of mutations that reduce accuracy in their coding genes in E. coli (Gorini 1974; Kurland et al. 1996) or their S. cerevisiae homologs (Alksne et al. 1993; Hendrick et al. 2001). Similar mutational results imply an error correction role for rRNA elements of the tunnel, Helix 18, and Helix 34, each of which includes nucleotides that can mutate to give hypoaccurate phenotypes (for review, see Kurland et al. 1996). The fact that the stimulator increases the frequency of +1 frameshift errors suggested that it might perturb the accuracy center of which these proteins and rRNA structures are a part. Furthermore, these same structures are very close to or, in the case off Helix 18, directly interact with initiation factor 1 (IF1) during initiation. This fact, among others, suggested to us that we should test the effect of the Ty3 stimulator on translation initiation accuracy.
Though the stimulator increases elongation errors, it does not affect initiation accuracy in the yeast S. cerevisiae. However, it does significantly alter initiation in E. coli, both increasing translation efficiency and decreasing initiation codon discrimination. The stimulator's effect on translation efficiency was very weak, only twofold. By comparison, the Shine-Dalgarno site stimulated translation almost 590-fold. The stimulator had a more significant effect on initiation codon discrimination in E. coli, resulting in a significant increase in use of UUG and CUG relative to AUG. The absolute efficiency of CUG initiation increased 10-fold and UUG initiation increased 7.7-fold. These changes corresponded to increases relative to AUG initiation of 4.5-fold and 3.5-fold, respectively. The increase in CUG initiation to 10% of the AUG reporter level is especially significant since we know of no evidence that this codon functions as an initiator in any gene in E. coli. In contrast, the Ty3 stimulator had little effect on initiation at GUG or at several weakly recognized non-AUG codons tested. In fact, the stimulator increased CUG initiation but had no effect on three stronger non-AUG codons (AUU, AUC, and AUA). These data show that the Ty3 stimulator effect does not simply increase non-AUG initiation in general but, rather, that it distinguishes between interactions of initiator tRNA with various non-AUG codons, apparently perturbing a ribosomal function that specifically interferes with recognition of UUG and CUG.
Stenstrom et al. (2001a) also found that certain sequences immediately downstream of the initiation codon could increase non-AUG initiation. Of five such regions tested in their study, two caused UUG initiation at approximately the same efficiency as AUG while having a much smaller effect on GUG initiation. This result resembles the effect of the Ty3 stimulator, but Stenstrom et al. did not test other non-AUG codons. However, the Stenstrom group attributed the majority of the effect to the codon immediately downstream of the initiation codon (the “+2 codon”). We held this codon constant in our experiments, as UCU, in our wild type and mutant constructs to eliminate this type of nearest neighbor effect. We suspect, therefore, that the effects we observed are distinct from those described by Stenstrom et al.
Other error-inducing mutations of the translational machinery affect initiation codon discrimination differently than does the Ty3 stimulator. For example, mutations in infC, which encodes initiation factor 3 (IF3), increased the use of several weakly utilized non-AUG codons—CUG, AUU, AUC, AUA, and ACG—while leaving expression from AUG, GUG, or UUG unchanged (Sacerdot et al. 1996; Sussman et al. 1996; O'Connor et al. 2001). IF3 directly modulates accuracy during initiation (Sacerdot et al. 1996; O'Connor et al. 2001). The very different spectrum of effects of the Ty3 stimulator suggests that IF3 does not mediate the stimulator's effect.
Mutations altering A-site rRNA nucleotides have an effect distinct from that of the IF3 mutations, increasing the use of all non-AUG codons tested, including UUG, GUG, CUG, AUC, AUA, and AUU (O'Connor et al. 1997). The more selective effect of the Ty3 stimulator specifically on UUG and CUG is also inconsistent with its modulating the effect of the A-site nucleotides.
The effect of the stimulator on initiation is quite insensitive to its distance downstream of the P-site, a result strikingly different from the effect of spacing on frameshifting. This flexibility in spacing resembles the flexibility of another translational stimulatory mRNA sequence, the downstream box, which can function whether 1 or 6 codons downstream of the initiation point (Etchegaray and Inouye 1999).
MATERIALS AND METHODS
Bacterial and yeast strains
The strains used for all experiments in this work were the E. coli strain DH5α (F−φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ− thi-1 gyrA96 relA1) (Hanahan 1983) and the S. cerevisiae strain FY1679-18B (Matα, his3-Δ200, leu2-Δ1, trp1-Δ63, ura3-52, GAL2) (Dujon et al. 1994). Assays of β-galactosidase were performed as previously described for E. coli (Miller 1972) and S. cerevisiae (Farabaugh et al. 1989). Triplicate assays were conducted on at least three independent transformants. In all cases, the standard error of the mean was <10%.
Media
All E. coli strains were grown on LB + Amp medium: 10 g of Bacto-tryptone (Difco), 5 g of Bacto yeast extract (Difco), 10 g of NaCl, and 100 mg of ampicillin per liter. Bacterial transformation was performed using the CaCl2 method as described (Miller 1972). All transformation experiments using S. cerevisiae strains were performed by the method of Ito et al. (1983) and involved introduction of plasmids carrying the URA3 gene and selection for complementation of the ura3-52 auxotrophic mutation. The transformants were grown on synthetic medium with 2% glucose (SD) supplemented with histidine, leucine, and tryptophan (Rose et al. 1990) but lacking uracil. Growth on this medium requires that a plasmid-borne URA3 gene complement the strain's ura3-52 auxotrophic mutation.
Reporter plasmid constructions
All plasmids used in this study are derivatives of the plasmid pMB38, a lacZ reporter plasmid that includes the bla gene of pBR322, conferring resistance to ampicillin and the URA3 gene of S. cerevisiae, to select for transformants in yeast. Expression of lacZ in S. cerevisiae from the pMB38 reporter vector depends on transcription and translation from the HIS4 gene, the first 33 codons of which have been fused to the beginning of the lacZ gene (Belcourt and Farabaugh 1990). Fusion between HIS4 and lacZ is through a BamHI–KpnI linker region. The plasmid was engineered for use in this study by deleting the region from the initiator codon of HIS4 to the KpnI site at the 5′-end of the lacZ segment and substituting a BamHI–KpnI fragment that included one of the several initiation regions used in the study. To make this construct, a polymerase chain reaction (PCR) experiment was performed with two primers. A primer (“5′ to Sal”) (see Belcourt and Farabaugh 1990) used in all reactions primes synthesis from a location 5′ to a unique SalI site, which is 714 bp upstream of the HIS4 gene. Each reaction used a specific primer that included the initiation region to be inserted at the beginning of the lacZ gene immediately upstream of a KpnI site to be used in subcloning the PCR fragment. Priming by these oligonucleotides was directed by a sequence complementary to the 15 nucleotides immediately 5′ of the HIS4 gene. The PCR fragment inserted between unique SalI and KpnI sites in pMB38 to create each clone. Each of these oligonucleotides introduced the sequence of a canonical Shine-Dalgarno (SD) interaction site (5′-AGGAGG-3′) separated from the initiator codon by a 6-nt BamHI site (5′-GGATCC-3′). The SD site directs ribosomes to recognize the initiation region (Shine and Dalgarno 1974). As a control for the effect of the SD, separate oligonucleotides were used to introduce a non-SD sequence (5′-ATCCCT-3′) to replace the wild-type SD. The non-SD sequence was chosen because it has been shown to not be recognized by normal E. coli ribosomes (Lee et al. 1996). In our hands, the non-SD sequence provides only very low levels of translation even with an AUG initiation codon. Table 2 provides the sequences of the initiation regions from the plasmids used in this work. Versions of each of these sequences with multiple initiation codons (shown as “XXX” in the table) have been constructed, as described in the text.
TABLE 2.
Plasmid constructs used in this work
ACKNOWLEDGMENTS
We thank Qiang Qian for technical assistance in early aspects of this work and to Gregory McCarty for his initial work on the Ty3 stimulator's effect in E. coli. This work was supported by Public Health Service grant GM 29480 from the National Institute of General Medical Sciences. Part of the work was performed while P.J.F. was a visiting scientist at the Université Paris XI, Orsay, France supported by the Centre Nationale de la Recherche Scientifique.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.13306.
REFERENCES
- Alksne L.E., Anthony R.A., Liebman S.W., Warner J.R. An accuracy center in the ribosome conserved over 2 billion years. Proc. Natl. Acad. Sci. 1993;90:9538–9541. doi: 10.1073/pnas.90.20.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: Translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Belcourt M.F., Farabaugh P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Clemons W.M., Jr., Brodersen D.E., Morgan-Warren R.J., Hartsch T., Wimberly B.T., Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Dujon B., Alexandraki D., Andre B., Ansorge W., Baladron V., Ballesta J.P., Banrevi A., Bolle P.A., Bolotin-Fukuhara M., Bossier P., et al. Complete DNA sequence of yeast chromosome XI. Nature. 1994;369:371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- Etchegaray J.P., Inouye M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli . J. Biol. Chem. 1999;274:10079–10085. doi: 10.1074/jbc.274.15.10079. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P., Liao X.-B., Belcourt M., Zhao H., Kapakos J., Clare J. Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae . Mol. Cell. Biol. 1989;9:4824–4834. doi: 10.1128/mcb.9.11.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J., Zhao H., Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: Frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. Streptomycin and misreading of the genetic code. In: Nomura M., et al., editors. Ribosomes. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1974. pp. 791–803. [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hendrick J.L., Wilson P.G., Edelman I.I., Sandbaken M.G., Ursic D., Culbertson M.R. Yeast frameshift suppressor mutations in the genes coding for transcription factor Mbf1p and ribosomal protein S3: evidence for autoregulation of S3 synthesis. Genetics. 2001;157:1141–1158. doi: 10.1093/genetics/157.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C., Hughes D., Ehrenberg M. Limitations of translational accuracy. In: Neidhardt F.C. III, et al., editors. Escherichia coli and Salmonella: Cellular and molecular biology. ASM Press; Washington, DC: 1996. pp. 979–1004. [Google Scholar]
- Lee K., Holland-Staley C.A., Cunningham P.R. Genetic analysis of the Shine-Dalgarno interaction: Selection of alternative functional mRNA–rRNA combinations. RNA. 1996;2:1270–1285. [PMC free article] [PubMed] [Google Scholar]
- Li Z., Stahl G., Farabaugh P.J. Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA. 2001;7:275–284. doi: 10.1017/s135583820100190x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- O'Connor M., Thomas C.L., Zimmermann R.A., Dahlberg A.E. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Gregory S.T., Rajbhandary U.L., Dahlberg A.E. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001;7:969–978. doi: 10.1017/s1355838201010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M., Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Pon C.L., Gualerzi C.O. Mechanism of protein biosynthesis in prokaryotic cells. Effect of initiation factor IF1 on the initial rate of 30 S initiation complex formation. FEBS Lett. 1984;175:203–207. doi: 10.1016/0014-5793(84)80737-1. [DOI] [PubMed] [Google Scholar]
- Ringquist S., Shinedling S., Barrick D., Green L., Binkley J., Stormo G.D., Gold L. Translation initiation in Escherichia coli: Sequences within the ribosome-binding site. Mol. Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Wintermeyer W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem. Sci. 2001;26:124–130. doi: 10.1016/s0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., Shin B.S., Dever T.E., Burley S.K. Engaging the ribosome: Universal IFs of translation. Trends Biochem. Sci. 2001;26:705–709. doi: 10.1016/s0968-0004(01)02024-2. [DOI] [PubMed] [Google Scholar]
- Rose M., Winston F., Hieter P. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- Sacerdot C., Chiaruttini C., Engst K., Graffe M., Milet M., Mathy N., Dondon J., Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli . Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M., Gomez-Lorenzo M.G., Grassucci R.A., Jorgensen R., Andersen G.R., Beckmann R., Penczek P.A., Ballesta J.P., Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenstrom C.M., Isaksson L.A. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3′ side. Gene. 2002;288:1–8. doi: 10.1016/s0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]
- Stenstrom C.M., Holmgren E., Isaksson L.A. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001a;273:259–265. doi: 10.1016/s0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- Stenstrom C.M., Jin H., Major L.L., Tate W.P., Isaksson L.A. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli . Gene. 2001b;263:273–284. doi: 10.1016/s0378-1119(00)00550-3. [DOI] [PubMed] [Google Scholar]
- Sussman J.K., Simons E.L., Simons R.W. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W., Peske F., Beringer M., Gromadski K.B., Savelsbergh A., Rodnina M.V. Mechanisms of elongation on the ribosome: Dynamics of a macromolecular machine. Biochem. Soc. Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]