Abstract
Although recent microarray studies have provided evidence of RNA interference (RNAi)-mediated off-target gene modulation, little is known about whether these changes induce observable phenotypic outcomes. Here we show that a fraction of randomly selected small inhibitory RNAs (siRNAs) can induce changes in cell viability in a target-independent fashion. The observed toxicity requires an intact RNAi pathway and can be eliminated by the addition of chemical modifications that reduce off-target effects. Furthermore, an analysis of toxic and nontoxic duplexes identifies a strong correlation between the toxicity and the presence of a 4-base-pair motif (UGGC) in the RISC-entering strand of toxic siRNA. This article provides further evidence of siRNA-induced off-target effects generating a measurable phenotype and also provides an example of how such undesirable phenotypes can be mitigated by addition of chemical modifications to the siRNA.
Keywords: siRNA, off-target effects, RNAi, toxicity
INTRODUCTION
Recent investigations have demonstrated that transcripts having <100% complementarity with an siRNA can be targeted for knockdown by the RNAi pathway (Jackson et al. 2003; Haley and Zamore 2004; Birmingham et al. 2006). This phenomenon, referred to as “off-targeting,” is concentration dependent, responsible for up to threefold suppression of dozens of genes, and mediated by either the sense or antisense strand of the siRNA. While off-target gene modulation is well documented (Lin et al. 2005), little is known about the possible effect of off-targeting on overall cellular physiology. Here we demonstrate that the phenomenon of off-target-derived phenotypes is much more extensive than originally anticipated and, depending on the specificity of the assay, can be responsible for as many as 30% of positives identified in a screen. Although these findings represent a significant obstacle to applying RNAi in high-throughput screening strategies, this article describes an siRNA chemical modification pattern (Jackson et al. 2006) that significantly diminishes off-target effects and the associated phenotypes with minimal (to no) effect on on-target gene silencing.
RESULTS
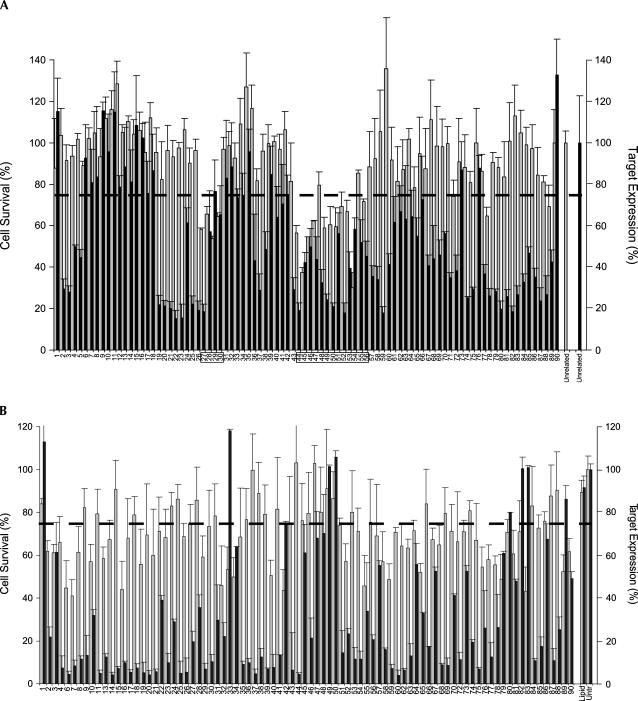
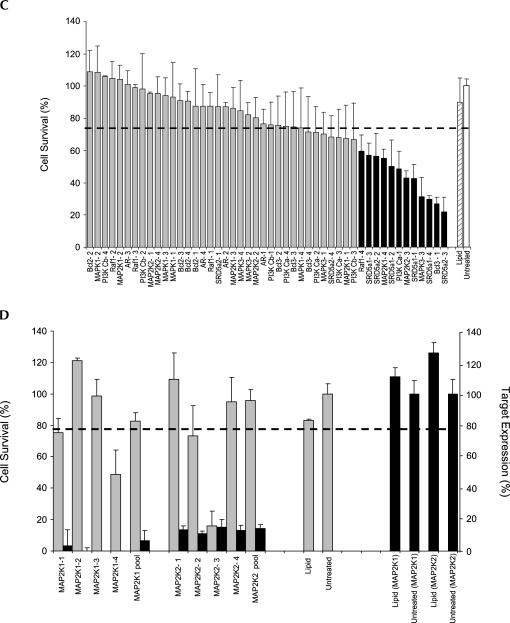
A population of 176 randomly selected siRNAs derived from two separate walks targeting diazepam-binding inhibitor (DBI NM_020548, position 202–291) and firefly luciferase (M15077, position 1434–1612) were evaluated to determine if a relationship existed between cell survival and lipid-mediated siRNA delivery. Using a cell viability cutoff of 75% to differentiate toxic from nontoxic duplexes, 51 siRNAs (29%) were identified as being capable of significantly reducing overall cell viability under the assay conditions (Fig. 1A,B). No direct correlation between siRNA specific activity and toxicity was observed, a conclusion that was independently confirmed by an analysis of a separate collection of 48 “functional” (>80% silencing) siRNAs targeting 12 additional genes (Fig. 1C,D). Additional studies demonstrated that the toxic phenotype was apoptotic in nature (data not shown), concentration dependent, and present in all of the cell lines tested. A dilution study performed on four separate siRNAs (MAP2K2-d3, SRD5A1-d1, SRD5A1-d3, and SRD5A2-d3) showed that the toxic effects steadily dissipated as concentrations were diminished, disappearing completely below 1 nM (Fig. 2A). Moreover, introduction of toxic siRNAs into three different cell lines (HeLa, MCF7, and DU145) led to similar but not identical levels of cell death, suggesting that some cell types were more sensitive than others (Fig. 2B).
FIGURE 1.
(Continued on next page)
FIGURE 1.
Target-independent, sequence-specific siRNA toxicity. (A) HeLa cells were transfected with 90 different siRNA targeting DBI (positions 202–291). Cellular viability and gene knockdown were determined. Data are displayed in a linear manner according to siRNA position in walk. The dotted line represents the 75% viability threshold arbitrarily chosen to delineate toxic from nontoxic siRNA. Toxicity data (gray bars) are superimposed on DBI mRNA expression data (black bars) for the same siRNA set. Boxed areas indicate toxic siRNA with sequence similarity. The values for toxicity represent the average of three independent experiments (each performed in triplicate). For mRNA levels, values represent the average of three independent experiments. Error bars depict standard deviation from the mean. (B) HeLa cells were transfected with one of 86 different siRNA (10 nM) targeting PPLUC (firefly luciferase, NM_15077, position 1434–1612), and cellular viability and gene knockdown were determined using an AlamarBlue assay and bDNA, respectively. Data are displayed according to siRNA position in walk. The dotted line represents the 75% viability threshold. Toxicity data (gray bars) are superimposed on luc mRNA expression data (black bars) for the same siRNA set. (C) HeLa cells were transfected with one of 48 rationally designed and functional (>80% silencing) siRNAs targeting 12 different genes (Bcl2, MAPK1, PI3K Cb, PI3K Ca, AR, Raf1, SRD5-1, SRD5-2, MAP2K1, MAP2K2, MAPK3, and Bcl3). Data are sorted based on the level of siRNA-induced toxicity and show the absence of a strong correlation between the level of target knockdown and toxicity. For all experiments, siRNAs were transfected at 10 nM concentrations, and cellular viability assays were performed 72 h after transfection. The values for toxicity represent the mean of three independent experiments (each performed in triplicate). For mRNA levels, the values represent the mean of three independent experiments. Error bars depict standard deviation from the mean. (D) A side-by-side comparison of toxicity and gene knockdown of eight siRNAs targeting two separate genes (MAP2K1 and MAP2K2). While all four duplexes in each cluster provide equivalent levels of gene silencing (>80%), only a single siRNA in each group is toxic.
FIGURE 2.
The toxic phenotype is apoptotic in nature, concentration dependent, and cell type independent. (A) Four toxic siRNAs targeting three different genes (MAP2K2, SRD5α1, or SRD5α2) were transfected into HeLa cells at concentrations ranging between 0.625 and 10 nM. Seventy-two hours after transfection, the effects of each duplex on cellular viability were determined using an Alamar Blue assay. (B) Toxic and nontoxic siRNAs targeting two different genes (MAP2K2 and SRD5α2) were transfected into HeLa (white bars), MCF7 (gray bars), or DU145 (black bars) cells at 10 nM concentrations and assayed for cell viability (72 h). For all experiments, the values for toxicity represent the mean of three independent experiments (each performed in triplicate).
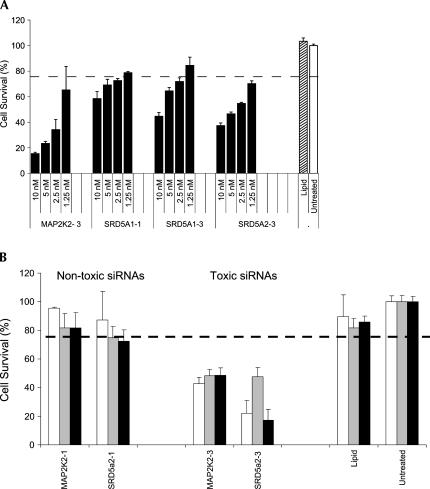
Three separate lines of experimentation led to the conclusion that the cell viability phenotype was dependent on RNAi. First, the ability of toxic siRNAs to induce cell death was severely compromised when a critical component of the RNAi mechanism, eIF2C2 (hAgo2), was obstructed. Previous studies have shown that eIF2C2 is responsible for RNAi-mediated mRNA cleavage and that knockdown of this gene product severely cripples the pathway (Meister et al. 2004). Control studies confirmed this finding, demonstrating that siRNA-mediated knockdown of eIF2C2 disabled subsequent siRNA-induced silencing of reporter genes (Fig. 3a–i). Surprisingly, when eIF2C2− cells were transfected with toxic siRNA, no decrease in cell viability was observed (Fig. 3j). As similar results were obtained with siRNAs targeting a second component of the RNAi pathway (eIF2C1) (data not shown), but not control siRNA, these findings strongly suggest that an uncompromised RNAi pathway is necessary for observance of the toxic phenotype.
FIGURE 3.
siRNA-induced toxicity is mediated through the RNAi pathway. (a) Bar graph showing the level of knockdown of eIF2C2 using a pool of siRNAs. The values represent the mean of three independent experiments. Error bars depict standard deviation from the mean. (b–i) Control experiments performed to demonstrate that eIF2C2 knockdown eliminates RNAi. Cells were transfected during Transfection 1 (T1) with either eIF2C2 or control (RISC-Free) siRNAs (10 nM). Forty-eight hours later, cells were recovered and replated at 5 × 103 cells per well. Twenty-four hours later, cells were transfected again (Transfection 2, T2) with the original siRNA from T1 plus (1) either the control siRNA or the EGFP-targeting siRNA, and (2) an EGFP expression cassette (20 ng/well). Twenty-four hours later, cells were assayed for EGFP expression at the mRNA level (branched DNA; data not shown) and the protein level (fluorescent microscopy). (b,d) T1 = control siRNA, T2 = control siRNA, EGFP, and Hoechst stain, respectively. (c,e) T1 = control siRNA, T2 = EGFP-targeting siRNA, EGFP, and Hoechst stain, respectively. (f,h) T1 = eIF2C2 targeting siRNA, T2 = control siRNA, EGFP, and Hoechst stain, respectively. (g,i) T1 = eIF2C2-targeting siRNA, T2 = EGFP-targeting siRNA, EGFP, and Hoechst stain, respectively. (j) Bar graph demonstrating the effects of eIF2C2 knockdown on siRNA toxicity. HeLa cells having normal (control) or low (eIF2C2) levels of eIF2C2 (Ago2) expression were transfected with toxic siRNA targeting PI3K-Ca, MAP2K2, or SRD5α1. (k) Bar graph showing the effect of 2-nt truncations (19-mer → 17-mer) on siRNA toxicity. Toxic siRNAs (and the equivalent 17-bp truncated forms) target seven different genes: PI3K-Ca, MAP2K2, SRD5α1, SRD5α2, PPY-Luc, GAPDH, and PPIB.
In a second approach, the ability of toxic siRNA to induce cell death was tested when the size of the duplex was reduced from 19 bp to 17 bp. Previous studies have shown that duplexes shorter than 19 bp target mRNA sequences inefficiently, suggesting that RISC fails to mediate RNAi when duplex length drops below a minimum length (Elbashir et al. 2001). When the length of 19-bp toxic siRNA was reduced by 2 bp (17 bp total), the level of toxicity was reduced dramatically (Fig. 3k), indicating that entry and/or processing by RISC was necessary for induction of toxicity.
Finally, chemical modifications that eliminate RNAi-mediated off-target effects (described in detail in our accompanying paper Jackson et al. 2006) were tested for the ability to abolish siRNA-induced toxicity. Recent studies have shown that (1) complementarity between the seed region (positions 2–7) of the siRNA and regions in the 3′-UTR is associated with off-targeting (Birmingham et al. 2006), and (2) addition of minimal 2′-O-methyl modification in the seed region of siRNA dramatically limits the number of off-targeted genes without altering target-specific knockdown (Fig. 4A,B; Jackson et al. 2006). When toxic siRNAs were modified with the 2′-O-methyl modification pattern, the cell viability phenotype was eliminated in the majority of cases, suggesting that suppression of off-targets negates the siRNA toxicity (Fig. 4C). Taken together, these three lines of evidence (eIF2C2 knockdown, duplex truncation, and chemical modification) strongly suggest that the observed siRNA-induced toxicity is RNAi-dependent and the result of off-target gene modulation.
FIGURE 4.
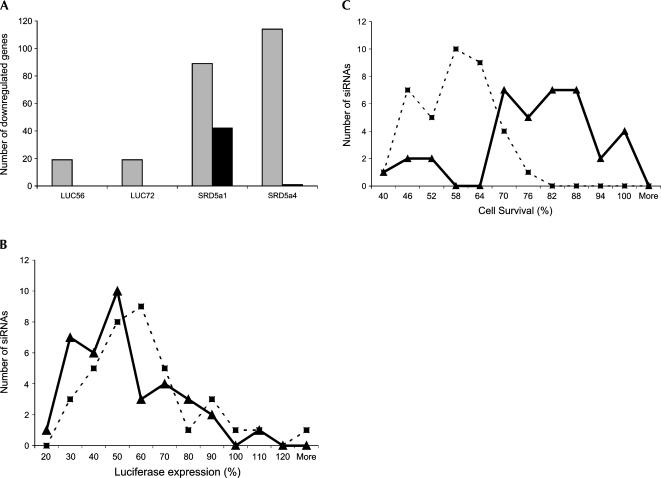
siRNA-induced toxicity is off-target effect-mediated. (A) Bar graph shows the relative numbers of off-targeted genes down-regulated by twofold or more (mock-transfected samples used as a reference) for four siRNAs (Luc-56, Luc-72, SRD5α1-1, and SRD5α1-4) in unmodified (gray bars) and modified (black bars) forms (two biological replicates for each sample). (B) Line graph showing the distribution of functionalities of 37 toxic sequences in unmodified (dotted line, squares) and modified (smooth line, triangles) forms. A χ2 test of independence comparing the functionality of modified and unmodified sequences shows that the difference is statistically insignificant (p = 0.245). (C) Line graph showing the distribution of toxicities of 37 toxic siRNAs in unmodified (dotted line, squares) and modified (smooth lines, triangles) forms. A χ2 test of independence comparing the viability of cells transfected with modified and unmodified sequences shows that the difference is statistically significant (p = 1.4 × 10−9). The values for toxicity represent the average of three independent experiments (each performed in triplicate).
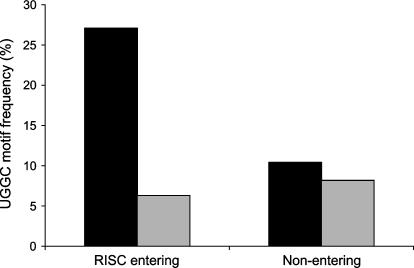
The clustered distribution of toxic siRNAs along the DBI walk (see positions 27–30 and 44–56, boxed areas) and the identification of AAA/UUU and/or GCCA/UGGC motifs in all 12 of the toxic siRNAs derived from the random functional set of duplexes (48 sequences) (Fig. 1C) suggested that the toxicity was associated with one or more motifs. To test this, 289 siRNAs containing a clear RISC-entry strand bias (as determined by thermodynamic bias rules previously established, Reynolds et al. 2004) were assessed for effects on cell viability. We predicted that any toxic motif should be overrepresented in the RISC-entering strand of toxic siRNAs (but not nontoxic siRNAs) and equally (or under-) represented in the non-RISC-entering strands of toxic and nontoxic duplexes. Ninety-eight nontoxic and 92 toxic siRNAs (190 total siRNAs out of the original 289 duplexes) exhibited clear toxic/nontoxic phenotypes. Subsequent motif analysis of this collection identified a single motif, 5′-UGGC, with the predicted properties (Fig. 5) (χ2 value of 15.40, p = 0.00009, adjusted p-value < 0.05 after applying the Bonferroni correction to account for multiple [256] comparisons), suggesting a strong correlation between the observed siRNA-induced toxicity and the UGGC motif. Interestingly, while several other motifs consisting of AAA and UUU trinucleotides were observed to be overrepresented in toxic versus nontoxic populations, these were not found to be statistically significant.
FIGURE 5.
Identification of UGGC motifs in the RISC-entering strand of toxic siRNA. Bar graph showing the relative frequency of the UGGC motif in the RISC-entering and RISC-nonentering strands of toxic and nontoxic siRNAs. Black bars—toxic siRNAs; gray bars—nontoxic.
DISCUSSION
The siRNA-induced toxicity described in this work is sequence-dependent, target-independent, reliant on the ability of the duplex to enter an intact RNAi pathway, and in most cases, sensitive to the addition of chemical modifications that minimize off-target effects. Based on these findings, we have concluded that off-target effects are capable of inducing strong, quantifiable phenotypes. While similar conclusions were recently reported by Lin and coworkers (Lin et al. 2005), the fraction of sequences that were capable of inducing off-target phenotypes in this study falls beyond previous expectations and has broad potential implications in all RNAi-based studies pertaining to functional genomics, gene mapping, drug target discovery and validation, and therapeutics.
Identification of a motif associated with a cell viability phenotype was unexpected. The differential frequency at which the UGGC motif is observed in the passive and RISC-entering strands of toxic siRNAs strongly suggests that the action of this sequence is mediated by RISC. This is in contrast to the two previously described siRNA motifs (GUCCUUCAA, and UGUGU) that induce IFN phenotypes through Toll-like receptor recognition (Hornung et al. 2005; Judge et al. 2005). As IFN induction can (in some cell types) lead to changes in cell viability, we examined our database of sequences to determine whether any contained the previously described motifs. Of the >300 sequences used in this study, none contains the GUCCUUCAA motif. The UGUGU motif was equally represented in toxic and nontoxic fractions (four out of 98 toxic and seven out of 192 nontoxic), suggesting either that (1) this motif does not generate a phenotype in the cell viability assay used in this study, or (2) the context of the motif in the surrounding sequence is of great importance for the appearance of a toxic phenotype.
Since the majority of toxic sequences evaluated in this study are responsive to the off-target minimizing chemical modification pattern (Fig. 4C) and microarray studies demonstrate that lipid transfection of toxic sequences under optimal conditions fails to induce an IFN response (data not shown), it is likely that the observed sequence-specific toxicity is unrelated to Toll-like receptor activation. However, as only 30% of the toxic sequences contained the UGGC motif (the only motif that passed the statistical significance threshold after Bonferroni correction for multiple comparisons), multiple mechanisms likely play a role in the induction of cellular toxicity. It is possible that in some cases, siRNA toxicity is not associated with knockdown of a single gene, but results from an overall cellular imbalance associated with cumulative cellular stress. Interestingly, visual examination of toxic siRNA sequences suggests that these motifs are highly enriched with AU-rich sequences. Although separately none of these motifs is overrepresented in the toxic population in a statistically significantly way, previous studies (Shimizu et al. 1991; Levine et al. 1993; Gao et al. 1994; King 2000; Jing et al. 2005) demonstrated that AU-rich elements (AREs) present in the 3′-UTR of transcripts play an important role in transcript stability. As 34% of the toxic siRNAs in our study contain one of six closely related AU-rich pentamers—AUUUG, GUUUU, AUUUU, CUUUU, UUUUU, GUUUG—(as compared to 17% of nontoxic siRNAs) and seven of nine (78%) of the duplexes containing both motifs were toxic, it is possible that the protein machinery that mediates AU-rich-element- and RNAi-dependent gene regulation may act additively or synergistically to enhance the levels of off-target gene modulation.
The clear differential distribution of the UGGC motif in RISC entering versus passive strands indicates that the motif correlated with toxicity is acting through the RNAi mechanism. Previous studies of microRNA (Lim et al. 2005) and siRNA target (Haley and Zamore 2004) recognition have demonstrated that RISC–target interaction involves the “seed region” (positions 2–7) of the guide strand. Although the position of the UGGC motif within the guide strand was predominantly localized to the 5′-end of the strand, the correlation is not absolute. Moreover, as (1) some UGGC-containing siRNAs induce toxicity while others do not, and (2) results from the DBI walk show that toxic siRNAs occur in clusters and share significant levels of sequence homology, it is likely that regions other than the seed areas contribute to siRNA off-target recognition.
In summary, we have demonstrated that a substantial number of siRNAs can induce off-target related phenotypes. If not adequately addressed, these effects can lead to false positives in broad RNAi-based phenotypic screens, thus clouding data interpretation. In this study, we demonstrate that chemical modification (in particular, modification of position 2 of the guide strand, described by Jackson et al. [2006]) can substantially limit off-target related phenotypes. Future application of these modifications to chemically synthesized siRNAs will expand the utility of RNAi by minimizing the potential for introduction of confounding experimental artifacts.
MATERIALS AND METHODS
siRNA synthesis
siRNA duplexes were synthesized using 5′-silyl-2′-ACE chemistry (Hartsel et al. 2004). All siRNAs were analyzed by mass spectrometry to ensure quality, and toxic siRNAs were re-synthesized and re-evaluated to ensure that the observed phenotypes were not the result of any impurities. For the 17-bp duplexes used in the deletion study, 2 bp was removed from the 5′ or 3′ sense end of the corresponding 19-bp siRNA. Chemically modified siRNAs were prepared according to the methods described by Jackson et al. (2006). “RISC-free” siRNAs are nontargeting duplexes containing chemical modifications that interfere with RISC uptake (see Jackson et al. 2006). The targeted genes, corresponding GenBank accession numbers, and siRNA sequences are presented in Supplemental Material (see http://www.dharmacon.com/Admin/tech/publications/Publication.aspx?id=166).
Transfection and gene expression analysis
HeLa, MCF7, and DU145 cell lines were obtained from ATCC and cultured using standard media conditions supplemented with penicillin (100 U/mL) and streptomycin (100 μg/mL). For transfection experiments, cells were seeded in antibiotic-free media at 5 × 103 cells per well in a 96-well plate, 24 h before the experiment. The cell density described for these experiments (∼50% confluency at the time of transfection) was critical for observing siRNA-induced toxicity. Cells were transfected with siRNAs (10 nM, 1 pmol/well) using Lipofectamine 2000 (0.1 μL/well; Invitrogen). For gene expression analysis (LUC walk), HEK293 cells were transfected as described previously (Reynolds et al. 2004). mRNA expression levels were assessed using Quantigene (Genospectra) branched DNA (bDNA) assays (Collins et al. 1997) according to the manufacturer's instructions. In all experiments, GAPDH (a housekeeping gene) was used as a reference. All graphical figures represent the average values obtained from three independent experiments, each performed in triplicate. Error bars represent the standard deviation.
Cell viability assay
Cell survival was determined by Alamar Blue (BioSource Int.) according to the manufacturer's instructions. Briefly, 72 h (HeLa) or 144 h (MCF7, DU145) after transfection, 25 μL of Alamar Blue dye was added to wells containing cells in 100 μL of media. Cultures were then incubated for 0.5 h (HeLa) or 2 h (MCF7 and DU145) at 37°C in a humidified atmosphere with 5% CO2. The data presented are an average of nine data points coming from three independent experiments performed in triplicate on different days. For the purpose of this study, siRNAs were defined as toxic when the average from nine different experiments (taking into account standard deviations) showed cell viability below 75%.
Microarray experiments
For each sample, 650 ng of total RNA isolated from siRNA-treated HeLa cells was amplified and Cy3-labeled (Cy-3 CTP; Perkin Elmer) using Agilent's Low Input RNA Fluorescent Linear Amplification Kit and hybridized against Cy5-labeled RNA derived from lipid-treated (control) samples. Hybridizations were performed using Agilent's Human 1A (V2) Oligo Microarrays (∼22,000 probes) and an Agilent Microarray Scanner (Model G2505B). The raw image was processed using Feature Extraction software (v6.1.1 or v7.5.1), Spotfire Decision Site 7.2 software, and the Spotfire Functional Genomics Module. A twofold cutoff (log ratio of >0.3 or <−0.3) and consistency between two biological replicates were applied to identify off-targeted genes. Outlier flagging was not used.
Microscopy
Live cells were stained with Hoechst 33,342 (2 μg/mL, 15 min at 37°C; Molecular Probes), and images were taken using a Leica DML fluorescent microscope InSight CCD camera and SPOT 3.5 software.
eIF2C2 knockdown experiments
HeLa cells were transfected (T1) with a pool of siRNAs directed against eIF2C2 (1 pmol/well) or with a control siRNA (1 pmol/well). Cells were then re-plated at 48 h (5 × 103 cells/well in 96-well plates) and transfected (T2) with a set of toxic siRNAs. The values for toxicity represent the average of three independent experiments (each performed in triplicate).
Statistical analysis of sequence motif distribution
Differential end stability was used to identify 289 siRNA sequences that had a strong RISC-entry bias. Each sequence was tested for toxicity and divided into toxic and nontoxic groups. Sequences whose toxicity/nontoxicity was within one standard deviation of the cutoff were removed, leaving a total of 190 duplexes (98 nontoxic, and 92 toxic) for the statistical analysis.
The occurrence of each possible 4-mer (256 possible combinations) was tabulated in both the RISC-entering and nonentering strands of toxic and nontoxic siRNAs, and a χ2 test for independence was performed to identify those motifs associated with toxicity. The associated p-value was produced using the CHIDIST worksheet function of Excel, and the Bonferroni correction was applied to these results to account for multiple comparisons.
ACKNOWLEDGMENTS
The EGFP-expressing plasmid pTK 590 was a generous gift from Dr. Tal Kafri, University of North Carolina Chapel Hill. We would like to thank the Dharmacon production team and especially Stephanie Hartsel for oligonucleotide synthesis. Elena Maksimova, Barbara Robertson, Queta Boese, and Alexey Wolfson provided helpful discussions. Julia Kendall was instrumental in manuscript preparation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.28106.
REFERENCES
- Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J., et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Collins M.L., Irvine B., Tyner D., Fine E., Zayati C., Chang C., Horn T., Ahle D., Detmer J., Shen L.P., et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F.B., Carson C., Levine T., Keene J. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc. Natl. Acad. Sci. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B., Zamore P. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004;11:559–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- Hartsel S.A., Kitchen D., Scaringe S., Marshall W.S. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods Mol. Biol. 2004;288:33–50. doi: 10.1385/1-59259-823-4:033. [DOI] [PubMed] [Google Scholar]
- Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Johnson J.M., Karpilow J., Nichols K., Marshall W., Khvorova A., et al. Position-specific chemical modification of siRNA increases specificity of gene silencing. RNA. 2006 doi: 10.1261/rna.30706. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Judge A.D., Sood V., Shaw J.R., Fang D., McClintock K., MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- King P.H. RNA-binding analyses of HuC and HuD with the VEGF and c-myc 3′-untranslated regions using a novel ELISA-based assay. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.7.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T., Gao F., King P.H., Andrews L.G., Keene J.D. Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin X., Ruan X., Anderson M.G., McDowell J.A., Kroeger P.E., Fesik S.W., Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Leake D., Scaringe S.A., Marshall W.S., Boese Q., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Shimizu N., Ohta M., Fujiwara C., Sagara J., Mochizuki N., Oda T., Utiyama H. Expression of a novel immediate early gene during 12-O-tetradecanoylphorbol-13-acetate-induced macrophagic differentiation of HL-60 cells. J. Biol. Chem. 1991;266:12157–12161. [PubMed] [Google Scholar]