Abstract
Cross-species DNA sequence comparison is the primary method used to identify functional noncoding elements in human and other large genomes. However, little is known about the relative merits of evolutionarily close and distant sequence comparisons. To address this problem, we identified evolutionarily conserved noncoding regions in primate, mammalian, and more distant comparisons using a uniform approach (Gumby) that facilitates unbiased assessment of the impact of evolutionary distance on predictive power. We benchmarked computational predictions against previously identified cis-regulatory elements at diverse genomic loci and also tested numerous extremely conserved human–rodent sequences for transcriptional enhancer activity using an in vivo enhancer assay in transgenic mice. Human regulatory elements were identified with acceptable sensitivity (53%–80%) and true-positive rate (27%–67%) by comparison with one to five other eutherian mammals or six other simian primates. More distant comparisons (marsupial, avian, amphibian, and fish) failed to identify many of the empirically defined functional noncoding elements. Our results highlight the practical utility of close sequence comparisons, and the loss of sensitivity entailed by more distant comparisons. We derived an intuitive relationship between ancient and recent noncoding sequence conservation from whole-genome comparative analysis that explains most of the observations from empirical benchmarking. Lastly, we determined that, in addition to strength of conservation, genomic location and/or density of surrounding conserved elements must also be considered in selecting candidate enhancers for in vivo testing at embryonic time points.
The majority of long-range cis-regulatory elements in the human genome have yet to be identified. These gene regulatory modules, which likely number in the tens or hundreds of thousands, are hard to detect, since they lack any obvious distinguishing features analogous to codon structure, splicing motifs, open reading frames, or other hallmarks of protein-coding genes. Furthermore, functional sequences of this class are mostly unique in the genome (Bejerano et al. 2004a), which largely rules out paralogy-based identification. The possibility that regulatory elements could lie more than 1 Mb from their target genes (Lettice et al. 2003; Nobrega et al. 2003) presents another challenge. Consequently, cross-species sequence comparisons, which rely upon the slow substitution rate of many categories of functional DNA relative to neutral sequence, have emerged as the pre-eminent means of identifying candidate cis-regulatory elements in large genomes such as human (Pennacchio and Rubin 2001; Nobrega et al. 2003; Ahituv et al. 2004; Chapman et al. 2004; de la Calle-Mustienes et al. 2005; Hughes et al. 2005; King et al. 2005; Woolfe et al. 2005).
Distant comparisons, such as human–fugu (Brenner et al. 1993; Nobrega et al. 2003; de la Calle-Mustienes et al. 2005; Woolfe et al. 2005), have proven especially powerful at high-specificity prediction of functional elements, since neutral sequences have had sufficient time to diverge beyond recognition. However, despite the similar gene content of the human and fugu genomes, even functional noncoding elements are likely to have diverged over such great distances, as demonstrated by the small number (thousands) of human–fugu conserved noncoding sequences (CNSs) in the genome. At the other extreme, primate sequence comparisons (Boffelli et al. 2003) are likely to capture most functional components of the human genome due to shared biology but suffer from low resolution due to insufficient neutral divergence among primate taxa (Eddy 2005). Mammalian genome comparisons have been proposed as a compromise between the requirements of sequence divergence and biological similarity (Cooper et al. 2003), and efforts are under way to sequence 16 additional mammalian genomes, albeit at low coverage (Margulies et al. 2005).
Here, we present an empirical and genomic evaluation of the relative merits of close and distant sequence comparisons at detecting functional noncoding regions in the human genome. Our study complements recent theoretical analyses of this problem (Eddy 2005, Stone et al. 2005) and is directly relevant to the choice of species for whole-genome sequencing and comparative analysis. Additionally, given that regulatory divergence is often proposed as a primary mechanism of phenotypic variation, it is important to characterize the rate of decline of noncoding sequence conservation with increasing evolutionary distance. To impartially assess the effect of evolutionary distance on the predictive power of noncoding conservation, we used a uniform computational approach (Gumby) to detect CNSs in primate, mammalian, and more distant sequence alignments. Close and distant comparisons were tested at three diverse genomic loci, for which numerous cis-regulatory elements have been characterized experimentally. To complement these empirical results, we performed a whole-genome meta-analysis of human–rodent, human–mouse–chicken, human–mouse–frog, and human–mouse–fish whole-genome CNS sets and uncovered a general principle linking shallow and deep evolutionary constraint. Finally, we performed systematic in vivo testing of extremely conserved human–rodent CNSs in an in vivo transgenic mouse enhancer assay and identified with high specificity developmental enhancers missed by human–fish comparative analysis, and in the process determined that genomic context is a critical factor in identifying such enhancers.
Results
Assessment of whole-genome noncoding conservation among mammals and more distant species
Coding exons are known to retain sequence similarity across great evolutionary distances, such as that between human and pufferfish (Aparicio et al. 2002). In contrast, intronic and intergenic conserved elements “evaporate” much more rapidly, thus limiting the sensitivity of distant noncoding sequence comparisons. To quantify the decay of noncoding sequence conservation with evolutionary distance, we generated whole-genome CNS sets through Gumby analysis (see Methods) of the following three-way genome alignments: human–mouse–rat (HMR), human–mouse–chicken (HMG), human–mouse–frog (HMX), and human–mouse–fish (HMF) (see Methods). In the human–rodent alignment, we identified 171,853 CNSs representing 2.2% of the human genome (Gumby P-value ≤ 1e-3). At the same P-value threshold, the corresponding CNS statistics were 40,033 and 0.37% for human–mouse–chicken, 14,568 and 0.13% for human–mouse–frog, and 5668 and 0.044% for human–mouse–fish. As expected, the more distantly related genomes exhibit markedly less conservation relative to human, suggesting a reduction in sensitivity that offsets their increased specificity in detecting functional noncoding regions.
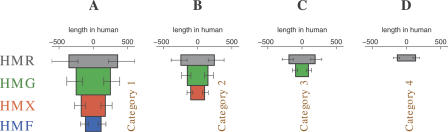
In order to correlate mammalian and more ancient noncoding conservation, we classified whole-genome CNSs into four primary categories. Category 1 consists of human–rodent CNSs that overlap human–mouse–chicken, human–mouse–frog, and human–mouse–fish CNSs. Category 2 extends only to human–mouse–frog, Category 3 to human–mouse–chicken, and Category 4 is restricted to human–rodent alone. Median length and −log(P-value) of CNSs within a given category and phylogenetic scope (for instance, Category 2 and human–mouse–frog) were represented by the dimensions of a single rectangular block. Building on such blocks, we define shapes generated by stacking blocks of the same category as evolutionary stacking patterns (ESPs, Fig. 1A–D). Although there is considerable variation within each category, the four ESPs characterizing the four CNS categories illustrate two general trends:
Figure 1.
Whole-genome noncoding conservation and the funnel principle: correlation between ancient and recent noncoding conservation (funnel principle), illustrated by four evolutionary stacking patterns (ESPs). HMR/G/X/F: human–mouse–rat/chicken/frog/fish. The four ESPs depict four sets of whole-genome HMR CNS, categorized by their most ancient overlapping nonmammalian CNS. Stacked below the rectangular blocks representing HMR CNSs are blocks depicting the corresponding ancient CNSs. (A) Category 1 CNSs extend to HMF, (B) Category 2 to HMX, (C) Category 3 to HMG, and (D) Category 4 is limited to HMR. Block width is proportional to median CNS length in human, and block area is proportional to the median of −log(P-value). Block height thus represents degree of evolutionary constraint at the basepair level. Error bars mark the range from the 25th to the 75th percentile of CNS length. The funnel principle takes its name from the funnel-like shape of ESPs.
CNSs shrink as evolutionary distance increases, with the tallest stacking pattern tapering from 712 bp at its phylogenetically “shallow” end (human–rodent) to 228 bp at its “deep” end (human–mouse–fish).
Close-species CNSs are longer and have stronger P-values when they are also conserved in distant species. For example, while human–rodent CNSs conserved in chicken, frog, and fish have median length 712 bp and P-value 4.1e-35, human–rodent CNSs with no significant nonmammalian conservation have median length 268 bp and P-value 2.2e-06. Since they arise from the characteristic funnel shape of ESPs, we refer to these two trends collectively as the “funnel principle” of noncoding conservation.
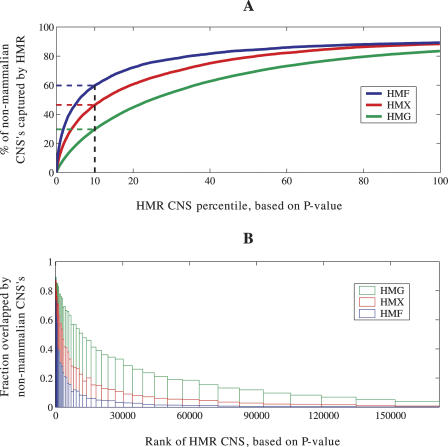
The funnel principle suggests that highly conserved human–rodent CNSs should be enriched for conservation in nonmammals. To quantify this enrichment, we sorted whole-genome human–rodent CNSs by P-value and calculated the fraction of human–mouse–chicken, human–mouse–frog, and human–mouse–fish CNSs overlapped by various quantiles of the human–rodent set (Fig. 2A). Remarkably, we see that the top 10% of the human–rodent CNSs overlap 60% of the CNSs in the human–mouse–fish set, 46% of the human–mouse–frog set, and 30% of the human–mouse–chicken set, indicating that a large fraction of deeply conserved CNSs can paradoxically be identified through shallow evolutionary comparisons. Similarly, almost half (47%) of the human–mouse–fish CNSs are contained in the top 5% of the human–rodent set. In terms of probabilities, the highest-scoring human–rodent CNSs have an 86% chance of being significantly conserved in chicken, while the lowest-scoring ones have less than a 4% chance of being in the human–mouse–chicken set (Fig. 2B). The corresponding likelihoods are 81% and 0.75% in human–mouse–frog and 64% and 0.2% in human–mouse–fish (see Supplemental Methods for analyses of potential artifacts).
Figure 2.
Whole-genome noncoding conservation and the funnel principle: strong enrichment for nonmammalian conservation in top-ranked (lowest-P-value) human–mouse–rat CNSs. (A) Cumulative fraction of nonmammalian CNSs overlapped by various quantiles of human–mouse–rat CNSs. The top 10% of human–rodent CNSs (by P-value) constitute a set of 17,185 sequences with a high degree of recent evolutionary conservation, that encompass 60% of whole-genome human–mouse–fish CNSs, 46% of human–mouse–frog CNSs, and 30% of human–mouse–chicken CNSs. (B) The 171,853 human–rodent CNSs are binned by P-value. Vertical bars represent the fraction of human–rodent CNSs in each bin that overlap more ancient CNSs.
Although we see a significant correlation between ancient and recent noncoding conservation, there are also important differences between the two phenomena. While half (2834) of the whole-genome human–mouse–fish CNSs have a human–rodent P-value < 1.9e-28, there are 7686 human–rodent CNSs that meet the same P-value criterion and yet lack significant conservation in fish. These data indicate that human–rodent analysis is capable of identifying a much larger set of unambiguously constrained noncoding elements than are obtainable from mammal–fish comparison.
It is conceivable that the funnel principle could merely reflect biases in the algorithm used to identify CNSs, as would indeed be the case if close and distant comparisons were not performed uniformly, or if the CNS sets were corrupted with significant numbers of false-positive predictions. We therefore performed evolutionary simulations and further statistical analyses, which confirmed that such artifacts are unlikely (see Supplemental Methods).
In vivo experimental validation and assessment: Four human-genome loci
DACH1: In vivo testing of the top-scoring human–rodent conserved elements
In order to empirically evaluate the power of extreme human–rodent conservation to identify developmental enhancers, we analyzed the locus of DACH1, a transcription-cofactor gene involved in limb, eye, and brain development (Davis et al. 1999). The 2-Mb genomic region containing DACH1 and most of its flanking intergenic DNA (human chr13:70,207,792–72,205,000; NCBI Build 35) contains 1084 human–mouse CNSs by the standard criterion of 70% sequence identity over 100 bp, thus necessitating some criterion for prioritizing the abundance of human–rodent elements. It has previously been demonstrated that human–fish conservation constitutes one such criterion for prioritizing human–rodent elements, in that restricting the analysis to human–fish CNSs facilitated the identification of transcriptional enhancers with a high true-positive rate (Nobrega et al. 2003). In the present study, we attempted to achieve a comparable true-positive rate and yet greater sensitivity by focusing instead on the human–rodent CNSs with the most extreme (i.e., very low) P-values.
We first assessed the overlap between the most conserved human–rodent CNSs and human–fish CNSs in the DACH1 locus. Consistent with the above-described funnel principle, we found that 22 of the 36 strongest human–mouse–rat CNSs in this locus (P-value ≤ 1e-50) are conserved in at least one of the three available fish genomes. These 22 include five of the seven human–fish CNSs previously validated through in vivo enhancer testing (Nobrega et al. 2003). Thus, in addition to having a high likelihood of ancient conservation in distant species, the 36 human–rodent CNSs with extreme P-values are also enriched for known developmental enhancers, relative to the entire set of 1084 human–mouse CNSs in the vicinity of DACH1.
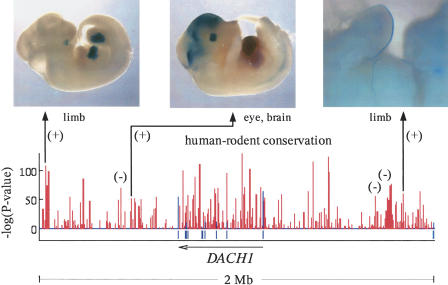
To demonstrate the independent predictive power of extreme human–rodent conservation, we focused on the 14 CNSs within the aforementioned set of 36 that exhibited no conservation in fish and randomly selected six of them to assay for transcriptional enhancer activity in vivo. Our assay fuses the human conserved element to a β-galactosidase reporter vector and assesses the ability of the conserved fragment to drive tissue-specific expression in transgenic mice at embryonic day 11.5–12.5 (e11.5–12.5) (Kothary et al. 1989) (see Supplemental Methods). For three of the six extreme human–rodent elements tested, we found reproducible β-galactosidase expression localized to the limbs, eyes, and forebrain, consistent with aspects of the endogenous developmental expression pattern of DACH1 (Caubit et al. 1999; Davis et al. 1999) (Fig. 3). β-galactosidase expression was not reproducibly localized to any other anatomical structure of the embryos.
Figure 3.
DACH1 locus: Identification of long-range embryonic enhancers by extreme human–rodent CNSs with no conservation in fish. The Gumby human–mouse–rat conservation plot in the lower half of the figure depicts 784 CNSs (red vertical bars) in a 2-Mb genomic region containing the DACH1 gene, many of which have extremely low P-values. Blue ticks below the line mark DACH1 exons. Six of the human–rodent CNSs with P-value < 1e-50 and no conservation in fish were tested for enhancer activity at embryonic day 11.5–12.5; the resulting positives and negatives are marked by (+) and (−) symbols, respectively. The identified enhancers drove reproducible β-galactosidase expression in limbs, eyes, and brain, consistent with the endogenous expression domains of DACH1.
To test the predictive power of human–rodent conservation P-values on a larger scale, we retrospectively analyzed 133 human–fugu and ultra-conserved (Bejerano et al. 2004b) CNSs that were tested for in vivo enhancer function as part of a separate whole-genome survey (L. Pennacchio, N. Ahituv, A. Moses, M. Nobrega, S. Prabhakar, M. Shoukry, S. Minovitsky, A. Visel, I. Dubchak, A. Holt, et al., in prep.). Each of the 133 CNSs was assigned a Gumby human–rodent conservation score (−log(P-value)). We found that positive enhancers had significantly higher conservation scores than negatives (t-test P-value = 0.0001), which further confirms the validity of using human–rodent P-values to prioritize candidate embryonic enhancers.
Human chromosome 16: Genomic distribution of enhancers active at embryonic day 11.5–12.5
Though reliable indicators of function, human–fish CNSs tend to be limited in number and strongly clustered in genomic regions containing a handful of developmental genes and transcription factors (Sandelin et al. 2004; Woolfe et al. 2005). For example, the human–mouse–fish CNSs on human chromosome 16 are highly skewed towards four gene-poor loci (Fig. 4) containing the developmentally regulated genes SALL1, IRX3, IRX5, IRX6, ATBF1, and WWOX. The density graph of human–rodent CNSs on this chromosome displays peaks at the same locations as human–mouse–fish and also additional peaks absent in human–mouse–fish. On the basis of positive results from the DACH1 pilot study, we hypothesized that human–rodent CNSs with extreme P-values could also identify developmental transcriptional enhancers in these additional loci, to compensate for their poor coverage by human–mouse–fish CNSs.
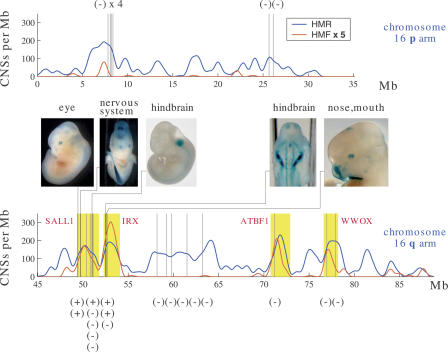
Figure 4.
Extreme human–rodent CNSs with no conservation in fish identify enhancers at e11.5–12.5 only in certain genomic regions. Upper and lower plots: human–rodent (blue) and human–mouse–fish (scaled by a factor of 5, red) CNS density on the p (upper) and q (lower) arms of human chromosome 16. We tested 24 human–rodent CNSs with P-value < 1e-40 and no conservation in fish (locations marked by vertical lines) for enhancer activity at embryonic day 11.5–12.5. The five enhancers thus identified were located exclusively in two of the four loci with the highest human–mouse–fish CNS density on the chromosome (yellow bands). These loci contain developmentally regulated genes, all but one of which are transcription factors.
To test this hypothesis, we focused on the 50 top-scoring “non-fish” human–mouse–rat CNSs on chromosome 16 (P-value < 1e-40), of which 36 were located outside the four developmental loci encompassed by human–mouse–fish conservation. We tested 11 of these 36 for in vivo enhancer function through our mouse enhancer transgenesis assay and found to our surprise that not a single one of these 11 CNSs drove a reproducible embryonic expression pattern, in contrast to our experience at the DACH1 locus. Excessive distance between the tested CNSs and their flanking genes is an unlikely explanation for the negative results, since 10 of the 11 CNSs lie within 1 Mb of a RefSeq transcription start site. It is possible that these CNSs are not transcriptional enhancers despite extreme human–rodent conservation, or that their activities are beyond the resolution/sensitivity of this assay or perhaps even that they are enhancers at a different embryonic time point.
To determine if extreme human–mouse–rat CNSs are better able to capture transcriptional enhancers flanking key developmental transcription factor and signaling genes, we tested 13 of the 14 extreme human–rodent CNSs (P-value < 1e-40) that lie within the four gene-poor regions on chromosome 16 containing the aforementioned developmental genes. Of these 13 CNSs, five (38%) drove reproducible β-galactosidase gene expression, a success rate comparable to the rate of 41% observed in systematic tests of human–fish CNSs on the same chromosome (L. Pennacchio, unpubl.; http://enhancer.lbl.gov/). The disparity in success rate among different loci on chromosome 16 suggests that genomic context must be considered in addition to conservation score in selecting candidate enhancers for testing at this embryonic time point.
SCL benchmark: Benefits of multiple-eutherian comparison
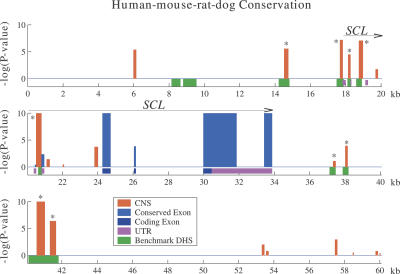
The human stem cell leukemia (SCL) locus provides an excellent benchmark for evaluating the effect of phylogenetic scope on comparative sequence analysis, based on the detailed experimental definition of nine nonexonic murine DNaseI-hypersensitive sites (DHSs) and one additional enhancer in the genomic region containing the SCL gene and its flanking intergenic segments (Chapman et al. 2004). Weak homology with chicken has been reported (Göttgens et al. 2000) for a subset of these functional elements in alignments generated using DIALIGN (Brudno et al. 2003a). However, with the exception of one of the SCL promoters, none of the 10 experimentally defined elements shows significant conservation in LAGAN alignments to nonmammalian species such as chicken, frog, fugu, tetraodon, or zebrafish, exemplifying the loss of sensitivity entailed by distant sequence comparisons (data not shown).
Given the poor sensitivity of distant sequence comparisons, we aligned the available human, mouse, rat, and dog sequences from this locus (human chr1:47,367,748–47,427,851; NCBI Build 35) using MLAGAN, which yielded a total divergence of 0.79 substitutions/site. At the default P-value threshold of 0.5, Gumby conservation analysis (see Supplemental Methods) detected eight of the 10 experimentally defined noncoding elements, with 11 new predictions (Fig. 5). Thus, as in the case of DACH1, P-value prioritization of CNSs in just a few eutherian genomes is sufficient to identify with acceptable true-positive rate functional elements missed by distant sequence comparison. Of the eight functional regions detected by human–mouse–rat–dog analysis, seven have P-value ≤ 1.3e-4, whereas only one of the new predictions meets the same criterion, suggesting that most of the known functional elements in this region are clearly distinguishable from neutrally evolving DNA. Indeed, when we reduced the statistical power of the sequence comparison by restricting our analysis to human and mouse alone, (branch length 0.47 substitutions/site), Gumby still identified only one new prediction within the P-value range of the seven prominent benchmark elements. Remarkably, even mouse–rat pairwise comparison (P-value ≤ 0.5) succeeded in identifying six of the benchmark elements with only three new predictions, despite the minimal neutral divergence (0.14 substitutions/site) between the two rodents. These results demonstrate that very low levels of neutral sequence divergence are sufficient for identification of well-conserved enhancers and DHSs, though functional elements marked by marginal levels of sequence conservation are better detected when total branch length is augmented by introduction of additional eutherians to the species set.
Figure 5.
SCL locus: Benchmarking eutherian sequence comparison against empirical data. The displayed 60-kb genomic region contains the SCL gene and its flanking intergenic regions. Human–mouse–rat–dog CNSs (red) and exonic (blue) conserved regions identified by Gumby (P-value ≤ 0.5) are shown as vertical bars. The nine CNSs marked with asterisks identify 8/10 benchmark sequence elements with empirical evidence of cis-regulatory function (green rectangles). Additionally, there are 11 new predictions at this P-value threshold.
α-globin benchmark: Simians, primates, mammals
The human α-globin locus is another well-characterized genomic region, with a recent synthesis of extensive computational and empirical analyses cataloging 17 functional noncoding DNA elements (Hughes et al. 2005). The elements comprise 11 promoters, four nonpromoter DHSs involved in transcriptional regulation, and two putative regulators of alternative splicing. In addition, this locus is well suited to evaluating the relative merits of close and distant sequence comparisons, due to the availability of sequence data from 22 vertebrate species spanning a broad range of evolutionary distances from old world monkeys to teleost fish. The analyzed genomic region (human chr16:48,339–219,839; NCBI Build 35) spans the block of conserved synteny telomeric to the α-globin genes, the α-globin genes themselves, and the LUC7L gene. To ensure accurate alignment, we split the locus into four subregions and aligned each subregion separately (see Supplemental Methods).
As was the case with the SCL locus, conservation of the benchmark functional elements in distant species is extremely limited; frog and fish have no apparent sequence homology with any of the 17 noncoding functional elements, and chicken shows homology with only two of the 17. Sequence conservation is also limited in opossum, and even hedgehog (which is the most diverged of the eutherians considered here) shows no similarity for five of the 17 benchmark sequence elements (see Hughes et al. [2005 for a detailed breakdown of conservation by species).
To assess the relative power of various eutherian comparisons at this locus, we selected the following three species sets: (1) simians (human, baboon, colobus, squirrel monkey, owl monkey, marmoset, dusky titi), (2) primates (simian group plus the prosimian galago), and (3) eutherians minus nonhuman primates and hedgehog (human, mouse, rat, cat, cow, pig). In the eutherian set, which had a total branch length ranging from 1.24 to 1.55 substitutions/site across the four subregions, Gumby identified 13/17 benchmark elements, with 22 new predictions (P-value ≤ 0.5). Primate and simian comparisons were performed with P-value thresholds adjusted to yield the same number of new predictions (22) as in the eutherian comparison, so as to fairly assess relative sensitivity of the three species sets. The resulting sensitivity of the primate comparison (0.45–0.69 substitutions/site) was 11/17, while the simian comparison (0.25–0.39 substitutions/site) had a sensitivity of 9/17, demonstrating that predictive power declines as evolutionary divergence decreases in closely related species. Although the simian comparison displayed the lowest statistical power, it is notable that a sensitivity of 53% (9/17) and a true-positive rate of at least 29% (9/(9 + 22)) were achieved by comparing no more than six simian genomes with human in any of the four subregions.
Discussion
Ancient human–fish noncoding conservation has been the mainstay of searches for enhancers of key developmental genes, whereas mammalian sequence comparisons have been considered insufficiently specific, especially in large, highly conserved intergenic regions harboring hundreds of human–rodent CNSs. This dichotomy disappears in light of the funnel principle established by whole-genome meta-analysis of close and distant sequence comparisons: The longer and more constrained a mammalian CNS, the deeper its evolutionary conservation in distant vertebrates (on average), and vice versa. Thus, extreme human–rodent P-values serve as a proxy for human–fish conservation, and most human–fish CNSs can be identified through human–rodent analysis alone. The correspondence between recent and ancient sequence conservation is likely to grow even stronger when more mammalian genomes are added to the human–mouse–rat trio (Margulies et al. 2005). A relationship between ancient and recent noncoding conservation consistent with the funnel principle has been reported earlier (Ovcharenko et al. 2004), though the correspondence described between the two evolutionary scales was significantly weaker than is evident from our results. Although human–fish sequence comparison identifies developmental enhancers with high specificity, only a small fraction of the expected tens or hundreds of thousands of functional noncoding elements in the human genome are conserved in fish. Extreme P-value mammalian CNSs form a superset covering the majority of human–fish as well as a much larger fraction of noncoding functional elements in the human genome, while still maintaining a low false-positive rate. More generally, in identifying candidates for experimental testing, the tradeoff between sensitivity and specificity can be tuned simply by adjusting the threshold conservation score (P-value), in contrast to the more common technique of varying the evolutionary distance between the compared species.
In vivo assays of 30 extreme human–rodent CNSs with no conservation in fish yielded eight positive enhancers at e11.5–12.5, close to the success rate of human–fish comparison. This experimentally confirms the theoretical prediction that extreme conservation in relatively close species is on a par with conservation between highly diverged species. In terms of genomic distribution, the success rate was 8/19 among human–rodent CNSs in the neighborhood of developmental genes such as DACH1, SALL1, IRX3, IRX5, IRX6, ATBF1, and WWOX, and it was 0/11 at other loci on human chromosome 16, most likely since the assay is limited to genes with complex tissue-specific expression at e11.5/12.5 and produces false negatives in loci that are silent or regulated in a relatively simple manner at this embryonic time point. Since such in vivo tests are expensive and time-consuming, one strategy for large-scale functional genomics would be to prioritize loci that are known to require tissue-specific regulation at the selected developmental stage. Alternatively, one could prioritize loci of unknown function that nevertheless have a high density of ancient (human–mouse–fish, or perhaps even human–mouse–frog or human–mouse–chicken) CNSs, since the loci on human chromosome 16 yielding high success rates in the embryonic enhancer assay were originally identified solely on the basis of their high human–mouse–fish CNS density (Fig. 4). For instance, the second-largest peak of human–mouse–fish CNS density in the human genome (data not shown) occurs between the first exons of the uncharacterized human genes ZNF503 and C10orf11, making this gene a candidate for deeply conserved and highly specific early developmental regulation.
Results from whole-genome conservation analysis, from benchmarking using pre-existing functional data sets from three diverse genomic loci and from systematic in vivo characterization of 30 new enhancer predictions, all indicate the considerable statistical power of sequence comparisons involving just a few (three to six) eutherian mammals. Another consistent theme is the loss of sensitivity when more distant species such as marsupials, chicken, frog, and fish are compared with human, either because they have diverged in their cis-regulatory programs or because of stabilizing selection that allows the regulatory sequence to diverge while retaining the same tissue and temporal specificity (Ludwig et al. 2000; Oda-Ishii et al. 2005). At the other extreme, simian sequence comparisons in the α-globin locus performed surprisingly well despite their low total divergence (0.25–0.39 substitutions/site), achieving a sensitivity of 53% and a true-positive rate of at least 29%. The funnel principle provides one possible explanation: Since the length of a CNS in human–mouse–chicken is on average greater than that of its equivalent in human–mouse–fish, and the corresponding mammalian CNS is longer still, the size of constrained blocks in simians should by extrapolation be greater than that in mammals, thus offering simian comparisons a larger target, and partially compensating for their lower neutral divergence.
It is common in analyses of the statistical power of sequence comparisons to fix the size of an individual constrained block as the total branch length (neutral divergence) is varied (Margulies et al. 2003; Eddy 2005; Stone et al. 2005). However, the funnel principle implies that the two variables are not independent, and that constrained blocks shrink when more distant species are compared. Thus, for a given total branch length, one could maximize block size, and consequently statistical power, by choosing multiple extremely close species (say, simian primates) over a few more diverged species (mammals).
In this study, we have focused on the identification of large sequence elements with empirical evidence of cis-regulatory function, such as enhancers, promoters, and DNaseI-hypersensitive sites, which have typical lengths of 100 bp or more. Previous theoretical studies (Cooper et al. 2005; Eddy 2005) have shown that higher-resolution functional prediction at the level of a transcription-factor binding site (6–12 bp), or even a single basepair, is likely to require sequence from more than ten mammals spread across the clade. However, a more accessible initial strategy might be to use the existing mammalian genome sequences for prediction of larger, higher-level functional elements, many of which show little or no sequence conservation in distant species. It should also be possible to use sequence data from multiple primates to identify distant regulatory elements that evolve too rapidly to be detected in mammalian sequence comparisons. We have demonstrated that such strategies are highly effective, based on systematic benchmarking of sequence comparisons across a broad range of phylogenetic scopes against empirical data from a diverse array of genomic loci. While the sequencing of additional mammalian genomes will incrementally facilitate identification of large regulatory modules in the human genome, it is likely that the greatest strength of deep mammalian genomic alignments will be in computationally dissecting their internal structure.
Methods
Development of a uniform statistical scoring scheme for sequence conservation (Gumby)
In order to uniformly evaluate the benefits and limitations of close versus distant sequence comparisons, we sought a computational algorithm general enough to process alignments at all evolutionary distances, identify conserved regions of any size, and, most importantly, quantify their statistical significance (P-value). For generality and convenience, we further stipulated that the method should require no training data, no prior estimates of evolutionary rates (branch lengths), and only one arbitrary parameter, which could remain fixed across all evolutionary distances. The Gumby program meets these design goals by making minimal assumptions about the statistical features of conserved noncoding regions and treating the sequence alignment as its own training set. Gumby takes its name from the Gumbel distribution, which is the extreme value distribution underlying Karlin-Altschul statistics. The input data are an alignment, a phylogenetic tree (topology only, without branch lengths), and annotations of coding regions (optional). The algorithm proceeds through five steps:
Noncoding regions in the input alignment are used to estimate the neutral mismatch frequency pN between each pair of aligned sequences. This is done simply by counting the number of mismatches in nonexonic positions and dividing by the number of aligned nonexonic positions. Failure to provide exon annotations introduces a bias in the mismatch frequency that is proportional to the fraction of genomic DNA contained in exons, which is generally small in vertebrates.
A log-odds scoring scheme for constrained versus neutral evolution is then independently initialized for each pair of sequences, based on the assumption that the mismatch frequency pC in constrained regions equals pN/R, where the ratio R is an arbitrary parameter. For example, if R = 3/2 (default value), constrained regions are expected to evolve at 2/3 times the neutral rate, until sequence divergence begins to saturate. The log-odds mismatch score for the sequence pair is then given by S0 = log((pN/R)/pN) = −log(R), and the match score is S1 = log((1 − pN/R)/(1 − pN)). The default R-ratio (1.5) was selected to optimize the sensitivity–specificity tradeoff in detecting empirically defined regulatory elements in the SCL locus (Supplemental Fig. 1). However, other values of R gave very similar results, perhaps because seven of the 10 benchmark regulatory elements are very significantly conserved and, therefore, robustly distinguishable from the rest of the locus. Gap characters in the alignment are assigned a weighted average of mismatch and match scores: SG = pNS0 + (1 – pN)S1. This gap score is guaranteed to be negative, and has the effect of lightly penalizing indels: a compromise between treating them as mismatches, which is the usual practice for algorithms implementing phylogenetic “shadowing,” and ignoring them altogether, as is common when the species set is sufficiently diverged. Missing data, i.e. “N” nucleotides, are given a zero score. However, one drawback of any scoring scheme that penalizes gaps is that failure to flag sequencing gaps with “N” characters results in spurious alignment gaps, which artificially lower the conservation score of the corresponding region in the alignment.
Each alignment column is scored as a sum of pairwise log-odds scores along a circular tour of the phylogenetic tree. For example, for the phylogenetic tree shown in Supplemental Figure 2, the circular-tour column score is S = S(human,mouse) + S(mouse,cow) + S(cow,dog) + S(dog,lemur) + S(lemur, human). This averaging scheme traverses each branch in the phylogenetic tree the same number of times, and thus permits simple phylogenetic scoring of multiple alignments while avoiding the drawbacks of sum-of-pairs and consensus schemes. The resulting conservation score fulfills the requirements of Karlin-Altschul statistics, in that positive column scores are possible, though the average column score is negative (Karlin and Altschul 1990).
Conserved regions appear as stretches of alignment columns with a high aggregate score. Gumby traverses the alignment from left to right, initiating a conserved segment at each new alignment column with a positive score and extending the segment until the aggregate score becomes negative, at which point the right edge of the segment is retraced to the alignment column yielding the maximal aggregate segment score. This procedure guarantees that both boundaries of each segment are maximal with respect to segment score, thus eliminating the need for setting arbitrary window sizes and allowing detection of long, weakly conserved regions as well as short, strongly conserved elements. This feature is also important in achieving generality across close and distant sequence comparisons, since short conserved elements are not likely to be statistically significant in sequence comparisons with low total neutral divergence (for example, primate shadowing).
The aggregate score of the alignment columns in each conserved region is translated into a P-value using Karlin-Altschul statistics. As is the case with the BLAST algorithm (Altschul et al. 1990), the P-value of a given conserved element varies with the size of the search space, since one is more likely to find a given degree of conservation by random chance in a long alignment than in a short alignment. To make the P-values comparable across alignments of different lengths, Gumby normalizes them to refer to a fictitious fixed-length alignment with the same statistical properties as the true alignment. In other words, the P-value answers the question, ′What is the likelihood of seeing such a high conservation score in a pseudo-alignment of length 10 kb that is generated by randomly selecting columns from the given alignment?′ The 10-kb P-value is related to the expected number of false positives in a 10-kb region (i.e. the 10-kb E-value) as follows: P = 1 – exp(−E). When P ≪ 1, P ≈ E. Thus, the P-value also doubles as an estimate of the false-positive rate.
One pitfall in applying Karlin-Altschul statistics to global alignments is the fact that column scores are not identically distributed. For example, the distribution of scores at positions that are aligned in, say, five of 10 species is different from that at positions that are unique to a single species. Also, the number of aligned species is highly correlated between neighboring alignment columns, due to the block-like structure generated by long indels. The fictitious randomly permuted alignments modeled by Karlin-Altschul statistics do not have this correlation structure and are, therefore, less likely to contain local high-scoring segments than are neutral regions in real alignments. Thus, a straightforward application of the permutation-based null model generates unrealistically strong P-values. To compensate for this effect, Gumby takes the conservative approach of estimating the Karlin-Altschul Κ and λ parameters only on the basis of columns that are aligned in at least k species (k ≥ 2). As k is increased, the number of columns in the null model decreases. Gumby sets k to the maximum value such that the number of columns in the null model is at least 40% of the length of the base sequence. Another source of spurious P-values is the suppression of null-model scores by large indels that are artifacts of missing sequence data. Gumby again takes the conservative approach of penalizing indels only after the null model has been generated.
Gumby availability
Gumby conservation analysis is automatically performed when DNA sequences are submitted to the VISTA server (Frazer et al. 2004) (http://genome.lbl.gov/vista), and conserved regions are graphically displayed using RankVISTA. Pre-computed whole-genome Gumby results based on pairwise alignments are available as RankVISTA tracks on the VISTA Browser (http://pipeline.lbl.gov). Gumby source code is available at http://pga.lbl.gov/gumby.
Generation of whole-genome CNS sets
Whole-genome CNS sets were generated as described in (Ahituv et al. 2005). Syntenic blocks between the compared genomes were defined by PARAGON, globally aligned using MLAGAN (Brudno et al. 2003b), and scanned for statistically significant conserved regions using Gumby. This procedure minimizes false alignments, since only syntenic conservation is allowed. As in previous large-scale analyses (Ahituv et al. 2005), we sought to improve alignment accuracy by performing three-way (human, mouse, nonmammal) instead of pairwise (human, nonmammal) genome alignments (Brudno et al. 2003b). Consequently, we analyzed alignments between the human and mouse genomes and those of rat (HMR), chicken (HMG), frog (HMX), and fish (HMF; union of human–mouse–fugu, human–mouse–tetraodon and human–mouse–zebrafish). CNSs were defined as conserved regions (P-value ≤ 1e-3) that do not overlap known genes, mRNAs, or spliced ESTs in any of the aligned genomes. This P-value threshold is a factor of 500 below the default threshold of 0.5 and is, therefore, not suitable for high-sensitivity identification of noncoding regulatory elements. However, it severely limits false-positive predictions and, therefore, facilitates reliable characterization of the relations between recent and ancient noncoding conservation.
DACH1 and human chromosome 16: Identifying candidate embryonic enhancers
Due to the extreme levels of noncoding constraint observed in the DACH1 locus, we identified human–mouse–rat CNSs in this genomic region using a Gumby R-ratio of 10.0 and a P-value threshold of 1e-50. However, in the larger subsequent analysis of extremely conserved human–mouse–rat CNSs on human chromosome 16, we retained the default R-ratio of 1.5 and relaxed the P-value threshold to 1e-40, so as to obtain a larger and more representative set of conserved elements. In addition to the aforementioned filters for transcriptional evidence, extreme human–rodent CNSs with more than 40 bp of overlap with human or nonhuman unspliced ESTs from more than one library were discarded, as were CNSs overlapping single unspliced ESTs with BLASTX matches (E-value ≤ 0.5) in peptide sequence databases (Altschul et al. 1990). CNSs within 50 bp of exons of known genes were also removed, so as to eliminate potential regulators of pre-mRNA splicing. The remaining human–rodent CNSs were filtered for overlap with Gumby human–mouse–fish CNSs, or with human–fugu, human–tetraodon or human–zebrafish “net” alignments in the UCSC Genome Browser (http://genome.ucsc.edu).
Acknowledgments
We thank Alexander Poliakov and Inna Dubchak for incorporating Gumby into the VISTA web server, Nadav Ahituv for the Gateway-HSP68-LacZ vector, Jim Hughes for feedback regarding α-globin sequence analysis, Qian-fei Wang, Jan-Fang Cheng, and Michael Brudno for numerous productive conversations during development of Gumby, and James Noonan, Nadav Ahituv, Qian-fei Wang, and Dario Boffelli for comments on the manuscript. F.P. was supported by a fellowship from the Canadian Institutes of Health Research. Research was conducted at the E.O. Lawrence Berkeley National Laboratory, supported by the Grant # HL066681, Berkeley-PGA, under the Programs for Genomic Application, funded by National Heart, Lung, and Blood Institute, USA, and performed under Department of Energy Contract DE-AC02-05CH11231, University of California.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4717506
References
- Ahituv N., Rubin E.M., Nobrega M.A., Rubin E.M., Nobrega M.A., Nobrega M.A. Exploiting human–fish genome comparisons for deciphering gene regulation. Hum. Mol. Genet. 2004;13:R261–R266. doi: 10.1093/hmg/ddh229. [DOI] [PubMed] [Google Scholar]
- Ahituv N., Prabhakar S., Poulin F., Rubin E.M., Couronne O., Prabhakar S., Poulin F., Rubin E.M., Couronne O., Poulin F., Rubin E.M., Couronne O., Rubin E.M., Couronne O., Couronne O. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum. Mol. Genet. 2005;14:3057–3063. doi: 10.1093/hmg/ddi338. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., Christoffels A., Rash S., Hoon S., Smit A., Rash S., Hoon S., Smit A., Hoon S., Smit A., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Haussler D., Blanchette M., Haussler D., Blanchette M., Blanchette M. Into the heart of darkness: Large-scale clustering of human non-coding DNA. Bioinformatics. 2004a;20(Suppl. 1):I40–I48. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004b;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko I., Pachter L., Rubin E.M., Pachter L., Rubin E.M., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Brenner S., Elgar G., Sandford R., Macrae A., Venkatesh B., Aparicio S., Elgar G., Sandford R., Macrae A., Venkatesh B., Aparicio S., Sandford R., Macrae A., Venkatesh B., Aparicio S., Macrae A., Venkatesh B., Aparicio S., Venkatesh B., Aparicio S., Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Brudno M., Chapman M., Göttgens B., Batzoglou S., Morgenstern B., Chapman M., Göttgens B., Batzoglou S., Morgenstern B., Göttgens B., Batzoglou S., Morgenstern B., Batzoglou S., Morgenstern B., Morgenstern B. Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics. 2003a;4:66. doi: 10.1186/1471-2105-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S., Davydov E., Green E.D., Sidow A., Batzoglou S., Green E.D., Sidow A., Batzoglou S., Sidow A., Batzoglou S., Batzoglou S. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003b;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caubit X., Thangarajah R., Theil T., Wirth J., Nothwang H.G., Ruther U., Krauss S., Thangarajah R., Theil T., Wirth J., Nothwang H.G., Ruther U., Krauss S., Theil T., Wirth J., Nothwang H.G., Ruther U., Krauss S., Wirth J., Nothwang H.G., Ruther U., Krauss S., Nothwang H.G., Ruther U., Krauss S., Ruther U., Krauss S., Krauss S. Mouse Dac, a novel nuclear factor with homology to Drosophila dachshund shows a dynamic expression in the neural crest, the eye, the neocortex, and the limb bud. Dev. Dyn. 1999;214:66–80. doi: 10.1002/(SICI)1097-0177(199901)214:1<66::AID-DVDY7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Chapman M.A., Donaldson I.J., Gilbert J., Grafham D., Rogers J., Green A.R., Göttgens B., Donaldson I.J., Gilbert J., Grafham D., Rogers J., Green A.R., Göttgens B., Gilbert J., Grafham D., Rogers J., Green A.R., Göttgens B., Grafham D., Rogers J., Green A.R., Göttgens B., Rogers J., Green A.R., Göttgens B., Green A.R., Göttgens B., Göttgens B. Analysis of multiple genomic sequence alignments: A web resource, online tools, and lessons learned from analysis of mammalian SCL loci. Genome Res. 2004;14:313–318. doi: 10.1101/gr.1759004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G.M., Brudno M., Green E.D., Batzoglou S., Sidow A., Brudno M., Green E.D., Batzoglou S., Sidow A., Green E.D., Batzoglou S., Sidow A., Batzoglou S., Sidow A., Sidow A. Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res. 2003;13:813–820. doi: 10.1101/gr.1064503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Green E.D., Batzoglou S., Sidow A., Batzoglou S., Sidow A., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.J., Shen W., Heanue T.A., Mardon G., Shen W., Heanue T.A., Mardon G., Heanue T.A., Mardon G., Mardon G. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev. Genes Evol. 1999;209:526–536. doi: 10.1007/s004270050285. [DOI] [PubMed] [Google Scholar]
- de la Calle-Mustienes E., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Allende M.L., Gomez-Skarmeta J.L., Gomez-Skarmeta J.L. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. A model of the statistical power of comparative genome sequence analysis. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I., Pachter L., Poliakov A., Rubin E.M., Dubchak I., Poliakov A., Rubin E.M., Dubchak I., Rubin E.M., Dubchak I., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttgens B., Barton L.M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Barton L.M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Mistry S., Grafham D., McMurray A., Vaudin M., Grafham D., McMurray A., Vaudin M., McMurray A., Vaudin M., Vaudin M., et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- Hughes J.R., Cheng J.F., Ventress N., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Cheng J.F., Ventress N., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Ventress N., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., De Gobbi M., de Jong P., Rubin E., Higgs D.R., de Jong P., Rubin E., Higgs D.R., Rubin E., Higgs D.R., Higgs D.R. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc. Natl. Acad. Sci. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Altschul S.F., Altschul S.F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Chiaromonte F., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary R., Clapoff S., Darling S., Perry M.D., Moran L.A., Rossant J., Clapoff S., Darling S., Perry M.D., Moran L.A., Rossant J., Darling S., Perry M.D., Moran L.A., Rossant J., Perry M.D., Moran L.A., Rossant J., Moran L.A., Rossant J., Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Goode D., Elgar G., Hill R.E., de Graaff E., Elgar G., Hill R.E., de Graaff E., Hill R.E., de Graaff E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M., Bergman C., Patel N.H., Kreitman M., Patel N.H., Kreitman M., Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Blanchette M., Haussler D., Green E.D., Blanchette M., Haussler D., Green E.D., Haussler D., Green E.D., Green E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Vinson J.P., Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Clamp M., Vinson J.P., Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Clamp M., Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Clamp M., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Clamp M., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Clamp M., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Clamp M., Green E.D., Lander E.S., Mullikin J.C., Clamp M., Lander E.S., Mullikin J.C., Clamp M., Mullikin J.C., Clamp M., Clamp M. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc. Natl. Acad. Sci. 2005;102:4795–4800. doi: 10.1073/pnas.0409882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M., Ovcharenko I., Afzal V., Rubin E.M., Afzal V., Rubin E.M., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Oda-Ishii I., Bertrand V., Matsuo I., Lemaire P., Saiga H., Bertrand V., Matsuo I., Lemaire P., Saiga H., Matsuo I., Lemaire P., Saiga H., Lemaire P., Saiga H., Saiga H. Making very similar embryos with divergent genomes: Conservation of regulatory mechanisms of Otx between the ascidians Halocynthia roretzi and Ciona intestinalis. Development. 2005;132:1663–1674. doi: 10.1242/dev.01707. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I., Stubbs L., Loots G.G., Stubbs L., Loots G.G., Loots G.G. Interpreting mammalian evolution using Fugu genome comparisons. Genomics. 2004;84:890–895. doi: 10.1016/j.ygeno.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Rubin E.M., Rubin E.M. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Bailey P., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Bailey P., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Wasserman W.W., Ericson J., Lenhard B., Ericson J., Lenhard B., Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E.A., Cooper G.M., Sidow A., Cooper G.M., Sidow A., Sidow A. Trade-offs in detecting evolutionarily constrained sequence by comparative genomics. Annu. Rev. Genomics Hum. Genet. 2005;6:143–164. doi: 10.1146/annurev.genom.6.080604.162146. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]