Abstract
Human and mouse genome sequences contain roughly 100,000 regions that are unalignable in primary sequence and neighbor corresponding alignable regions between both organisms. These pairs are generally assumed to be nonconserved, although the level of structural conservation between these has never been investigated. Owing to the limitations in computational methods, comparative genomics has been lacking the ability to compare such nonconserved sequence regions for conserved structural RNA elements. We have investigated the presence of structural RNA elements by conducting a local structural alignment, using FOLDALIGN, on a subset of these 100,000 corresponding regions and estimate that 1800 contain common RNA structures. Comparing our results with the recent mapping of transcribed fragments (transfrags) in human, we find that high-scoring candidates are twice as likely to be found in regions overlapped by transfrags than regions that are not overlapped by transfrags. To verify the coexpression between predicted candidates in human and mouse, we conducted expression studies by RT-PCR and Northern blotting on mouse candidates, which overlap with transfrags on human chromosome 20. RT-PCR results confirmed expression of 32 out of 36 candidates, whereas Northern blots confirmed four out of 12 candidates. Furthermore, many RT-PCR results indicate differential expression in different tissues. Hence, our findings suggest that there are corresponding regions between human and mouse, which contain expressed non-coding RNA sequences not alignable in primary sequence.
Approximately half of the ∼3 billion nucleotides in the human genome represent repetitive sequences. Roughly two-thirds of the remaining nucleotides can be aligned with the mouse genome (http://genome.ucsc.edu/). This leaves about one-third of the nonrepetitive human genome unalignable with the mouse. It has generally been assumed that these large fractions of the genomes are not conserved between human and mouse because of lack of sequence similarity. This, however, is not necessarily true, since it is possible that they are conserved, just not at the sequence level, but at the structural level. This applies to structural DNA motifs, non-coding RNAs (ncRNAs), and specific proteins, where maintaining the structure is of more importance than maintaining the sequence. This has been observed for many functional classes of RNA molecules, including, tRNA, rRNA, RNAse P, and SRP RNA.
Recently Cheng et al. (2005) mapped the sites of polyadenylated [poly(A)+], cytosolic, RNA transcription for chromosomes, 6, 7, 13, 14, 19, 20, 21, 22, X, and Y at a 5-bp resolution in eight cell lines using tiling microarrays. For one of the cell lines, HepG2, they also mapped nonpolyadenylated [poly(A)−] and nuclear transcripts. Based on their data, we estimate that at least 32% of the human genome is transcribed. Combining their data with known and predicted transcripts, including all introns, ∼58% of the human genome is transcribed on either strand. Considerable amounts (60%–84%) of these transcripts do not overlap with known coding genes and are therefore possibly non-coding (Cheng et al. 2005). It has furthermore been suggested that at least half of all transcripts in mammals do not encode proteins (Ravasi et al. 2005).
Owing to computational limitations, the focus has mainly been on regions that are conserved in sequence, where many ncRNAs have been found (Griffiths-Jones et al. 2003; Washietl et al. 2005). Recently, an approach based on the Sankoff algorithm (Sankoff 1985), FOLDALIGN (Havgaard et al. 2005), was updated to conduct searches on regions with low sequence similarity. While conducting a mutual scan of two sequences, FOLDALIGN simultaneously aligns and folds regions compared between the two sequences, thereby making it possible to search for conserved local structural sequences, such as ncRNAs, in previously unalignable regions.
The question we seek to answer is: Are there places in the assumed nonconserved regions of mammals that have evolutionary constraints on maintaining their RNA structure?
Results
Data set
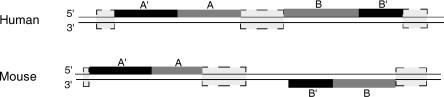
We chose to focus on the regions in human that could not be aligned with mouse and vice versa. Altogether the human genome contains 1,369,651 non-gap, non-repeat, >100-bp regions constituting 493,754,093 bp that cannot be aligned with the mouse. The longest region is 12,680 bp, and the average length is 360 bp. The mouse contains 1,225,106 such regions, constituting 388,573,892 bp, the longest being 11,103 bp and the average length being 317. Since it has been concluded that the large majority of ncRNAs, analyzed by the FANTOM consortium, display positional conservation across species (The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group 2005), we generated a list of 101,563 human–mouse pairs that were adjacent to a matching alignment (see Fig. 1). FOLDALIGN was run on 36,970 pairs, on both strands, from the same 10 chromosomes for which Cheng et al. made transcriptional maps. Transcribed fragments (transfrags) were used to denote array-detected regions of transcription (Cheng et al. 2005). Using BLAT (Kent 2002), we did not find any known, corresponding ncRNAs in our pairs, searching all of Rfam (Griffiths-Jones et al. 2003).
Figure 1.
Pair generation. Example of the generation of two pairs, pair A and pair B. The black boxes are matching alignments, the light gray boxes are repeats, gaps, or alignments, and the dark gray boxes are the pairs, A and B. Pair A is downstream of the A′ +/+ alignment, and both regions end at a repeat, gap, or another alignment. Pair B is upstream of the B′ +/− alignment; therefore, both regions are placed at the 5′-end of the alignment; again, the length of these pairs is limited by repeats, gaps, or another alignment. The remaining nonboxed lines are nonconserved regions, which are not adjacent to matching alignments.
Model chromosome and randomizations
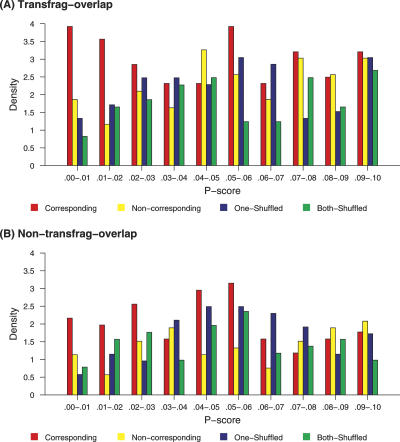
Human chromosome 20 was chosen as a model chromosome. For chromosome 20, we performed three different randomizations, maintaining the dinucleotide content (Altschul and Erickson 1985; Workman and Krogh 1999), and ran FOLDALIGN on these. We randomized (1) both sequences in the pairs, (2) only the human sequences and then only the mouse sequences, and (3) randomized the pairs themselves, comparing probable noncorresponding pairs. In this study, we only consider predicted FOLDALIGN alignments, which are longer than 59 nt and show >40% base-pairing, as candidates (see Methods). This resulted in 2260 candidates on chromosome 20 that were compared to the transfrags from the transcriptional maps for chromosome 20. In total, 1176 showed an overlap with a transfrag. Intriguingly, the 1176 transfrag-overlapping candidates show significantly better scores than the 1084 that did not overlap with a transfrag (Fig. 2).
Figure 2.
Probability density histograms. The densities of the P-scores, in chromosome 20, for the corresponding pairs, noncorresponding pairs, pairs in which one sequence is shuffled, and pairs in which both sequences are shuffled. The histograms depict (A) the candidates that overlap transfrags and (B) the candidates that do not.
In order to estimate a false-positive rate, we compared the probability densities of the original pairs to the three different sets of randomized pairs. The P-score values represent the contrast of the candidate to the rest of the region in the pairs. These are calculated in a manner similar to BLAST, although whereas BLAST estimates the scores from random alignments, FOLDALIGN, because of the computational complexity, estimates the scores from the surrounding regions of the best hit (Havgaard et al. 2005). The P-scores were considerably lower for the candidates derived from the original pairs. On average, the probability density of candidates with a P-score below 0.03 was twice as high for candidates from original pairs (Fig. 2). This indicates that approximately half of the candidates with P-scores below 0.03 show higher structure conservation than what would be expected by chance. Throughout this paper, we refer to our candidates with a P-score below 0.03 as our top candidates.
More genomes
To further increase the significance of our candidates, we searched for corresponding regions in rat, dog, and chicken that were not conserved in sequence with either mouse or human genomes, and ran FOLDALIGN on these pairs. We did this by analyzing the multiple alignments between these organisms (http://genome.ucsc.edu/) in a search for regions, in the vicinity of our original pairs, that were alignable between human or mouse, but not both, and a third organism. In practice, this often involved finding corresponding regions between mouse and rat, or human and dog, and then pairing human and rat, or dog and mouse. It should be noted that the fact that two regions are adjacent to alignable sequences does not necessarily imply that they correspond to each other, since deletions or rearrangements might have occurred. Using this approach, we could find a potential corresponding region in at least one additional organism for half of all our candidates.
In addition to this, we also used BLAST (Altschul et al. 1990) to look for hits to our predicted candidates that were conserved in sequence in chimp, rhesus monkey, dog, cow, rat, or chicken. This resulted in 1290 hits whereof 1288 were hits between the closely related human–chimp, human–rhesus monkey, or mouse–rat, with an average sequence similarity above 95%. Because of this high sequence similarity, we could not use these hits to provide further information to help verify the structure prediction.
The top candidates
In the 10 chromosomes analyzed, 1297 candidates score below 0.03. Applying the results from the analysis of chromosome 20, we estimate that approximately half of these candidates cannot be explained by random events. We were able to assign a potential corresponding region in at least one additional organism to 500 of these top candidates. In 17% (83 of 500) of the third organism scans, the P-score was lower than 0.03, whereas this fraction is 5% (399 of 7844) for all the candidates. We have noticed some similarities between human–mouse structures and the corresponding structures in the third organism by manual comparison, but since we perform a local scan with a maximum motif length of 200, it is very difficult to compare these structures reliably without using a local multiple alignment program that considers unaligned sequences and secondary structure. To date, no such reliable program exists for a bifurcated structure for a small number of sequences.
These 1297 candidates have, on average, 45% sequence identity, between the human and the mouse sequences, and 51% of the nucleotides are involved in base-pairing. The fraction of intronic and intergenic candidates is approximately the same. In the top candidates, 3.8% of the mouse sequences overlap with FANTOM3 transcripts (The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group 2005), whereas 3.2% of all the candidates have such overlaps. To search for structural similarities to known ncRNA families, we ran Ravenna (Weinberg and Ruzzo 2004a, b), for every model in Rfam (Griffiths-Jones et al. 2003), on our candidates and found no significant hits.
We have created a database (http://genome.kvl.dk/resources/hm_ncrna_scan) with all candidates that were longer than 59 nt and have >40% base-pairing. This database contains different information concerning the candidates.
Experimental verification
We have designed primers for reverse-transcription (RT) PCRs of 36 mouse candidates that overlapped with human transfrags from chromosome 20. For RT-PCRs, we have used cDNAs generated with random hexamers or oligo(dT)s. In total, 32 and 23 ncRNA candidates gave positive results in cDNAs made with random hexamers and oligo(dT)s, respectively. Twenty-seven of these 36 candidates were tested in five different tissues. We found differential expression of 10 sequences in poly(A)+ cDNAs, and of 15 sequences in cDNAs made with random hexamers.
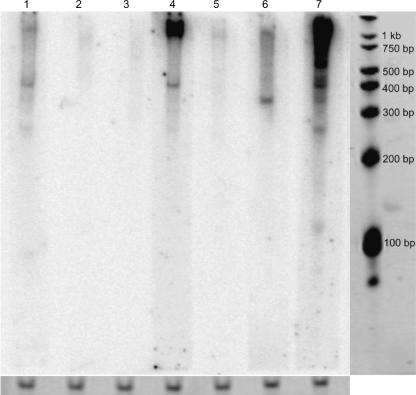
In addition to the RT-PCRs, we have made Northern blots for 12 differentially expressed mouse candidates, chosen based on the RT-PCR results. Four candidates were confirmed using this approach (Fig. 3).
Figure 3.
Northern blots. The results of Northern blotting for seven mouse candidates, where lanes 1, 4, 6, and 7 show positive results with size estimates of ∼400, 400, 350, and 400 bp, respectively. A 100-bp RNA marker as a size standard and 5.8S rRNAs as a loading control are shown.
Discussion
We have revealed that the assumed nonconserved regions between human and mouse show higher secondary structure conservation than what would be expected by chance. Interestingly, the probability density of the best scoring candidates is twice as high for candidates that overlap transfrags compared to those that do not. We have scanned 36% of all human–mouse pairs, which resulted in 1297 good scoring candidates. Based on our analysis of these 36% (10 chromosomes), we estimate that genome wide, ∼3600 candidates score below 0.03. Half of these, or 1800 candidates, would not be explainable by random events. The question that remains unanswered is why this is. Some of the regions with conserved secondary structure are probably structural ncRNAs, having the same or related function, where secondary structure is of more importance than primary sequence. We find that 89% of our tested candidates are transcribed using sensitive RT-PCR. Performing Northern blots (a less sensitive method) on 12 of these, we detected four candidates in mouse.
Whether transcription implies function remains an open question. Recently Wyers et al. (2005) discovered unexpected transcribed regions in Saccharomyces cerevisiae that were rapidly degraded by a novel mechanism. They noted that this mechanism was also responsible for degradation of several Pol I and Pol III transcripts, and named these transcripts CUTs for cryptic unstable transcripts, which supports the existence of a post-transcriptional quality control mechanism. Davis and Ares Jr. (2006) later discovered that several such transcripts originated near known promoter elements and hypothesized that these transcripts reflect important features of RNA polymerase activity at the promoter rather than being uninformative transcriptional “noise.” In addition, Babak et al. (2005) designed microarrays, based on QRNA predictions (Rivas and Eddy 2001), for high-throughput screening of highly conserved intergenic and intronic sequences in human, mouse, and rat. They performed Northern blots on 55 good candidates, where only eight were confirmed to be transcribed in mouse, and none of those was found in human. These results are intriguing: What is the explanation for such a high sequence and structural conservation between human and mouse if they are not transcribed and functional in both organisms? It is possibly caused by a recent inactivation of the human orthologs, different temporal and/or spacial transcription, or low transcript abundance in human. Still further experimental investigation of this phenomenon is needed.
There are two probable reasons why we do not detect our selected confirmed RT-PCR candidates with Northern blots. One is that our candidates are, in fact, CUTs that are rapidly degraded and are therefore not detectable. The other reason is that they are low-level transcripts. In fact, the studies by Holland (2002) reveal that transcript abundance in yeast varies over six orders of magnitude, where important transcription factors are expressed at levels as low as one-thousandth transcript per cell. These low-level transcripts would more likely be detected by sensitive RNA-probed RT-PCR than with less sensitive Northern blots. It is also interesting to note that despite the major efforts of the FANTOM consortium, their data still only covers ∼40% of the known RNAs (The FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group 2005).
Methods
Data set
From the regions that could not be aligned in primary sequence, we constructed a set of pairs for each chromosome. Each pair contains one region from each genome if and only if the two regions were immediately upstream or downstream from a matching alignment, as shown in Figure 1. Since the alignments are of double-stranded DNA, a +/+ alignment is equivalent to a −/− alignment. Therefore, we have also made a data set where we found the complementary strand to each region in each pair. This is because the structural alignment is not necessarily the same on the complementary strand because of G-U base pairs. All sequence data and alignments were obtained from the UCSC Genome Browser (http://genome.ucsc.edu/). The following tracks for human (assembly hg17) and mouse (assembly mm5) were used in the making of the nonconserved pairs:
axtNet tracks for human versus mouse and vice versa, which contain chained and netted alignments, that is, the best chains in the genome, with gaps in the best chains filled in by next best chains where possible. The alignments are produced by the BLASTZ alignment program (http://www.bx.psu.edu/miller_lab/).
RepeatMasker track, made using the RepeatMasker program (http://www.repeatmasker.org), which screens DNA for interspersed repeats and low complexity DNA sequences.
Simple repeat track, produced by the TandemRepeatFinder (http://tandem.biomath.mssm.edu/trf/trf.html), which displays simple, possibly imperfect, tandem repeats.
Gap track, which holds information about the gaps in the assemblies.
Transcriptional fragments
The list of transfrags was obtained from http://cgap.nci.nih.gov/Info/2005.1, and the coordinates were updated to assembly hg17 (May 2004) using UCSC’s liftOver tool (http://www.genome.ucsc.edu/cgi-bin/hgLiftOver).
Filtering
Studying every consensus structure in the Rfam database (http://www.sanger.ac.uk/Software/Rfam/) indicated an average of ∼41% nucleotides involved in the base-pairing within the structures. The well-defined ncRNAs like 5S RNAs, SRP, and tRNA had base-pairing ranging from 47% to 57%. In addition, we did not want to include simple short structures like a single short hairpin. From these observations, an initial filtering of sequences above 59 nt, to include pre-miRNAs, and base-pairing above 40%, to include all the known and well characterized ncRNAs but not necessarily difficult ncRNAs like snoRNAs, was performed.
More genomes
The multiple alignments and the genome sequences for the chimp (panTro1), rhesus monkey (rheMac2), rat (rn3), dog (canFam2), cow (bosTau2), and chicken (galGal2) were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/). We inspected all BLAST alignments of comparable length (<20 nt difference) and with >50% sequence identity. All of our 1290 hits had >75% sequence identity. We used the multiple alignments of human with six other organisms. These were the chimp (panTro1), rat (rn3), dog (canFam1), chicken (galGal2), zebrafish (danRer1), and fugu (fr1).
Running FOLDALIGN
The data sets were run for ∼5 mo on 70 2-GB-RAM nodes in a linux cluster. These were run with a motif length (λ) 200 and length difference (δ) 15, using the default score matrix.
RT-PCRs
RNAs from mouse liver, kidney, testicle, embryo, and thymus were transcribed to cDNA with ImProm-II Reverse Transcription System (Promega) following the manufacturer’s protocol, using oligo(dT)15 primer or random hexamers, respectively, in each RT reaction. Additionally, negative–no template, and negative–no reverse transcriptase controls were performed. Primer pairs were designed within 200 nt of potentially novel RNA coding sequences using the Primer3 Web-based software at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www_slow.cgi (Rozen and Skaletsky 2000). Two microliters of each RT reaction were used as a template in PCR amplification with sequence-specific primers for each candidate. The PCR components were 0.05 U of Ampliqon III TEMPase DNA polymerase (Ampliqon), 0.35 μM each primer, and 2 mM Mg2+ (25 mM) in a total volume of 16 μL. PCR reactions were performed on a PTC-200 thermocycler (MJ Research) in 35 cycles of the Touchdown 60°C PCR program with denaturation at 95°C, annealing with lowering temperature from 60°C by 1°C after each cycle in the first 10 cycles, 25 cycles at 50°C, 72°C for 1 min, and a final elongation at 72°C for 5 min. Amplification products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining.
Northern blots
Total mouse liver RNA (6 μg) (Ambion) and total human liver RNA (1 μg) (Ambion) were resolved on denaturing 5% TBE–urea polyacrylamide gels (Bio-Rad). Gels were stained with ethidium bromide to visualize 5.8S rRNAs as loading controls. RNAs were transferred by electroblotting to a Hybond-N nylon membrane (Amersham Biosciences) using the Bio-Rad semidry blotting apparatus (Trans-blot SB; Bio-Rad). After immobilizing of RNAs by baking at 80°C for 15 min, we prehybridized the membranes for 1 h at 65°C in standard Northern buffer (6× SSC, 0.2× SDS, 10× Denhardt’s solution). Antisense RNA probes complementary to potentially novel RNAs were prepared by in vitro transcription with [α-32P]dUTP, and T7 RNA Polymerase (mirVana miRNA Probe Construction Kit; Ambion) following the manufacturer’s protocol. The blots were hybridized with a 5′-32P phosphorylated RNA probe (in 6× SSC, 0.2× SDS, 5× Denhardt’s solution), overnight at 42°C. Washes were performed at room temperature, with a final wash at 42°C. The blots were visualized in a STORM840 PhosphorImager (Molecular Dynamics).
Acknowledgments
This work was supported by the Danish research councils SJVF and STF and the Danish Center for Scientific Computing. We thank the anonymous reviewers for valuable comments.
Footnotes
[Supplemental material and database access is available online at http://genome.kvl.dk/resources/hm_ncrna_scan.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5226606
References
- Altschul S.F., Erickson B.W., Erickson B.W. Significance of nucleotide sequence alignments: A method for random sequence permutation that preserves dinucleotide and codon usage. Mol. Biol. Evol. 1985;2:526–538. doi: 10.1093/oxfordjournals.molbev.a040370. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Babak T., Blencowe B.J., Hughes T.R., Blencowe B.J., Hughes T.R., Hughes T.R. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genomics. 2005;6:104. doi: 10.1186/1471-2164-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucelotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Davis C.A., Ares M., Jr., Ares M., Jr. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium. RIKEN Genome Exploration Research Group. Genome Science Group The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Bateman A., Marshall M., Khanna A., Eddy S.R., Bateman A., Marshall M., Khanna A., Eddy S.R., Marshall M., Khanna A., Eddy S.R., Khanna A., Eddy S.R., Eddy S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havgaard J.H., Lyngsø R.B., Stormo G.D., Gorodkin J., Lyngsø R.B., Stormo G.D., Gorodkin J., Stormo G.D., Gorodkin J., Gorodkin J. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40%. Bioinformatics. 2005;21:1815–1824. doi: 10.1093/bioinformatics/bti279. [DOI] [PubMed] [Google Scholar]
- Holland M.J. Transcript abundance in yeast varies over six orders of magnitude. J. Biol. Chem. 2002;277:14363–14366. doi: 10.1074/jbc.C200101200. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Fukuda S., Ru K., Frith M.C., Gongora M.M., Ru K., Frith M.C., Gongora M.M., Frith M.C., Gongora M.M., Gongora M.M., et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2005;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E., Eddy S.R., Eddy S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., et al., editors. Bioinformatics methods and protocols. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sankoff D. Simultaneous solution of the RNA folding, alignment and protosequence problems. SIAM J. Appl. Math. 1985;45:810–825. [Google Scholar]
- Washietl S., Hofacker I.L., Lukasser M., Hüttenhoffer A., Stadler P., Hofacker I.L., Lukasser M., Hüttenhoffer A., Stadler P., Lukasser M., Hüttenhoffer A., Stadler P., Hüttenhoffer A., Stadler P., Stadler P. Genome-wide mapping of conserved RNA secondary structures predicts thousands of functional non-coding RNAs in human. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Weinberg Z., Ruzzo W.L., Ruzzo W.L. Exploiting conserved structure for faster annotation of non-coding RNAs without loss of accuracy. Bioinformatics. 2004a;20(suppl. 1):i334–i342. doi: 10.1093/bioinformatics/bth925. [DOI] [PubMed] [Google Scholar]
- Weinberg Z., Ruzzo W.L., Ruzzo W.L. Proceedings Eighth Annual International Conference on Computational Molecular Biology (RECOMB), ASM Press; Washington, DC: 2004b. Faster genome annotation of non-coding RNA families without loss of accuracy. pp. 243–251. [Google Scholar]
- Workman C., Krogh A., Krogh A. No evidence that mRNA have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J.-C., Dufour M.-E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Rougemaille M., Badis G., Rousselle J.-C., Dufour M.-E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Badis G., Rousselle J.-C., Dufour M.-E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Rousselle J.-C., Dufour M.-E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Dufour M.-E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Régnault B., Devaux F., Namane A., Séraphin B., Devaux F., Namane A., Séraphin B., Namane A., Séraphin B., Séraphin B., et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]