Abstract
Pre-mRNA processing often occurs in coordination with transcription thereby coupling these two key regulatory events. As such, many proteins involved in mRNA processing associate with the transcriptional machinery and are in proximity to DNA. This proximity allows for the mapping of the genomic associations of RNA binding proteins by chromatin immunoprecipitation (ChIP) as a way of determining their sites of action on the encoded mRNA. Here, we used ChIP combined with high-density microarrays to localize on the human genome three functionally distinct RNA binding proteins: the splicing factor polypyrimidine tract binding protein (PTBP1/hnRNP I), the mRNA export factor THO complex subunit 4 (ALY/THOC4), and the 3′ end cleavage stimulation factor 64 kDa (CSTF2). We observed interactions at promoters, internal exons, and 3′ ends of active genes. PTBP1 had biases toward promoters and often coincided with RNA polymerase II (RNA Pol II). The 3′ processing factor, CSTF2, had biases toward 3′ ends but was also observed at promoters. The mRNA processing and export factor, ALY, mapped to some exons but predominantly localized to introns and did not coincide with RNA Pol II. Because the RNA binding proteins did not consistently coincide with RNA Pol II, the data support a processing mechanism driven by reorganization of transcription complexes as opposed to a scanning mechanism. In sum, we present the mapping in mammalian cells of RNA binding proteins across a portion of the genome that provides insight into the transcriptional assembly of RNA–protein complexes.
RNA binding proteins regulate many stages of RNA metabolism to help determine gene expression programs (for reviews, see Keene and Tenenbaum 2002; Hieronymus and Silver 2004). To prepare mRNA for export to and translation in the cytoplasm, many events including splicing, cleavage of the 3′ end, and editing occur cotranscriptionally (Kornblihtt et al. 2004; Aguilera 2005a). After transcription, the mRNA–protein complex may reorganize as it is exported to the cytoplasm and prepared for translation (Fairman et al. 2004). Thus, significant efforts have been made to determine how RNA binding proteins interact at genes and/or mRNAs at various stages of mRNA metabolism (Tenenbaum et al. 2000; Brown et al. 2001; Hieronymus and Silver 2003; Ule et al. 2003; Yu et al. 2004). These early genomic efforts suggested that, similar to DNA binding proteins, functional groupings of genes are being bound by RNA binding proteins perhaps as post-transcriptional operons (Keene and Tenenbaum 2002). However, it is not known at which stage of RNA processing these operons are established.
Much of pre-mRNA processing is coupled to transcription both physically and functionally, potentially through interactions with the C-terminal domain (CTD) of RNA polymerase II (RNA Pol II) (Mortillaro et al. 1996; Yuryev et al. 1996; Kim et al. 1997). The CTD is phosphorylated as transcription begins. Following initiation, RNA Pol II transcribes at varying rates and undergoes conformational changes during transcript elongation that may result in pausing at particular loci (for review, see Arndt and Kane 2003). Many connections between transcription and RNA processing have been found to occur during transcription elongation (Howe et al. 2003; Lindstrom et al. 2003). A recent study suggested cotranscriptional specificity for RNA binding proteins; some splicing factors were constitutively recruited while others depended on the splicing outcome (Mabon and Misteli 2005). RNA binding proteins also have critical roles in maintaining genomic stability (Aguilera 2005b; Li and Manley 2005).
Three mammalian proteins known to modulate nuclear RNA processing include the mRNA export factor ALY, the splicing factor polypyrimidine tract binding protein (PTBP1), and the 3′ end cleavage factor CSTF2. ALY is an RNA binding protein that links splicing to mRNA export as a component of the exon junction complex (EJC) (Zhou et al. 2000). PTBP1 represses alternative exons like those of FGFR1 and FGFR2, where PTBP1’s influence yields receptors with higher affinities for fibroblast growth factor (Carstens et al. 2000; Wagner and Garcia-Blanco 2002; Jin et al. 2003). CSTF2 is the mammalian homolog of the yeast protein, Rna15p, and is a 3′ end cleavage factor that can modulate alternative 3′ end cleavage of transcripts (Takagaki and Manley 1998). Interestingly, some of the same proteins are involved in the regulation of splicing and 3′ end processing suggesting a link between the two processes (Lin and Patton 1995; Castelo-Branco et al. 2004). However, little is known about which mRNAs are the substrates for these RNA binding proteins.
RNA binding proteins also influence transcription initiation (Kornblihtt et al. 2004). In mammalian cells, numerous RNA binding proteins are associated with hypo-phosphorylated RNA Pol II or transcription factors (Michelotti et al. 1995; Calvo and Manley 2001; Stains et al. 2005). Additionally, RNA splicing factors can influence transcription in a hormone dependent manner at promoters (Dowhan et al. 2005). The mechanism of action for RNA binding proteins at promoters remains unclear.
Chromatin immunoprecipitation (ChIP) is a powerful approach to localize proteins to genomes. A variety of RNA binding proteins including splicing, nuclear export, and 3′ processing factors associate with the yeast genome to reveal different distributions across genes and gene sets (Komarnitsky et al. 2000; Lei et al. 2001; Lei and Silver 2002; Yu et al. 2004; Lacadie and Rosbash 2005). Genomic localization of RNA binding proteins identified sites that are consistent with known functions. A yeast spliceosome component, the U1 snRNP, localized preferentially to intron-containing genes, while the RNA exonuclease Rat1p localized to the 3′ ends of genes where it is involved in transcription termination as well as mRNA degradation (Kotovic et al. 2003; Kim et al. 2004; Lacadie and Rosbash 2005). These studies underscore the potential for the genomic localization approach to identify functional targets of RNA binding proteins and improve our understanding of the early assembly and organization of mRNPs.
Results
PTBP1 localizes to actively transcribed genes
Since much of pre-mRNA processing is physically and functionally coupled to transcription, we postulated that ChIP could be used to localize RNA binding proteins to chromatin in mammalian cells. To determine the distribution of RNA binding proteins across DNA, we performed ChIP with an antibody against PTBP1 to isolate both direct and indirect associations with chromatin in HeLa cells (Fig. 1). We first examined two genes known to be regulated by PTBP1 during splicing, fibroblast growth factor receptor 1 (FGFR1), which is expressed in HeLa cells, and fibroblast growth factor receptor 2 (FGFR2), which is not expressed in HeLa cells (Fig. 2A, RNA expression array data) (Jin et al. 2000, 2003; Brodsky et al. 2005). As a reference and control, we designed primers for the poly-A polymerase testis specific gene (PAPOLB) that is intron-less and not expressed in HeLa cells (RNA expression array data) (Brodsky et al. 2005). Enrichment values were calculated using the input-normalized signal of the target in the ChIP relative to the input-normalized signal of PAPOLB (see Methods).
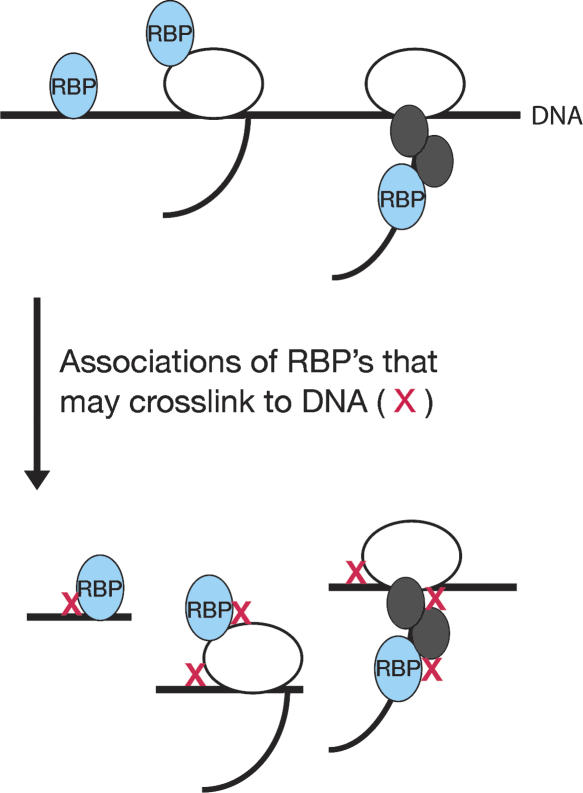
Figure 1.
Cross-linking events that may yield enriched DNA fragments by chromatin immunoprecipitation of an RNA binding protein (RBP). HeLa cells were cross-linked with 10 mM DMA and 1% formaldehyde (cross-linking events depicted as red x’s). This approach should increase the frequency of protein–protein and protein–DNA cross-linking, enabling the detection of both direct and indirect protein–DNA interactions.
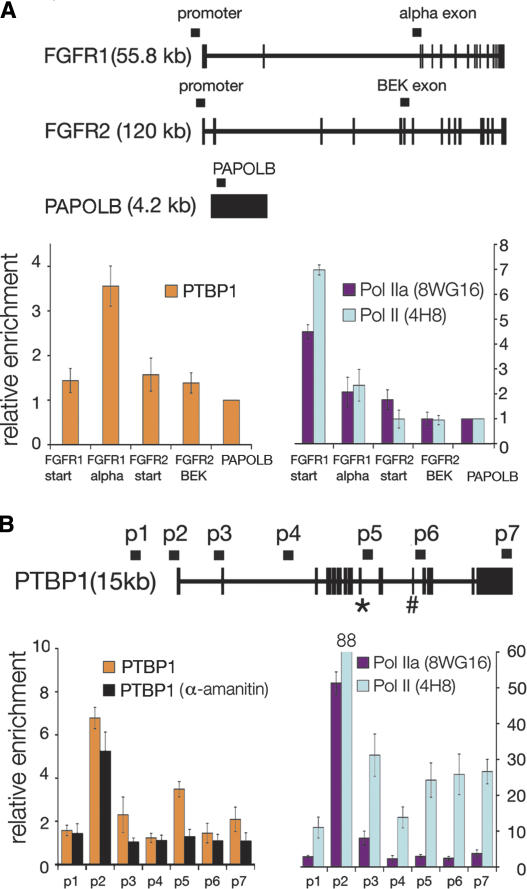
Figure 2.
PTBP1 and RNA polymerase II are enriched at sites within transcribed genes. (A) Schematic representation of the FGFR1, FGFR2, and PAPOLB genes with primers depicted above genes; thick segments represent exons, and thin segments represent introns. PCR analysis of ChIP’s targeting PTBP1, promoter associated RNA polymerase II (Pol IIa, 8WG16), and total RNA polymerase II (non-phosphospecific Pol II, 4H8). Error bars represent the standard deviation calculated from three to seven biological repeats. (B) PTBP1, Pol IIa, and Poll II are enriched at PTBP1’s promoter and alternative exons. Starting upstream of the transcription start site, eight primer sets (denoted p1–p7) were designed to target sites across PTBP1. Alternative exons include exon 9 (*) and exon 11 (#). HeLa cells were grown in 40 μg/mL a-amanitin, and then cross-linked and sonicated for the α-amanitin ChIP experiments.
We observed enrichment of PTBP1 at FGFR1’s α-exon relative to PAPOLB, but significant enrichment was not observed at the previously identified PTBP1-regulated FGFR2 exon nor at the promoters of FGFR1 and FGFR2 (Fig. 2A; primers in Supplemental data 1). The absence of PTBP1 enrichment at FGFR2 correlates with the lack of FGFR2 expression in HeLa cells, suggesting a transcriptional bias for localization. Typically, we considered a value >2 significant enrichment above background and within the precision of the experiment.
To investigate the relationship between transcription and splicing, we performed ChIP with two antibodies against RNA Pol II. We had previously shown that hypo-phosophorylated RNA Pol II (Pol IIa) localized predominately to promoter regions while RNA Pol II (Pol II) localized at promoters, internal and terminal exons (Brodsky et al. 2005). Thus, the enrichment patterns for RNA Pol II outlined active promoters, elongating polymerase, and sites of accumulation that likely reflect slower transcription elongation. ChIP analysis across FGFR1 and FGFR2 with antibodies against RNA Pol II reflected the expression of FGFR1 and the lack of FGFR2 expression (Fig. 2A, RNA expression array data) (Brodsky et al. 2005).
To examine enrichment patterns across another gene, we designed a series of primers across the PTBP1 gene itself. Many RNA binding proteins exhibit auto-regulation, as has been shown for PTBP1 (Wollerton et al. 2004). PTBP1 localized to a number of regions across the PTBP1 gene including the promoter (Fig. 2B, p2) and around alternative exon 9 (Fig. 2B, p5). Thus, the ChIP approach effectively identified PTBP1 target regions. Similar to patterns observed previously, we observed Pol IIa mostly at the promoter and Pol II across the PTBP1 gene (Fig. 2B). In comparison, we observed PTBP1 sites overlapping with RNA Pol II accumulation within the gene.
PTBP1 associations in gene regions are sensitive to reductions in transcription
To determine if PTBP1’s association with genes depends on transcription, we treated cells with the transcription elongation inhibitor, α-amanitin, before performing the ChIP assay. The drug α-amanitin blocks the translocation of RNA Pol II and enabled us to test the dependence of genomic association on transcription elongation (Reid et al. 2003; Gong et al. 2004). For the α-amanitin experiments we again used PAPOLB for background correction because α-amanitin treatment did not induce its expression (absence of expression not shown). Our data show that α-amanitin treatment reduced internal PTBP1 enrichment across PTBP1 (Fig. 2B). Enrichment at the promoter, while reduced, remained significant for PTBP1 after drug treatment. Therefore, the localization of PTBP1 within a gene is sensitive to either the reduction of active elongation complexes or RNA synthesis.
Genome location analyses identify many RNA binding protein association sites
To determine sites of interaction and the specificity of cotranscriptional association with the genome, we combined ChIP with tiled oligonucleotide microarrays (ChIP-chip). We hybridized amplified DNA to tiled microarrays spanning the Encyclopedia of DNA Elements (ENCODE) regions (The ENCODE Project Consortium 2004; Brodsky et al. 2005). We compared DNA from chromatin immunoprecipitations of PTBP1, ALY, and CSTF2, to differentiate how RNA binding proteins with different basal functions in pre-mRNA processing may be recruited to different locations reflecting their roles in mRNA processing. Additional expression information came from total HeLa RNA that was reverse-transcribed with random primers and probed on the tiled ENCODE arrays (RNA tract in Fig. 3).
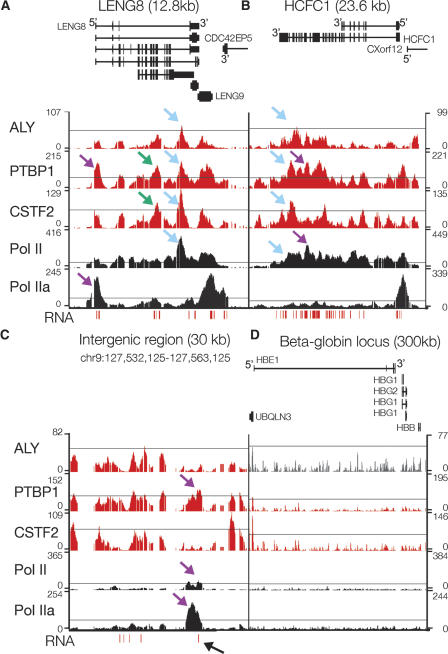
Figure 3.
High resolution mapping of ALY, PTBP1, CSTF2, Pol II, Pol IIa, and RNA shows accumulation at distinct sites. Pol II sites were determined using an antibody, 4H8, that recognizes both phosphorylated and unphosphorylated RNA polymerase II. Pol IIa sites were determined using an antibody, 8WG16, which recognizes primarily unphosphorylated RNA polymerase II. RNA was reverse-transcribed with random primers and the resulting cDNA was probed on the tiled ENCODE arrays. Representative plots of –10 log (P-value) across LENG8 (A), HCFC1 (B), an intergenic region (C), and the β-globin locus (D). Schematics of the genes from the known genes database are shown and labeled at the 5′ end to indicate the direction of transcription of each gene. The thin line in each plot marks a P-value of 10−6 except for ALY, where it marks a P-value of 10−5. Blue arrows highlight sites where RNA binding proteins were similar to other RNA binding proteins as well as RNA Pol II, purple arrows highlight sites where RNA binding proteins were different than one another, green arrows highlight sites where there was RNA binding protein enrichment in the absence of significant RNA Pol II accumulation, and the black arrow highlights an unannotated loci where PTBP1, RNA Pol II, and RNA signal were observed.
We examined enrichment patterns across individual genes and found distinct patterns for RNA binding proteins relative to RNA Pol II. Figure Figure 3 shows representative plots of −10 log (P-value) for probes across four genomic regions. While ALY, PTBP1, and CSTF2 exhibited significant enrichment on LENG8 and HCFC1, their localization patterns were different (Fig. 3A,B). In Figure Figure 3, blue arrows highlight sites where gene associations were similar between RNA binding proteins as well as RNA Pol II, purple arrows highlight sites where RNA binding proteins were different from one another, and green arrows highlight sites where there was RNA binding protein enrichment in the absence of significant RNA Pol II accumulation. LENG8 and HCFC1 represented typical genes, with PTBP1 and RNA Pol II showing similar patterns across the genes. In comparison, CSTF2 and ALY appeared at the 3′ ends of LENG8 and HCFC1.
Interestingly, RNA binding proteins were present at unannotated regions where PTBP1 enrichment overlapped with RNA Pol II as well as RNA (indicated with a black arrow, Fig. 3C). These regions may represent novel small genes. At loci where there was no apparent transcription, such as the β-globin locus, there was often no significant enrichment for the RNA binding proteins (Fig. 3D, RNA expression array data) (Brodsky et al. 2005).
ALY, CSTF2, and PTBP1 are biased to particular exons reflecting their functions
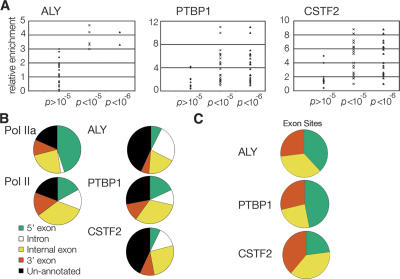
To determine sites of enrichment for ALY, CSTF2, and PTBP1, we used a nonparametric approach generalizing the Mann-Whitney U-test (Brodsky et al. 2005). The P-values reported here represent comparisons between the ChIP and control samples of probe intensities in each 1000-base pair window. As shown in Figure 4A, we found that a P-value of 10−6 minimized the false-positive rate for PTBP1 and CSTF2. For ALY, a P-value <10−5 was justified by the PCR data.
Figure 4.
Analysis of array data showed P < 10−6 were significant and annotation of these sites revealed unique patterns and biases. (A) PCR validation of array data, performed with PCR assay used in Figure Figure 1. Each point in the scatter plots represents a PCR validation. (B) RefSeq site annotation of sites enriched for RNA polymerase II, ALY, PTBP1, and CSTF2. Sites were annotated as being 5′ exons, introns, internal exons, 3′ exons, and unannotated as described in Methods. Numbers for each annotation are in Table 1. (C) Exon biases of RNA binding proteins. The percent of each protein’s exons that are 5′, 3′, or internal (by default) was determined and plotted on a pie chart to illustrate factor biases.
To search for general patterns amongst ALY’s, PTBP1’s, and CSTF2’s sites of enrichment, we classified sites using the RefSeq database for their location relative to 5′ exons, introns, internal exons, and 3′ exons of annotated genes as well as unannotated, intergenic regions (Fig. 4B; Table 1; Supplemental data 2). The RNA binding proteins exhibited unique distribution patterns. ALY, while having fewer significant sites (189 total sites), was most often at introns (51 sites) and internal exons (36 sites). Statistical analysis using the two-sided Fisher’s exact test demonstrated that ALY intron sites coincided less often then expected by chance with the other factors and locations (P-values 2.2 × 10−29 to 0.17, Supplemental data 3). PTBP1 had 748 significant sites and many were at 5′ (215 sites) and internal exons (295 sites). The distribution of PTBP1 sites resembled the distribution of Pol II sites (Fisher’s exact test P-values 0.07–1, Supplemental data 3). CSTF2 had an intermediate number of sites (481 total sites) and, compared with PTBP1, had a greater portion of its sites within introns and regions not annotated in the RefSeq gene database. The most significant differences between PTBP1 and CSTF2 occurred between 5′ exon-intron and 5′ exon-internal exon sites (respective Fisher’s exact test P-values are 3.2 × 10−7 and 2.5 × 10−4, Supplemental data 3).
Table 1.
A summary of RefSeq site annotations for ALY, PTBP1, CSTF2, RNA polymerase II, and RNA polymerase IIa illustrated protein specific patterns
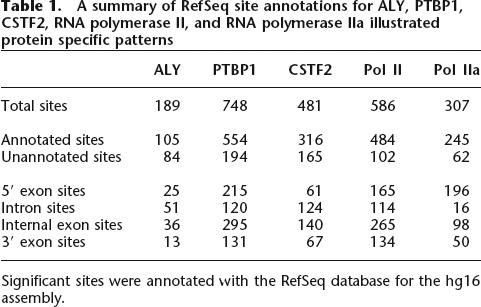
Significant sites were annotated with the RefSeq database for the hg16 assembly.
The RNA binding proteins had different exon biases reflecting their different known functions during pre-mRNA splicing, 3′ end processing, and mRNA export. Generally, ∼57% of PTBP1 sites were at annotated exons while ∼40% of CSTF2 and ∼30% of ALY sites were at annotated exons. Amongst exon positions, PTBP1 was biased for 5′ exons, ALY was biased for 5′ and internal exons, and CSTF2 was biased for internal and 3′ exons (Fig. 4C). We did not find significant motifs within the promoters bound by PTBP1 or CSTF2. Additionally, we did not identify significant correlations with alternatively spliced exons or splice site donor strength.
To examine the patterns across individual genes, we sorted genes with enrichment into categories defined by the location(s) of factor enrichment (Table 2). With this analysis, we found that 54% of genes with ALY enrichment sites only had ALY sites at introns and/or internal exons. We also found that 73% of genes with PTBP1 enrichment sites had PTBP1 sites at the 5′ exon combined with various patterns downstream within the gene. There were two predominant categories of CSTF2-enriched genes: The first category had 38% of the CSTF2 genes with sites only within the gene at introns and/or internal exons, the second category had 41% of CSTF2 genes with sites at the 3′ exon and in various patterns upstream within the gene. These gene specific patterns could reflect trends in different functional roles during pre-mRNA splicing, 3′ end processing, and mRNA export.
Table 2.
Classification of genes for localization patterns highlighted the distinct association patterns of ALY, PTBP1, and CSTF2 across individual genes
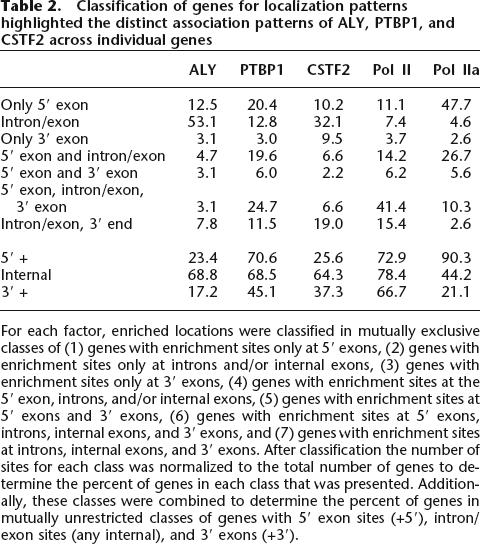
For each factor, enriched locations were classified in mutually exclusive classes of (1) genes with enrichment sites only at 5′ exons, (2) genes with enrichment sites only at introns and/or internal exons, (3) genes with enrichment sites only at 3′ exons, (4) genes with enrichment sites at the 5′ exon, introns, and/or internal exons, (5) genes with enrichment sites at 5′ exons and 3′ exons, (6) genes with enrichment sites at 5′ exons, introns, internal exons, and 3′ exons, and (7) genes with enrichment sites at introns, internal exons, and 3′ exons. After classification the number of sites for each class was normalized to the total number of genes to determine the percent of genes in each class that was presented. Additionally, these classes were combined to determine the percent of genes in mutually unrestricted classes of genes with 5′ exon sites (+5′), intron/exon sites (any internal), and 3′ exons (+3′).
ALY, PTBP1, and CSTF2 localizations reveal exon specificity
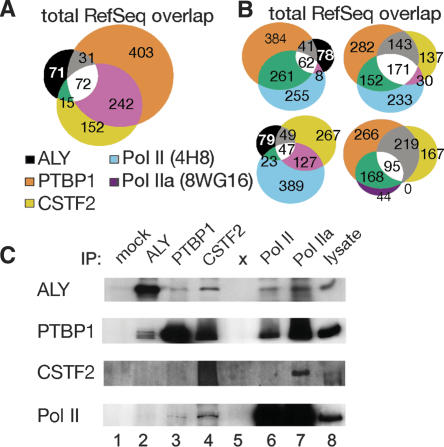
To further characterize the specificity of the observed protein associations, we analyzed the overlap in genomic association as diagrammed in Figure 5A. We identified 986 sites bound by ALY, PTBP1, and/or CSTF2. Over the annotated regions, we observed that two or more of the RNA binding proteins shared 36% of these sites. We determined that the overlaps are statistically significant, with P-values <10−4 as determined by simulations with 10,000 random sites. To examine whether there was any bias for sites where RNA binding proteins overlap, we classified these sites for 5′ exon, introns, internal exon, 3′ exon, and unannotated (analysis not shown). We found multiple RNA binding proteins coincided more often at internal exons and 3′ exons, (39% and 47% shared by two or more), while there was less overlap at promoter regions (23% shared by two or more).
Figure 5.
Analysis of ChIP-enriched sites showed that most RNA binding protein sites are unique to ALY, PTBP1, or CSTF2. Additionally, many RNA binding protein sites overlapped with RNA Pol II and this is consistent with protein complex association between RNA binding proteins and RNA Pol II. (A) Overlap analysis of RNA binding proteins within the RefSeq-annotated regions. Numbers in each set indicate the number of enrichment sites that fell into that overlap category. (B) Overlap analysis of ALY, PTBP1, CSTF2, Pol II, and Pol IIa within RefSeq-annotated regions. (C) RNA binding proteins associated with one another as well as with RNA polymerase II. α-ALY (lane 2), α-PTBP1 (lane 3), α-CSTF2 (lane 4), and α-RNA polymerase II (lane 5 is 4H8, and lane 6 is 8WG16) antibodies were used to immunoprecipitate complexes from HeLa cell lysates. Mock IPs lacked primary antibody (lane 1).
Sites of RNA binding protein enrichment overlap with RNA Pol II enrichment sites
RNA binding protein sites overlapped significantly with RNA Pol II sites (Fig. 5B). PTBP1 sites coincided with RNA Pol II sites more than CSTF2 and ALY (Fig. 5B). PTBP1 associated at many RNA Pol II sites across genes (65%) but slightly more often at 5′ (86%) and 3′ exons (75%, Fig. 5B, location analysis in Supplemental data 4). Less overlap between PTBP1 and RNA Pol II occurred at internal exons (62%) and introns (50%). Thus, not all PTBP1 and RNA Pol II internal exon sites were the same, suggesting that there is some disconnect between the transcription machinery and PTBP1’s sites of accumulation. For CSTF2, increased overlap with RNA Pol II occurred at 5′ exons (61%) and terminal exons (63%) as opposed to all gene sites (47%). In contrast, CSTF2 shared fewer sites with RNA Pol II at internal exons (50%) and introns (40%). Overall, CSTF2 sites coincided to a lesser extent with RNA Pol II sites than PTBP1. With only 39% of its gene sites overlapping with RNA Pol II, ALY coincided to a lesser extent with sites of RNA Pol II accumulation. Biochemical experiments substantiated these shared sites as the different RNA binding proteins co-immunoprecipitated with one another and with different phosphorylation states of RNA Pol II (Fig. 5C).
RNA binding protein locations significantly coincided with RNA signal (Table 3). RNA was reverse-transcribed with random primers and the resulting cDNA was probed on the tiled ENCODE arrays. As summarized in Table 3, PTBP1 overlapped with RNA signal 49% of the time, similar to RNA Pol II at 57%. Additional analysis of RNA expression array data demonstrated that PTBP1 bound to the 5′ exons of highly expressed genes (Supplemental data 5). Meanwhile, ALY and CSTF2 coincided much less with RNA signal, at 35% and 28%, respectively. This may reflect ALY and CSTF2 localization to lowly transcribed and intergenic regions as well as intronic sites normally absent in mature mRNA (Fig. 4; Supplemental data 5).
Table 3.
The percent overlap between ChIP enrichment sites and RNA-tiled array signal
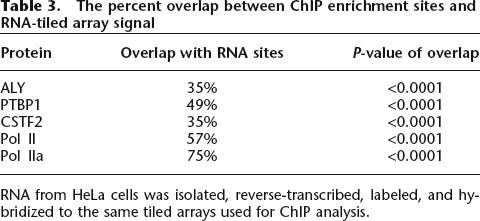
RNA from HeLa cells was isolated, reverse-transcribed, labeled, and hybridized to the same tiled arrays used for ChIP analysis.
RNA binding proteins are recruited to exons
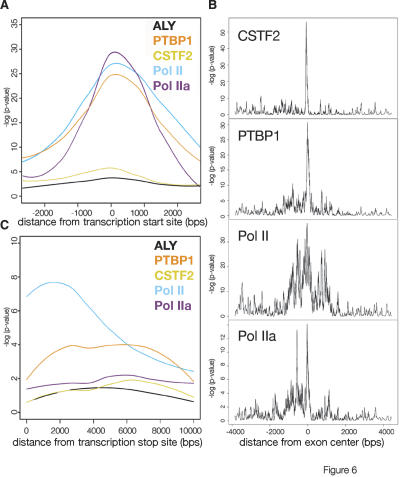
To extract more information from the high density of probes available on the Affymetrix arrays, we mapped the maximum enrichment sites (the top 10%) at different annotations for each factor to determine the approximate order of recruitment. We defined locations based on RefSeq annotations and exon centers that are halfway between annotated 5′ and 3′ ends of RefSeq exons, excluding the 5′ and 3′ exons. P-values were calculated for the strongest 10% sites for each location using a 25-base pair window. In Figure 6A the P-values at promoters were compared for RNA Pol II and RNA binding proteins. As expected, the hypo-phosphorylated RNA Pol II (Pol IIa) appeared slightly earlier than RNA Pol II (Pol II). Like elongating RNA Pol II (Pol II), PTBP1 signal was spread broadly in the promoter region around the 5′ exon and was distinguished from the tight promoter signal of hypo-phosphorylated RNA Pol II (Pol IIa).
Figure 6.
Location analysis of strongest enrichment at particular classes of sites shows factor-specific association patterns. (A) 5′ exon signal analysis of each factor examined. The P-value patterns at the top 10% sites for 5′ and 3′ exons were smoothed and plotted as curves to show signal patterns relative to the beginning or end of genes. (B) For each factor, enrichment around internal exons was averaged but not smoothed to highlight minor peaks up and down stream of exons. (C) Location analysis performed as in A for 3′ exon signal of each factor examined.
At internal exons, PTBP1 and CSTF2 showed relatively homogenous, strong enrichment around the center of internal exons (Fig. 6B). In contrast, elongating RNA Pol II had a broad location pattern while Pol IIa signal was stronger at the 5′ ends of internal exons. At the 3′ end of genes RNA Pol II (Pol II) enrichment extended beyond the 3′ ends of genes, consistent with the synthesis of larger pre-mRNAs (Fig. 6C). Interestingly, PTBP1 enrichment increased as the RNA Pol II enrichment tailed off further from the 3′ exon, suggesting some PTBP1 may be globally involved in transcription termination. ALY was not enriched at sufficient numbers of locations to provide meaningful signal for global signal analysis.
Discussion
This study illustrated that RNA binding proteins bind a variety of sites across genes as well as putative intergenic regions in the human genome. We used a genomic tiling array for nonrepetitive sequences in the ENCODE regions to determine the distribution of three different RNA binding proteins across the human genome. We observed hundreds of sites (748 for PTBP1, 481 for CSTF2, and 189 for ALY), mostly near known annotated genes that are actively expressed. These data identified genes that may be regulated by these RNA binding proteins.
Localization of RNA binding proteins to different sites across genes suggests that RNA binding proteins may be critical parts of the transcription machinery during both initiation and elongation phases of RNA synthesis. Previously it had been proposed that RNA binding proteins regulate groups of genes during transcription and/or post-transcriptionally through various mechanisms to act as operons (Keene and Tenenbaum 2002). Our findings focus on one stage of this combinatorial control of RNA metabolism as this approach characterizes how RNA binding proteins interact with the genome during the earliest phases of mRNA metabolism. These data also indicate that some RNA binding proteins associate with many, but not all, active genes. Interestingly, we observed across genes being transcribed distinct and unique binding patterns as the RNA binding proteins interacted at particular regions. Furthermore, not all RNA Pol II-enriched locations had significant RNA binding protein levels, suggesting instances of decoupling between transcriptional pausing and RNA binding protein accumulation, even on actively transcribed genes Figs. 3, 5). These findings provide evidence that RNA binding proteins of various functions localize to genes in mammalian cells and the myriad gene locations are reminiscent of their known mRNA processing functions.
RNA binding proteins have distinct interaction patterns across genes
Interestingly, all locations with significant RNA Pol II did not have PTBP1 enrichment, even when PTBP1 was observed elsewhere along the same gene (see Figs. 2, 3, 5B). This suggests a difference between RNA Pol II accumulation and RNA binding protein association. Moreover, at internal exons, RNA binding proteins had a narrower distribution than RNA Pol II, suggesting that the RNA binding proteins are not always acting together with RNA Pol II. Thus, RNA binding proteins associate with particular exons during transcription and may be key components of paused transcription complexes at exons. During this time, the transcription machinery may be reorganizing as different RNA binding proteins, and other factors move on and off the complex. Thus, our ChIP-chip data are consistent with a reorganization model for cotranscriptional pre-mRNA processing.
The association patterns we observed shed new light on past evidence that indicated that RNA processing as well as RNA processing factors can alter transcription elongation rates (Enriquez-Harris et al. 1991; Cramer et al. 1999; Kadener et al. 2002; de la Mata et al. 2003; Howe et al. 2003; Nogues et al. 2003; Proudfoot 2003; Robson-Dixon and Garcia-Blanco 2004; Ujvari and Luse 2004). On internal exons, PTBP1 and CSTF2 exhibited relatively homogeneous and strong enrichment around the center of exons while RNA Pol II had a broader enrichment both 5′ and 3′ from the center of the exon. In contrast, hypo-phosphorylated RNA Pol II (Pol IIa) was biased to the 5′ ends of internal exons, possibly marking alternative 5′ exons. We speculate that RNA binding protein accumulation at the center of exons may represent maximal transcription pausing where the RNA binding protein may be recruited to bind to the growing mRNA during transcription. Likewise, RNA Pol II movement may slow down both upstream of and downstream from exons. Interestingly, both PTBP1 and CSTF2 had some significant enrichment at distinct distances 5′ of the exon, consistent with recruitment of the RNA binding protein to the transcription machinery before the exon is synthesized. Consistent with this interpretation are past findings that PTBP1 binds to intronic sequences both upstream of and downstream from exons (Wagner and Garcia-Blanco 2002).
RNA binding proteins interact with promoter regions
Although much of the connection between splicing and transcription is thought to occur during the elongation phase of mRNA synthesis, a number of RNA binding proteins have been found to be interacting with promoters during transcription initiation (Dowhan et al. 2005; Stains et al. 2005). Each of the RNA binding proteins examined herein, PTBP1, CSTF2, and ALY, associated with 5′ ends of genes near promoter regions. This suggests that cotranscriptional assembly of RNA–protein complexes may begin during transcription initiation for some genes.
Additionally, promoters can affect alternative splicing decisions. Our data reveal a correlation between the association of alternative splicing factor, PTBP1, and RNA polymerase II. There may be a kinetic mechanism where transcription complexes containing RNA binding proteins assemble at the promoter and then reorganize during transcription in an elongation-rate sensitive manner (Dowhan et al. 2005; Stains et al. 2005). An attractive extension of this interpretation is that RNA binding proteins associate at promoters to modulate the elongation rate and downstream splicing decisions. Thus, cotranscriptional alternative splicing decisions may be the sum of both promoter and local interaction effects (Enriquez-Harris et al. 1991; Cramer et al. 1999; Fong and Zhou 2001; de la Mata et al. 2003).
Transcription termination
Many RNA binding proteins, including PTBP1 and CSTF2, regulate 3′ pre-mRNA processing. Consistent with this coordination, all three factors examined were found at numerous 3′ exons and in some cases many kilobases past the mature 3′ end of the mRNA. RNA Pol II tailed off past the 3′ end but PTBP1 enrichment increased a few kilobases past the mature 3′ end. These data suggest that PTBP1 may play a role in the coupling between transcription termination and 3′ processing. CSTF2 was also found at a variety of locations across genes but gene-by-gene accounting demonstrated that it was most often at the 3′ ends of genes. This is consistent with its known roles in 3′ end pre-mRNA processing. The yeast ALY homolog, Yra1p, is 3′ biased in its association with genes. However, in mammalian cells, we observed that ALY is predominantly associated at introns. Thus, we found different trends for each RNA binding protein at the 3′ end, potentially reflecting different roles of each factor in 3′ processing.
Significance of genomic localization of RNA binding proteins
The connections between transcription and pre-mRNA processing provide a potential mechanism for a number of RNA binding proteins to affect genetic stability, potentially through transcription-activated recombination (Aguilera 2005b; Li and Manley 2005; Luna et al. 2005). Our observation that RNA binding proteins, especially PTBP1, are common parts of transcription initiation, elongation, and termination complexes provides many opportunities for a role similar to SF2 in destabilizing R-loops. Additionally, other RNA binding proteins have been found or are thought to interact directly with transcription machinery such as PGC1-α, CoAA, and hnRNP K (Kornblihtt et al. 2004; Moumen et al. 2005).
In sum, these data demonstrated that RNA binding proteins localize to actively transcribing genes during both initiation and elongation phases of transcription. These data, combined with other studies, suggest a fundamental role for RNA binding proteins in the regulation of transcription and transcription-RNA processing coupled reactions. We expect the ChIP strategy presented herein to be a valuable tool to uncover the specificity and function of RNA binding proteins’ roles in transcription and early stages of pre-mRNA processing. Future whole-genome localization may provide sufficient information to uncover sequence motifs. Additionally, there is a need for new assays to probe the connections between promoter regulation and pre-mRNA processing decisions. As RNA Pol II–RNA binding protein complexes may be reorganizing during mRNA synthesis, these data are consistent with models in which cotranscriptional assembly of RNPs is a dynamic process (Mortillaro et al. 1996; Wetterberg et al. 2001; Custodio et al. 2004; Ujvari and Luse 2004; Yu et al. 2004).
Methods
Preparation of chromatin
The chromatin immunoprecipitation procedure was optimized and adapted from previously described methods (Shang et al. 2000). For the α-amanitin experiments, cells were grown in the presence of 40 μg/mL α-amanitin (Roche) for 4 h prior to cross-linking; 1 × 108 suspension HeLa cells were cross-linked in the presence of 10 mM DMA (Pierce) for 10 min at room temperature (Kurdistani et al. 2002). Cross-linked cells were then washed three times in 25 mL PBS and subsequently treated with 1% formaldehyde for 10 min at room temperature. Extracts were prepared for sonication as described previously (Ren et al. 2002). The sonication program was optimized for a Fisher 550 Sonic Dismembrator with a Fisher 1/4″ Microtip: 10–25-sec pulses with 3-min rests at 4°C to yield chromatin fragments that average 1 kb in size. The quality of our sonicated chromatin was monitored after protease treatment (20 μg of pronase/100 μL chromatin), reverse cross-linking (65°C, 16 h), and RNase treatment.
Chromatin immunoprecipitation
Chromatin from 1.5 × 107 cells was adjusted to 1% Triton, 0.1% deoxycholate and pre-cleared with 60 μL protein A/protein G sepharose bead mix, salmon sperm DNA, and protease inhibitors. We used the following monoclonal antibodies: Bb7 anti-PTBP1 (ATCC, help from D. Black), 8WG16 anti-Pol IIa (Covance), and 4H8 anti-Pol II (Abcam). Antibody concentrations were determined empirically and antibody was added at 4°C (>16 h). IPs were performed with protein A/protein G 1:1 sepharose bead mix (Pharmacia) for 2 h followed by sequential 10-min washes as previously described (Shang et al. 2000). To increase IP specificity, 3 M urea (final concentration) was included in all wash buffers for the chromatin IPs. Samples were eluted in TE and 1% SDS for 15 min at 65°C, vortexing every 2 min. Protein samples were set aside for IP evaluation by immunoblotting and elutes were treated with 20 μg Pronase (Roche) and then reverse cross-linked overnight at 65°C. The DNA was extracted and washed with phenol-chloroform and ethanol-precipitated using 20 mM NaCl to increase solubility of SDS.
Real-time PCR analysis
Primers used in real-time PCR were designed to amplify 100 bp regions using ABI’s Primer Express 1.0. Each real-time PCR reaction contained 50 nM of primers, 1× ABI SYBR Green PCR Reaction Mix, and processed chromatin (∼1/200th of IP, ∼1 ng of input). The log-linear regression analysis of amplification curves calculates a fluorescence value proportional to the initial quantity of target DNA (Ostermeier et al. 2003). We normalized this value to an input chromatin sample, then normalized this ratio to a reference PAPOLB ratio to calculate a relative enrichment value for the target [(TargetIP) / (Targetinput)] / [(PAPOLBIP) / (PAPOLBinput)]. Experiments were performed in technical quadruplicate and the error bars represent the standard deviation from three to seven biological replicates.
Immunoprecipitation
HeLa cells were grown in DMEM, 10% FBS, and lysates were made from cells that were 80% confluent with lysis buffer (50 mM HEPES, pH7.5, 200 mM NaCl, 0.5% Triton X-100, 5% glycerol, 0.25 mM Na3VO4, RNAsin [Roche] and protease inhibitors [Roche]). Cleared lysate from 8 × 106 cells was combined with antibody against each factor or no antibody for 2 h at 4°C. Each IP was then added to 15 μL of 1:1 protein A:protein G sepharose bead mix for 2 h at 4°C. Beads were then washed 6× 10 min at 4°C with lysis buffer and then once in TE. Samples were eluted in sample loading buffer.
DNA amplification and array handling
Isolated DNA was amplified isothermally using random nonamer primers and Klenow polymerase (Invitrogen) for more than 4 h, yielding ∼2 g of DNA per IP. DNA was prepared and hybridized on Affymetrix ENCODE oligonucleotide tiled arrays using the fragmentation, hybridization, staining, and scanning procedures described previously (Kennedy et al. 2003; Brodsky et al. 2005). Affymetrix ENCODE microarrays have interrogating 25mer oligonucleotide probes tiled every 20 bp on average. A sample of chromatin was set aside before IP and used to represent the input DNA.
Identification of ChIP-enriched regions on tiled array
Identifying the genomic locations of ChIP-enriched regions from raw microarray data involves four steps: background subtraction off microarray signal, normalization of intensities between arrays, redundant probe filtering, and statistical testing for differences in probe intensities between ChIP and control groups.
On the Affymetrix ENCODE microarray, for each 25mer PM probe that matches a genomic locus, there is a mismatch (MM) probe that is identical to the PM probe except for a mismatch at the 13th nucleotide. We take an approach related to that of Wu and colleagues to estimate target specific signal y from PM and MM intensities:
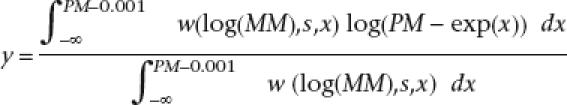 |
w(log(MM),s,x)is a normal distribution function with mean log(MM) and standard error s which is estimated from the log intensity of all probes on the array (Wu et al. 2004).
Much of the difference in signal intensity between arrays may be attributed to factors unrelated to the primary experimental objective, including sample preparation, array manufacture, and processing. Quantile normalization is used to remove these sources of variation between arrays and make the distribution of probe intensities the same across all the arrays (Bolstad et al. 2003).
The statistical test to find ChIP-enriched regions assumes probe intensities to be independent. Therefore, redundant probes were filtered to ensure that no 25mer is mapped to two genomic locations within 1000 bp and no two probes map to the same genomic location.
We use a generalization of the nonparametric Mann-Whitney test to identify ChIP-enriched regions (Lehman 1975). This test slides across all 1000-bp windows of genomic DNA covered by the microarray and compares all the probes in the ChIP and control samples to determine whether the 1000-bp region is ChIP-enriched. Within each window, ChIP and control probe intensity was centered by subtracting the mean of that probe in all samples. Then all the mean-subtracted probes in ChIPs and controls within the window are ranked together, and a statistic U is calculated as the sum of ranks for the ChIP probes. The window P-value is based on a permutation distribution of U, in which the ChIP and control ranks are randomly permuted within the same probe.
Enrichment of annotated regions
The significance of the overlap between ChIP enriched regions and annotated gene regions is computed by simulation. First the number of ChIP enriched regions that intersect with annotated regions and the total number of base pairs in this overlap are counted. The ChIP regions are then randomly assigned to genomic locations that are tiled on the Affymetrix ENCODE microarray and the intersection and base-pair count is recorded. The random assignment process is repeated 10,000 times and the intersection number and base-pair counts are used as a null distribution from which P-values for the enrichment of annotated regions may be estimated.
Genomic annotation
Sites were determined to be near a genomic annotation if they were within the apparent 1000-bp resolution. Sites shorter than 1000 bp were scaled to 1000 bp around the center of the site. Sites that were longer than 1000 bp used the data-determined length for their resolution size. Databases were downloaded from the University of California at Santa Cruz (UCSC) Golden Path Genome Browser and loaded into a local MySQL database. Exons were compared and classified as one or more of the following: start, terminal, alternatively spliced, constitutive, or cassette.
Acknowledgments
We thank E. Fox, G. Hall, P. Hollasch, M. Berkeley, and the DFCI Affymetrix core for all their assistance. We thank J. Carroll for introduction to mammalian ChIP. We thank J. Parvin, M. Greenberg, and M. Meyerson for helpful discussions and the suggestion of the α-amanitin experiments. We thank A. McKee, D. Drubin, O. Johnstone, and M. Yu for helpful discussions and critical reading of the manuscript. We also thank D. Black for advice on the α-PTBP1 antibody and J. Manley for providing the α-CSTF2 antibody. This work was supported by grants from the Department of Defense, the National Institutes of Health (P.A.S.), the Claudia Adams Barr Young Investigator program, a NHGRI K22 grant (A.S.B.), the Claudia Adams Barr Program for Innovative Basic Cancer Research (X.S.L.), T90 DK070078-01 (C.A.M.), and by an NIH training grant in Cellular and Developmental Biology (I.A.S.).
Footnotes
[Supplemental material is available online at www.genome.org and http://silver.med.harvard.edu/brodsky/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5211806
References
- Aguilera A. Cotranscriptional mRNP assembly: From the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005a;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Aguilera A. mRNA processing and genomic instability. Nat. Struct. Mol. Biol. 2005b;12:737–738. doi: 10.1038/nsmb0905-737. [DOI] [PubMed] [Google Scholar]
- Arndt K.M., Kane C.M., Kane C.M. Running with RNA polymerase: Eukaryotic transcript elongation. Trends Genet. 2003;19:543–550. doi: 10.1016/j.tig.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P., Irizarry R.A., Astrand M., Speed T.P., Astrand M., Speed T.P., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brodsky A.S., Meyer C.A., Swinburne I.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Meyer C.A., Swinburne I.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Swinburne I.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Hall G., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Keenan B.J., Liu X.S., Fox E.A., Silver P.A., Liu X.S., Fox E.A., Silver P.A., Fox E.A., Silver P.A., Silver P.A. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V., Jin P., Ceman S., Darnell J.C., O’Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Jin P., Ceman S., Darnell J.C., O’Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Ceman S., Darnell J.C., O’Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Darnell J.C., O’Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., O’Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Jin X., Feng Y., Wilkinson K.D., Keene J.D., Feng Y., Wilkinson K.D., Keene J.D., Wilkinson K.D., Keene J.D., Keene J.D., et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Calvo O., Manley J.L., Manley J.L. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- Carstens R.P., Wagner E.J., Garcia-Blanco M.A., Wagner E.J., Garcia-Blanco M.A., Garcia-Blanco M.A. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco P., Furger A., Wollerton M., Smith C., Moreira A., Proudfoot N., Furger A., Wollerton M., Smith C., Moreira A., Proudfoot N., Wollerton M., Smith C., Moreira A., Proudfoot N., Smith C., Moreira A., Proudfoot N., Moreira A., Proudfoot N., Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Caceres J.F., Cazalla D., Kadener S., Muro A.F., Baralle F.E., Kornblihtt A.R., Caceres J.F., Cazalla D., Kadener S., Muro A.F., Baralle F.E., Kornblihtt A.R., Cazalla D., Kadener S., Muro A.F., Baralle F.E., Kornblihtt A.R., Kadener S., Muro A.F., Baralle F.E., Kornblihtt A.R., Muro A.F., Baralle F.E., Kornblihtt A.R., Baralle F.E., Kornblihtt A.R., Kornblihtt A.R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- Custodio N., Carvalho C., Condado I., Antoniou M., Blencowe B.J., Carmo-Fonseca M., Carvalho C., Condado I., Antoniou M., Blencowe B.J., Carmo-Fonseca M., Condado I., Antoniou M., Blencowe B.J., Carmo-Fonseca M., Antoniou M., Blencowe B.J., Carmo-Fonseca M., Blencowe B.J., Carmo-Fonseca M., Carmo-Fonseca M. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA. 2004;10:622–633. doi: 10.1261/rna.5258504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R., Cramer P., Bentley D., Kornblihtt A.R., Bentley D., Kornblihtt A.R., Kornblihtt A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Dowhan D.H., Hong E.P., Auboeuf D., Dennis A.P., Wilson M.M., Berget S.M., O’Malley B.W., Hong E.P., Auboeuf D., Dennis A.P., Wilson M.M., Berget S.M., O’Malley B.W., Auboeuf D., Dennis A.P., Wilson M.M., Berget S.M., O’Malley B.W., Dennis A.P., Wilson M.M., Berget S.M., O’Malley B.W., Wilson M.M., Berget S.M., O’Malley B.W., Berget S.M., O’Malley B.W., O’Malley B.W. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERα and CAPERβ. Mol. Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Enriquez-Harris P., Levitt N., Briggs D., Proudfoot N.J., Levitt N., Briggs D., Proudfoot N.J., Briggs D., Proudfoot N.J., Proudfoot N.J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991;10:1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman M.E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E., Gollnick P., Nilsen T.W., Jankowsky E., Nilsen T.W., Jankowsky E., Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Fong Y.W., Zhou Q., Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Gong X.Q., Nedialkov Y.A., Burton Z.F., Nedialkov Y.A., Burton Z.F., Burton Z.F. α-amanitin blocks translocation by human RNA polymerase II. J. Biol. Chem. 2004;279:27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- Hieronymus H., Silver P.A., Silver P.A. Genome-wide analysis of RNA–protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Hieronymus H., Silver P.A., Silver P.A. A systems view of mRNP biology. Genes & Dev. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- Howe K.J., Kane C.M., Ares M., Jr., Kane C.M., Ares M., Jr., Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., McCutcheon I.E., Fuller G.N., Huang E.S., Cote G.J., McCutcheon I.E., Fuller G.N., Huang E.S., Cote G.J., Fuller G.N., Huang E.S., Cote G.J., Huang E.S., Cote G.J., Cote G.J. Fibroblast growth factor receptor-1 α-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res. 2000;60:1221–1224. [PubMed] [Google Scholar]
- Jin W., Bruno I.G., Xie T.X., Sanger L.J., Cote G.J., Bruno I.G., Xie T.X., Sanger L.J., Cote G.J., Xie T.X., Sanger L.J., Cote G.J., Sanger L.J., Cote G.J., Cote G.J. Polypyrimidine tract-binding protein down-regulates fibroblast growth factor receptor 1 α-exon inclusion. Cancer Res. 2003;63:6154–6157. [PubMed] [Google Scholar]
- Kadener S., Fededa J.P., Rosbash M., Kornblihtt A.R., Fededa J.P., Rosbash M., Kornblihtt A.R., Rosbash M., Kornblihtt A.R., Kornblihtt A.R. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. 2002;99:8185–8190. doi: 10.1073/pnas.122246099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D., Tenenbaum S.A., Tenenbaum S.A. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Kennedy G.C., Matsuzaki H., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Matsuzaki H., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Dong S., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Liu W.M., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Huang J., Liu G., Su X., Cao M., Chen W., Zhang J., Liu G., Su X., Cao M., Chen W., Zhang J., Su X., Cao M., Chen W., Zhang J., Cao M., Chen W., Zhang J., Chen W., Zhang J., Zhang J., et al. Large-scale genotyping of complex DNA. Nat. Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- Kim E., Du L., Bregman D.B., Warren S.L., Du L., Bregman D.B., Warren S.L., Bregman D.B., Warren S.L., Warren S.L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S., Nedea E., Greenblatt J.F., Buratowski S., Greenblatt J.F., Buratowski S., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P., Cho E.J., Buratowski S., Cho E.J., Buratowski S., Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A.R., de la Mata M., Fededa J.P., Munoz M.J., Nogues G., de la Mata M., Fededa J.P., Munoz M.J., Nogues G., Fededa J.P., Munoz M.J., Nogues G., Munoz M.J., Nogues G., Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotovic K.M., Lockshon D., Boric L., Neugebauer K.M., Lockshon D., Boric L., Neugebauer K.M., Boric L., Neugebauer K.M., Neugebauer K.M. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani S.K., Robyr D., Tavazoie S., Grunstein M., Robyr D., Tavazoie S., Grunstein M., Tavazoie S., Grunstein M., Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- Lacadie S.A., Rosbash M., Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lehman E.L. Nonparametrics: Statistical methods based on ranks. Holden Day; San Francisco: 1975. [Google Scholar]
- Lei E.P., Silver P.A., Silver P.A. Intron status and 3′ end formation control cotranscriptional export of mRNA. Genes & Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Krebber H., Silver P.A., Krebber H., Silver P.A., Silver P.A. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Manley J.L., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lin C.H., Patton J.G., Patton J.G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Emigh C.A., McCleery J.A., Yates J.R., III, Hartzog G.A., McCleery J.A., Yates J.R., III, Hartzog G.A., Yates J.R., III, Hartzog G.A., Hartzog G.A. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R., Jimeno S., Marin M., Huertas P., Garcia-Rubio M., Aguilera A., Jimeno S., Marin M., Huertas P., Garcia-Rubio M., Aguilera A., Marin M., Huertas P., Garcia-Rubio M., Aguilera A., Huertas P., Garcia-Rubio M., Aguilera A., Garcia-Rubio M., Aguilera A., Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Mabon S.A., Misteli T., Misteli T. Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo. PLoS Biol. 2005;3:e374. doi: 10.1371/journal.pbio.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti E.F., Tomonaga T., Krutzsch H., Levens D., Tomonaga T., Krutzsch H., Levens D., Krutzsch H., Levens D., Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- Mortillaro M.J., Blencowe B.J., Wei X., Nakayasu H., Du L., Warren S.L., Sharp P.A., Berezney R., Blencowe B.J., Wei X., Nakayasu H., Du L., Warren S.L., Sharp P.A., Berezney R., Wei X., Nakayasu H., Du L., Warren S.L., Sharp P.A., Berezney R., Nakayasu H., Du L., Warren S.L., Sharp P.A., Berezney R., Du L., Warren S.L., Sharp P.A., Berezney R., Warren S.L., Sharp P.A., Berezney R., Sharp P.A., Berezney R., Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumen A., Masterson P., O’Connor M.J., Jackson S.P., Masterson P., O’Connor M.J., Jackson S.P., O’Connor M.J., Jackson S.P., Jackson S.P. hnRNP K: An HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Nogues G., Munoz M.J., Kornblihtt A.R., Munoz M.J., Kornblihtt A.R., Kornblihtt A.R. Influence of polymerase II processivity on alternative splicing depends on splice site strength. J. Biol. Chem. 2003;278:52166–52171. doi: 10.1074/jbc.M309156200. [DOI] [PubMed] [Google Scholar]
- Ostermeier G.C., Liu Z., Martins R.P., Bharadwaj R.R., Ellis J., Draghici S., Krawetz S.A., Liu Z., Martins R.P., Bharadwaj R.R., Ellis J., Draghici S., Krawetz S.A., Martins R.P., Bharadwaj R.R., Ellis J., Draghici S., Krawetz S.A., Bharadwaj R.R., Ellis J., Draghici S., Krawetz S.A., Ellis J., Draghici S., Krawetz S.A., Draghici S., Krawetz S.A., Krawetz S.A. Nuclear matrix association of the human β-globin locus utilizing a novel approach to quantitative real-time PCR. Nucleic Acids Res. 2003;31:3257–3266. doi: 10.1093/nar/gkg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J. Dawdling polymerases allow introns time to splice. Nat. Struct. Biol. 2003;10:876–878. doi: 10.1038/nsb1103-876. [DOI] [PubMed] [Google Scholar]
- Reid G., Hubner M.R., Metivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F., Hubner M.R., Metivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F., Metivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F., Manu D., Beaudouin J., Ellenberg J., Gannon F., Beaudouin J., Ellenberg J., Gannon F., Ellenberg J., Gannon F., Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Volkert T., Terragni J., Young R.A., Dynlacht B.D., Terragni J., Young R.A., Dynlacht B.D., Young R.A., Dynlacht B.D., Dynlacht B.D. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson-Dixon N.D., Garcia-Blanco M.A., Garcia-Blanco M.A. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 2004;279:29075–29084. doi: 10.1074/jbc.M312747200. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu X., DiRenzo J., Lazar M.A., Brown M., Hu X., DiRenzo J., Lazar M.A., Brown M., DiRenzo J., Lazar M.A., Brown M., Lazar M.A., Brown M., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Stains J.P., Lecanda F., Towler D.A., Civitelli R., Lecanda F., Towler D.A., Civitelli R., Towler D.A., Civitelli R., Civitelli R. Heterogeneous nuclear ribonucleoprotein K represses transcription from a cytosine/thymidine-rich element in the osteocalcin promoter. Biochem. J. 2005;385:613–623. doi: 10.1042/BJ20040680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y., Manley J.L., Manley J.L. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- Tenenbaum S.A., Carson C.C., Lager P.J., Keene J.D., Carson C.C., Lager P.J., Keene J.D., Lager P.J., Keene J.D., Keene J.D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari A., Luse D.S., Luse D.S. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J. Biol. Chem. 2004;279:49773–49779. doi: 10.1074/jbc.M409087200. [DOI] [PubMed] [Google Scholar]
- Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B., Ruggiu M., Mele A., Ule A., Darnell R.B., Mele A., Ule A., Darnell R.B., Ule A., Darnell R.B., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A., Garcia-Blanco M.A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- Wetterberg I., Zhao J., Masich S., Wieslander L., Skoglund U., Zhao J., Masich S., Wieslander L., Skoglund U., Masich S., Wieslander L., Skoglund U., Wieslander L., Skoglund U., Skoglund U. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20:2564–2574. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Garcia-Blanco M.A., Smith C.W., Smith C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A., Gentleman R., Murillo F.M., Spencer F, Irizarry R.A., Gentleman R., Murillo F.M., Spencer F, Gentleman R., Murillo F.M., Spencer F, Murillo F.M., Spencer F, Spencer F. A model based background adjustment of oligonucleotide expression arrays. Johns Hopkins University, Department of Biostatistics Working Papers; Baltimore, MD: 2004. [Google Scholar]
- Yu M.C., Bachand F., McBride A.E., Komili S., Casolari J.M., Silver P.A., Bachand F., McBride A.E., Komili S., Casolari J.M., Silver P.A., McBride A.E., Komili S., Casolari J.M., Silver P.A., Komili S., Casolari J.M., Silver P.A., Casolari J.M., Silver P.A., Silver P.A. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes & Dev. 2004;18:2024–2035. doi: 10.1101/gad.1223204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A., Patturajan M., Litingtung Y., Joshi R.V., Gentile C., Gebara M., Corden J.L., Patturajan M., Litingtung Y., Joshi R.V., Gentile C., Gebara M., Corden J.L., Litingtung Y., Joshi R.V., Gentile C., Gebara M., Corden J.L., Joshi R.V., Gentile C., Gebara M., Corden J.L., Gentile C., Gebara M., Corden J.L., Gebara M., Corden J.L., Corden J.L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo M.J., Straesser K., Katahira J., Hurt E., Reed R., Luo M.J., Straesser K., Katahira J., Hurt E., Reed R., Straesser K., Katahira J., Hurt E., Reed R., Katahira J., Hurt E., Reed R., Hurt E., Reed R., Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]