Abstract
Heparanase is a mammalian endo-β-D-glucuronidase that cleaves heparan sulfate (HS) side chains at a limited number of sites. Heparanase enzymatic activity is thought to participate in degradation and remodeling of the extracellular matrix (ECM) and to facilitate cell invasion associated with tumor metastasis, angiogenesis and inflammation. Traditionally, heparanase activity was well correlated with the metastatic potential of a large number of tumor-derived cell types. More recently, heparanase up regulation was detected in an increasing number of primary human tumors, correlating, in some cases, with poor postoperative survival and increased tumor vascularity. The present study was undertaken to develop a highly sensitive ELISA assay suitable for the determination and quantification of human heparanase in tissue extracts and body fluids. The assay preferentially detects the 8+50 kDa active heparanase heterodimer vs. the latent 65 kDa pro-enzyme and correlates with immunoblot analysis of heparanase containing samples. It detects heparanase at concentrations as low as 200 pg/ml, and is suitable for quantification of heparanase in tissue extracts and urine.
Keywords: heparanase, ELISA, antibody
Introduction
Heparanase is an endoglycosidase that specifically cleaves heparan sulfate (HS) side chains of heparan sulfate proteoglycans (HSPG) [1-3]. HSPG consist of a protein core to which HS side chains are covalently attached. These complex macromolecules are highly abundant in the extracellular matrix (ECM) and are thought to play an important structural role, contributing to ECM integrity and insolubility [4]. In addition, HS side chains can bind to a variety of biological mediators such as growth factors, cytokines and chemokines, thus providing a readily available reservoir of active molecules that can be liberated upon local or systemic cues [5]. Moreover, HSPG on the cell surface participates directly in signal transduction cascades by potentiating the interaction between certain growth factors and their receptors [6-8]. HS-degrading activity is thus expected to affect several fundamental aspects of cell behavior under normal and pathological settings. Traditionally, heparanase activity was implicated in cellular invasion associated with angiogenesis, inflammation and cancer metastasis [9-12]. This notion recently gained further support by employing siRNA and ribozyme technologies, clearly depicting heparanase-mediated HS cleavage and ECM remodeling as critical requisites for metastatic spread [13]. Since the cloning of the heparanase gene and the availability of specific molecular probes, heparanase upregulation was documented in an increasing number of primary human tumors, correlating with reduced postoperative survival, enhanced local and distant metastasis and increased microvessel density [14-21]. The enzyme has also been implicated in diabetic nephropathy [22, 23] and immune responses [2, 11, 12, 24]. Collectively, these studies provide compelling evidence for the clinical relevance of the enzyme, making it an attractive target for drug development. Heparanase gene induction in human malignancies, as well as in several other pathologies such as cirrhosis, nephrosis and diabetes [22, 23, 25] further implies the enzyme as a valuable clinical diagnostic marker. Several assays have been reported for measuring heparanase enzymatic activity, utilizing its HS degrading capacity [26-31]. However, a method for the detection and quantification of small amounts of heparanase in tissue extracts and body fluids has not been reported. Here, we report the development of a sensitive ELISA method suitable for determination and quantification of human heparanase. The assay preferentially detects the 8+50 kDa active heparanase heterodimer vs. the 65 kDa latent proenzyme. It correlates with immunoblot analysis of heparanase containing samples, detects heparanase at concentrations as low as 200 pg/ml and issuitable for quantification of heparanase in tissue extracts, plasma and urine samples. A 4-5 fold average increase in heparanase levels was found in urine collected from cancer and diabetes patients vs. healthy donors, further supporting the notion that heparanase may be considered as a diagnostic and prognostic marker, and a valid target for drug development.
Materials and Methods
Antibodies and reagents. Monoclonal anti-heparanase antibody 1E1 was generated by immunizing Balb/C mice with the entire 65 kDa heparanase protein. Hybridomas were obtained by routine procedure and were selected by an ELISA screen using the 65 kDa heparanase for coating. Several hybridomas that reacted positively with recombinant human heparanase were selected for further characterization. Anti-heparanase1453 polyclonal antibody was raised in rabbit against the entire 65 kDa heparanase precursor isolated from the conditioned medium of heparanase-transfected 293 cells [32], and has been shown to recognize both the latent and active forms of heparanase [32-34]. HRP-conjugated goat anti-rabbit antibody was purchased from Jackson ImmunoResearch (West Grove, PA). Microtiter 96-well plates (Maxisorp) were from Nunc™(Roskilde, Denmark). HRP colorimetric substrate 3,3′,5,5′-tetramethylbenzidine (TMB) was purchased from Dako (Glostrup, Denmark). Bovine serum albumin (BSA) was from Biological Industries (Beit Haemek, Israel). Single chain (GS3) active heparanase, comprised of the 8 kDa and 50 kDa heparanase heterodimer gene construct, was kindly provided by Dr. Christian Steinkuhler (IRMB/Merck Research Laboratories, Pomezia, Italy) [30], and the protein was purified from the conditioned medium of baculovirus infected insect cells [30].
ELISA procedure. Wells of microtiter plates were coated (18 h, 4°C) with 2 μg/ml of 1E1 monoclonal anti-heparanase antibody in 50μl of coating buffer (0.05 M Na2CO3, 0.05 M NaHCO3, pH 9.6) and were then blocked with 2% BSA in PBS for 1h at 37°C. Samples (200 μl) were loaded in duplicates and incubated for 2 h at room temperature, followed by the addition of 100 μl antibody 1453 (1 μg/ml) for additional 2 h at room temperature. HRP-conjugated goat anti-rabbit IgG (1:20,000) in blocking buffer was added (1 h, room temperature) and the reaction was visualized by the addition of 50 μl chromogenic substrate (TMB) for 30 min. The reaction was stopped with 100 μl H2SO4and absorbance at 450 nm was measured with reduction at 630 nm using ELISA plate reader. Plates were washed five times with washing buffer (PBS, pH 7.4, containing 0.1% (v/v) Tween 20) after each step. As a reference for quantification, a standard curve was established by a serial dilution of recombinant 8+50 GS3 active heparanase enzyme (390 pg/ml-25 ng/ml).
Sample preparation and immunoblotting. Liver and lung tissues were harvested from control and heparanase transgenic mice and homogenized by Polytron homogenizer (Kinamatica, Luzerne, Switzerland) in 10 volumes of PBS supplemented with protease inhibitors. Lysate samples were centrifuged and the supernatant was applied on to 35S-labeled ECM to evaluate heparanase activity (see below), or pre-absorbed with concanavalin A (Con A)-Sepharose beads to concentrate the samples and reduce non-specific reactivity, and subjected to SDS-PAGE under reducing conditions. Following electrophoresis, proteins were transferred to PVDF membrane (BioRad) and reacted with the appropriate antibody followed by HRP-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) and an enhanced chemiluminescent substrate (Pierce, Rockford, IL), as described [35-37].
Heparanase activity assay. Preparation of ECM-coated 35 mm dishes and determination of heparanase activity were performed as described in detail elsewhere [27, 34, 38]. To evaluate heparanase activity in tissue extracts, fresh liver and lung tissues derived from heparanase over-expressing transgenic (hpa-tg) mice [41] and control mice were homogenized, as described above, and the supernatant fractions were incubated (18 h, 37°C, pH 6.0) with35S-labeled ECM. The incubation medium (1 ml) containing sulfate labeled degradation fragments was subjected to gel filtration on a Sepharose CL-6B column. Fractions (0.2 ml) were eluted with PBS and their radioactivity counted in a β-scintillation counter. Degradation fragments of HS side chains are eluted at 0.5< Kav<0.8 (peak II, fractions 15-30) and represent heparanase generated degradation products [33, 37].
Statistics. Data are presented as mean + SE. Statistical significance was analyzed by two-tailed Student’s t-test. The value of P<0.05 is considered as significant.
Results
Establishment of quantitative ELISA
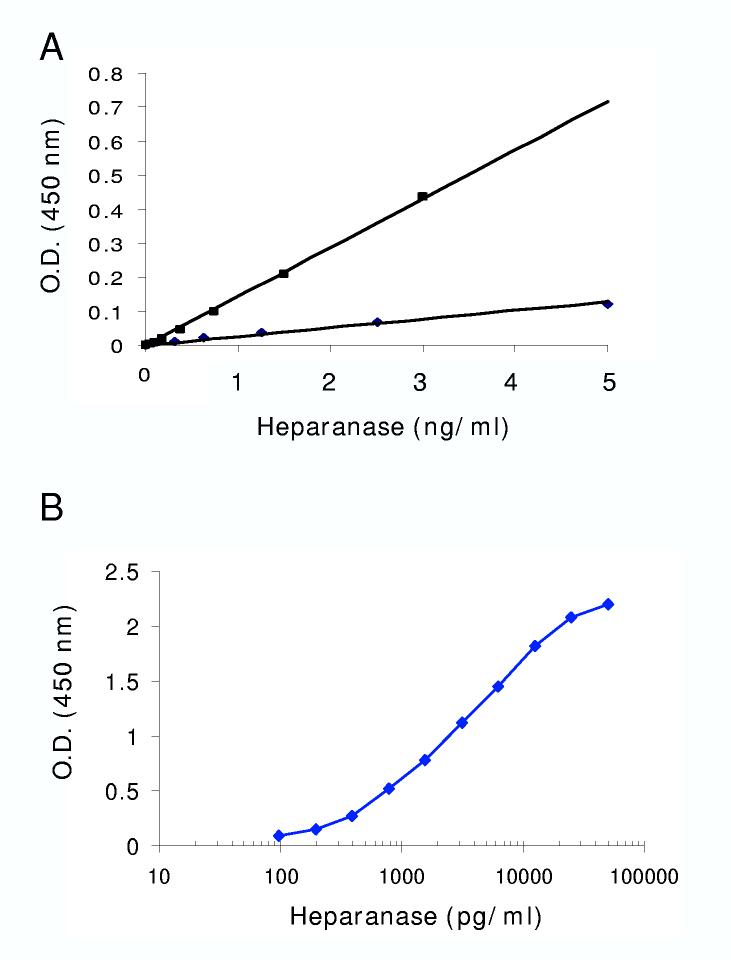
Heparanase up regulation in several pathological settings, including cancer, and the secreted nature of the enzyme predict elevated heparanase levels in patients’ body fluids. In order to detect and quantify heparanase in multiple samples, we set to establish an ELISA method suitable for the detection of low protein levels. Several anti-heparanase antibodies from different species (mouse, rabbit, goat) were tested for their ability to detect heparanase in a sandwich ELISA. The antibodies were tested reciprocally, and the best pair selected for further characterization was monoclonal antibody 1E1, applied for coating, and polyclonal antibody 1453 used for detection of the bound enzyme. Similar to several other classes of enzymes, heparanase is first synthesized as a latent enzyme that appears as a 65 kDa protein when analyzed by SDS-PAGE [1, 2]. The protein undergoes proteolytic processing yielding an 8 kDa peptide at the N-terminus and a 50 kDa polypeptide at the C-terminus that heterodimerize to form the active heparanase enzyme [38-40]. Thus, we first compared the sensitivity of our assay for the 65 kDa latent heparanase vs. the 8+50 kDa active heterodimer. As demonstrated in figure 1A, ELISA reactivity was significantly higher with the 8+50 kDa active enzyme than with the 65 kDa proenzyme. Applying increasing amounts of the 8+50 kDa heparanase further revealed the linearity of the assay, up to 30 ng/ml. Plotting the data in a semi logarithmic scale revealed sensitivity of 200 pg/ml (Fig. 1B), providing a linearity range exceeding 2 orders of magnitude.
Figure 1.
Sensitivity and linearity of heparanase ELISA method. A. Anti-heparanase 1E1 monoclonal antibody preferentially recognizes the 8+50 active heparanase heterodimer. Microtiter plate was coated with 1E1 monoclonal antibody. Latent 65 kDa heparanase (◆) and active 8+50 kDa heparanase (■) were added at the indicated amounts, and the ELISA procedure was carried out as described in “Materials and Methods”. Note that the 1E1 antibody preferentially recognizes the 8+50 kDa heterodimer. B. Dose response. Sensitivity of the heparanase ELISA method was evaluated by applying the indicated amounts (pg/well) of the 8+50 kDa active heparanase and plotting the O.D. results on a semi logarithmic scale.
Quantification of heparanase in tissue extracts
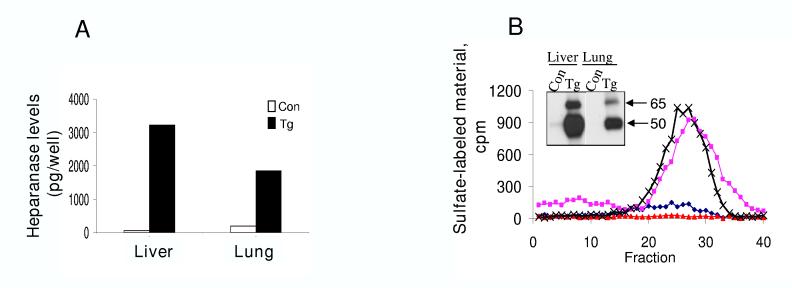
We have recently established and characterized transgenic mice (hpa-tg) over expressing heparanase in most tissues [41], and used this system to study the involvement of the enzyme in tissue function, morphogenesis, and vascularization [41]. In order to further validate the ELISA method, we determined and quantified heparanase levels in tissue extracts of control vs. transgenic mice (Fig. 2). Only trace amounts of heparanase were detected by the ELISA method in tissue extracts derived from control mice (Fig. 2A). In contrast, high levels of heparanase were detected in tissue extracts derived from heparanase transgenic mice (Fig. 2A), and concentrations of 75 and 50 ng heparanase/gram tissue were calculated for liver and lung tissues, respectively. We next correlated the ELISA results with the more common heparanase immunoblotting and enzymatic activity analyses. Under the current experimental conditions, no protein bands (Fig. 2B, inset) or enzymatic activity (Fig. 2B) were detected in the liver and lung tissue extracts derived from control mice. Heparanase was highly abundant in the hpa-tg liver tissue (Fig. 2B, inset). Lower levels of heparanase were detected by immunoblotting in the corresponding lung tissue (Fig. 2B, inset), similar, but not identical to the trend depicted by the ELISA method. Both samples exhibited similar levels of heparanase enzymatic activity (Fig. 2B), in agreement with the semi-quantitative feature of this assay.
Figure 2.
Detection and quantification of heparanase in tissue extracts. Liver and lung tissues were harvested from control and heparanase transgenic mice and homogenized by Polytron homogenizer and the supernatant fractions subjected to ELISA, heparanase activity assay and immunoblotting. A. ELISA. Heparanase levels were determined by the ELISA method in 40 μg of total tissue extracts. B. Enzymatic activity. Similar amounts of tissue extracts were applied onto sulfate-labeled ECM for determination of heparanase enzymatic activity. Samples were also subjected to immunoblot analysis with anti-heparanase 1453 antibody (inset).
Heparanase ELISA as a diagnostic tool
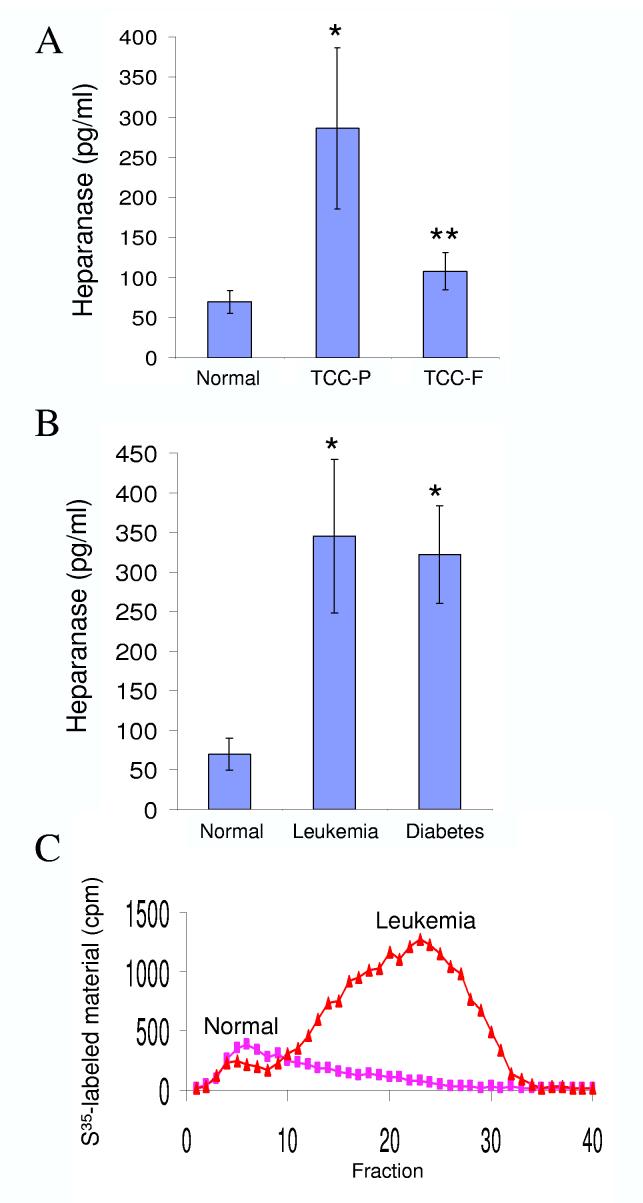
Traditionally correlated with the metastatic potential of cancer cells, heparanase up regulation has been demonstrated in an increasing number of primary human tumors, correlating with reduced post operative survival rates [14-21]. Heparanase is also up regulated in diabetic nephropathy [22, 23]. Thus, a reliable method capable of quantifying heparanase in body fluids may serve as a valuable diagnostic tool. We approached this task by evaluating heparanase levels in urine derived from normal human volunteers vs. urine collected from cancer and diabetes patients (Fig. 3). The average concentration of heparanase in urine samples collected from 27 normal individuals was 69±20 pg/ml (Fig. 3A). In contrast, heparanase levels in urine samples obtained from primary transitional cell carcinoma (TCC-P) patients (n=10) was 286±100 pg/ml, differences that are statistically highly significance (p=0.0009). Moreover, urinary heparanase concentrations were markedly reduced in TCC patients following surgical resection (n=19, TCC-F; 107±23 pg/ml, Fig. 3A), decrease that is statistically significance compared with heparanase levels in the urine of TCC-P patients (p=0.03), further supporting the notion that urinary heparanase originates primarily from the tumor mass. Similarly, elevated heparanase levels were measured in the urine of leukemia patients (n=8, 345 ± 97 pg/ml, Fig. 3B), differences that are statistically highly significance (p=0.00009), indicating systemic spread of the enzyme from solid and hematological tumors. Furthermore, heparanase levels in urine samples obtained from 34 diabetes patients with kidney complications was 322 ±61 pg/ml (Fig. 3B; p=0.0006), in agreement with previous studies demonstrating elevated heparanase enzymatic activity in the urine of diabetic patients [22]. Applying urine sample collected from a representative healthy donor on labeled ECM yielded no heparanase activity (Fig. 3C). In contrast, heparanase enzymatic activity was readily detected in urine collected from leukemia patient (Fig. 3C), correlated with the ELISA results (Fig. 3B). Al together, these results indicate that the newly developed ELISA method is suitable for the quantification of heparanase in tissue extracts and urine samples and is therefore of experimental and clinical significance.
Figure 3.
Detection and quantification of heparanase in urine samples. A. ELISA. Urine samples were collected from normal human volunteers (n=27) and from transitional bladder cell carcinoma prior to (A, TCC-P, n=10), and following surgical resection of the primary tumor (A, TCC-F, n=19). Urine samples were also collected from leukemia (B, n = 8: 4 AML, 2 ALL, 2 CLL) and diabetic (n = 34) patients with kidney complications. Heparanase levels were determined in duplicates by the ELISA method and the results are expressed as mean + SE. It should be noted that heparanase levels in the urine of healthy donors was often below the assay sensitivity, and the exact amounts above baseline could not be determined. Actual heparanase levels in the urine of healthy donors may be, thus, lower than those calculated here. C. Enzymatic activity. Urine collected from representative healthy volunteer and leukemia patient was incubated (18 h, 37°C, pH 6.0) with sulfate-labeled ECM and heparanase activity was determined as described in “Materials and Methods”.
Discussion
Heparanase up regulation documented in an increasing number of human malignancies [14-21], as well as in several other pathologies [22-24], and the cloning of the same human heparanase cDNA sequence by various laboratories [42-45] suggest that a single prominent HS-degrading endoglycosidase is expressed by mammalian cells, positioning heparanase as a potentially new and promising drug target. Several assays have been developed for quantifying heparanase enzymatic activity, detecting the degradation of fluorescent or radiolabeled HS chains [26, 29, 31, 46], or the degradation of intact, metabolically sulfate labeled ECM deposited by cultured cells [27]. These assays require gel filtration to separate degradation products, are time and labor consuming and semi-quantitative in nature. Some assays were developed specifically as a high-throughput screen for heparanase inhibitors, utilizing recombinant enzyme and are less adequate for the detection of heparanase in biological samples [46]. The ELISA method described in the present study is highly sensitive, capable of detecting heparanase at amounts as low as 200 pg/ml, and exhibits linearity up to 30 ng/ml (Fig. 1). Notably, the monoclonal anti-heparanase antibody 1E1 used for coating the microtiter wells, preferentially reacts with the active 8+50 kDa heterodimer compared with the 65 kDa latent heparanase (Fig. 1A). Thus, the ELISA results are expected to reflect heparanase activity. Indeed, a positive correlation was obtained between the ELISA, immunoblotting and enzymatic activity results, utilizing tissue extracts derived from heparanase transgenic mice vs. control mice (Fig. 2). Yet, while differences in heparanase levels in the liver and lung tissue were clearly evident by ELISA, and even more so by immunoblotting analyses, differences were not evident by the activity assay employed (Fig. 2), clearly demonstrating the quantitative advantage of the ELISA method.
The ability to detect and quantify heparanase levels in urine samples supports a clinical application of the ELISA method. Indeed, a significant increase in heparanase levels was observed in the urine of bladder and leukemia patients, suggesting a systemic spread of heparanase in both solid and hematological malignancies. A marked reduction in urinary heparanase was detected following resection of the primary tumor, indicting a correlation with tumor mass. Moreover, elevated levels of heparanase were found in the urine of diabetic patients suffering from renal complications. In fact, results obtained initially by us [22] and then by other groups [23, 25], suggesting that heparanase plays a role in the generation of albuminuria in diabetic patients.
Taken together, we describe, for the first time, a highly sensitive and reliable ELISA method capable of quantifying heparanase in tissue extracts and urine samples. Our preliminary results indicate that the same is true for other body fluids such as plasma and pleural effusions. The significant increase in heparanase levels found in the urine of cancer and diabetic patients provides a basis for a large scale screen evaluating heparanase as a diagnostic and prognostic marker for several disorders such as solid and hematological malignancies, diabetes, inflammation, autoimmunity, and kidney disorders.
Acknowledgments
This work was supported by grants from the Israel Science Foundation (grant 532/02); National Cancer Institute, NIH (grant RO1-CA106456); the Israel Cancer Research Fund; and the Rappaport Family Institute Fund
References
- [1].Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- [3].Dempsey LA, Brunn GJ, Platt JL. Heparanase, a potential regulator of cell-matrix interactions. Trends Biochem Sci. 2000;25:349–351. doi: 10.1016/s0968-0004(00)01619-4. [DOI] [PubMed] [Google Scholar]
- [4].Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- [5].Esko M. M. F. a. J. D. The sweet and sour of cancer: glycans as novel therapeutic targets. Nature Review Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- [6].David G. Integral membrane heparan sulfate proteoglycans. FASEB J. 1993;7:1023–1030. doi: 10.1096/fasebj.7.11.8370471. [DOI] [PubMed] [Google Scholar]
- [7].Ornitz DM. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. Bioessays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- [8].Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- [9].Nakajima M, Irimura T, DiFerrante D, DiFerrante N, Nicolson GL. Heparan sulfate degradation: relation to tumor invasion and metastatic properties of Mouse B 16 Melanoma sublines. Science (Wash. DC) 1983;220:611–613. doi: 10.1126/science.6220468. [DOI] [PubMed] [Google Scholar]
- [10].Vlodavsky I, Fuks Z, Bar-Ner M, Ariav Y, Schirrmacher V. Lymphoma cells mediated degradation of sulfated proteoglycans in the subendothelial extracellular matrix: relation to tumor cell metastasis. Cancer Research. 1983;43:2704–2711. [PubMed] [Google Scholar]
- [11].Naparstek Y, Cohen IR, Fuks Z, Vlodavsky I. Activated T lymphocytes produce a matrix-degrading heparan sulphate endoglycosidase. Nature. 1984;310:241–244. doi: 10.1038/310241a0. [DOI] [PubMed] [Google Scholar]
- [12].Matzner Y, Bar-Ner M, Yahalom J, Ishai-Michaeli R, Fuks Z, Vlodavsky I. Degradation of heparan sulfate in the subendothelial extracellular matrix by a readily released heparanase from human neutrophils. Possible role in invasion through basement membranes. J Clin Invest. 1985;76:1306–1313. doi: 10.1172/JCI112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase Gene Silencing, Tumor Invasiveness, Angiogenesis, and Metastasis. J Natl Cancer Inst. 2004;96:1219–123. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- [14].Koliopanos A, Friess H, Kleeff J, Shi X, Liao Q, Pecker I, Vlodavsky I, Zimmermann A, Buchler MW. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 2001;61:4655–4659. [PubMed] [Google Scholar]
- [15].Kim AW, Xu X, Hollinger EF, Gattuso P, Godellas CV, Prinz RA. Human heparanase-1 gene expression in pancreatic adenocarcinoma. J Gastrointest Surg. 2002;6:167–172. doi: 10.1016/s1091-255x(01)00087-7. [DOI] [PubMed] [Google Scholar]
- [16].Rohloff J, Zinke J, Schoppmeyer K, Tannapfel A, Witzigmann H, Mossner J, Wittekind C, Caca K. Heparanase expression is a prognostic indicator for postoperative survival in pancreatic adenocarcinoma. Br J Cancer. 2002;86:1270–1275. doi: 10.1038/sj.bjc.6600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gohji K, Hirano H, Okamoto M, Kitazawa S, Toyoshima M, Dong J, Katsuoka Y, Nakajima M. Expression of three extracellular matrix degradative enzymes in bladder cancer. Int J Cancer. 2001;95:295–301. doi: 10.1002/1097-0215(20010920)95:5<295::aid-ijc1051>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- [18].Tang W, Nakamura Y, Tsujimoto M, Sato M, Wang X, Kurozumi K, Nakahara M, Nakao K, Nakamura M, Mori I, Kakudo K. Heparanase: a key enzyme in invasion and metastasis of gastric carcinoma. Mod Pathol. 2002;15:593–598. doi: 10.1038/modpathol.3880571. [DOI] [PubMed] [Google Scholar]
- [19].Maxhimer JB, Quiros RM, Stewart R, Dowlatshahi K, Gattuso P, Fan M, Prinz RA, Xu X. Heparanase-1 expression is associated with the metastatic potential of breast cancer. Surgery. 2002;132:326–333. doi: 10.1067/msy.2002.125719. [DOI] [PubMed] [Google Scholar]
- [20].Takaoka M, Naomoto Y, Ohkawa T, Uetsuka H, Shirakawa Y, Uno F, Fujiwara T, Gunduz M, Nagatsuka H, Nakajima M, Tanaka N, Haisa M. Heparanase expression correlates with invasion and poor prognosis in gastric cancers. Lab Invest. 2003;83:613–622. doi: 10.1097/01.lab.0000067482.84946.bd. [DOI] [PubMed] [Google Scholar]
- [21].Kelly T, Miao H-Q, Yang Y, Navarro E, Kussie P, Huang Y, MacLeod V, Casciano J, Joseph L, Zhan F, Zangari M, Barlogie B, Shaughnessy J, Sanderson RD. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63:8749–8756. [PubMed] [Google Scholar]
- [22].Katz A, Van-Dijk DJ, Aingorn H, Erman A, Davies M, Darmon D, Hurvitz H, Vlodavsky I. Involvement of human heparanase in the pathogenesis of diabetic nephropathy. Isr Med Assoc J. 2002;4:996–1002. [PubMed] [Google Scholar]
- [23].Levidiotis V, Freeman C, Tikellis C, Cooper ME, Power DA. Heparanase is involved in the pathogenesis of proteinuria as a result of glomerulonephritis. J Am Soc Nephrol. 2004;15:68–78. doi: 10.1097/01.asn.0000103229.25389.40. [DOI] [PubMed] [Google Scholar]
- [24].Vlodavsky I, Eldor A, Haimovitz-Friedman A, Matzner Y, Ishai-Michaeli R, Lider O, Naparstek Y, Cohen IR, Fuks Z. Expression of heparanase by platelets and circulating cells of the immune system: possible involvement in diapedesis and extravasation. Invasion Metastasis. 1992;12:112–12. [PubMed] [Google Scholar]
- [25].Maxhimer JB, Somenek M, Rao G, Pesce CE, Baldwin D, Jr., Gattuso P, Schwartz MM, Lewis EJ, Prinz RA, Xu X. Heparanase-1 gene expression and regulation by high glucose in renal epithelial cells: a potential role in the pathogenesis of proteinuria in diabetic patients. Diabetes. 2005;54:2172–2178. doi: 10.2337/diabetes.54.7.2172. [DOI] [PubMed] [Google Scholar]
- [26].Freeman C, Parish CR. A rapid quantitative assay for the detection of mammalian heparanase activity. Biochem J. 1997;325:229–237. doi: 10.1042/bj3250229. ( Pt 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vlodavsky I. Preparation of extracellular matrices produced by cultured corneal endothelial and PF-HR9 endodermal cells., in Protocols in Cell Biology. In: Bonifacino JS, J MD, Hartford B, Lippincott-Schwartz J, Yamada KM, editors. John Wiley & Sons; New York: 1999. pp. 10.14.11–10.14.14. [DOI] [PubMed] [Google Scholar]
- [28].Behzad F, Brenchley PE. A multiwell format assay for heparanase. Anal Biochem. 2003;320:207–213. doi: 10.1016/s0003-2697(03)00358-0. [DOI] [PubMed] [Google Scholar]
- [29].Huang KS, Holmgren J, Reik L, Lucas-McGady D, Roberts J, Liu CM, Levin W. High-throughput methods for measuring heparanase activity and screening potential antimetastatic and anti-inflammatory agents. Anal Biochem. 2004;333:389–398. doi: 10.1016/j.ab.2004.06.023. [DOI] [PubMed] [Google Scholar]
- [30].Nardella C, Lahm A, Pallaoro M, Brunetti M, Vannini A, Steinkuhler C. Mechanism of activation of human heparanase investigated by protein engineering. Biochemistry. 2004;43:1862–1873. doi: 10.1021/bi030203a. [DOI] [PubMed] [Google Scholar]
- [31].Tsuchida S, Podyma-Inoue KA, Yanagishita M. Ultrafiltration-based assay for heparanase activity. Anal Biochem. 2004;331:147–152. doi: 10.1016/j.ab.2004.04.033. [DOI] [PubMed] [Google Scholar]
- [32].Zetser A, Levy-Adam F, Kaplan V, Gingis-Velitski S, Bashenko Y, Schubert S, Flugelman MY, Vlodavsky I, Ilan N. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci. 2004;117:2249–2258. doi: 10.1242/jcs.01068. [DOI] [PubMed] [Google Scholar]
- [33].Gingis-Velitski S, Zetser A, Kaplan V, Ben-Zaken O, Cohen E, Levy-Adam F, Bashenko Y, Flugelman MY, Vlodavsky I, Ilan N. Heparanase uptake is mediated by cell membrane heparan sulfate proteoglycans. J. Biol. Chem. 2004;279:44084–44092. doi: 10.1074/jbc.M402131200. [DOI] [PubMed] [Google Scholar]
- [34].Levy-Adam F, Abboud-Jarrous G, Guerrini M, Beccoti D, Vlodavsky I, Ilan N. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J. Biol. Chem. 2005;280:20457–20466. doi: 10.1074/jbc.M414546200. [DOI] [PubMed] [Google Scholar]
- [35].Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J. Biol. Chem. 2004;279:23536–23541. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- [36].Levy-Adam F, Miao HQ, Heinrikson RL, Vlodavsky I, Ilan N. Heterodimer formation is essential for heparanase enzymatic activity. Biochem. Biophys. Res. Commun. 2003;2003:885–891. doi: 10.1016/s0006-291x(03)01478-5. [DOI] [PubMed] [Google Scholar]
- [37].Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- [38].Levy-Adam F, Miao HQ, Heinrikson RL, Vlodavsky I, Ilan N. Heterodimer formation is essential for heparanase enzymatic activity. Biochem Biophys Res Commun. 2003;308:885–891. doi: 10.1016/s0006-291x(03)01478-5. [DOI] [PubMed] [Google Scholar]
- [39].Fairbanks MB, Mildner AM, Leone JW, Cavey GS, Mathews WR, Drong RF, Slightom JL, Bienkowski MJ, Smith CW, Bannow CA, Heinrikson RL. Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. J Biol Chem. 1999;274:29587–29590. doi: 10.1074/jbc.274.42.29587. [DOI] [PubMed] [Google Scholar]
- [40].McKenzie E, Young K, Hircock M, Bennett J, Bhaman M, Felix R, Turner P, Stamps A, McMillan D, Saville G, Ng S, Mason S, Snell D, Schofield D, Gong H, Townsend R, Gallagher J, Page M, Parekh R, Stubberfield C. Biochemical characterisation of the active heterodimer form of Human Heparanase (Hpa1) protein expressed in insect cells. Biochem J. 2003;373:423–435. doi: 10.1042/BJ20030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zcharia E, Metzger S, Chajek-ShaulL T, Aingorn H, Elikn M, Friedmann Y, Weinstein T, Jin-Ping L, Lindahl U, Vlodavsky I. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. FASEB J. 2004;18:252–263. doi: 10.1096/fj.03-0572com. [DOI] [PubMed] [Google Scholar]
- [42].Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, Spector L, Pecker I. Mammalian heparanase: gene cloning. expression and function in tumor progression and metastasis, Nat Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- [43].Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- [44].Kussie PH, Hulmes JD, Ludwig DL, Patel S, Navarro EC, Seddon AP, Giorgio NA, Bohlen P. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun. 1999;261:183–187. doi: 10.1006/bbrc.1999.0962. [DOI] [PubMed] [Google Scholar]
- [45].Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274:24153–24160. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- [46].Nardella C, Steinkuhler C. Radiolabeled heparan sulfate immobilized on microplate as substrate for the detection of heparanase activity. Anal Biochem. 2004;332:368–375. doi: 10.1016/j.ab.2004.05.050. [DOI] [PubMed] [Google Scholar]