Abstract
The shoot apical meristem comprises an organized cluster of cells with a central region population of self-maintaining stem cells providing peripheral region cells that are recruited to form differentiated lateral organs. Leaves, the principal lateral organ of the shoot, develop as polar structures typically with distinct dorsoventrality. Interdependent interactions between the meristem and developing leaf provide essential cues that serve both to maintain the meristem and to pattern dorsoventrality in the initiating leaf. A key component of both processes are the class III HD–ZIP genes. Current findings are defining the developmental role of members of this family and are identifying multiple mechanisms controlling expression of these genes.
Introduction
A hallmark of land plant evolution has been development of the leaf. Leaves are the principal organ for capture of energy from sunlight and conversion, through photosynthesis, into organic components for growth. In angiosperms, leaves are typically planar, dorsoventrally flattened structures. Dorsoventrality is specified early in development of primordia. In the initiating leaf, the dorsal, or adaxial, side is immediately adjacent to the shoot apical meristem, whereas the ventral, or abaxial, side is farther from the shoot meristem (Figure 1A and 1B). In the mature leaf, the adaxial side is usually the upper sun-exposed side of the leaf and the abaxial side is the lower shaded side of the leaf.
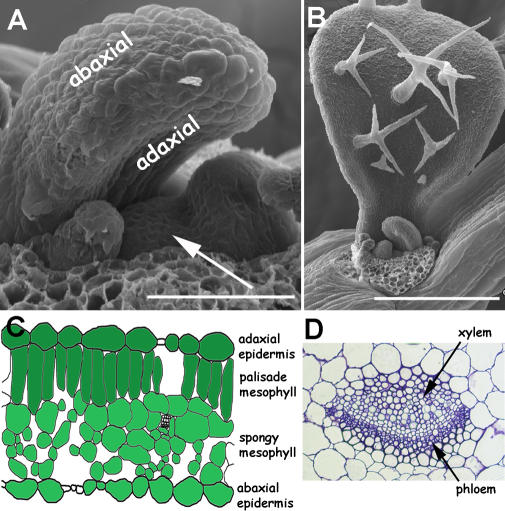
Figure 1. Leaves Arise on the Flanks of the Shoot Meristem.
(A) Vegetative apex of Arabidopsis. The adaxial side of the leaf is adjacent to the central shoot apical meristem, whereas the opposite, abaxial, side of the leaf is farther from the shoot meristem. Dorsoventrality is established early in development and is clearly evident in the leaf on the left, which arches over the meristem due to differential growth on each side of the leaf. Scale bar is 50 μm.
(B) Developing vegetative leaf of Arabidopsis. Adaxial trichomes on the larger leaf are one marker distinguishing dorsoventrality. Scale bar is 250 μm.
(C) Diagrammatic representation of a leaf cross section with adaxial and abaxial outer epidermal and inner mesophyll marked.
(D) Cross section of leaf midvein with adaxial xylem and abaxial phloem cells marked.
A series of surgical experiments carried out in the 1950s, and elaborated upon more recently, were the initial key to mechanisms that establish leaf dorsoventral patterning [1–3]. Separation of initiating primordia from the meristem by surgical incision generated a radial, abaxial leaf. This suggested, firstly, lateral organ patterning required an interaction between the initiating organ and the shoot apical meristem and, secondly, that in the absence of this interaction loss of dorsoventrality resulted in radial organs. Thus positional information in the context of the apical meristem specifies dorsoventral patterning and development of a leaf as a planar structure.
The first hint of a molecular basis for dorsoventral patterning came from the aptly named phantastica (phan) mutant in Antirrhinum [4,5]. Severely affected leaves in phan mutants are abaxial and fully radial, whereas weakly affected leaves have abaxial sectors on the adaxial leaf surface surrounded by ectopic lamina. The phenotypes of phan are entirely consistent with a requirement for dorsoventrality in lamina development. Subsequent to the work in Antirrhinum, there has been a relative explosion of information in the field, with identification and description of many additional genes and potential networks that combine to pattern adaxial and abaxial fate in the leaf [6–8]. To provide a basic primer to this field, this review will focus entirely on the role of one family of genes in leaf patterning and meristem function. These genes are the Class III HD–ZIP family. Members of this gene family are both necessary and sufficient for adaxial leaf fate and they potentially represent a pivotal component for leaf patterning–shoot meristem interactions. Class III HD–ZIP genes also provide a good example of the role of miRNAs in plant development.
Markers of Dorsoventrality
The extent to which adaxial and abaxial sides of a mature leaf can be distinguished varies between species; however, in developmental model species such as the dicot Arabidopsis or the monocot maize, many cell types differentiate top from bottom [9–11]. In Arabidopsis, epidermal cells on both sides of the leaf are jigsaw-shaped, but adaxial cells are larger and uniform in size relative to variable abaxial cell size. Trichome density is a useful marker on early juvenile leaves, where adaxial trichomes are much more frequent than abaxial trichomes. In subepidermal cell layers, closely aligned elongate palisade mesophyll cells lie juxtaposed to the adaxial epidermis. Less closely spaced, larger spongy mesophyll cells form abaxial internal tissue (Figure 1C). The palisade and spongy mesophyll tissues are optimized for light capture and gas exchange, respectively. Vasculature is also patterned in the dorsoventral dimension with xylem, the water-conducting tissue, adaxial to the organic nutrient–conducting phloem tissue (Figure 1D). Vascular bundles within the stem are also patterned with xylem more central to peripheral phloem. Conceptually, the dorsoventral vascular patterning of the leaf can be translated into a collateral central–peripheral pattern within the stem.
Class III HD–ZIP Genes—The Arabidopsis Family
Class III HD–ZIP transcription factors have in common a homeodomain DNA binding motif and a leucine zipper dimerization motif (HD-ZIP), and are a subset of a much larger group of plant proteins that also include a sterol/lipid binding (START) domain [12,13] (Figure 2A). Although lipid ligands for a small number of START domain proteins have been identified in animals, none to date have been found for plant START proteins. There are five Class III HD–ZIP genes in Arabidopsis, each encoding a protein in the range of 833–852 amino acids, and sharing between 60% to 85% amino acid homology (Figure 2B). PHABULOSA (PHB) and PHAVOLUTA (PHV) are most closely related to one another, sharing 85% amino acid identity [14]. Likewise, ATHB8 and ATHB15 form a relatively closely related pair, sharing 75% amino acid identity [15]. ATHB15 has also been published under the name CORONA (CNA) [16,17]. REVOLUTA (REV), in some previous work published as INTERFASICULAR FIBERLESS1 (IFL1), shares between 60% and 66% amino acid identity with other members of the group [18–20].
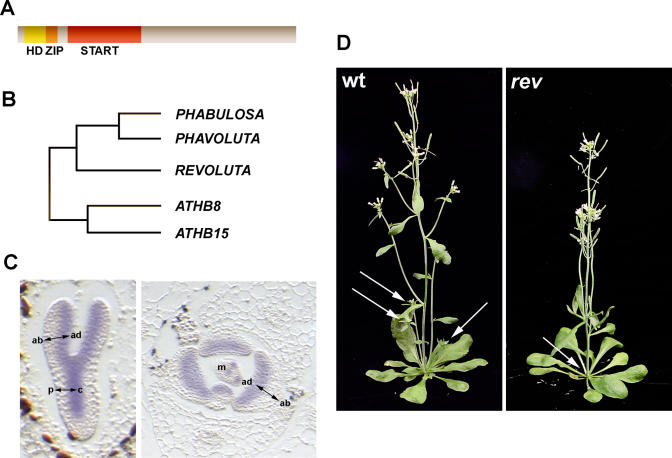
Figure 2. Class III HD–ZIP Genes in Arabidopsis .
(A) Class III HD–ZIP genes encode 833–852 amino acid proteins with main domains indicated; an N-terminal HD-ZIP domain, and a 213–218 amino acid START domain.
(B) Relationship between five Arabidopsis Class III HD–ZIP genes.
(C) Representation of expression pattern of PHB in longitudinal section of embryo (left) and transverse section of shoot apex (right). In the embryo, expression is adaxial in cotyledons and in central provasculature. In the shoot apex, expression is adaxial in developing leaves and in the meristem.
ad, adaxial; ab, abaxial; p, peripheral; c, central; m, meristem
(D) Phenotype of rev mutants. In wild-type, axillary meristems in axils of leaves give rise to lateral branches (arrows). In rev mutants, axillary meristems are frequently absent, and fewer or no lateral shoots are produced.
Patterned Expression
All five Class III HD–ZIP genes in Arabidopsis have well-defined tissue-specific expression patterns within the embryo and shoot. There are several principal reports describing tissue specific expression patterns, as determined by in situ hybridization, for REV [15,18,20], PHB [14], and PHV [15]. For comparative analysis, expression patterns in embryo and shoot for all five HD–ZIP genes have also been reported [17].
PHB and REV are expressed early in embryogenesis and appear throughout the 16-cell embryo. As development proceeds, expression becomes confined to the adaxial domain of the cotyledons and the central region of the embryo, including the shoot apical meristem and provasculature (Figure 2C). The expression of PHV and ATHB15 reiterate this pattern although expression initiates later and is more restricted than that of PHB and REV. The adaxial expression pattern is also in the developing organs of the shoot. However, in the shoot apical meristem PHB is expressed throughout very early stages of organ initiation as well as in regions that predict sites of successive organs. Discrete bands of expression extend from these presumptive organ sites to the very central region of the shoot apical meristem. The significance of this expression pattern is still to be determined. Potentially, expression of genes within these bands may contribute to coordinating central zone stem cell activity with peripheral zone lateral organ cell recruitment. In the shoot, REV is expressed adaxially in lateral organs, as well as in vasculature, and has a complex pattern in the shoot apical meristem, but, like PHB, predicts the site of organ initiation. ATHB15 has high levels of expression in the vasculature of the shoot and in axillary and floral meristems. In contrast to other members, expression of ATHB8 is more limited, being restricted to procambial cells of the embryo and vasculature of developing organs [17,21–23]. Some of the differences in expression patterns of these genes are consistent with differential contributions to development, as described below.
Loss-of-Function Effects
A visible gross plant phenotype for loss-of-function mutations in Class III HD–ZIP genes has only been described for REV, as redundancy seems to mask the role of other family members in development [18–20,24]. REV mutants have a diverse range of phenotypes. One of the most prominent defects is a failure in initiation and development of secondary meristems in the axils of vegetative rosette and cauline leaves, resulting in plants with few branches (Figure 2C). Flowers display variable phenotypes and may lack organs or form only rudimentary tapered filaments. In the inflorescence stem, there is a loss of xylem and interfasicular layers interconnecting vascular bundles. Despite this loss, there is no defect in vascular dorsoventral patterning.
Loss of PHB and PHV, either as single mutants or together in a double mutant, has no evident phenotypic consequences. However, mutations in both genes enhance rev defects, indicating PHB and PHV are redundant with each other and also with REV [15,17]. Depending on the background, loss of all three genes results in either partially abaxial cotyledons without a shoot apical meristem or a single abaxial cotyledon, or, in the extreme, failure to correctly pattern embryo development. Additional loss of ATHB15 in this background increases the frequency of the more severe phenotype. These patterning defects coincide with the expression pattern of these genes and are consistent with loss of both organ adaxial patterning and shoot central patterning within the embryo.
All HD–ZIP genes function in postembryonic development. Mutations in both PHB and PHV enhance shoot phenotypes of rev [17]. Like PHB and PHV, loss of either ATHB8 or ATHB15 or combined loss of both of these genes has no gross phenotypic effect, although vascular development is slightly perturbed in athb15 mutants. However, mutations in ATHB8 and ATHB15 suppress axillary and floral meristem defects of rev mutants. Together, genetic interactions suggest overlapping redundant as well as competitive interactions between these genes in development [17].
Dominant Effects
Mutations in PHB were first reported as temperature-sensitive, semidominant mutants with radial, adaxialized leaves and enlarged meristems [25]. Ectopic meristems surround the adaxial leaf as would be expected if adaxial fate promotes meristem formation or, alternatively, if PHB independently promotes meristem formation. Like PHB, gain-of-function mutations in REV, as in rev-10d and the allele amphivasal vascular bundle 1 (avb1), result in stem fasciation indicative of an increase in meristem size, adaxialized leaves, and conversion of normally collateral vasculature of the stem, where xylem is central to peripheral phloem to amphivasal vasculature with the xylem surrounding phloem [15,26]. All of these phenotypes are not always evident even with identical mutations and genetic background, suggesting that growth conditions may influence expressivity of the phenotype.
All dominant mutations in Class III HD–ZIP genes occur within a defined region around the 3′ end of the fourth exon and 5′ end of the fifth exon (Figure 3). An allelic series of phb-d mutants revealed either a point mutation at a splice site, resulting in a small percentage of transcripts with a short peptide sequence insertion, or point mutations resulting in an amino acid change [14]. Multiple dominant alleles of phv and rev also have single nucleotide changes within the same region as found in phb-d alleles [14,15,26]. However, as discussed below, changes in amino acid sequence in the dominant mutants do not appear to be significant as the site of all these mutations is within the predicted binding site for the small regulatory microRNAs, miR165 and miR166 [27,28].
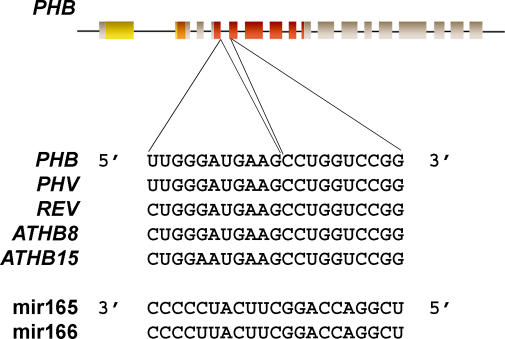
Figure 3. MicroRNAs and Class III HD–ZIP Target Sequences.
Diagrammatic representation of the PHB gene. Exons are represented as boxes and introns as lines. The three structural domains in the coding sequence are the HD-ZIP (yellow-orange) and the START (red) domains. Below them is the sequence of the microRNA binding site in the five Class III HD–ZIP genes and miR165 and miR166 sequences.
Regulation by microRNAs
MicroRNAs are approximately 21 nucleotides in length and are generated from longer precursor transcripts. The precursor transcripts notably form a hairpin loop structure that is recognized and cleaved into a double stranded form carrying the microRNA and a complementary sequence. Ultimately, the microRNA as a single strand is guided to the target transcript, a process involving the small RNA binding proteins of the ARGONAUTE family. Subsequently, target transcripts are either cleaved within the region binding the microRNA or are subject to translational inhibition. Much more detail on this pathway and the genes involved can be found in reviews on the subject [29–31].
The Arabidopsis genome encodes two copies of miR165 and seven copies of miR166, which have near perfect match with a sequence conserved within the transcript of all Arabidopsis Class III HD–ZIP genes [27] (Figure 3). So far, regulation of Class III HD–ZIP genes by microRNAs appears to occur by transcript cleavage, as is typical for many plant genes targeted by microRNAs. Several lines of evidence support this model [15,26,32,33]. Firstly, REV and PHB cDNA constructs that alter the microRNA binding site but not the corresponding amino acid sequence induce the same phenotypes as the gain-of-function mutations in these genes. Secondly, transcript cleavage products can be generated in in vitro assays and can be detected in plant extracts. Thirdly, in phb-1d mutants, the mutant PHB transcript is ectopically expressed and occurs throughout the leaf, whereas constitutive expression of wild-type PHB, using a 35S promoter, does not, in general, induce a phenotype. Although constitutive expression of the Class III HD–ZIP genes does not result in a phenotype, there are two interesting exceptions. Overexpression of PHB did result in adaxialization of early leaves in a small proportion of transformants [33] and overexpression of ATHB8 induces a phenotype with an increase in vascular xylem [34]. Occasional overexpression phenotypes may reflect dosage-dependent effects between microRNAs and target transcripts.
As might be expected, overexpression of microRNAs results in a reduction in Class III HD–ZIP gene transcripts. Two semidominant mutants, meristem enlarged1 (men1) and jabba-1D (jab-1D), isolated from activation tagged lines of Arabidopsis, have increased levels of miR166a and miR166g, respectively [35,36]. Although homozygous men1 are seedling lethal, men1/+ plants have a range of phenotypes including an enlarged shoot apical meristem and slight downcurling of leaves suggesting weak abaxialization of lateral organs [35]. Meristem, leaf patterning, and vascular defects also appear in the jab-1D mutant, and the severity of these phenotypes is stronger in homozygous compared with heterozygous plants [36]. In both men1/+ and jab-1D, overexpression of different miR166 genes has differential effects on expression of individual members of the Class III HD–ZIP genes. In jab-1D mutants, PHB, PHV, and ATHB15 are downregulated. However, REV is upregulated and this accounts for adaxialization of leaves but not meristem defects. Again, differential effects of miR165/166 on Class III HD–ZIP genes may be a consequence of dosage-dependent interactions between microRNA and target and the degree to which expression patterns of these two overlap. This in turn is potentially influenced by regulatory interactions between the different Class III HD–ZIP genes.
Regulation of Regulators
One gene involved in microRNA-mediated regulation of PHB is ARGONUATE1 (AGO1). AGO1 is a key component of RNA-mediated gene silencing. In plants, AGO1 binds microRNAs and is sufficient to mediate cleavage of target transcripts [37,38]. Mutations in ago1 disrupt many aspects of development, including organ dorsoventral patterning. In plants carrying strong ago1 mutant alleles, lateral organ development is severely affected, leading to a number of disparate interpretations of the phenotype [39–41]. It is possible that both adaxial and abaxial fates are affected by ago1 such that leaves in severely affected plants have lost polarity. However, in strong ago1 mutants PHB expression is expanded throughout the leaf and weak ago1 alleles have leaf development defects consistent with adaxialization of the leaf, indicating that one role of AGO1 is regulation of PHB via miRNAs [41].
Another component implicated in microRNA-mediated regulation of PHB is SERRATE (SE). Mutations in SE have a number of effects on shoot development including time to flowering, meristem size, leaf serrations, and dorsoventral patterning defects [42–45]. In severe mutants, leaves and leaf vasculature are adaxialized. se mutants have increased levels of expression of several Class III HD–ZIP genes. In the case of PHB, the domain of expression is expanded to the abaxial side of the leaf and the level of PHB is increased in the abaxial side of the leaf, similar to the pattern of misexpression in the dominant phb-1D mutants. The Class III HD–ZIP genes are likely secondarily affected by SE because the level of miR166 is greatly reduced in se mutants [45]. Concomitant with a reduction in miR166, the level of the precursor transcript for miR166 is increased in se mutants indicating that SE may function in processing miRNA precursor transcripts. SE encodes a protein with a domain sharing limited similarity to a zinc finger, although the precise function is still to be determined [44].
Dominant mutations in PHB and mutations in AGO1 and SE all result in PHB misexpression with increased levels of transcript in the adaxial domain and ectopic expression in the abaxial domain of young leaf primordia. Changes in adaxial and abaxial expression in these backgrounds suggest miRNAs regulating PHB expression are expressed throughout early leaf primordia. One report shows miR165/166 expression throughout the Arabidopsis shoot [46]. However two other reports, in Arabidopsis and in maize, indicate that these microRNAs are spatially restricted in the shoot apex and may function in patterning [41,47]. A further report examining expression in the embryo indicates dynamic expression throughout development, but in early embryogenesis miR166 is initially abaxial and at the distal tips of initiating cotyledons, an expression pattern closely complementary to that of the HD–ZIP genes [36]. Resolution of the degree to which miR165/166 and HD–ZIP target genes overlap may come from analysis of the expression pattern of sensor constructs where a cell-autonomous reporter gene carries the miRNA target binding site [48]. To add to this picture, Class III HD–ZIP tissue-specific expression appears to be directed simply by the promoter [21,22,49], in which case microRNAs may serve to modulate expression levels, particularly at boundaries where regulator and target expression overlap.
The LOB-domain gene ASYMMETRIC LEAVES2, which is required in leaf patterning, appears to negatively regulate miR165. Levels of miR165 are further increased in mutants lacking both ASYMMETRIC LEAVES2 and the RNA silencing pathway gene RDR6, an RNA-dependant RNA polymerase [46]. However, regulation of miR165 by ASYMMETRIC LEAVES2 and RDR6 is likely to be indirect as RDR6 acts in a pathway with three genes, AGO7, SGS3, and DCL4, known to regulate production of a specific class of small RNAs known as trans-acting siRNA [50–55]. Downstream targets of these trans-siRNA pathway genes are ETTIN and ARF3, two AUXIN RESPONSE FACTOR genes required for abaxial fate [50,56,57].
Linking to Chromatin
Aside from cleavage, microRNA targeting to the PHB transcript also influences the methylation status of the PHB locus [58]. High levels of methylation are usually associated with transcriptionally inactive chromatin, and, conversely, low methylation levels are typically associated with transcriptionally active chromatin. In wild-type plants the PHB locus is methylated at the 3′ end of the gene, although methylation levels are low in meristem-enriched tissues where PHB is expressed. The dominant allele, which may no longer effectively bind miR165/166, fails to mediate methylation of the PHB gene. Thus miR165/166 may function, directly or indirectly, in transcriptional as well as posttranscriptional regulation of PHB. The PHV gene is also methylated in the 3′ region indicating Class III HD–ZIP genes as a whole may be subject to multiple levels of regulation. The significance of this regulatory system is yet to be established, but multiple levels of regulation may serve several purposes. Conceivably, microRNA-directed cleavage of transcripts acts as an efficient mechanism for rapid inactivation of transcripts within a cell. Simultaneous or subsequent methylation of the locus would then maintain a stable repressed state in cells where gene expression is no longer required.
Conclusions
The Class III HD–ZIP genes play multiple, possibly interdependent, roles in plant development. Conservation of these genes and expression patterns throughout land plants, in particular in lower land plant species, highlight a critical role in development of the basic plant body [59]. Spatial, temporal, and quantitative regulation of expression appears to involve a number of mechanisms including posttranscriptional and transcriptional gene silencing mediated by microRNAs. The importance of microRNAs as regulators of this gene family is reflected in conservation of miR166 and conservation of Class III HD–ZIP gene function in divergent plant species [27,47,60,61]. An additional layer of regulation may involve modulation of function via a sterol-type ligand. Evaluation of the contribution and interplay of these regulatory mechanisms and the degree to which components of regulation are conserved are clearly going to be subjects of much future research.
Supporting Information
Accession Numbers
Accession numbers from the Genbank genebank (http://www.ncbi.nlm.nih.gov/Genbank) are for: PHABULOSA (PHB), 2G34710; PHAVOLUTA (PHV), 1G30490; ATHB8, 4G32880; ATHB15, 1G52150; and REVOLUTA (REV), 5G60690.
Acknowledgments
I thank Catherine Kidner and Robert Martienssen for critical reading of the manuscript and Peter Etchells for the figure of vasculature. My apologies to colleagues whose research I failed to adequately cite due to space limitations, in particular to those whose work moves beyond the bounds of Arabidopsis.
Abbreviations
- HD–ZIP
homeodomain–leucine zipper
- phan
phantastica
- se and SE
SERRATE
- START
sterol/lipid binding domain
Footnotes
Competing interests. The author has declared that no competing interests exist.
Funding. MEB is a recipient of a Royal Society Wolfson Merit Award, and her laboratory is funded by the Biotechnology and Biological Sciences Research Council, United Kingdom.
References
- Sussex IM. Morphogenesis in Solanum tuberosum L.: Experimental investigation of leaf dorsiventrality and orientation in the juvenile shoot. Phytomorph. 1955;5:286–300. [Google Scholar]
- Snow M, Snow R. The dorsiventrality of leaf primordia. New Phytol. 1959;58:188–207. [Google Scholar]
- Reinhardt D, Frenz M, Mandel T, Kuhlemeier C. Microsurgical and laser ablation analysis of leaf positioning and dorsoventral patterning in tomato. Development. 2005;132:15–26. doi: 10.1242/dev.01544. [DOI] [PubMed] [Google Scholar]
- Waites R, Hudson A. phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus . Development. 1995;121:2143–2154. [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum . Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- Engstrom EM, Izhaki A, Bowman JL. Promoter bashing, microRNAs, and Knox genes. New insights, regulators, and targets-of-regulation in the establishment of lateral organ polarity in Arabidopsis. Plant Physiol. 2004;135:685–694. doi: 10.1104/pp.104.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME. Networks in leaf development. Curr Opin Plant Biol. 2005;8:59–66. doi: 10.1016/j.pbi.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Kepinski S. Integrating hormone signaling and patterning mechanisms in plant development. Curr Opin Plant Biol. 2005;9:28–34. doi: 10.1016/j.pbi.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants. New York: John Wiley and Sons; 1977. 550. p. [Google Scholar]
- Freeling M. A conceptual framework for maize leaf development. Dev Biol. 1992;153:44–58. doi: 10.1016/0012-1606(92)90090-4. [DOI] [PubMed] [Google Scholar]
- Tefler A, Poethig RS. Leaf development in Arabidopsis . In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. New York: Cold Spring Harbor Press; 1994. pp. 379–401. [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD–ZIP proteins. Plant Mol Biol. 1998;38:609–622. doi: 10.1023/a:1006016319613. [DOI] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, et al. Radial patterning of Arabidopsis shoots by class III HD–ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Green KA, Prigge MJ, Katzman RB, Clark SE. CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell. 2005;17:691–704. doi: 10.1105/tpc.104.026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, et al. Class III homeodomain–leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. IFL1, a gene-regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain–leucine zipper protein. Plant Cell. 1999;11:2139–2152. doi: 10.1105/tpc.11.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Riechmann JL, Zhang JZ. INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA . Plant Cell. 2000;12:315–317. doi: 10.1105/tpc.12.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, et al. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana . Development. 1995;121:4171–4182. doi: 10.1242/dev.121.12.4171. [DOI] [PubMed] [Google Scholar]
- Kang J, Dengler N. Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis . Planta. 2002;216:212–219. doi: 10.1007/s00425-002-0847-9. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H. HD–zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 2003;44:1350–1358. doi: 10.1093/pcp/pcg164. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana . Development. 1995;121:2723–2735. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis . Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 2004;45:369–385. doi: 10.1093/pcp/pch051. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Curr Opin Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones–Rhoades MW, Tang G, Zamore PD, et al. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, et al. The Arabidopsis ATHB-8 HD–zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001;126:643–655. doi: 10.1104/pp.126.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, et al. microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 2005;42:84–94. doi: 10.1111/j.1365-313X.2005.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD–ZIP target genes. Development. 2005;132:3657–3668. doi: 10.1242/dev.01942. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, et al. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, et al. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1 . Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Clarke JH, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis . Plant J. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis . Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- Li H, Xu L, Wang H, Yuan Z, Cao X, et al. The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell. 2005;17:2157–2171. doi: 10.1105/tpc.105.033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker NP, Bowman JL. Roles for Class III HD–Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 2004;135:2261–2270. doi: 10.1104/pp.104.040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis . Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci U S A. 2005;102:9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana . Proc Natl Acad Sci U S A. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis . Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Current Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD–ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL. Evolution of Class III Homeodomain-leucine zipper genes in streptophytes. Genetics 173. 2006. In press. [DOI] [PMC free article] [PubMed]
- Floyd SK, Bowman JL. Gene regulation: Ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- McHale NA, Koning RE. MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell. 2004;16:1730–1740. doi: 10.1105/tpc.021816. [DOI] [PMC free article] [PubMed] [Google Scholar]