Abstract
An evolutionarily ancient mechanism is used for intracellular membrane fusion events ranging from endoplasmic reticulum–Golgi traffic in yeast to synaptic vesicle exocytosis in the human brain. At the heart of this mechanism is the core complex of N-ethylmaleimide-sensitive fusion protein (NSF), soluble NSF attachment proteins (SNAPs), and SNAP receptors (SNAREs). Although these proteins are accepted as key players in vesicular traffic, their molecular mechanisms of action remain unclear. To illuminate important structure–function relationships in NSF, a screen for dominant negative mutants of yeast NSF (Sec18p) was undertaken. This involved random mutagenesis of a GAL1-regulated SEC18 yeast expression plasmid. Several dominant negative alleles were identified on the basis of galactose-inducible growth arrest, of which one, sec18-109, was characterized in detail. The sec18-109 phenotype (abnormal membrane trafficking through the biosynthetic pathway, accumulation of a membranous tubular network, growth suppression, increased cell density) is due to a single A-G substitution in SEC18 resulting in a missense mutation in Sec18p (Thr394→Pro). Thr394 is conserved in most AAA proteins and indeed forms part of the minimal AAA consensus sequence that serves as a signature of this large protein family. Analysis of recombinant Sec18-109p indicates that the mutation does not prevent hexamerization or interaction with yeast α-SNAP (Sec17p), but instead results in undetectable ATPase activity that cannot be stimulated by Sec17p. This suggests a role for the AAA protein consensus sequence in regulating ATP hydrolysis. Furthermore, this approach of screening for dominant negative mutants in yeast can be applied to other conserved proteins so as to highlight important functional domains in their mammalian counterparts.
INTRODUCTION
A major recent development in cell biology is the realization that fundamental cellular processes are controlled by similar mechanisms in all eukaryotes. An excellent example of this is intracellular membrane traffic. Yeast genetics (Pryer et al., 1992), in vitro biochemistry (Rothman, 1994), and molecular cloning (Sudhof, 1995) have converged to indicate that vesicle formation and consumption are governed by the same ubiquitous protein machinery (Ferro-Novick and Jahn, 1994). An early clue that this might be the case was provided when the gene encoding N-ethylmaleimide-sensitive fusion protein (NSF), the first protein identified as being required for reconstituted mammalian intracellular membrane fusion (Block et al., 1988), was cloned. It was observed (Wilson et al., 1989) that NSF displayed both sequence and functional homology to Sec18p, a protein required for endoplasmic reticulum (ER) to Golgi transport in yeast (Eakle et al., 1988). NSF and Sec18p are ATPases required for interorganelle transport at multiple stages of the biosynthetic and endocytic pathways (Graham and Emr, 1991; Rothman, 1994). Furthermore, NSF and Sec18p form a 20 S complex with soluble NSF attachment proteins (SNAPs; Sec17p in yeast) and SNAP receptors (SNAREs), and abundant evidence suggests that these proteins are also required for most intracellular membrane trafficking events (Woodman, 1997). Taken together, this strongly suggests that NSF and Sec18p are orthologues that function at the heart of a universal mechanism of membrane fusion.
Although the importance of NSF is unquestioned, the precise role of this key protein in the membrane fusion process has been intensely debated (Morgan and Burgoyne, 1995; Woodman, 1997). Suggestions for its function range from a chaperone (Morgan and Burgoyne, 1995) that acts either at an early predocking priming stage to enable subsequent docking/fusion (Mayer et al., 1996) or at a postfusion stage to recycle used SNARE complexes (Littleton et al., 1998; Weber et al., 1998), through to a membrane fusogen (Otter-Nilsson et al., 1999). For many proteins, mutational studies, in particular the generation and analysis of dominant-acting mutants, have been invaluable in gaining insight into their cellular and molecular functions (Herskowitz, 1987; Feig, 1999). However, because of its large size and essential multidomain structure (Whiteheart et al., 1994), NSF is not well suited to structure–function studies using conventional deletion/alanine-scanning mutagenesis. Indeed, the only published mutagenic analyses of NSF/Sec18p have targeted residues in the Walker A and B boxes known to be required for ATP binding or hydrolysis (Sumida et al., 1994; Whiteheart et al., 1994; Nagiec et al., 1995; Colombo et al., 1996; Matveeva et al., 1997; Steel and Morgan, 1998; Steel et al., 1999). Although these studies have emphasized the critical requirement for D1 domain ATPase activity, the function of regions beyond the Walker boxes has not been examined.
To redress this situation and shed light on important functional amino acids in NSF, we took advantage of its orthology with Sec18p. We reasoned that dominant negative mutants of SEC18 (i.e., genes encoding mutant polypeptides that disrupt the function of the wild-type gene product in the cell [Herskowitz, 1987]) might represent generally useful tools. We therefore used random mutagenic approaches using the Saccharomyces cerevisiae SEC18 gene and screened for dominant negative mutations active in vivo. Because Sec18p and NSF are members of the ATPases associated with a variety of cellular activities (AAA) protein family (Confalonieri and Duguet, 1995), any mutants that were isolated could potentially also provide information on this large, evolutionarily conserved family. Here we describe the strategies used and the isolation and characterization of one such dominant inhibitory mutant: sec18-109.
MATERIALS AND METHODS
Materials
Affinity-purified anti-Sec18p antiserum was a generous gift from Dr. A. Haas (Lehrstuhl fur Mikrobiologie, University of Wurzburg, Germany). Monoclonal anticarboxypeptidase Y antibody was a generous gift from Dr. T. Stevens (University of Oregon, Eugene, OR). The pSEY8 plasmid was a generous gift from Dr. S. Emr (University of California, San Diego, CA). The sec18-1 yeast strains, HMSF176 and RSY271, were generous gifts from Dr. R. Schekman (University of California, Berkeley, CA). INVSc1 cells, pYES2 vectors, and yeast transformation kits were purchased from Invitrogen (Groningen, The Netherlands); pQE vectors and nitrilotriacetic acid agarose were purchased from Qiagen (Dorking, UK). Unless specified otherwise, all other reagents were of analytical grade and obtained from Sigma (Poole, UK).
Yeast Strains
JRY188 (MATα; leu2-3, 112; ura3-52; trp1; his4; sir3; rme) was used for generation and analysis of dominant negative SEC18 alleles. This strain was crossed with HMSF176 to create the temperature-sensitive sec18-1 strain, CHY01 (MATα; leu2-3, 112; trp1; his4; sec18-1), used to score mutagenic efficiency and for morphological analysis. The pep4 strain, BJ5464 (MATα; ura3-52; leu2Δ1; trp1; prb1Δ1.6R; can1; his3Δ200; pep4::HIS3), was used to calibrate carboxypeptidase Y (CPY) processing. The diploid INVSc1 strain (leu2; ura3-52; trp1-289; his3Δ1) was used for expression of epitope-tagged SEC18 constructs. RSY271 (MATa; ura3-52; his4; sec18-1) was used for assessment of dominant lethality using pSEY8-derived plasmids.
Plasmids
A 685-bp EcoRI/BamHI fragment containing the GAL1/GAL10 divergent promoter was excised from pBM150 and ligated into the corresponding sites in YCplac22 (Gietz and Sugino, 1988) to produce the centromeric, inducible yeast expression vector, YCpGAL. The wild-type SEC18 gene was then excised from pSEY8 (Eakle et al., 1988) as a 3-kbp BamHI/HindIII fragment and ligated into the same sites in YCpGAL to create YCpGAL–SEC18. The pYES2 plasmid carrying His6–V5 epitope-tagged SEC18 was obtained from Invitrogen. The bacterial expression plasmids encoding His6-tagged Sec18p and Sec17p have been described previously (Steel et al., 1999).
Mutagenesis
Random mutagenesis using PCR
PCR mutagenesis uses suboptimal cation and nucleotide conditions for Taq polymerase, thus decreasing the fidelity of replication (Muhlrad et al., 1992). For this strategy, the oligonucleotides 5′-GCTCACTCATTAGGCACC-3′ and 5′-CCGCACAGATGCGTAA-3′ were used to anneal to YCpGAL–SEC18 and produce a PCR product of ∼4.1 kbp containing the GAL–SEC18 cassette. After first optimizing the MgCl2 concentration, a ratio of either 8:1 or 12:1 of Mg2+ to Mn2+ ions, respectively, was used in the PCR reaction to induce mutations. The YCpGAL vector was then digested with BamHI and HindIII and cotransformed along with the PCR products into JRY188 cells. This resulted in the insertion of the PCR products into the gapped plasmid by homologous recombination in vivo and allowed direct screening for mutants within the yeast strain.
Random mutagenesis using mutD Escherichia coli
The rifampicin-resistant mutD strain of E. coli contains a mutant ε subunit of DNA polymerase III, resulting in a deficient DNA proofreading ability (Echols et al., 1983). This strain was transformed with YCpGAL–SEC18, and the resulting pool of mutagenized plasmids was recovered using standard DNA procedures (Sambrook et al., 1989) and used to transform JRY188 yeast cells.
Site-directed mutagenesis
For epitope-tagging studies in yeast, the sec18-109 mutation was introduced into pYES2–SEC18 using the “Quickchange”' site directed mutagenesis kit (Stratagene, La Jolla, CA). The mutagenic primers that were used were as follows: sense 5′-GGTTATTGGTATGCCCAATCGTAAAGATCTAATAGACAGTGC-3′, antisense 5′-GCACTGTCTATTAGATCTTTACGATTGGGCATACCAATAACC-3′. For assessment of dominant lethality using the pSEY8 plasmid (hereafter referred to as pSEY8–SEC18), the same approach and primers were used to create pSEY8–sec18-109, whereas pSEY8–sec18E350Q was generated using the following primers: sense 5′-CATATTATTATTTTCGATCAGCTGGATTCTG-3′, antisense 5′-CAGAATCCAGCTGATCGAA-AATAATAATATG-3′. For bacterial expression of recombinant His6-tagged Sec18-109p, the coding sequence was PCR amplified from YCpGAL–sec18-109 and ligated into the BamHI and HindIII sites of pQE-30 (Qiagen). All constructs were checked by either manual or automated DNA sequencing (Oswel, Southampton, UK).
Isolation of Dominant Negative Mutants
JRY188 cells transformed directly by the PCR mutagenesis approach, or indirectly with a mutD-mutagenized YCpGAL–SEC18 preparation, were plated onto minimal media supplemented with 3% glucose at 25°C. These were then replica-plated onto media containing either glucose or galactose (3%). Plasmids were recovered from five colonies exhibiting galactose-sensitive growth, and the mutant SEC18 genes were subcloned as BamHI/HindIII fragments back into the YCpGAL vector, before the dominant negative phenotype was rechecked by transforming fresh JRY188 cells. Mutations in the confirmed dominant negative alleles were identified by manual sequencing.
Density Shift Analysis
JRY188 yeast cells transformed with either YCpGAL–SEC18 or YCpGAL–sec18-109 were inoculated into 10 ml minimal media supplemented with raffinose (3%) and grown overnight at 25°C until the OD600 reached 0.2. The cultures were then split into two 5-ml cultures, and galactose was added to one culture to a final concentration of 3%. These cultures were grown for a further 5 h at 25°C, at which point the cells were pelleted by centrifugation, resuspended in 500 μl of 10% Percoll in TE buffer (10 mM Tris–HCl, pH 7.4, 1 mM EDTA) and fractionated on a 20–100% Percoll/TE buffer continuous gradient (10 ml). The gradients were centrifuged at 2000 × g for 10 min at room temperature, and the OD600 of 0.5-ml fractions was measured.
Analysis of CPY Processing
Untransformed JRY188 and BJ5464 cells and JRY188 cells transformed with either YCpGAL–SEC18 or YCpGAL–sec18-109 were inoculated into 50 ml minimal media supplemented with glucose (2%) and grown overnight at 30°C until the OD600 reached 0.5. Cells were harvested, washed with water, and resuspended in minimal media supplemented with 2% galactose and incubated further. Cells were then spheroplasted by incubation in lyticase solution (500 U/ml lyticase in 1 M sorbitol, 14 mM β-mercaptoethanol, 0.1 M EDTA, pH 8), resuspended in Laemmli buffer, and boiled before they were loaded onto 10% polyacrylamide gels and transferred onto nitrocellulose. CPY was visualized after immunoblotting using a monoclonal CPY antibody and enhanced chemiluminescence (ECL) detection. The position of p2 CPY was established by comparing the mobility of a pep4 strain, BJ5464, on the same gel (pep4 strains are deficient in proteinase B and therefore cannot cleave the p2 form into mature CPY).
β-Lactamase Secretion Assay
JRY188 cells were cotransformed with pYJS50, a plasmid carrying a prepro-α-factor/β-lactamase fusion gene under the control of the α-factor promoter, and either YCpGAL–SEC18 or YCpGAL–sec18-109. Cotransformants were inoculated into 200 ml minimal media supplemented with glycerol/ethanol (3%) and grown at 25°C until the OD600 reached ∼0.2. After 2 h of growth at 25°C, galactose was added to 3%, and the cultures were grown further. Duplicate 5-ml samples were removed at intervals, and low-speed supernatants were prepared from glass bead homogenates. β-Lactamase activity was assayed spectrophotometrically by adding 50 μl of sample to 950 μl nitrocefin solution (0.5 mg/ml nitrocefin in 100 mM sodium phosphate, pH 7) and monitoring the change in absorbance at 490 nm. Triton X-100–releasable β-lactamase activity was assayed as a measure of intracellular protein accumulation and expressed as a percentage of the total β-lactamase activity present in the samples.
Electron Microscopy
JRY188 cells transformed with either YCpGAL–SEC18 or YCpGAL–sec18-109 were inoculated into 200 ml minimal media supplemented with glycerol/ethanol (3%) and grown overnight at 25°C until the OD600 reached 0.2. Galactose was then added to 3%, and the cultures were grown further. For comparison, the temperature-sensitive sec18-1 strain, CHY01, was grown similarly overnight and then shifted to the restrictive temperature (37°C). Samples of 50 ml were immediately fixed in solution by the addition of 1:10 volume of 10× prefixative solution (10% glutaraldehyde, 2% methanol-free formaldehyde, 0.4 M potassium phosphate, pH 7). After 5 min, the samples were pelleted and processed before embedding in LR white. Sections cut for electron microscopy were stained sequentially with 2% uranyl acetate for 1–5 min and then with Reynolds lead citrate for 30 s.
Molecular Mass Estimation of Epitope-tagged Sec18p
INVSc1 yeast cells were transformed with pYES2–SEC18 or pYES2–sec18-109. These plasmids encode forms of Sec18p tagged with His6 and the V5 epitope at the C terminus. Transformants were inoculated into 200 ml minimal media supplemented with glucose (2%) and grown overnight at 25°C until the OD600 reached 0.6. Cells were harvested, washed with water, and resuspended in minimal media supplemented with 2% galactose and incubated further. Cells were then spheroplasted and lysed using a French press. Soluble proteins were isolated by centrifugation at 100,000 × g and separated on the basis of native molecular mass by gel filtration chromatography on a Superdex 200 column (Pharmacia, Uppsala, Sweden; 16:60, 120 ml bed volume) in buffer A (20 mM HEPES, pH 7.0, 200 mM KCl, 2 mM 2-mercaptoethanol, 0.5 mM ATP, 10% [wt/vol] glycerol, 50 mM imidazole) collecting 2-ml fractions. Column fractions were run on 10% polyacrylamide gels, transferred onto nitrocellulose, and immunoblotted for the V5 epitope using ECL to specifically detect recombinant forms of Sec18p.
Purification of Recombinant Proteins
Recombinant His6-tagged proteins were extracted using a French press from XL-1Blue E. coli resuspended in breaking buffer and purified using Ni-Nitrilotriacetic acid agarose as previously described (Steel et al., 1999). Further purification of His6–Sec18p and His6–Sec18-109p was achieved by gel filtration chromatography on the Superdex 200 column in buffer A (Steel et al., 1999). All chromatography was performed at room temperature using a Pharmacia FPLC system.
Sec18p Binding Assay
This was performed in polypropylene tubes as described previously (Steel et al., 1999). Bound samples were run on 10% polyacrylamide gels, and proteins were detected by immunoblotting followed by ECL detection.
Sec18p ATPase Assay
ATPase assays were performed in flat-bottomed 96-well microtiter plates as described previously (Steel et al., 1999).
RESULTS
SEC18 is an essential gene in S. cerevisiae, and so to identify dominant loss-of-function mutants it was necessary to achieve conditional expression of mutant Sec18p proteins. This was accomplished by placing the wild-type SEC18 gene downstream of the galactose-inducible GAL1 promoter in a low copy number centromeric plasmid (YCpGAL–SEC18). Two random mutagenic strategies were then used: in vitro PCR mutagenesis and in vivo mutagenesis using a mutD strain of E. coli. Mutagenic efficiency was estimated by assaying complementation of the temperature-sensitive phenotype of a sec18-1 yeast strain grown on galactose. It was found that the proportion of temperature-sensitive, i.e., mutant, colonies was higher using PCR mutagenesis (20–45%, depending on PCR conditions) compared with the mutD approach (∼5%). To reveal potential dominant mutations, the same pool of mutagenized DNA was transformed into the wild-type JRY188 yeast strain, which contains a normal chromosomal copy of the SEC18 gene. Of 3000 transformed colonies screened, 5 (2 from the mutD strategy, 3 from mutagenic PCR) exhibited the desired inducible dominant negative phenotype, i.e., normal growth on glucose-containing media and no growth on galactose-containing media. These five dominant loss-of-function mutant alleles were named sec18-108, -109, -110, -111, and -112. Of these, the only allele characterized extensively thus far is the sec18-109 mutant, which was produced using the mutD approach.
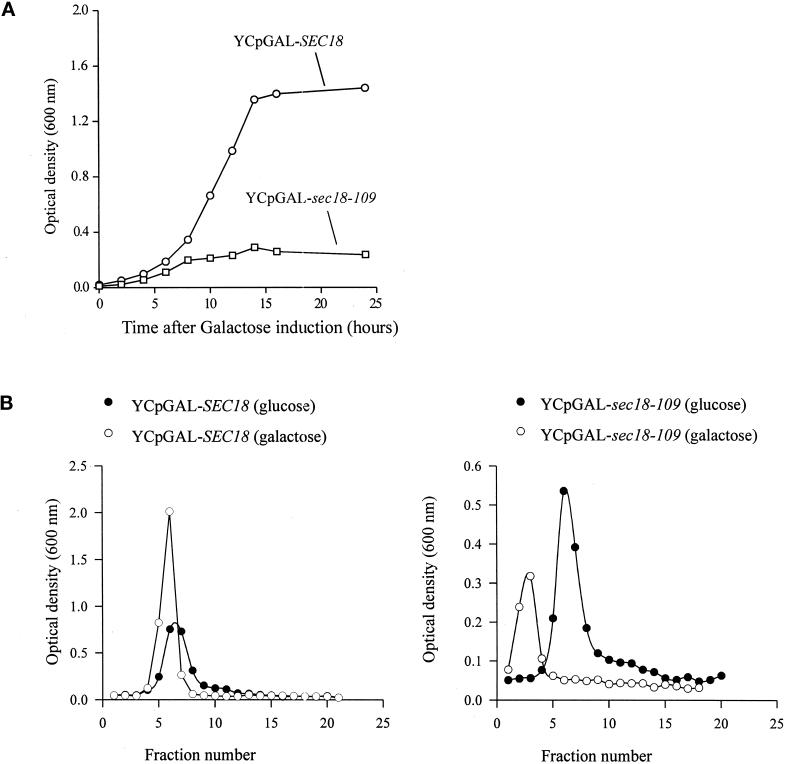
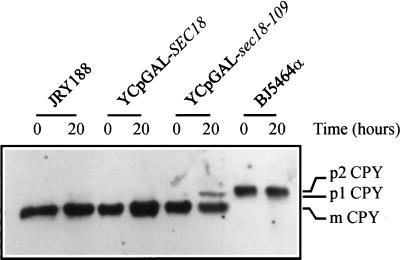
The galactose-inducible growth arrest exhibited by the dominant negative mutants that were isolated is typified by sec18-109. It can be seen that galactose-induced expression of wild-type Sec18p in JRY188 cells allowed normal exponential growth but that this was prevented by expression of Sec18-109p (Figure 1A). The pioneering work of Novick et al. (1980) established that yeast secretion (sec) mutants, including the sec18-1 mutant, undergo a density increase when shifted to the restrictive temperature caused by the accumulation of intracellular organelles. Similarly, we observed that wild-type yeast transformed with YCpGAL–sec18-109 exhibited a marked galactose-induced density increase, whereas those transformed with YCpGAL–SEC18 showed no such change (Figure 1B). To determine whether this density increase was the result of a block in membrane traffic, two independent assays of transport through the biosynthetic pathway were performed. Transport through the exocytotic pathway was monitored by assaying intracellular β-lactamase activity after transformation with pYJS50. This plasmid encodes a β-lactamase–prepro-α-factor fusion protein that is correctly processed and secreted via the exocytotic pathway (A. Boyd, unpublished data). Cotransformation with YCpGAL–sec18-109 caused a time-dependent accumulation of intracellular β-lactamase activity, whereas cotransformation with YCpGAL–SEC18 was without effect (Table 1), indicating a block at one or more stages of the exocytotic pathway by Sec18–109p. Transport of CPY to the vacuole was determined by following its processing from the core glycosylated p1 form through the mannosylated p2 form to the proteolyzed mature m form (Graham and Emr, 1991). In untransformed cells and cells transformed with YCpGAL–SEC18, only the mature form of CPY was accumulated internally (Figure 2), consistent with the normal trafficking and processing of CPY en route to the vacuole. In contrast, transformation with YCpGAL–sec18-109 caused internal accumulation of p1 CPY (Figure 2), indicating that this dominant negative mutant interferes with Sec18p-dependent ER–Golgi membrane traffic.
Figure 1.
YCpGAL–sec18-109 causes galactose-induced growth arrest and increased cell density. (A) JRY188 yeast cells transformed with either YCpGAL–SEC18 or YCpGAL–sec18-109 were inoculated into minimal media supplemented with glycerol/ethanol (3%) and grown overnight. Galactose was then added to 3%, and the cultures were grown for a further 24 h. At the time points indicated, samples were removed, and the OD600 was measured. (B) JRY188 yeast cells transformed with either YCpGAL–SEC18 or YCpGAL–sec18-109 were inoculated into minimal media supplemented with raffinose (3%) and grown overnight. The cultures were then split into two 5-ml cultures, control (●) and induced (galactose was added to a final concentration of 3%, ○). These cultures were grown for a further 5 h, at which point the cells were pelleted, resuspended, and fractionated on a 20–100% Percoll continuous gradient before the OD600 of 0.5-ml fractions was measured.
Table 1.
YCpGAL–sec18-109 causes intracellular accumulation of a secreted protein
| Time after galactose addition (h) | % Intracellular β-lactamase activity
|
|
|---|---|---|
| YCpGAL–SEC18 | YCpGAL–sec18-109 | |
| 2 | 3.077 | 5.704 |
| 4 | 2.288 | 8.520 |
| 6 | 2.086 | 12.810 |
| 8 | 2.642 | 12.940 |
| 10 | 2.53 | 18.98 |
JRY188 yeast cells were cotransformed with pYJS50, a plasmid carrying a prepro-α-factor/β-lactamase fusion gene under the control of the α-factor promoter, and either YCpGAL–SEC18 or YCpGAL–sec18-109. Cotransformants were inoculated into minimal media supplemented with glycerol/ethanol (3%) and grown until the OD600 reached ∼0.2. After 2 h growth, galactose was added to 3%, and the cultures were grown for a further 10 h. Duplicate 5-ml samples were removed from these cultures at 2-h intervals, and low speed supernatants were prepared from glass bead homogenates. Triton X-100–releasable β-lactamase activity was assayed as a measure of intracellular protein accumulation and expressed as a percentage of the total β-lactamase activity present in the samples.
Figure 2.
YCpGAL–sec18-109 causes aberrant processing of CPY. Untransformed JRY188 and BJ5464 cells and JRY188 cells transformed with either YCpGAL–SEC18 or YCpGAL–sec18-109 were inoculated into minimal media supplemented with glucose (2%) and grown overnight. Cells were harvested, washed with water, and resuspended in minimal media supplemented with 2% galactose and incubated overnight. Samples were removed from these cultures immediately after galactose addition (0 h) and after overnight incubation (20 h), spheroplasted, and loaded onto 10% polyacrylamide gels before transfer onto nitrocellulose. Transformed and untransformed JRY188 cells were loaded at 2 μg protein per track, whereas BJ5464 cells were loaded at 10 μg per track. CPY was visualized after immunoblotting and ECL detection. The position of p2 CPY is defined by its mobility in the pep4 strain, BJ5464, which selectively accumulates the p2 form. (p1, p2, and mCPY represent core-glycosylated, mannose-linked, and mature CPY, respectively).
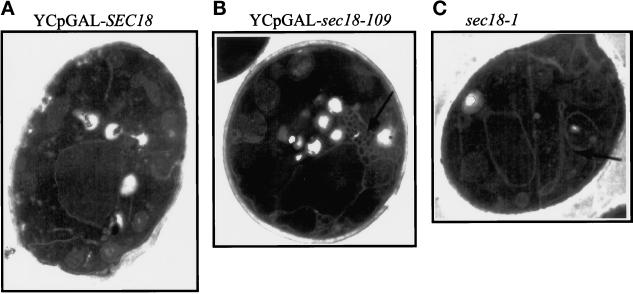
Although all yeast sec mutants exhibit a characteristic temperature-induced block in secretion, electron microscopy analysis has revealed differential organelle accumulation by individual sec strains (Novick et al., 1980). The sec18-1 strain exhibits accumulation of ER membranes and small vesicles (Novick et al., 1980), consistent with the requirement for Sec18p in multiple stages of membrane traffic through the biosynthetic pathway (Graham and Emr, 1991). To study the effect of Sec18-109p expression on cell morphology, electron microscopy was performed on wild-type JRY188 yeast transformed with either YCpGAL–sec18-109 or YCpGAL–SEC18. Galactose induction was performed after growing the cells to early log phase in glycerol/ethanol-supplemented minimal medium. It can be seen that expression of Sec18–109p caused the accumulation of a fenestrated membranous structure (Figure 3B). Exaggerated membranous tubules were also seen in the sec18-1 strain at the restrictive temperature (Figure 3C) (Novick et al., 1980), and so presumably they represent ER-derived membrane. In contrast, expression of wild-type Sec18p did not cause the formation of such tubular networks, indicating that the membrane defects observed were solely due to the mutation in sec18-109.
Figure 3.
YCpGAL–sec18-109 induces the formation of membranous tubules. JRY188 yeast cells transformed with either YCpGAL–SEC18 (A) or YCpGAL–sec18-109 (B) were inoculated into minimal media supplemented with glycerol/ethanol (3%) and grown overnight. Galactose was then added to 3%, and the cultures were grown for a further 5 h. For comparison, a temperature-sensitive sec18-1 strain (C) was grown similarly overnight and then shifted to the restrictive temperature (37°C) for 3 h. Samples were immediately fixed in solution, and the cells were prepared for electron microscopy. Arrows denote the exaggerated tubular network seen in B and C but not in A.
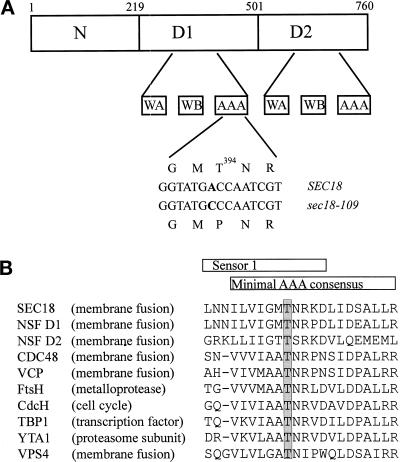
To identify the sec18-109 mutation, both YCpGAL–SEC18 and YCpGAL–sec18-109 were sequenced. This revealed an additional codon (GTG) in the wild-type gene between GAT1679 and GTT1682, as defined in the original SEC18 gene sequence cloned by Eakle et al. (1988). This extra codon is confirmed in the Saccharomyces Genome Database ORF YBR080c and predicts an additional glycine residue. Sequencing of the sec18-109 mutant allele revealed that the only change from the wild-type gene was a single nucleotide substitution, A1721→C, resulting in a single predicted amino acid substitution, Thr394→Pro. Figure 4A shows the position of Thr394 in Sec18p, located in the D1 domain beyond the Walker A and B boxes involved in ATP binding and hydrolysis. Thr394 is within the minimal AAA consensus sequence that defines members of the large and functionally diverse AAA protein family (Patel and Latterich, 1998) and is highly conserved in most, but not all, AAA proteins (Figure 4B).
Figure 4.
The sec18-109 mutation is within the AAA consensus sequence. (A) Schematic representation of Sec18p illustrating the position of the sec18-109 mutation in the minimal AAA consensus sequence of the D1 domain. Walker A and B boxes are denoted by WA and WB, respectively, and the AAA consensus sequence is denoted by AAA. Sequencing of the sec18-109 mutant allele revealed that the only change from the wild-type gene was a single nucleotide substitution, A1721→C (in bold), resulting in a single predicted amino acid substitution: Thr394→Pro. (B) Alignment of the amino acid sequences of selected members of the AAA family. The position of the highly conserved Thr residue is shown (shaded box).
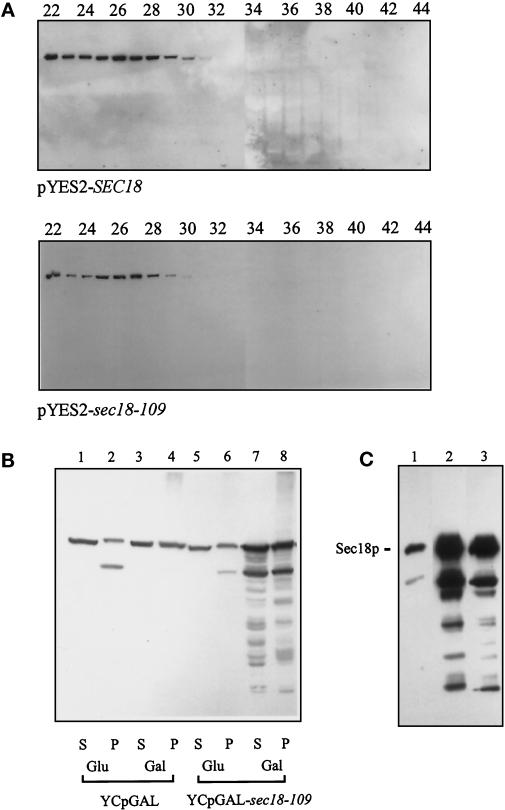
NSF is known to be a homohexameric protein (Fleming et al., 1998), and its multisubunit structure is thought to be essential for its function in membrane traffic (Whiteheart et al., 1994). Sec18p can also form homohexamers (Hanson et al., 1997; Steel et al., 1999), so if Sec18–109p induced the formation of aberrant oligomeric forms, this could potentially explain the dominant negative phenotype observed. To address this issue, V5 epitope-tagged versions of Sec18p and Sec18-109p were expressed in wild-type INVSc1 cells using the GAL1-regulated, 2-μm–based pYES2 plasmid. Cytosolic (100,000 × g supernatant) fractions were then subjected to gel filtration chromatography, and the native molecular masses of the recombinant proteins were estimated by Western blotting using an anti-V5 epitope antibody. It can be seen that the distribution of epitope-tagged Sec18p and Sec18-109p is similar, with both proteins peaking in fractions 26–27 (Figure 5A). From comparison with molecular weight standards run on the same column, this corresponds to a molecular mass of ∼640 kDa, which is identical to the estimated molecular mass of the putative hexameric pool of bacterially expressed recombinant His6–Sec18p (Steel et al., 1999). Thus, it appears that the dominant negative phenotype of the sec18-109 mutant is not due to gross structural changes in the oligomeric state of Sec18p.
Figure 5.
Oligomeric state and extent of overexpression of Sec18-109p. (A) JRY188 yeast cells expressing V5 epitope-tagged Sec18p or Sec18-109p from a 2-μm–based plasmid (pYES2) were inoculated into minimal media supplemented with glucose (2%) and grown overnight. Cells were harvested, washed with water, and resuspended in minimal media supplemented with 2% galactose and incubated for a further 2 h. Cells were then spheroplasted and lysed using a French press, and 100,000 × g supernatants were applied to a Superdex 200 gel filtration column for separation on the basis of native molecular mass (the void volume of this column accounts for 21 fractions, or 42 ml). Column fractions were run on 10% polyacrylamide gels, transferred onto nitrocellulose, and immunoblotted for the V5 epitope using ECL to specifically detect only recombinant forms of Sec18p. (B) JRY188 yeast cells transformed with either the YCpGAL vector or YCpGAL–sec18-109 were inoculated into minimal media supplemented with glucose (2%) and grown overnight. Samples were then split into two halves, and one half was reserved (Glu). The remaining cells were harvested, washed with water, and resuspended in minimal media supplemented with 2% galactose, and the cultures were incubated overnight (Gal). All samples were then spheroplasted, lysed using a French press, and centrifuged at 100,000 × g, and pellet and supernatant were fractions run on 10% polyacrylamide gels at equal protein loading per track. Both native and recombinant Sec18p were detected by immunoblotting with a Sec18p antibody and visualized by ECL. Quantification of Sec18p levels was performed by stripping and reprobing the same blot using 125I-labeled protein G. Phosphorimage analysis using ImageQuant software yielded the following relative densities: track 1, 3.99; track 2, 2.71; track 3, 2.69; track 4, 5.34; track 5, 3.95; track 6, 4.73; track 7, 23.24; track 8, 25.38. (C) Untransformed JRY188 cells (track 1) or JRY188 cells transformed with either YCpGAL–SEC18 (track 2) or YCpGAL–sec18-109 (track 3) were inoculated into minimal media supplemented with glucose (2%) and grown to mid log phase. Cells were harvested, washed with water, and resuspended in minimal media supplemented with 2% galactose and incubated for a further 20 h before being spheroplasted and loaded onto 10% polyacrylamide gels at equal protein loadings per track and transferred onto nitrocellulose. Both native and recombinant Sec18p were detected with a Sec18p antibody and visualized by ECL.
To estimate the degree of overexpression required to observe the strong phenotype of sec18-109, JRY188 cells transformed with either YCpGAL–sec18-109 or YCpGAL (i.e., an empty vector) were Western-blotted and probed with a Sec18p antibody. Sec18p was found to partition approximately equally to the 100,000 × g pellet and supernatant fractions (Figure 5B), as observed previously (Eakle et al., 1988). As expected, the presence of glucose or galactose had little effect on the expression of Sec18p in YCpGAL-transformed cells, because this represented the endogenous pool of Sec18p. A similar amount of Sec18p was detected in YCpGAL–sec18-109-transformed cells grown in the presence of glucose, indicating that expression of recombinant Sec18-109p was tightly repressed under these conditions; however, when YCpGAL–sec18-109-transformed cells were switched to galactose, a clear increase in Sec18p levels was observed, presumably caused by expression of recombinant Sec18–109p driven from the GAL1 promoter (Figure 5B). Quantification of 125I-labeled protein G binding by phosphorimager analysis using ImageQuant software (Pharmacia, Uppsala, Sweden) revealed an approximately sixfold increase in total cellular Sec18p levels in response to overnight incubation in galactose-containing media. Expression levels from the YCpGAL–SEC18 and YCpGAL–sec18-109 plasmids were approximately equal after the overnight inductions that were used (Figure 5C). Furthermore, the dominant negative effect of YCpGAL–sec18-109 is not due to overexpression of proteolytic fragments of Sec18p, because similar levels of breakdown products were seen in YCpGAL–SEC18-transformed cells (Figure 5C). The relatively modest level of overexpression of sec18–109p suggests that mixed hexamers containing both wild-type and mutant protomers would predominate in the cell and may be mainly responsible for the dominant lethal phenotype.
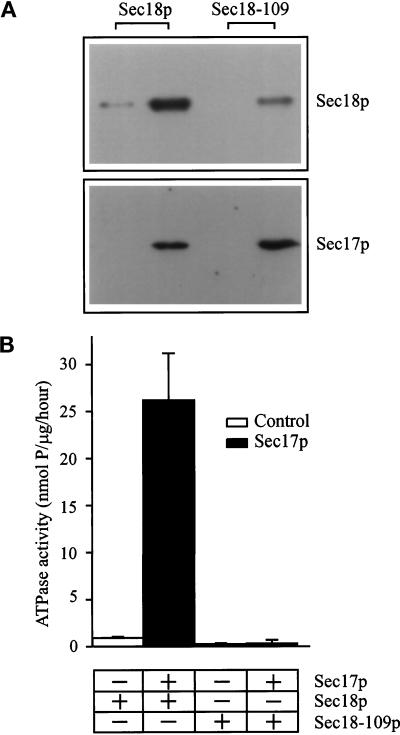
The function of NSF in membrane traffic is widely accepted as being dependent on binding to its cofactor, α-SNAP (Sec17p in yeast). To assess whether the sec18-109 phenotype was due to impairment of this protein–protein interaction, a Sec17p binding assay was performed using bacterially expressed, his-tagged Sec18p. It can be seen that the previously reported binding of Sec18p to immobilized Sec17p (Griff et al., 1992; Steel et al., 1999) is also exhibited by Sec18-109p (Figure 6A), albeit at slightly reduced efficiency, suggesting that the molecular defect in this mutant lies downstream of Sec17p binding. Once NSF has bound via α-SNAP to the SNARE complex, SNAP stimulation of NSF ATPase activity is thought to be essential for disassembly of the complex and the consequent priming of membrane fusion (Sumida et al., 1994; Whiteheart et al., 1994; Barnard et al., 1997). Sec18p has low but detectable ATPase activity that can be stimulated by Sec17p (Xu et al., 1998; Steel et al., 1999), and so this activity was investigated to determine whether it could explain the dominant negative sec18-109 phenotype. It can be seen that the ATPase activity of Sec18p was stimulated greatly by Sec17p, whereas Sec18-109p ATPase activity was undetectable in the presence or absence of Sec17p (Figure 6B). Sec18-109p ATPase activity remained negligible even when a concentration 50-fold that used for wild-type Sec18p was used, suggesting that the sec18-109 phenotype is a consequence of a profound block in ATPase activity.
Figure 6.
Sec18–109p can bind to Sec17p but has negligible ATPase activity. (A) Sec17p (100 μg/ml) was immobilized onto polypropylene tubes for 20 min. After washing, Sec18p or Sec18-109p (100 μg/ml) were added for 10 min. Bound proteins remaining after a wash step were solubilized in Laemmli buffer and analyzed on 10% polyacrylamide gels. Sec17p was detected using an anti–His-tag antibody, and Sec17p-bound proteins were detected using a Sec18p antibody followed by ECL. (B) Standard ATPase reactions were performed using Sec18p and Sec18-109p at 1 μg/ml final concentration in the assay. Incubations were performed at 37°C for 2 h. The proteins were incubated after preincubation with buffer A (open bars) or 400 μg/ml Sec17p in buffer A (filled bars). ATPase activity was assayed spectrophotometrically as release of Pi, and values were corrected by subtracting appropriate protein-free controls. The data shown represent means ± SD (n = 4) from a representative experiment.
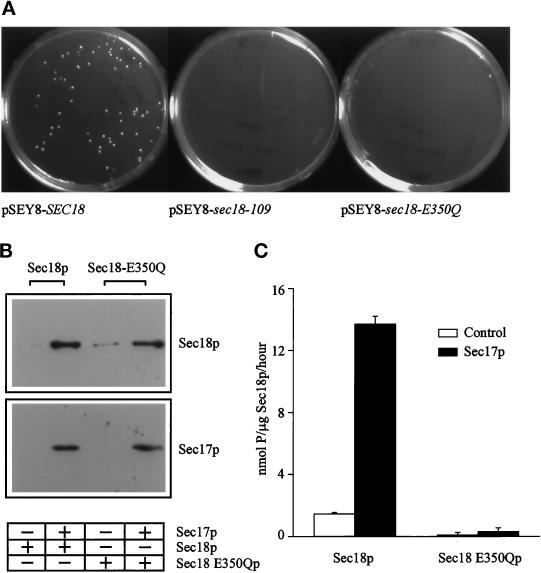
If the dominant negative effect of sec18-109 was really due to reduced ATPase activity in the mutant protein, then other ATPase-defective sec18 mutants should exhibit a similar phenotype. To test this hypothesis, we analyzed the sec18–E350Q mutant, in which a crucial Glu residue in the D1 domain Walker B box is replaced by Gln. The corresponding mutation in NSF (E329Q) has no effect on ATP binding but greatly inhibits hydrolysis of this bound ATP (Whiteheart et al., 1994). When the sec18–E350Q and sec18-109 mutations were introduced into the 2-μm–based pSEY8–SEC18 plasmid, this resulted in complete dominant lethality, whereas cells transformed with unaltered pSEY8–SEC18 remained viable (Figure 7A). In addition, recombinant Sec18–E350Q protein was able to bind to immobilized Sec17p (Figure 7B), as was the case for Sec18-109p (Figure 6A). Furthermore, Sec18–E350Q displayed negligible ATPase activity that could not be stimulated by Sec17p (Figure 7C), as also shown for Sec18-109p (Figure 6A). Because sec18-109 and sec18–E350Q are phenotypically indistinguishable at both the cellular and biochemical level, this strongly supports the conclusion that the sec18-109 phenotype described here is the result of a block in D1 domain ATPase activity.
Figure 7.
sec18-109 is phenotypically similar to a D1 ATP hydrolysis-defective mutant. (A) RSY271 yeast cells were transformed with pSEY8–SEC18, pSEY8-sec18–E350Q or pSEY8–sec18-109 and inoculated onto minimal plates. After 3 d growth at 25°C, the plates were photographed to assess viability. (B) Sec17p (100 μg/ml) was immobilized onto polypropylene tubes for 20 min. After washing, Sec18 or Sec18–E350Q proteins (100 μg/ml) were added for 10 min. Bound proteins remaining after a wash step were solubilized in Laemmli buffer and analyzed on 10% polyacrylamide gels. Sec17p was detected using an anti–His-tag antibody, and Sec17p-bound proteins were detected using a Sec18p antibody followed by ECL. (C) Standard ATPase reactions were performed using Sec18 and Sec18–E350Q proteins at 1 μg/ml final concentration in the assay. Incubations were performed at 37°C for 2 h. The proteins were incubated after preincubation with buffer A (open bars) or 400 μg/ml Sec17p in buffer A (filled bars). ATPase activity was assayed spectrophotometrically as release of Pi, and values were corrected by subtracting appropriate protein-free controls. The data shown represent means ± SD (n = 4) from a representative experiment.
DISCUSSION
In this work we have used a generally applicable approach to identify dominant negative mutants of the S. cerevisiae SEC18 gene. The mutant described here, sec18-109, has a phenotype similar to the well characterized temperature-sensitive sec18-1 mutant in terms of exaggerated ER morphology (Novick et al., 1981) and defective membrane traffic to the vacuole and cell surface (Graham and Emr, 1991) but has revealed new insights into the function of the AAA consensus sequence. All of the sec mutations are recessive (Novick et al., 1980), and so the phenotype of sec18-1 at the restrictive temperature can be assumed to mimic a null mutation in this essential gene. The similarity of the sec18-109 phenotype therefore suggests that this dominant mutation inhibits an essential function required for all Sec18p-dependent cellular processes. Because sec18-109 is phenotypically indistinguishable from the sec18–E350Q mutant, it is likely that this essential function is ATPase activity in the D1 domain. Significantly, the recessive nature of the temperature-sensitive sec18-1 mutant means that the (unknown) mutation in this allele cannot be used episomally and so investigations of Sec18p function have had to be performed in a sec18-1 background. The sec18-109 mutant described here will allow conditional inhibition of membrane traffic in any genetic background in vivo, and the isolated recombinant Sec18-109p could also be used in more recently developed yeast Sec18p-dependent in vitro reconstitution systems (Haas and Wickner, 1996; Barlowe, 1997). In addition, four further dominant negative alleles await full characterization. Preliminary work indicates that the sec18-112 mutant, despite displaying a galactose-induced density shift, does not exhibit the block in membrane traffic through the biosynthetic pathway exhibited by sec18-109 and sec18-1 (our unpublished observations). Thus, it may be that the remaining uncharacterized mutants selectively interfere with only a subset of Sec18p-dependent processes, making them potentially useful tools for the molecular dissection of membrane traffic.
The function of NSF in membrane traffic is thought to require hexamerization, followed by binding to SNAP, which stimulates NSF ATPase activity, resulting in SNARE disassembly and priming (Burgoyne and Morgan, 1998). If this model is correct, the ATPase-defective mutants, sec18-109 and sec18–E350Q (which have a normal oligomeric structure and ability to interact with Sec17p), would be predicted to result in the in vivo accumulation of unprimed SNARE complexes and hence the dominant lethal phenotype that is observed. This interpretation is consistent with the observation that an NSF D1 domain ATP-hydrolysis mutant acts as a dominant negative inhibitor of mammalian intra-Golgi transport and endosome fusion (Sumida et al., 1994; Whiteheart et al., 1994; Colombo et al., 1996) and can form, but not disassemble, 20 S SNARE complexes (Nagiec et al., 1995).
NSF and Sec18p belong to the AAA family of ATPases associated with various cellular activities (Confalonieri and Duguet, 1995). Members of this family contain one or two copies of a 230-amino acid AAA module and function in diverse cellular processes ranging from membrane traffic to proteolysis (Confalonieri and Duguet, 1995). The AAA module contains the Walker A and B boxes involved in nucleotide binding and hydrolysis, and a distinct AAA consensus sequence of unknown function (Patel and Latterich, 1998). Thr394, the residue mutated in Sec18-109p, is highly conserved in AAA proteins and indeed forms part of the minimal consensus sequence that defines the AAA family (Figure 4B). The undetectable ATPase activity of purified Sec18-109p is strikingly similar to that of the D1 domain ATP hydrolysis mutant, Sec18–E350Q, suggesting that the minimal AAA consensus sequence controls D1 domain ATPase activity in Sec18p.
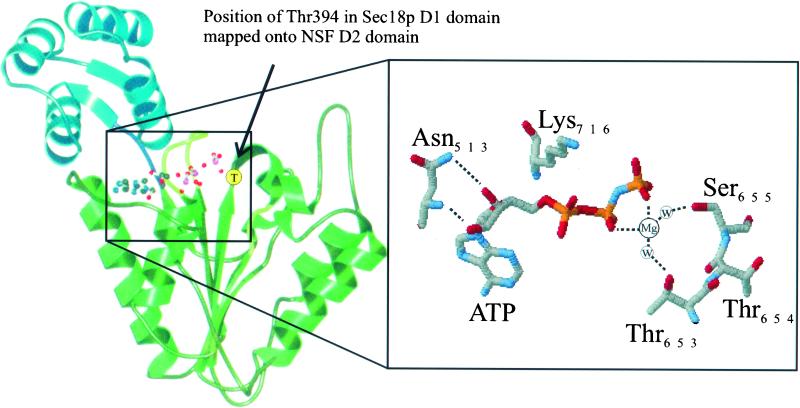
The structure of the D1 domain of Sec18p or NSF is unknown, but superimposing Thr394 onto the corresponding residue (Thr654,) in the recently solved crystal structure of the NSF D2 domain (Lenzen et al., 1998; Yu et al., 1998) reveals it to be at the base of a loop connecting β-sheet 4 to α-helix 5 (numbered according to Lenzen et al., 1998). This loop occupies a position between the γ-phosphate of ATP and the DExx (Walker B) box and so is in an ideal position to influence nucleotide hydrolysis (Figure 8) (Lenzen et al., 1998; Yu et al., 1998). Indeed, the two adjacent amino acids, Thr653 and Ser655, both serve to coordinate the Mg2+ ion essential for nucleotide hydrolysis (Figure 8). Although we cannot rule out an effect on ATP binding, these crystal structures strongly suggest that the defective ATPase phenotype observed in Sec18-109p is a consequence of a block in ATP hydrolysis. Furthermore, the high conservation of this Thr in the minimal AAA consensus sequence may indicate that this domain is important for ATP hydrolysis in AAA proteins generally.
Figure 8.
Modeling the sec18-109 mutation using the NSF D2 domain crystal structure. Ribbon diagram of the D2 domain of NSF based on the crystal structure determined by Yu et al. (1998). The amino acid corresponding to Thr394 of Sec18p (Thr694 in NSF) is indicated (T in circle) and is present between β sheet 4 and α helix 5 (numbered according to Lenzen et al., 1998). The position of Thr694 relative to the γ-phosphate of ATP is illustrated in the expanded box.
More recent sequence analyses have suggested that the AAA family is a subset of a larger family of ATPases, termed the AAA+ family (Neuwald et al., 1999). Members of the AAA+ family are thought to function as molecular chaperones (Neuwald et al., 1999), as was originally suggested for NSF and the AAA proteins (Morgan and Burgoyne, 1995). Thr394 is within the AAA+ Sensor 1 domain that has been suggested to detect nucleotide binding or hydrolysis (Neuwald et al., 1999). Alternatively, the direct contact between Sensor 1 and the γ-phosphate of ATP (Neuwald et al., 1999), taken together with the data presented here, may suggest a more active role for this domain in regulating ATP hydrolysis.
Finally, the mutD strategy used here to create sec18-109 can be applied to any essential yeast gene and requires no subcloning or PCR steps, only the E.coli mutator strain and the plasmid of interest. Indeed, a similar approach using hydroxylamine mutagenesis to successfully isolate dominant lethal BiP mutants has recently been described (McClellan et al., 1998). If the gene chosen has mammalian homologues, this may provide important structural/functional information. Most importantly, if the mutated residue(s) is conserved, this allows the generation of dominant negative mutants of the mammalian protein to be used in more complex systems. The informative power of such dominant negative mutants is illustrated by the hundreds of articles in which dominant inhibitory Ras or Ras-related GTPases were used (Feig, 1999). We hope that the simple, relatively rapid approach described here will allow new dominant-acting mutations of important genes to be identified.
ACKNOWLEDGMENTS
We thank Drs. Albert Haas, Tom Stevens, Scott Emr, and Randy Schekman for generous provision of materials, Dr. Reinhard Jahn for suggesting the modeling of the sec18-109 mutation on the NSF D2 crystal structure, and Drs. Wally Whiteheart and Richard Newmann for generous help with structural analysis. We thank Bob Burgoyne, Albert Haas, and Wally Whiteheart for helpful discussions and comments on the manuscript. This work was supported by a Wellcome Trust Project Grant (to A.M.) and a Wellcome Prize PhD Studentship (to C.H.).
REFERENCES
- Barlowe C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J Cell Biol. 1997;139:1097–1108. doi: 10.1083/jcb.139.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard RJO, Morgan A, Burgoyne RD. Stimulation of NSF ATPase activity by a-SNAP is required for SNARE complex disassembly and exocytosis. J Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block MR, Glick BS, Wilcox CA, Weiland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci USA. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD, Morgan A. Analysis of regulated exocytosis in adrenal chromaffin cells: insights into NSF/SNAP/SNARE function. BioEssays. 1998;20:328–335. doi: 10.1002/(SICI)1521-1878(199804)20:4<328::AID-BIES9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Colombo MI, Taddese M, Whiteheart SW, Stahl PD. A possible predocking attachment site for N-ethylmaleimide-sensitive fusion protein. J Biol Chem. 1996;271:18810–18816. doi: 10.1074/jbc.271.31.18810. [DOI] [PubMed] [Google Scholar]
- Confalonieri F, Duguet M. A 200-amino acid ATPase module in search of a basic function. BioEssays. 1995;17:639–650. doi: 10.1002/bies.950170710. [DOI] [PubMed] [Google Scholar]
- Eakle KA, Bernstein M, Emr SD. Characterization of a component of the yeast secretory machinery: identification of the SEC18 gene product. Mol Cell Biol. 1988;8:4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H, Lu C, Burgers PMJ. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1983;80:2189–2192. doi: 10.1073/pnas.80.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Fleming KG, Hohl TM, Yu RC, Muller SA, Wolpensinger B, Engel A, Enelhardt H, Brunger AT, Söllner TH, Hanson PI. A revised model for the oligomeric state of the N-ethylmaleimide-sensitive fusion protein, NSF. J Biol Chem. 1998;273:15675–15681. doi: 10.1074/jbc.273.25.15675. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griff IC, Schekman RW, Rothman JE, Kaiser CA. The yeast SEC17 gene product is functionally equivalent to mammalian alpha-snap protein. J Biol Chem. 1992;267:12106–12115. [PubMed] [Google Scholar]
- Haas A, Wickner W. Homotypic vacuole fusion requires Sec17p (yeast a-SNAP) and Sec18p (yeast NSF) EMBO J. 1996;15:3296–3305. [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexed visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- Lenzen CU, Steinmann D, Whiteheart SW, Weis WI. Crystal structure of the hexamerisation domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94:525–563. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Chapman ER, Kreber R, Garment MB, Carlson SD, Ganetzky B. Temperature-sensitive paralytic mutations demonstrate that synaptic exocytosis requires SNARE complex assembly and disassembly. Neuron. 1998;21:401–413. doi: 10.1016/s0896-6273(00)80549-8. [DOI] [PubMed] [Google Scholar]
- Matveeva EA, He P, Whiteheart SW. N-Ethylmaleimide-sensitive fusion protein contains high and low affinity ATP-binding sites that are functionally distinct. J Biol Chem. 1997;272:26413–26418. doi: 10.1074/jbc.272.42.26413. [DOI] [PubMed] [Google Scholar]
- Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (a-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Endres JB, Vogel JP, Palazzi D, Rose MD, Brodsky JL. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol Biol Cell. 1998;9:3533–3545. doi: 10.1091/mbc.9.12.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A, Burgoyne RD. Is NSF a fusion protein? Trends Cell Biol. 1995;5:335–339. doi: 10.1016/s0962-8924(00)89059-5. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Nagiec EE, Bernstein A, Whiteheart SW. Each domain of the N-ethylmaleimide-sensitive fusion protein contributes to its transport activity. J Biol Chem. 1995;270:29182–29188. doi: 10.1074/jbc.270.49.29182. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman RW. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman RW. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Otter-Nilsson M, Hendriks R, Pecheur-Huet E-I, Hoekstra D, Nilsson T. Cytosolic ATPases, p97 and NSF, are sufficient to mediate rapid membrane fusion. EMBO J. 1999;18:2074–2083. doi: 10.1093/emboj/18.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- Pryer NK, Wuestehube LJ, Schekman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–62. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Steel GJ, Laude AJ, Boojawan A, Harvey DJ, Morgan A. Biochemical analysis of the Saccharomyces cerevisiae Sec18 gene product: implications for the molecular mechanism of membrane fusion. Biochemistry. 1999;38:7764–7772. doi: 10.1021/bi990315v. [DOI] [PubMed] [Google Scholar]
- Steel GJ, Morgan A. Selective stimulation of the D1 ATPase domain of N-ethylmaleimide-sensitive fusion protein (NSF) by soluble NSF attachment proteins. FEBS Lett. 1998;423:113–116. doi: 10.1016/s0014-5793(98)00072-6. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Sumida M, Hong RM, Tagaya M. Role of two nucleotide-binding regions in a N-ethylmaleimide-sensitive factor involved in vesicle-mediated protein transport. J Biol Chem. 1994;269:20636–20641. [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachi M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. N-Ethylmalemide-sensitive fusion protein: A trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DW, Wilcox CA, Flynn GC, Chen E, Kuang W, Henzel WJ, Block MR, Ullrich A, Rothman JE. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- Woodman PG. The roles of NSF, SNAPs and SNAREs during membrane fusion. Biochim Biophys Acta. 1997;1357:155–172. doi: 10.1016/s0167-4889(97)00039-6. [DOI] [PubMed] [Google Scholar]
- Xu Z, Sato K, Wickner W. LMA1 binds to vacuoles at Sec18p (NSF), transfers upon ATP hydrolysis to a t-SNARE (Vam3p) complex, and is released during fusion. Cell. 1998;93:1125–1134. doi: 10.1016/s0092-8674(00)81457-9. [DOI] [PubMed] [Google Scholar]
- Yu RC, Hanson PI, Jahn R, Brunger AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat Struct Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]