Abstract
Obligate intracellular parasites of the phylum Apicomplexa exhibit gliding motility, a unique form of substrate-dependent locomotion essential for host cell invasion and shown to involve the parasite actin cytoskeleton and myosin motor(s). Toxoplasma gondii has been shown to express three class XIV myosins, TgM-A, -B, and -C. We identified an additional such myosin, TgM-D, and completed the sequences of a related Plasmodium falciparum myosin, PfM-A. Despite divergent structural features, TgM-A purified from parasites bound actin in an ATP-dependent manner. Isoform-specific antibodies revealed that TgM-A and recombinant mycTgM-A were localized right beneath the plasma membrane, and subcellular fractionation indicated a tight membrane association. Recombinant TgM-D also had a peripheral although not as sharply defined localization. Truncation of their respective tail domains abolished peripheral localization and tight membrane association. Conversely, fusion of the tails to green fluorescent protein (GFP) was sufficient to confer plasma membrane localization and sedimentability. The peripheral localization of TgM-A and of the GFP-tail fusion did not depend on an intact F-actin cytoskeleton, and the GFP chimera did not localize to the plasma membrane of HeLa cells. Finally, we showed that the specific localization determinants were in the very C terminus of the TgM-A tail, and site-directed mutagenesis revealed two essential arginine residues. We discuss the evidence for a proteinaceous plasma membrane receptor and the implications for the invasion process.
INTRODUCTION
The phylum of Apicomplexa comprises numerous pathogens of medical and veterinary significance. Among them, Plasmodium falciparum is the most virulent of the Plasmodium species responsible for malaria in human. The opportunistic pathogen Toxoplasma gondii causes diseases in immunocompromised patients and in congenitally infected infants. Although members of the Apicomplexa differ significantly in their host range and cell type specificity, they do share a similar mechanism for host cell entry. Host cell penetration is a prerequisite for the survival of these obligate intracellular parasites and does not occur by induced phagocytosis but via an active process from the parasites (Sibley, 1995). In the absence of locomotive organelles, apicomplexan parasites have developed an unusual mode of substrate-dependent gliding locomotion that is necessary for invasion (Sibley et al., 1998). The inhibitory functions of cytochalasins on invasion by Plasmodium (Miller et al., 1979) and Eimeria (Russell, 1983) have been reported and argue for a participation of actin filaments. More recently, a direct involvement of T. gondii actin has been demonstrated, establishing that motility is essential for invasion and depends on the parasite's own cytoskeleton (Dobrowolski and Sibley, 1996). An actin-based motor is predicted to generate the force during invasion by Apicomplexa, and this hypothesis is corroborated by the inability of parasites to glide and to invade in the presence of the inhibitor of myosin heavy chain ATPase butanedione monoxime (Dobrowolski et al., 1997a; Pinder et al., 1998). Similarly, tachyzoites treated with jasplakinolide, a membrane-permeable actin-polymerizing and filament-stabilizing drug, reversibly inhibit host cell invasion (Shaw and Tilney, 1999). Members of the myosin superfamily are mechanoenzymes that convert chemical energy stored in ATP into a directed force along actin filaments (Spudich, 1994). In the past few years evidence has emerged for roles played by actin-based molecular motors in a wide range of membrane movements (Hasson and Mooseker, 1995; Mermall et al., 1998). In the case of host cell invasion by apicomplexan parasites, a capping model proposes that gliding motility shall be driven by the redistribution of transmembrane proteins from the apical to the posterior pole of the parasite along the subcortical actomyosin system. Correspondingly, the molecular motor powering gliding locomotion and capping is expected to localize right beneath the plasma membrane. Recently, three myosins were identified in T. gondii (Heintzelman and Schwartzman, 1997) and shown to build a 14th phylogenetic and structural class of myosins (Mermall et al., 1998). Antibodies cross-reacting with heterologous myosins were used to immunolocalize a myosin at the anterior pole of T. gondii tachyzoites (Schwartzman and Pfefferkorn, 1983), and more recent work from the same group potentially identified this myosin as TgM-A (Heintzelman and Schwartzman, 1999). In addition, antibodies raised against the conserved peptide LEAF revealed one or more myosins localized beneath the plasma membrane and also scattered throughout the cytosol of T. gondii tachyzoites (Dobrowolski et al., 1997a). Similarly, antibodies generated against a small fragment of the motor domain of Pf-myo1 showed labeling concentrated under the plasma membrane of P. falciparum merozoite but absent from the apical prominence itself (Webb et al., 1996). The P. falciparum myosin recognized by these antibodies migrates slightly above 100 kDa in SDS-PAGE, is expressed in mature schizonts and merozoites, and localizes predominantly around the periphery of the cell (Pinder et al., 1998).
Here, we report of the identification of a 91-kDa T. gondii myosin of class XIV, TgM-D, and of the completion of the PfM-A sequence, the likely P. falciparum homologue of TgM-A. The exquisite amenability of T. gondii to molecular genetics allowed us to investigate the determinants of subcellular localization of two class XIV myosins. A pair of basic residues is essential to target TgM-A to the periphery, defining its cargo-binding site down to the amino acid level. Such precise mapping has only been achieved for two other myosins, NINAC, a Drosophila myosin III interacting with INAD (Wes et al., 1999), and Myo2, a Saccharomyces cerevisiae myosin V binding to the vacuole (Catlett and Weisman, 1998). In addition, the first purification of such a myosin is presented, allowing us to show that, despite its structural divergences, TgM-A had the ability to bind F-actin in an ATP-dependent manner. Evidence is presented that the peripheral localization is largely independent of the actin cytoskeleton and is likely due to specific and saturable interaction with a proteinaceous component of the plasma membrane.
MATERIALS AND METHODS
Strains and Reagents
The bacterial strains for recombinant DNA techniques were Escherichia coli XL1-Blue and XLOR. The helper phage ExAssist, from Stratagene (La Jolla, CA), was used for the in vivo excision of the phagemid vectors from the λZAPII clones. Restriction enzymes were purchased from New England Biolabs (Beverly, MA). The secondary antibodies for Western blotting were from Bio-Rad (Hercules, CA), and those for immunofluorescence were from BioTrend (Cologne, Germany).
Growth of Parasites and Isolation of DNA and RNA
T. gondii tachyzoites (RH strain wild-type and RHhxgprt−) were grown in human foreskin fibroblasts (HFFs) or in vero cells (African green monkey kidney cells) maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 2 mM glutamine, and 25 μg/ml gentamicin. Parasites were harvested after complete lysis of the host cells and purified by passage through 3.0-μm filters and centrifugation in PBS. Genomic DNA was isolated from purified parasites by SDS-proteinase K lysis followed by phenol-chloroform, chloroform extractions and ethanol precipitation (Sibley and Boothroyd, 1992). Total RNAs were prepared using RNA Clean from AGS (Heidelberg, Germany) according to the manufacturer's instructions. P falciparum total RNA was purified from the isolate FCBR, Columbia grown in vitro (Knapp et al., 1989) and kindly provided by Dr. H. del Portillo (University of Sao Paulo, Sao Paulo, Brazil).
PCR Screening
PCR was performed on T. gondii genomic DNA and yielded two distinct myosin fragments. PCR reactions were carried out in a GeneAmp PCR System 2400 instrument (Perkin-Elmer, Norwalk, CT). Primers myo1 and myo5 (corresponding to the conserved motifs GESGAGKT and LEAFGNAKT, respectively) and the parameters used for the PCR reaction are described elsewhere (Schwarz et al., 1999). The PCR products were purified over a spin column (QIAquick PCR purification kit; Qiagen, Hilden, Germany) and the whole sample was cloned into PCR Script (Stratagene) as described by the manufacturer. Double-stranded DNA sequencing of the products was performed by the dideoxy-termination method using Sequenase version 2.0 (United Stated Biochemical, Cleveland, OH) with T3 and T7 primers.
Toxoplasma cDNA and Genomic Library Screening
The digoxigenin system (Boehringer Mannheim, Mannheim, Germany) for nonradioactive labeling and detection of nucleic acids was used to screen a T. gondii cDNA library. The 300-bp myosin fragment was digoxigenin-dUTP labeled by PCR and used as a probe to screen the RH (Elmer Pfefferkorn) cDNA library in λZAPII from National Institutes of Health AIDS reagents (kindly provided by D.S. Roos, University of Pennsylvania, Philadelphia, PA). The detection of positive clones was achieved by chemiluminescence with CSPD (Boehringer Mannheim) according to the manufacturer. Plaques from 15 × 104 phage were screened, and positive clones were identified on both duplicates. After two additional cycles of hybridization, positive clones were excised in vivo, and the insert sizes were determined by restriction digests. One clone contained a full-length cDNA corresponding to the previously described TgM-A and predicting a protein of 93 kDa (Heintzelman and Schwartzman, 1997). The second clone encoded another myosin lacking a start codon (TgM-D). Further screening of cDNA libraries failed to identify full-length TgM-D cDNA clones. The missing 5′ sequences of TgM-D were finally obtained by sequencing genomic cosmid clones encompassing the TgM-D locus. The cosmid library used the SuperCos vector modified with an SAG1/ble Toxoplasma selection cassette inserted into its HindIII site. The library was prepared from a Sau3AI partial digestion of RH genomic DNA ligated into the BamHI cloning site and was kindly provided by D. Sibley and D. Howe (Washington University, St. Louis, MO). The gene coding for TgM-D was interrupted by 11 introns of various sizes (our unpublished results). Southern blotting analysis of T. gondii genomic DNA digested with several restriction enzymes yielded patterns of bands matching those predicted by the restriction map of the cosmid clone, indicating that TgM-D is a single-copy gene.
We also obtained and sequenced a cosmid clone encompassing the TgM-B/C locus (GenBank accession number AF202585). This confirmed that the two proteins, which have a common N terminus and differ only in their C terminus, are really translated from two transcripts with alternatively spliced 3′ ends (as suggested by Heintzelman and Schwartzman, 1997). This also revealed that the reported common 5′ termini were cDNA library cloning artifacts, resulting from an in frame fusion at an EcoRI site with the 3′ end of GRA7. The correct cDNA could then be amplified by reverse transcription (RT)-PCR, cloned, and sequenced.
Identification of a Full-Length P. falciparum Myosin
The sequences of the short and conserved, positively charged tail domains of TgM-A and TgM-D showed a significant homology with an expressed sequence tag derived from the closely related organism P. falciparum (BLAST software search). An RT-PCR strategy using an antisense primer corresponding to the tail and a sense primer corresponding to a conserved motif in the head domain allowed us to amplify, clone, and sequence a large fragment of the myosin. The sequence corresponded to the partial sequence of a P. falciparum myosin present in the database (Pfmyo-1). The progress on the genome sequencing project allowed us to complete and to confirm the sequence of this myosin (PfM-A, accession number AF105118). The PfM-A gene contains only two introns clustered immediately downstream of the AUG, suggesting a possible regulatory function of the splicing. The deduced amino acid sequence of PfM-A predicts a protein of 90 kDa similar to TgM-A and TgM-D (Figure 1). The full cDNA coding for PfM-A was amplified by RT-PCR using the Titan RT-PCR system (avian myeloblastosis virus and Expand High Fidelity; Boehringer Mannheim) according to the manufacturer's instructions. The primers used in the PCR reaction were 5-′ccatccatgcatgctgttacaaatgaagaaataaaaacggc-3′ and 5′-ggatccttgagctaccattttttttcttatatgagc-3′, and the product of amplification was cloned into pBluescript (Stratagene).
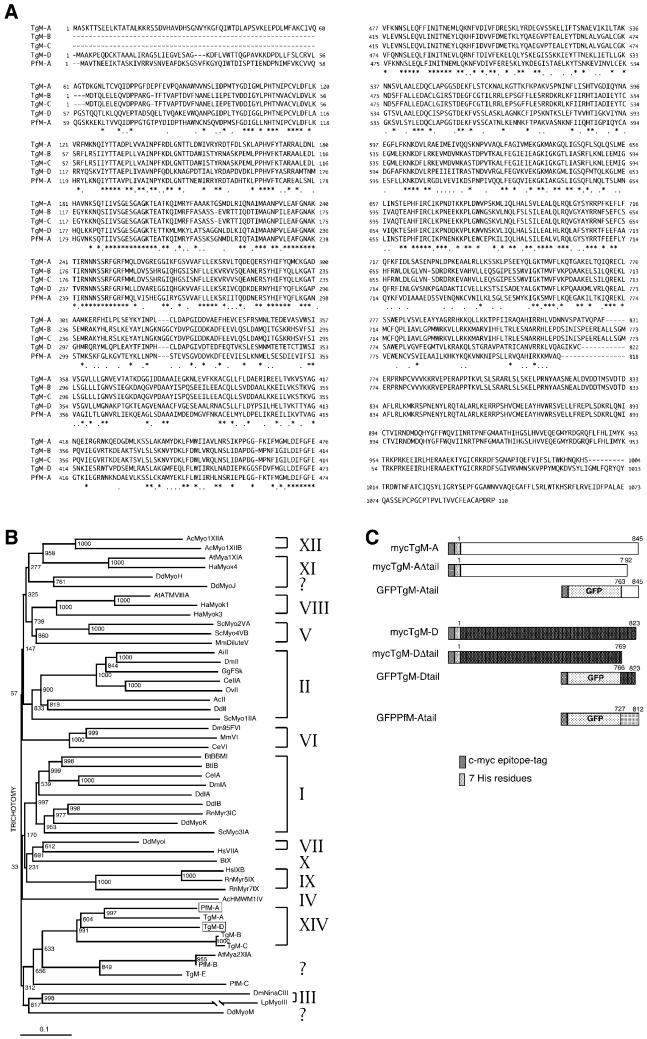
Figure 1.
TgM-D is a member of the class XIV myosins. (A) Sequence alignment of the class XIV myosins. Identical amino acids are indicated by a star; homologies are indicated by a dot. The TgM-A glutamine 419 corresponds to the TEDS site, and the serine 693 is found instead of the conserved glycine proposed to act as a pivot point of the lever arm. (B) Phylogenetic tree with 51 myosins representative of each known class. TgM-D and the completed PfM-A belong to the class XIV. (C) Schematic presentation of the constructs used in this study. The numbers on top of the bars indicate the corresponding residues from each myosin (see A). Most accession numbers can be found at http://www.mrc-lmb.cam.ac.uk/myosin/trees/accession.html, and some can be found in an article by Schwarz et al. (1999). Accession numbers for the apicomplexan myosins: TgM-A, AF006626; TgM-B, AF006627; TgM-C, AF006628; a genomic fragment with the correct 5′end of TgM-B and -C cDNAs, AF202585; TgM-D, AF105118; TgM-E, AF221131; PfM-A, AF105117; PfM-B, AF222716; and PfM-C, AF222717.
Construction of T. gondii Expression Vectors
TgM-A and TgM-D were amplified by PCR to introduce a SacI site after the start codon and a BamHI site before the stop codon, using the sense oligonucleotide 5′-gagctcgcgagcaaggaccacgtct-3′ combined with reverse oligonucleotide primer 5′-ggatccgaacgccggctgaacagtcg-3′ for TgM-A and 5′-gagctcggcacgagcctcttcagt-3′ combined with 5′-ggatccgcaaaccttgatgcccgctt-3′, for TgM-D. The PCR products were cloned into bacterial expression vectors (pET·TgM-A and pET·TgM-D) and analyzed for expression of the recombinant protein in BL21 (DE31, expressing T7 polymerase) after isopropyl-1-thio-β-d-galactopyranoside induction. Polypeptides of ∼93 kDa for TgM-A and 91 kDa for TgM-D were produced (our unpublished results). Vectors used to express TgM-A and TgM-D in T. gondii were derived from the pS214SCAT vector containing 5′ and 3′ flanking sequences of the SAG1 gene and pT5RCAT containing 5′flanking sequence of the TUB1 gene described previously (Soldati and Boothroyd, 1995). pS·TgM-A was generated by subcloning TgM-A from pET·TgM-A into pS/4R between NsiI and PacI sites (Soldati and Boothroyd, 1995). The vector pT·TgM-D was obtained by cloning TgM-D coding sequence into NsiI and PacI sites of pT/5R230 (Soldati and Boothroyd, 1995). In these constructs, both myosins are fused at the N terminus to a c-myc epitope tag and seven His residues (these sequences were imported from a modified pET vector [MQEQKLISEEDLAMAMHHHHHHH]), giving rise to the mycTgM-A and mycTgM-D proteins. The construct pS·TgM-AΔtail is identical to pS·TgM-A except that sequences corresponding to the last 53 amino acids of TgM-A are lacking. This deletion was obtained by PCR amplification using the sense primer described above and the complementary oligonucleotide 5′-ttaattaagaggatccgcattctctctgaatctgc-3′. To express full-length TgM-D, a fragment of the genomic clone was amplified by PCR using oligonucleotides 5′-atgcatgagctcgcggcaaaaccggag-3′ and 5′-cagccaaaaccgaagttcc.3′and cloned into the NsiI sites of pTc·TgM-D to generate pT·TgM-D. In construct pT·TgM-DΔtail, the last 54 amino acids of TgM-D have been deleted. The EcoRV–PacI fragment of 1091 bp corresponding to the C terminus of the protein has been replaced by the fragment corresponding to the C terminus lacking the tail, obtained by PCR amplification using 5′-ttaattaaagccgcaccaacatctttgct-3′ as a complementary oligonucleotide.
The plasmid pT·GFP was constructed by replacement of chloramphenicol acetyltransferase (CAT) by the green fluorescent protein (GFP) coding sequence between the NsiI and PacI sites in the vector pT5RCAT. An NsiI site was introduced at the N terminus of the GFP coding sequence using the oligonucleotides 5′-ggcgatgcatagtaaaggagaagaacttttc-3′, and PstI and PacI sites were introduced at the C terminus using the complementary oligonucleotides 5′-gggcttaattaagcaccgcctgcagctttgtatagttca-3′. The GFP mutant used in this work is fluorescent as nonfusion protein in tachyzoites (M. Soete, C. Hetlmann, and D. Soldati, unpublished data) and has been extensively modified in the codon usage (Haseloff et al., 1997). The constructs pT·GFP·TgM-Atail, pT·GFP·TgM-Dtail, and pT·GFP·PfM-Atail were generated by cloning the PCR products corresponding to the last 82 amino acids of TgM-A (as in Figures 7 and 9, or 62, as in Figures 1A, 5, and 8), the last 57 amino acids of TgM-D, and the last 85 amino acids of PfM-A, respectively, using PstI and PacI sites. The site-specific mutagenesis of TgM-Atail was achieved by PCR amplification using the following primers: 5′-gcgtactacgctggcatactccacgcggcgcag-ctgctgaaaaag-3′, 5′-aagaagcagctgctggcagcgacccccttcatcatt-3′, 5′-gcccaggctcacatcgccgcacacctggtggacaac-3′, 5′-cggttaattaaaccaggtgtctgcggatgtg-3′, and 5′-gccttaattaagcgcgaatgatgaaggggg-3′ for TgM-Atail-mut I, -mut II, -mut III, -Δ14, and -Δ22, respectively. The PCR products of the mutated tails were cloned in pT·GFPdhfrtsHXGPRT between PstI and PacI sites. The fragment coding for GFP fused to TgM-Atail was cut from pT·GFP·TgM-Atail and introduced into pUHD15-1 (Gossen and Bujard, 1992), using EcoRI and BamHI as cloning sites to generate pUHD15-1·GFP·TgM-Atail. The vector for nonfusion GFP expression, pUHD15-1GFP, was kindly provided by R. Löw (ZMBH, Heidelberg, Germany).
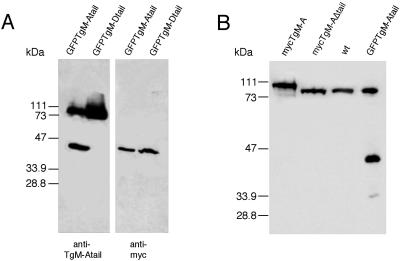
Figure 7.
The tails of TgM-A and TgM-D are sufficient to confer to GFP both peripheral localization and association with the particulate fraction. (A) Alignment of three highly related apicomplexan myosin tails. The basic residues are boxed. (B) Localization of GFP (a), GFP·TgM-Atail (b), GFP·TgM-Dtail (c), and GFP·PfM-Atail (d). Fluorescence was observed directly (a and b) or after indirect immunofluorescence with anti-myc antibodies (c and d). (C) Recombinant parasites expressing GFP, GFP·TgM-Atail, GFP·TgM-Dtail, and GFP·PfM-Atail were lysed in PBS and separated by high-speed centrifugations in soluble (S) and particulate (P) fraction. The partitioning of GFP and the tail chimeras reflected their respective intracellular localization observed by imunofluorescence microscopy.
Figure 9.
A dibasic motif in the tail of TgM-A is essential for the cell membrane localization of GFP-tail chimera. (A) The wild-type sequence of the tail of TgM-A (wt) is shown aligned with the site-specific mutants (I–III) and the two deletion mutants (Δ14 and Δ22). The mutated residues are indicated in red. (B) Confocal microscopic analysis of parasites stably transformed with the respective mutant tail constructs fused to GFP. Indirect immunofluorescence was performed using the anti-myc antibodies. (C) Anti-myc immunoblot analysis of recombinant parasites expressing the different constructs presented in A.
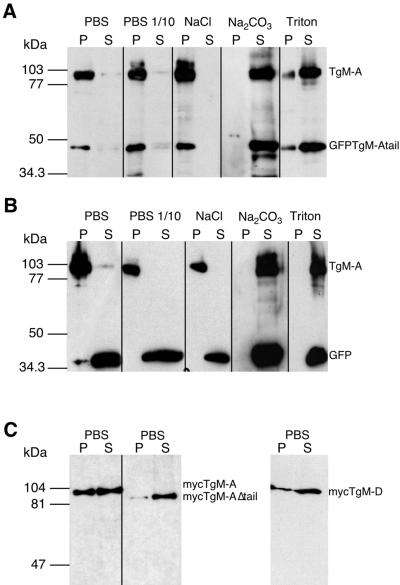
Figure 5.
Distribution of endogenous and recombinant myosins studied by subcellular fractionation. Cells expressing GFP (B) or GFP·TgM-Atail (A) were lysed in the presence of 5 mM ATP under different conditions and separated by high-speed centrifugations in soluble (S) and particulate (P) fractions. Cells were lysed in PBS, in a low-osmolarity buffer (PBS diluted 1:10), in PBS with 1 M NaCl, in 0.1 M Na2CO3, pH 11.5, or in PBS with 1% Triton X-100. The distribution of endogenous TgM-A and either GFP·TgM-Atail or GFP was determined simultaneously by immunoblotting with antibodies against the tail of TgM-A either alone (A) or combined with anti-myc antibodies (B). (C) The distribution of mycTgM-A, mycTgM-AΔtail, and mycTgM-D was detected by anti-myc antibody after lysis in PBS.
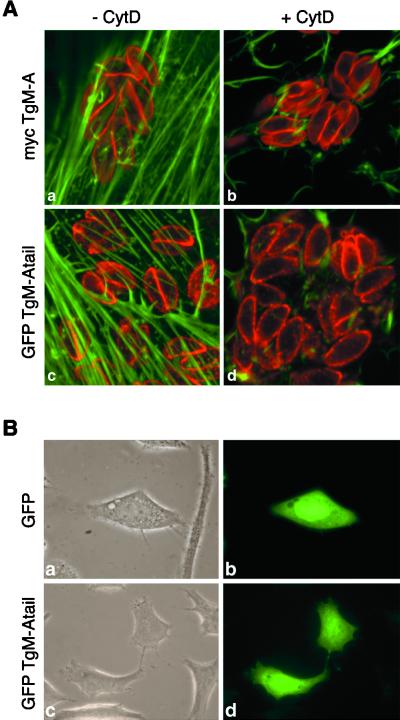
Figure 8.
Cytochalasin treatment did not influence the peripheral localization of mycTgM-A and GFP·TgM-Atail. GFP·TgM-Atail did not localize to the plasma membrane of HeLa cells. (A) MycTgM-A (a and b) and GFP·TgM-Atail (c and d) localized at the periphery of parasites after incubation in the presence (b and d) or absence (a and c) of 10 μg/ml cytochalasin D for 3 h, indicating that it did not depend on an intact actin cytoskeleton. To optimally present the host actin cytoskeleton and the peripheral staining in the parasites, one confocal section was recorded in the ventral part of the fibroblasts where the actin stress fibers are most prominent, and another confocal section was recorded through the middle of the parasites, 1 μm higher in the cell. Only the overlay is shown here. (B) Contrary to what was seen in T. gondii, GFP·TgM-Atail did not interact with the plasma membrane of an animal cell (d) and had a distribution similar to GFP (b). (a and c) Phase-contrast pictures corresponding to b and d.
Generation of a Serum Specific for the Tail of TgM-A
Two peptides covering the tail of TgM-A (CVLEAYYAGRRHKKQLLKKTP and AHIRRHLVDNNVSPATVQPAFC) were synthesized and coupled to keyhole limpet hemocyanin according to the instructions of the manufacturer (Pierce, Rockford, IL). Two rabbits were first immunized with 400 μg of both peptides with the complete Freund's adjuvant. Four successive subcutaneous injections with 200 μg of both peptides in incomplete adjuvant LQ (Gerbu, Heidelberg, Germany) were performed at 24-d intervals. The serum was immunoaffinity purified against the peptides previously coupled to Affi-Gel 10 according to the manufacturer (Bio-Rad). Four milligrams of each peptide were used to affinity purify 2 ml of sera.
Parasite and HeLa Cell Transfection and Selection of Transformants
T. gondii tachyzoites (RHhxgprt−) were transfected by electroporation as previously described. Selection of stable transformants expressing pS·TgM-A and pT·GFP·TgM-Atail were achieved by cotransfection of the expression vector with the selectable plasmid (pminHXGPRT). Restriction enzyme-mediated integration was applied as previously described (Black et al., 1995). The hypoxanthine-xanthine-guanine-phosphoribosyltransferase (HXGPRT) was used as a positive selectable marker gene in the presence of mycophenolic acid and xanthine as described (Donald et al., 1996). Freshly released parasites (5 × 107) of the RHhxgprt− strain were resuspended in cytomix buffer in the presence of 100 U of BamHI and were cotransfected with 10–20 μg of the plasmid pminHXGPRT, which carries the HXGPRT gene, and 80–100 μg of the plasmids expressing GFP fusions or myosins. Parasites expressing pT·TgM-D, pT·GFP·TgM-Dtail, and pT·GFP·PfM-Atail could not be obtained by cotransfection; therefore, the selectable marker gene dhfrtsXHGPRTdhfrts was introduced into the expression vector in a unique SacII site before selection. HeLa cells were transfected with pUHD15-1·GFP·TgM-Atail and by a calcium phosphate method as described previously (Graham and Eb, 1973).
Cytochalasin D Treatment
Cytochalasin D (Sigma, St. Louis, MO) was stored as DMSO stock at −20°C. Intracellular parasites grown for 24 h in HFFs on glass coverslips were incubated with 10 μg/ml cytochalasin D in medium at 37°C for up to 3 h. At the indicated time, the coverslips were taken out and rinsed twice with PBS, and cells were fixed as described below.
Indirect Immunofluorescence Microscopy and Detection of GFP in T. gondii
All manipulations were carried out at room temperature. Intracellular parasites grown for 24 h in HFFs on glass slides were fixed with 4% paraformaldehyde and 0.05% glutaraldehyde for 20 min. After fixation, slides were rinsed in PBS and 0.1 M glycine. Cells were then permeabilized in PBS and 0.2% Triton X-100 for 20 min and blocked in the same buffer with 2% FCS. Slides were incubated for 60 min with primary antibodies diluted in PBS and 1% FCS, washed, and incubated for 60 min with Alexa488- or FITC-labeled goat anti-mouse immunoglobulin Gs diluted in PBS and 1% FCS. After cytochalasin D treatment, stainings were carried out with the following modifications. Fixation was with 4% paraformaldehyde only, all subsequent buffers contained 5 mM EGTA to chelate Ca2+ and avoid disruption of actin filaments by gelsolin-like activities, and 2% BSA was used as blocking reagent. Staining of F-actin was performed with Oregon Green-phalloidin (Molecular Probes, Eugene, OR) for 60 min. The rapid freezing combined with fixation and permeabilization in ultracold methanol was performed as described (Neuhaus et al., 1998). Slides were mounted in Vectashield (Vector Laboratories, Burlingame, CA) and kept at 4°C in the dark. The mAb anti-myc was an ascites preparation of 9E10 used at the dilution 1:1000. The mAb DG52, recognizing SAG1, coupled to biotin was generously provided by Dr. J.F. Dubremetz (Institut Pasteur de Lille). Intracellular parasites expressing GFP were fixed according to the above protocol and mounted immediately for microscopic analysis. Confocal images were collected with a Leica (Nussloch, Germany) laser scanning confocal microscope (TCS-NT DM/IRB) using a 100× Plan-Apo objective, numerical aperture 1.30. Single optical sections were recorded with an optimal pinhole of 1.0 (according to Leica's instructions) and 16 times averaging. All other micrographs were obtained with a Zeiss (Thornwood, NY) Axiophot microscope with a camera (CH-250; Photometrics, Tucson, AZ). Adobe Photoshop (Adobe Systems, Mountain View, CA) was used for image processing.
Western Analysis of Parasite Lysates
SDS-PAGE was performed using standard methods (Leammli, 1970). Crude extracts from T. gondii tachyzoites were separated by SDS-PAGE and transferred to nitrocellulose. Western blot analysis was carried out essentially as described previously (Soldati et al., 1998), using 8–12% polyacrylamide gels run under reducing condition with 144 mM β-mercaptoethanol or 100 mM DTT in the loading samples. After electrophoresis, proteins were transferred to Hybond ECL nitrocellulose (Amersham, Arlington Heights, IL). For detection, the membranes were incubated with the mAb 9E10 (mouse ascites fluid diluted 1:1000 in PBS and 0.5% Tween 20) and then with the affinity-purified horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (1:2000), and bound antibodies were visualized using the ECL system (Amersham). Direct recording of chemoluminescent signals and densitometry by an LAS-1000 luminescent image analyser (Fuji, Tokyo, Japan) allowed quantification of signal intensities within a broad linear range.
Cell Fractionation
Freshly released parasites (108) from infected cells were resuspended into 300 μl of buffer and lysed by freezing and thawing three times followed by sonification (four times for 30 s on ice). Pellet and soluble fractions were separated by ultracentrifugation 1 h at 55,000 rpm at 4°C. The buffers were PBS, PBS and 1 M NaCl, PBS and 0.1 M Na2CO3, pH 11.5, PBS and 2% Triton X-100, and 3 M urea. In addition, all buffers contained 5 mM ATP to minimize the interaction of myosins with F-actin, and 5 mM EDTA.
Purification of Recombinant Myosin from T. gondii
Freshly lysed transgenic parasites expressing TgM-AΔtail (1010 parasites) were lysed under native conditions by freezing and thawing followed by sonification. The soluble fraction was separated by ultracentrifugation at 65,000 rpm, and the supernatant was applied to an Ni-nitrilotriacetic acid resin column and eluted according to the manufacturer (The QIAexpressionist; Janknecht et al., 1991); determination of protein concentration was performed by a Bradford assay (Bio-Rad).
Actin Binding Assays
The binding of TgM-AΔtail to F-actin was measured using an F-actin sedimentation assay as described previously (Jung and Hammer, 1994). F-actin from rabbit skeletal muscle was purified according to the method of Pardee and Spudich (1982). Binding assays were performed using a 4 μM final concentration of F-actin and 0.5 μg of TgM-AΔtail (corresponding to ∼0.06 μM), The buffer (plus ATP) contained 10 mM Tris-HCl, pH 7.5, 130 mM KCl, 2 mM MgCl2, 2 mM ATP, and 0.1 mM DTT. The washing steps were performed by resuspending the pellet in washing buffer and were followed by centrifugation at 50,000 rpm. Pellets and supernatants were analyzed by SDS-PAGE and Coomassie blue staining.
RESULTS
Identification and Cloning of Two Members of the Class XIV Myosins
To identify myosins in T. gondii, we designed a PCR screening strategy taking advantage of the highly conserved motifs present in the head domain (Schwarz et al., 1999). The approach identified TgM-A, a myosin already described by Heintzelman and Schwartzman (1997), and TgM-D, an additional member of the apicomplexan-specific class XIV of myosins (Figure 1B). The entire TgM-D gene was sequenced. The protein predicted from the open reading frame has 823 amino acid residues (Figure 1A) with a mass of 91 kDa (accession number AF105117).
The sequences of the short and conserved, positively charged tail domains of TgM-A and TgM-D showed a significant homology with an expressed sequence tag derived from the closely related organism P. falciparum. This allowed us to identify and subsequently clone a complete myosin corresponding to a partial P. falciparum sequence present in the database (PfMyo-1; see MATERIALS AND METHODS). We named this myosin PfM-A (Figure 1A) because its predicted sequence of 90 kDa has highest homology to TgM-A (Figure 1B). The degree of conservation at the amino acid level between the myosins is high and extends throughout the entire coding region. There is 55.7% identity between TgM-A and TgM-D, 63.1% between TgM-A and PfM-A, and 52.0% between TgM-D and PfM-A. Pairwise comparisons between any of these myosins with the other two, alternatively spliced, class XIV myosins from T. gondii, TgM-B and -C, give lower identity scores of ∼47–49%.
Like the other apicomplexan myosins TgM-D and PfM-A have a very short tail, do not contain the strictly conserved glycine residue at the proposed fulcrum point of the lever arm, and appear to lack conserved “IQ” motifs that bind calmodulin and calmodulin-related proteins. Among their particularities, and contrary to the P. falciparum molecule, all the T. gondii myosins do not follow the TEDS rule (Heintzelman and Schwartzman, 1997; Figure 1A), which describes the presence of an acidic or phosphorylatable residue at a precise position close to the actin-binding region (originally mapped by Brzeska et al., 1989, 1990; for review, see Bement and Mooseker, 1995). In lower eukaryotes, phosphorylation of this conserved serine or threonine was shown to be crucial for the stimulation of the ATPase activity of class I myosins (Bement and Mooseker, 1995; Carragher et al., 1998; Novak and Titus, 1998). It is unclear at the moment whether and how these class XIV motors are activated and how conformational changes in the molecules occur.
The different recombinant constructs of TgM-A and TgM-D used in this study are presented schematically in Figure 1C.
Antibodies to the Tail of TgM-A Are Isoform Specific
To investigate the expression and distribution of TgM-A, we raised polyclonal sera against two peptides covering its entire tail. The specificity of the antibodies was assessed by Western blotting of parasites expressing GFP·TgM-Atail and GFP·TgM-Dtail (Figure 2A). The anti-myc antibodies detected both GFP chimeras, but despite high overall sequence homologies, the anti-tail antibodies recognized only the TgM-A tail (Figure 2A). In wild-type parasites, the affinity-purified antibody recognized a single band migrating above 90 kDa, the predicted size of TgM-A (Figure 2B). Recombinant mycTgM-A was detected by the anti-tail antibodies and was slightly bigger than endogenous TgM-A. Confirming the antibody specificity, only the endogenous protein was detectable in parasites expressing mycTgM-AΔtail (Figure 2B).
Figure 2.
Immunoblot analysis of wild-type and recombinant parasites. (A and B) The antibody against the tail of TgM-A does not recognize the tail of TgM-D. Extracts of parasites expressing GFP·TgM-Atail or GFP·TgM-Dtail were blotted with anti-myc antibodies (mAb 9E10) and with anti-TgM-A tail antibodies (A). (B) The expression of mycTgM-A down-regulates the level of endogenous protein. Detection of endogenous and recombinant TgM-A with an antibody directed against its tail is shown. The lanes were loaded with similar amounts of freshly lysed wild-type parasites or parasites expressing mycTgM-A, mycTgM-AΔtail, or GFP·TgM-Atail.
The Expression of Endogenous TgM-A Is Down-regulated in the Transgenic Parasites Expressing Full mycTgM-A
Surprisingly, in recombinant parasites expressing full-length mycTgM-A, the endogenous TgM-A was barely detectable (Figure 2B). To obtain a quantitative impression of the phenomenon, equal numbers of wild-type and different recombinant parasites were analyzed (Figure 2B). MycTgM-A appeared to be expressed at approximately two- to threefold the level of endogenous protein found in wild-type parasites and parasites expressing mycTgM-AΔtail. We conclude that expression of full-length mycTgM-A, but not of the head domain alone, led to down-regulation of the endogenous TgM-A protein and expression. In parasites expressing GFP·TgM-Atail, the anti TgM-A tail antibodies simultaneously detected the 40-kDa GFP chimera and the endogenous TgM-A. Expression of the endogenous protein appeared slightly reduced compared with wild-type cells, indicating that the tail alone mimicked the down-regulation effect observed with the full-length myosin. Because the signal for the GFP chimera was approximately half as intense as for mycTgM-A (Figure 2B, first lane), we suggest that the effect is partial probably because of a lower expression level.
The Presence of Its Tail Influences the Expression of TgM-D
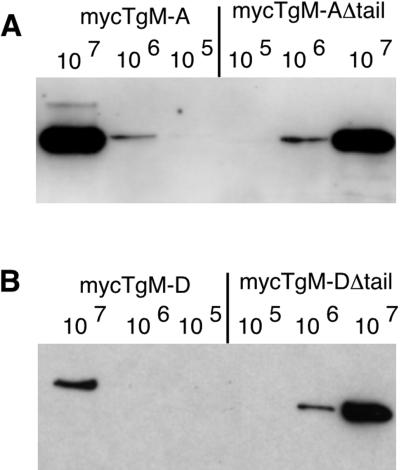
TgM-D was well expressed transiently, but our initial efforts to obtain stable transformants were unsuccessful, potentially because mycTgM-D overexpression was not well tolerated by the parasites. To circumvent this problem, we introduced the selectable marker directly in the expression vector and obtained a few stable cell lines expressing mycTgM-D. Expression of transgenic mycTgM-D (Figure 3B) was significantly reduced compared with mycTgM-A (Figure 3A), although the promoter used to drive its expression was previously shown to be stronger (Soldati and Boothroyd, 1995). The construct pT·TgM-DΔtail could readily be introduced into the parasites by simple cotransfection with the selection marker plasmid. Immunoblot analysis of total cell lysates with an antibody against the c-myc epitope showed that parasites transformed with pS·TgM-A or pT·TgM-D expressed a single protein of the expected sizes of 93 and 91 kDa (Figure 3). Deletion of their tails resulted in slightly higher mobility. A semiquantitative analysis was performed by comparing the signal intensities resulting from loading lanes with a precise number of parasites. This revealed that mycTgM-A and mycTgM-AΔtail were expressed at comparable levels (Figure 3A), whereas mycTgM-DΔtail was expressed at an ∼5- to 10-fold higher level than mycTgM-D (Figure 3B). These results provided evidence that the presence of the tail ofTgM-D limits the expression level of the recombinant protein.
Figure 3.
The presence of its tail limits the expression level of recombinant TgM-D. Western blot analysis used the anti-myc antibody on parasites expressing recombinant mycTgM-A and mycTgM-AΔtail (A) or mycTgM-D and mycTgM-DΔtail (B). The respective expression levels of each full-length and truncated myosin were judged by comparing the signals resulting from the loading of 105, 106, or 107 parasites, respectively. MycTgM-A and mycTgM-AΔtail,as well as mycTgM-DΔtail, were expressed at comparable levels, whereas full-length mycTgM-D was expressed at an approximately 10-fold reduced level.
TgM-A Localization Is Confined to the Parasite Periphery
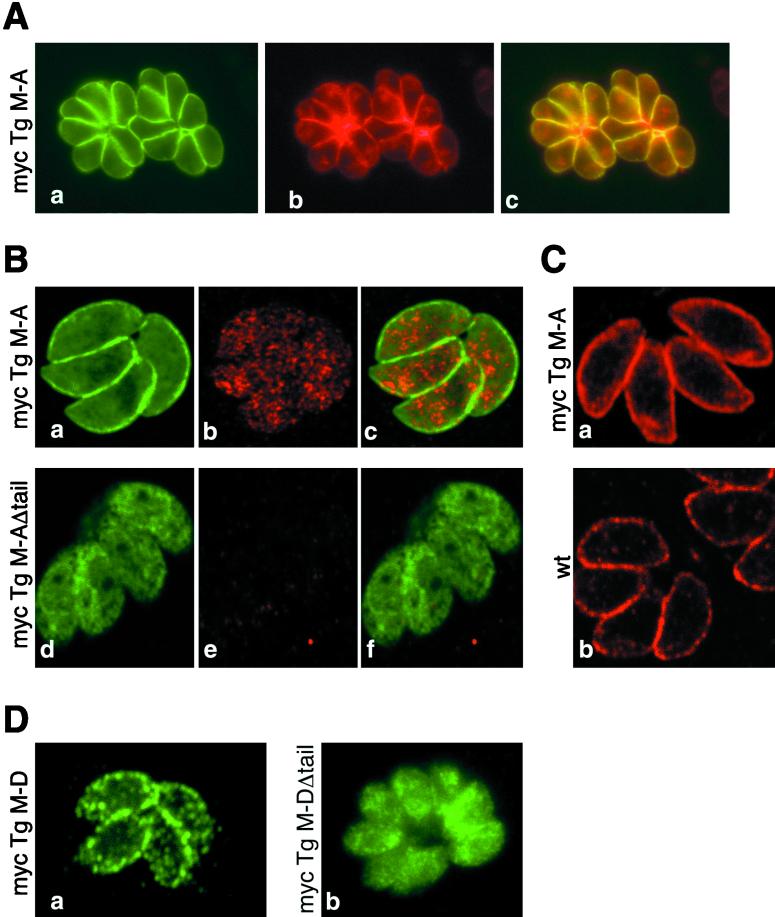
TgM-A and TgM-D share a high degree of homology, and it is not yet known whether T. gondii expresses additional similar myosins (in addition to the related TgM-B and -C, which harbor longer and different tail domains). To determine unambiguously, without risk of cross-reaction, the subcellular localization of TgM-A, TgM-D and mutants thereof, two approaches were used. First we generated isoform-specific antibodies. Second, we used an epitope-tagging strategy, taking advantage of the easy accessibility of T. gondii to genetic manipulation. The anti-myc tag antibody revealed that, aside from a relatively diffuse cytoplasmic distribution, a proportion of mycTgM-A localized precisely beneath the plasma membrane (Figure 4A, a), as visualized by the almost perfect colocalization with SAG1, the major surface antigen of T. gondii tachyzoites (Figure 4A, b). Because SAG1 is anchored at the plasma membrane via a glycosylphosphatidylinositol, the staining suggests that TgM-A associates closely with this membrane, although the resolution of light microscopy cannot exclude that the myosin interacts with the inner membrane complex. This latter structure is composed of flattened membrane cisternae found in close apposition with the plasma membrane of all Apicomplexa. It covers the elaborate basket of microtubules and contributes to the maintenance of cell shape and polarity (Morrissette et al., 1997).
Figure 4.
Immunofluorescence localization of TgM-A and TgM-D and their respective tail-less constructs. Classical (A and D) and confocal (B and C) immunofluorescence microscopic analysis of wild-type parasites (C, b) or parasites expressing mycTgM-A (A, B, a–c, and C, a), mycTgM-AΔtail (B, d–f), mycTgM-D (D, a), or mycTgM-DΔtail (D, b) is shown. Fixation was performed either by paraformaldehyde-glutaraldehyde (A, B, and D) or ultracold methanol (C). (A) MycTgM-A, visualized by anti-myc antibodies (a), colocalized at the parasite periphery (c) with SAG1, the major glycosylphosphatidylinositol-anchored surface antigen of T. gondii (b). (B and C) The antibody against the tail of TgM-A did not detect TgM-A (B, e) or mycTgM-A (B, b) at the plasma membrane when cells were fixed by paraformaldehyde-glutaraldehyde, even though mycTgM-A was detected at the periphery by anti-myc staining (B, a). The rapid freezing and fixation in ultracold methanol allowed peripheral localization of endogenous TgM-A (C, b) and mycTgM-A (C, a). Myc-TgM-AΔtail was mainly found in the cytoplasm (B, d). The anti-tail antibodies detected a cytosolic pool of mycTgM-A (B, b). (D) As detected by anti-myc antibodies, mycTgM-D was enriched at the parasite periphery (a) but in a way distinct from TgM-A (compare with A–C). MycTgM-DΔtail was mainly cytoplasmic (b).
Surprisingly, despite strong and specific signals obtained on Western blots (Figure 2), imunofluorescence using the anti-TgM-A tail antibodies failed to detect endogenous TgM-A in wild-type parasites (our unpublished results) and parasites expressing mycTgM-AΔtail (Figure 4B, e) and also failed to corroborate the peripheral localization of mycTgM-A (Figure 4B, compare a and b). Nevertheless, in the latter case a signal was clearly perceived and was similar in intensity (but not in staining pattern) to the cytoplasmic signal obtained for mycTgM-AΔtail (Figure 4B, compare d and b). It is reasonable to assume that the tail of TgM-A, being very rich in arginine and lysine residues, aldehyde cross-linking to plasma membrane proteins, may lead to complete epitope masking. Therefore, we used an alternative fixation method involving rapid freezing fixation and permeabilization in ultra-cold methanol, an ideal method to preserve both structure and antigenicity (Neuhaus et al., 1998). Figure 4C illustrates that both the endogenous TgM-A (Figure 4C, b) and the myc-tagged protein (Figure 4C, a) were now detected by the anti-tail antibody. As expected, the signal for TgM-A was slightly lower in wild-type parasites than in recombinants expressing the myc-tagged protein. The cytoplasmic staining for mycTgM-A (Figure 4B, b) was stronger than for endogenous TgM-A (Figure 4B, e). Finally, irrespective of the expression level, both proteins localized similarly all around the parasite periphery, confirming the epitope-tagging data.
The Short Tail Domains Are Necessary for Plasma Membrane Localization of the T. gondii Myosins
Myosin molecules are modular motors made up of three domains. The N-terminal domain is the actin binding motor unit per se. The middle neck domain bears the light chains and acts as a lever arm. The tail domain is exceptionally divergent and reflects the diversity in myosin functions. The tail is thought to target a given myosin to its cargo or site of action and thereby to determine the specific task of the motor. To assess the role of the short tail domains in the subcellular distribution of the two proteins, we constructed vectors expressing myosins lacking their tail. Recombinant parasites expressing mycTgM-AΔtail and mycTgM-DΔtail were analyzed by confocal microscopy (Figure 4, B and D). As already observed above, absence of tail in mycTgM-AΔtail caused the disappearance of the membrane-associated pool (Figure 4B, d).
In a way similar to mycTgM-A, mycTgM-D appeared to distribute predominantly at the parasite periphery (Figure 4D, a), even though not as sharply as mycTgM-A. The subtle difference in distribution may indicate a different mode or mechanism of localization. As in the case of mycTgM-A, the absence of tail caused an apparent redistribution of mycTgM-D to the cytoplasm (Figure 4D, b), and, as mentioned above (Figure 3B), it also led to a significant increase in the level of the recombinant product compared with the full-length protein.
The Tail of TgM-A Is Necessary for Distribution in the Particulate Fraction
To corroborate the localization data with biochemical fractionation, cells were first lysed in PBS, separated into soluble and sedimentable fractions by high-speed centrifugation, and analyzed by Western blotting. A comprehensive analysis was undertaken on cells expressing GFP·TgM-Atail, allowing for simultaneous investigation of endogenous TgM-A and of the GFP tail chimera. In agreement with the imunofluorescence, TgM-A partitioned with the particulate fraction (Figure 5A). The nature of this interaction was further investigated. TgM-A behaved as a strongly associated peripheral membrane protein, because it was resistant to solubilization by high salt. Full solubilization was achieved only by carbonate treatment (pH 11.5) and extraction by detergent (1% Triton X-100) or 3 M urea (our unpublished results).
Even though mycTgM-AΔtail appeared cytoplasmic (Figure 4B, d), it is formally possible that TgM-A localized at the periphery and, despite inclusion of ATP during fractionation, interacted with the particulate fraction through binding of its head domain to the actin cytoskeleton. Therefore, the role of the tail domain was investigated for recombinant TgM-A (Figure 5C). When parasites were lysed in PBS, about one-third to one-half of mycTgM-A sedimented with the particulate, membrane fraction whereas mycTgM-AΔtail partitioned essentially with the soluble cytosolic fraction, in perfect agreement with the imunofluorescence data. This indicated that the sedimentation of TgM-A was not due to interactions through the motor domain but was likely mediated by the tail domain (also see Figure 7 and accompanying text). Similar results were obtained for mycTgM-D (Figure 5C). Moreover, whereas endogenous TgM-A is quantitatively recovered with the particulate fraction (Figure 5, A and B), approximately half of mycTgM-A, which is overexpressed two- to threefold, is found in the soluble fraction (Figure 5C), possibly because a plasma membrane receptor is saturated.
TgM-A Binds to F-actin in an ATP-dependent Manner
The peripheral targeting of TgM-A appears not to depend on the interaction of the head domain with the actin cytoskeleton. This could potentially be caused by the fact that the motor domains of class XIV myosins are predicted to exhibit important structural divergences. Therefore, it was crucial to assess experimentally whether these proteins truly act as ATP-driven, actin-dependent motors. In addition, according to the capping model of invasion, the myosin involved would have to be able to grab onto F-actin and exert ATP-dependent traction. The complete solubility of recombinant mycTgM-AΔtail in tachyzoites allowed us to purify biochemical quantities of the protein under native conditions. The degree of purity of the recombinant mycTgM-AΔtail was examined by SDS-PAGE and Coomassie blue staining (Figure 6A). The identity of two low-molecular-mass bands (∼30 kDa) has not been fully determined yet (our unpublished results), but they were distinct from degradation products of TgM-A and could potentially represent copurifying light chain(s). To gain first insights into the biochemical characteristics of this mycTgM-AΔtail, a cosedimentation assay with F-actin was performed (Figure 6B). The results showed that mycTgM-AΔtail coprecipitated with actin only in the absence of ATP and that the sedimented myosin was subsequently, even though only incompletely, released by the addition of 10 mM ATP, demonstrating that this purified myosin was able to reversibly bind actin in an ATP-dependent manner. Similar results have been obtained with full-length mycTgM-A (our unpublished results). Together, our results also demonstrated the functionality of both recombinant TgM-A constructs.
Figure 6.
MycTgM-AΔtail sediments with F-actin in an ATP-dependent manner. (A) SDS-PAGE and Coomassie blue staining of mycTgM-AΔtail purified from an extract of recombinant parasites. (B) SDS-PAGE analysis and Coomassie blue staining of the supernatant (S) and pellets (P) obtained after cosedimentation of mycTgM-AΔtail with F-actin in the presence (+) or absence (−) of ATP. MycTgM-AΔtail was completely soluble in the absence of F-actin (Control). MycTgM-AΔtail could be released from F-actin by subsequent incubation with ATP. Rabbit muscle F-actin was used at 4 μM and ATP at 10 mM in the presence of 130 mM KCl.
The Short TgM-A and TgM-D Tail Domains Are Sufficient to Target a Reporter Protein to the Cell Periphery
The short basic tails of TgM-A, TgM-D and PfM-A are very similar in length and in amino acid composition. A comparison of the myosin tails is depicted in Figure 7A. The conserved spots of basic residues are highlighted. Analysis of the predicted secondary structure of these domains also revealed a conserved overall organization in three helices interrupted by short loops. To determine whether the short tail domains are sufficient to confer specific cellular distribution to the myosins, the tails of TgM-A (last 82 amino acids) TgM-D (last 57 amino acids), and PfM-A (last 85 amino acids) were fused to GFP. The plasmids pT·GFP and pT·GFP·TgM-Atail were stably integrated into tachyzoites by cotransfection with HXGPRT expression vector. We had to introduce the selectable marker gene on pT·GFP·TgM-Dtail and pT·GFP·PfM-Atail to obtain stable transformants. The transgenic parasites expressing GFP and GFP·TgM-Atail were analyzed by direct GFP fluorescence, whereas GFP·TgM-Dtail and GFP·PfM-Atail required indirect immunofluorescence analysis using anti-myc antibodies to definitely visualize the recombinant protein (Figure 7B). The nonfusion GFP was abundantly expressed, homogeneously distributed in the cytosol with a significant accumulation in the nucleus (Figure 7B, a). In contrast, GFP·TgM-Atail was found almost exclusively closely associated with the plasma membrane, with barely detectable cytosolic staining (Figure 7B, b). In a way reminiscent of the respective localization of the full-length proteins, GFP·TgM-Dtail showed a slightly more diffuse and speckled peripheral signal than the GFP·TgM-Atail chimera (Figure 7B, c). The difficulty reported above to express high amounts of mycTgM-D appeared indeed to be dependent of the tail domain, because parasites expressing GFP·TgM-Dtail were also difficult to obtain, and the fusion was expressed at a very reduced level compared with both GFP and GFP·TgM-Atail. This was visible on the immunofluorescence stainings and was confirmed by Western blotting (our unpublished results). Similarly, the GFP·PfM-Atail fusion was expressed at a low level and localized essentially to the parasite cytosol (Figure 7B, d).
The GFP·TgM-Atail and GFP·TgM-Dtail Chimeras Partition with the Membrane Fraction
The transgenic parasites expressing GFP and chimeras were lysed in PBS, separated into soluble and particulate fractions, and subsequently analyzed by Western blot. As illustrated in Figure 7C, the migration on SDS-PAGE of the GFP chimera was in agreement with their predicted sizes. GFP·TgM-Atail and GFP·TgM-Dtail completely partitioned with the particulate fraction, whereas GFP and GFP·PfM-Atail were entirely recovered in the supernatant. The quantitative association of GFP with the particulate fraction was comparable with the behavior of endogenous TgM-A. In contrast, GFP·TgM-Dtail sedimented despite the fact that the main part of mycTgM-D was soluble. This could potentially indicate that both GFP chimeras were expressed at levels that did not saturate their respective peripheral binding sites. In addition, it is important to note that both chimeras were fully solubilized by carbonate treatment, demonstrating that they were not simply aggregated in the cell (Figure 7C). In fact, the solubilization characteristics of GFP·TgM-Atail were extremely similar to the ones of endogenous TgM-A (see Figure 5A).
The Localization of TgM-A Is Not Due to Interactions with the Actin Cytoskeleton
Altogether, the data indicate a strong interaction of the tail of TgM-A with components of the parasite cortex, likely with proteins associated peripherally with the plasma membrane, even though an interaction with the closely apposed inner membrane complex cannot be excluded. To definitively eliminate the potential targeting role of the cortical actin cytoskeleton, F-actin was severely compromised by cytochalasin D incubation. It has been previously reported that phalloidin fails to stain the actin filaments in T. gondii (Dobrowolski et al., 1997b). Therefore, as indication of the treatment efficacy, the peripheral localizations of mycTgM-A (Figure 8A, a and b) and GFP·TgM-Atail (Figure 8A, c and d) are shown simultaneously with a phalloidin staining of the host actin cytoskeleton. No noticeable change of localization of both mycTgM-A and GFP·TgM-Atail was observed despite severe disturbance of the actin cytoskeleton (Figure 8A, compare b with a and d with c).
The Tail of TgM-A Does Not Localize to the Plasma Membrane of HeLa Cells
Stretches of basic residues in the tail of class I myosins have been shown to contain high-affinity phospholipid binding sites (Doberstein and Pollard, 1992). If TgM-A were to interact with the plasma membrane solely through unspecific interaction with lipids, the peripheral localization should be observed in unrelated cellular systems. When expressed in HeLa cells (Figure 8B), GFP·TgM-Atail is homogeneously distributed in the cytoplasm, confirming that the association with the particulate fraction in T. gondii was not due to improper folding. More importantly, the chimera showed no sign of membrane association. Indeed, its cytoplasmic localization (Figure 8B, d) was indistinguishable from the one of GFP (Figure 8B, b).
Identification of the Molecular Determinants of Peripheral Membrane Localization
To define precisely, at the amino acid level, the determinants of membrane localization, deletion analysis and site-specific mutagenesis of the basic residues in the tail of TgM-A fused to GFP were undertaken (Figure 9A). Faithful expression and stability of the respective constructs were confirmed by immunoblotting (Figure 9C). The GFP fusions with mutants of TgM-A tail were also examined by indirect immunofluorescence (Figure 9B). Deletion of the C-terminal extension composed of the last 14 amino acids (TgM-AtailΔ14) did not alter the association of mycTgM-A with the cell membrane. In contrast, a further deletion encompassing the last 22 amino acids (TgM-AtailΔ22) caused a complete loss of membrane localization. The mutagenesis of two arginine residues within the last 22 amino acids into alanine residues (TgM-Atail mut III) completely abolished localization. However, the conversion of the three other sets of basic residues into neutral amino acids (TgM-Atail mut I and TgM-Atail mut II) did not alter significantly the plasma membrane distribution. These data indicate that the localization of TgM-A is directed by two precise residues rather than by the overall positive charge of the tail domain.
DISCUSSION
A Family of Apicomplexan Myosins Presenting Divergent Structural Features
T. gondii and P. falciparum are both obligate intracellular parasites sharing a similar mechanism of host cell penetration. According to the capping model for host cell invasion, a myosin is necessary to mediate the ATP-dependent actin-based motility (Sibley et al., 1998). The molecular motor mediating this process has not been identified so far. In the present study, we have cloned and characterized two apicomplexan myosins that share a high degree of homology with the previously identified class XIV of myosins (Heintzelman and Schwartzman, 1997). With a molecular mass ranging between 90 and 93 kDa, these myosins are the smallest molecular motors identified so far. Each of the T. gondii myosins exhibit either a glutamine or an asparagine at the TEDS site, which is neither acidic nor phosphorylatable, potentially implying either different structural requirements for activation or a different post-translational modification involved in the regulation of these myosins. We are exploring the speculative hypothesis that a deamidase might convert asparagine or glutamine into acidic residues at the TEDS site. Among the >100 known myosins, there are few others making exception to the TEDS rule, such as two Acetabularia class XIII myosins, myosin IB of Entamoeba histolytica (Vargas et al., 1997), and a Drosophila class III myosin (Montell and Rubin, 1988). Like some T. gondii myosins, the Drosophila myosin IA has an Asn (Strom Morgan et al., 1994). Another striking feature of all apicomplexan myosins is the presence of a serine instead of the highly conserved glycine corresponding to Gly699 in the chicken myosin II (Kinose et al., 1996). This structural feature has only been reported for a plant myosin (HaMyok3; D. Menzel, unpublished data) and a Caenorhabditis elegans class XII myosin (Baker and Titus, 1997). The conserved glycine residue likely plays an essential role in the conformational changes around the actin- and nucleotide-binding sites, maybe functioning as a pivot point for the lever arm. Mutation of this glycine into an alanine residue dramatically alters the motor activity of the skeletal muscle myosin and the Dictyostelium discoideum myosin II, inhibiting the velocity of actin filament movement by 100-fold (Kinose et al., 1996; Patterson and Spudich, 1996; Patterson et al., 1997). Finally, class XIV myosins do not carry classical IQ motifs for the binding of light chains but harbor a conserved stretch of amino acids that might represent a very divergent form of this motif.
Because of these important structural divergences, it was crucial to assess experimentally whether the apicomplexan myosins truly act as ATP-driven, actin-dependent motors. Also, a requirement of the capping model of invasion is that the motor involved has to grab onto F-actin and exert ATP-dependent traction. The ability to express and purify recombinant myosins from T. gondii offers a unique opportunity to undertake combined in vivo and in vitro structure–function analyses of this novel class of myosins. Taking advantage of the solubility of mycTgM-AΔtail, we purified the recombinant protein from parasites under native conditions. This material was used in pilot experiments aiming at the biochemical characterization of TgM-A. Indeed, F-actin sedimentation assay showed that TgM-A binds actin in an ATP-dependent manner. Preliminary transient ATPase kinetics and motility experiments indicate further that TgM-A purified from parasites is fully functional (our unpublished results).
TgM-A Localizes at the Plasma Membrane via Its Tail Domain
By using a reverse genetic approach and by generating a tail-specific antibody, we have determined the localization of the T. gondii myosin TgM-A. It distributed beneath the plasma membrane of tachyzoites, an ideal position to transmit mechanical energy in forward motion and thus to propel cell invasion. At this point, we have to mention that Heintzelman and Schwartzman (1999) recently reported similar data concerning the association of TgM-A with the particulate fraction of T. gondii, but contrary to our results, they documented a punctate localization of TgM-A near the apical pole of tachyzoites, possibly associated with some intracellular structure or organelle such as the rhoptries. The discrepancy is probably related to our problems in detecting peripheral localization after aldehyde fixation associated with the spurious detection of a cross-reactive rhoptry antigen.
TgM-D appears enriched in the peripheral region of the parasite but not as sharply defined as for TgM-A. The loss of membrane targeting of TgM-A and TgM-D resulting from deletion of their tail supports the idea that this domain mediates interaction with cargo and brings the myosin to its site of action. Conversely, fusion of the TgM-A tail to GFP confers membrane association to that otherwise soluble cytosolic protein. These data strongly support that the tail domain is the necessary and sufficient determinant mediating specific interactions of the myosin with integral or peripheral membrane constituents.
Expression of mycTgM-A Down-regulates the Level of Endogenous TgM-A
MycTgM-A and GFP·TgM-Atail transgenes were well expressed in the parasites. Whereas the GFP chimera was essentially associated with the periphery and the particulate fraction, only approximately half of mycTgM-A behaved that way, suggesting that the specific membrane “receptor” is saturable. Intriguingly, drastic down-regulation of endogenous TgM-A was observed upon expression of recombinant mycTgM-A. This phenomenon appeared to be associated with overexpression of the full TgM-A and at least partially also of GFP·TgM-Atail but was not seen with mycTgM-AΔtail. We hypothesize that the parasites autoregulate the level of TgM-A in accordance with the abundance of a receptor present in limiting amounts at the cell periphery.
The difficulties encountered in the generation of recombinant parasites expressing TgM-D and the very low level of protein produced suggest a similar phenomenon. The high expression levels of TgM-DΔtail contrast with the low expression levels of GFP·TgM-Dtail and argue for a regulatory effect of the tail domain. Analogously, the tail of PfM-A seems to induce a comparable effect. It is conceivable that the limited expression of TgM-D, GFP·TgM-Dtail, and GFP·PfM-Atail are due to their inappropriate presence in cells lacking either a stage- or species-specific partner, respectively. The potential role of stage-specific regulation is not well understood yet. Preliminary studies on the developmental pattern of T. gondii myosin gene expression suggest that TgM-A is constitutively expressed in tachyzoites and bradyzoites, whereas TgM-D transcripts accumulate predominantly in bradyzoites (F. Delbac, A. Sänger, C. Toursel, S. Tomavo, and D. Soldati, unpublished results).
Note that in all cases expression is limited, but we did not notice deleterious effects on the morphology and proliferation of parasites. This contrasts with observations reported for the expression of other myosins. Overexpression of entire unconventional myosin myoIB from D. discoideum is an example of deleterious effects leading to a phenotype as severe as observed in null cells (Novak and Titus, 1997). Similarly, the overexpression of MYO4 or MYO2 in S. cerevisiae led to morphological abnormalities (Haarer et al., 1994).
The exact nature of the interaction between TgM-A tail and the cell membrane is not elucidated yet; however, in addition to the down-regulation phenomenon discussed above, the following evidence further points toward a parasite-specific proteinaceous receptor. First, the fractionation experiments with mycTgM-A as well as GFP·TgM-Atail argue for a very strong interaction of the tail with the constituents of the plasma membrane. Second, the tails of TgM-A and PfM-A have similar overall charges but markedly differ in their association with the periphery. Third, the mutagenesis experiments indicate that two specific residues and not simply the overall charge of the tail domain are important for localization, rendering it unlikely that it interacts nonspecifically with phospholipids. This is corroborated by the fact that the GFP·TgM-Atail had an exclusively cytoplasmic distribution in HeLa cells. Finally, after aldehyde fixation, the failure to detect the tail antigen specifically when TgM-A is at the plasma membrane suggests that it is engaged in strong interactions with a protein component.
Is TgM-A or TgM-D Powering Gliding Locomotion?
To accommodate the capping model for invasion, the myosin shall lie beneath the plasma membrane and interact with transmembrane proteins exposed at the cell surface at the time of invasion. The motility shall then be driven by the concerted redistribution of these transmembrane proteins toward the posterior pole of the parasite. The TRAP protein (trombospondin-related adhesive protein) expressed in the sporozoites of Plasmodium species and its homologue MIC2 in T. gondii (Wan et al., 1997) are exported at the surface of the parasite and relocalize to the posterior pole during invasion (Carruthers et al., 1999). This protein is an excellent candidate to interact directly or indirectly with the actomyosin system. Disruption of the TRAP gene in Plasmodium berghei demonstrates that this protein is necessary not only for sporozoite infection of the mosquito salivary glands and the rat liver but also for gliding motility of the sporozoites in vitro (Sultan et al., 1997). The TgMIC2 C-terminal domain has recently been shown to functionally complement TRAP mutants lacking their C-terminal domain in P. berghei (Kappe et al., 1999). Although MIC2 constitutes an attractive partner for TgM-A during invasion, it is only transiently present at the surface of the parasite during host cell penetration and therefore is unlikely to serve as receptor for TgM-A at the plasma membrane during the intracellular replicative phase of the life cycle (Carruthers and Sibley, 1997). It is conceivable that an abundant protein serves as a docking partner, ideally positioning the myosin close to its later site of action. Once at the plasma membrane, the myosin could respond immediately to an activation signal and trigger invasion by establishing a connection with proteins such as MIC2.
A recent study provides evidence for a myosin II responsible for the capping of surface receptors in E. histolytica (Arhets et al., 1998). In light of such a finding, it is rather surprising that no conventional myosin of type II has been identified in Apicomplexa so far. In comparison, despite its simplicity, even S. cerevisiae relies on the function of five myosins from three distinct classes (Brown, 1997). Our most recent studies revealed the presence of a fifth myosin of class XIV in T. gondii (our unpublished results). Nevertheless, because our screening is not exhaustive, we cannot yet exclude the presence of other classes of myosins in T. gondii. The genome sequencing project for P. falciparum is still in progress, and the current status reveals that at least two additional myosin genes are present in this parasite, both of which appear to be closest to class XIV. Although not yet confirmed by functional data, TgM-A is definitely guilty by localization. Besides unraveling its potentially crucial role in invasion, the investigation of the biochemical details of enzymatic and motile properties of this very divergent class of unconventional myosins holds the promise of shedding light on the fundamental requirements of myosin function.
Note added in proof. A fragment corresponding to the complete PfM-A cDNA reported here was previously described as Pfmyo-1 (Pinder et al., 1998). We named all the class XIV myosins according to the nomenclature introduced by Heintzelman and Schwartzman (1997). To minimize the confusion, Pinder and colleagues now kindly agree to rename Pfmyo-1 Pfmyo-A.
ACKNOWLEDGMENTS
We are indebted to C. Kistler for assistance and expertise in the actin sedimentation assays and to Dr. J. Ajioka for assistance in the screening of the T. gondii genomic libraries. We are very grateful to Dr. J.F. Dubremetz for providing the biotinylated anti-SAG1 antibody and to Dr. D. Chakabarti for kindly and promptly providing us with the P. falciparum cDNA clone Pf1550C. Dr. D Lawson at the Sanger Center was instrumental in completing the PfM-A gene sequence. The PfM-B and -C coding sequences were assembled from sequence data of chromosomes 5 and 13 obtained from the Sanger Center web site (http://www.sanger.ac.uk/Projects/P_falciparum/). Sequencing of P. falciparum chromosomes 5 and 13 was accomplished as part of the Malaria Genome Project with support by The Wellcome Trust. We thank Dr. J. Haseloff for sending us the modified GFP (mgfp-5ER). This work was supported by Deutsche Forschungsgemeinschaft grants SO 366/1-1 and SO366/1-2. E.S. was supported by Deutsche Forschungsgemeinschaft grant SFB 352 (to T.S.).
REFERENCES
- Arhets P, Olivo JC, Gounon P, Sansonetti P, Guillen N. Virulence and functions of myosin II are inhibited by overexpression of light meromyosin in Entamoeba histolytica. Mol Biol Cell. 1998;9:1537–1547. doi: 10.1091/mbc.9.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JP, Titus MA. A family of unconventional myosins from the nematode Caenorhabditis elegans. J Mol Biol. 1997;272:523–535. doi: 10.1006/jmbi.1997.1232. [DOI] [PubMed] [Google Scholar]
- Bement WM, Mooseker MS. TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil Cytoskeleton. 1995;31:87–92. doi: 10.1002/cm.970310202. [DOI] [PubMed] [Google Scholar]
- Black M, Seeber F, Soldati D, Kim K, Boothroyd JC. Restriction enzyme-mediated integration elevates transformation frequency and enables co-transfection of Toxoplasma gondii. Mol Biochem Parasitol. 1995;74:55–63. doi: 10.1016/0166-6851(95)02483-2. [DOI] [PubMed] [Google Scholar]
- Brown SS. Myosins in yeast. Curr Opin Cell Biol. 1997;9:44–48. doi: 10.1016/s0955-0674(97)80150-0. [DOI] [PubMed] [Google Scholar]
- Brzeska H, Lynch TJ, Martin B, Corigliano-Murphy A, Korn ED. Substrate specificity of Acanthamoeba myosin I heavy chain kinase as determined with synthetic peptides. J Biol Chem. 1990;265:16138–16144. [PubMed] [Google Scholar]
- Brzeska H, Lynch TJ, Martin B, Korn ED. The localization and sequence of the phosphorylation sites of Acanthamoeba myosins I. An improved method for locating the phosphorylated amino acid. J Biol Chem. 1989;264:19340–19348. [PubMed] [Google Scholar]
- Carragher BO, Cheng N, Wang ZY, Korn ED, Reilein A, Belnap DM, Hammer JA, III, Steven AC. Structural invariance of constitutively active and inactive mutants of Acanthamoeba myosin IC bound to F-actin in the rigor and ADP-bound states. Proc Natl Acad Sci USA. 1998;95:15206–15211. doi: 10.1073/pnas.95.26.15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers VB, Gidding OK, Sibley LD. Secretion of micronemal proteins is associated with Toxoplasma invasion of host cells. Cell Microbiol. 1999;1:225–235. doi: 10.1046/j.1462-5822.1999.00023.x. [DOI] [PubMed] [Google Scholar]
- Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–123. [PubMed] [Google Scholar]
- Catlett NL, Weisman LS. The terminal tail region of a yeast myosin-V mediates its attachment to the vacuole membranes and sites of polarized growth. Proc Natl Acad Sci USA. 1998;95:14799–14804. doi: 10.1073/pnas.95.25.14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein SK, Pollard TD. Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J Cell Biol. 1992;117:1241–1249. doi: 10.1083/jcb.117.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski JM, Carruthers VB, Sibley LD. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol Microbiol. 1997a;26:163–173. doi: 10.1046/j.1365-2958.1997.5671913.x. [DOI] [PubMed] [Google Scholar]
- Dobrowolski JM, Niesman IR, Sibley LD. Actin in the parasite Toxoplasma gondii is encoded by a single copy gene, ACT1 and exists primarily in a globular form. Cell Motil Cytoskeleton. 1997b;37:253–262. doi: 10.1002/(SICI)1097-0169(1997)37:3<253::AID-CM7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Dobrowolski JM, Sibley LD. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- Donald R, Carter D, Ullman B, Roos DS. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, Eb AJvd. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Petzold A, Lillie SH, Brown SS. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994;107:1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson T, Mooseker MS. Molecular motors, membrane movements and physiology: emerging roles for myosins. Curr Opin Cell Biol. 1995;7:587–594. doi: 10.1016/0955-0674(95)80017-4. [DOI] [PubMed] [Google Scholar]
- Heintzelman MB, Schwartzman JD. A novel class of unconventional myosins from Toxoplasma gondii. J Mol Biol. 1997;271:139–146. doi: 10.1006/jmbi.1997.1167. [DOI] [PubMed] [Google Scholar]
- Heintzelman MB, Schwartzman JD. Characterization of myosin-A and myosin-C: two class XIV unconventional myosins from Toxoplasma gondii. Cell Motil Cytoskeleton. 1999;44:58–67. doi: 10.1002/(SICI)1097-0169(199909)44:1<58::AID-CM5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Janknecht R, de Martynoff G, Lou J, Hipskind RA, Nordheim A, Stunnenberg HG. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc Natl Acad Sci USA. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Hammer JA., III The actin binding site in the tail domain of Dictyostelium myosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology region 2) FEBS Lett. 1994;342:197–202. doi: 10.1016/0014-5793(94)80500-8. [DOI] [PubMed] [Google Scholar]
- Kappe S, Bruderer T, Gantt S, Fujioka H, Nussenzweig V, Ménard R. Conservation of a gliding motility and cell invasion machinery in Apicomplexan parasites. J Cell Biol. 1999;147:937–944. doi: 10.1083/jcb.147.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinose F, Wang SX, Kidambi US, Moncman CL, Winkelmann DA. Glycine 699 is pivotal for the motor activity of skeletal muscle myosin. J Cell Biol. 1996;134:895–909. doi: 10.1083/jcb.134.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp B, Hundt E, Kupper HA. A new blood stage antigen of Plasmodium falciparum transported to the erythrocyte surface. Mol Biochem Parasitol. 1989;37:47–56. doi: 10.1016/0166-6851(89)90101-1. [DOI] [PubMed] [Google Scholar]
- Leammli UK. Cleavage of structural protein during the assembly of the head of bacteriophageT4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- Miller LH, Aikawa M, Johnson JG, Shiroishi T. Interaction between cytochalasin B-treated malarial parasites and erythrocytes. Attachment and junction formation. J Exp Med. 1979;149:172–184. doi: 10.1084/jem.149.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C, Rubin GM. The Drosophila ninaC locus encodes two receptor cell-specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell. 1988;52:757–772. doi: 10.1016/0092-8674(88)90413-8. [DOI] [PubMed] [Google Scholar]
- Morrissette NS, Murray JM, Roos DS. Subpellicular microtubules associate with an intramembranous particle lattice in the protozoan parasite Toxoplasma gondii. J Cell Sci. 1997;110:35–42. doi: 10.1242/jcs.110.1.35. [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Horstmann H, Almers W, Maniak M, Soldati T. Ethane-freezing/methanol-fixation of cell monolayers: a procedure for improved preservation of structure and antigenicity for light and electron microscopies. J Struct Biol. 1998;121:326–342. doi: 10.1006/jsbi.1998.3971. [DOI] [PubMed] [Google Scholar]
- Novak KD, Titus MA. Myosin I overexpression impairs cell migration. J Cell Biol. 1997;136:633–647. doi: 10.1083/jcb.136.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Titus MA. The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol Biol Cell. 1998;9:75–88. doi: 10.1091/mbc.9.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- Patterson B, Ruppel KM, Wu Y, Spudich JA. Cold-sensitive mutants G680V and G691C of Dictyostelium myosin II confer dramatically different biochemical defects. J Biol Chem. 1997;272:27612–27617. doi: 10.1074/jbc.272.44.27612. [DOI] [PubMed] [Google Scholar]
- Patterson B, Spudich JA. Cold-sensitive mutations of Dictyostelium myosin heavy chain highlight functional domains of the myosin motor. Genetics. 1996;143:801–810. doi: 10.1093/genetics/143.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder JC, Fowler RE, Dluzewski AR, Bannister LH, Lavin FM, Mitchell GH, Wilson RJ, Gratzer WB. Actomyosin motor in the merozoite of the malaria parasite, Plasmodium falciparum: implications for red cell invasion (in process citation) J Cell Sci. 1998;111:1831–1839. doi: 10.1242/jcs.111.13.1831. [DOI] [PubMed] [Google Scholar]
- Russell DG. Host cell invasion by Apicomplexa: an expression of the parasite's contractile system? Parasitology. 1983;87:199–209. doi: 10.1017/s0031182000052562. [DOI] [PubMed] [Google Scholar]
- Schwartzman JD, Pfefferkorn ER. Immunofluorescent localization of myosin at the anterior pole of the coccidian Toxoplasma gondii. J Protozool. 1983;30:657–661. doi: 10.1111/j.1550-7408.1983.tb05339.x. [DOI] [PubMed] [Google Scholar]
- Schwarz EC, Geissler H, Soldati T. A potentially exhaustive screening strategy reveals two novel divergent myosins in Dictyostelium. Cell Biochem Biophys. 1999;30:413–435. doi: 10.1007/BF02738122. [DOI] [PubMed] [Google Scholar]
- Shaw MK, Tilney LG. Induction of an acrosomal process in Toxoplasma gondii: visualization of actin filaments in a protozoan parasite. Proc Natl Acad Sci USA. 1999;96:9095–9099. doi: 10.1073/pnas.96.16.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley LD. Invasion of vertebrate cells by Toxoplasma gondii. Trends Cell Biol. 1995;5:129–132. doi: 10.1016/s0962-8924(00)88964-3. [DOI] [PubMed] [Google Scholar]
- Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature. 1992;359:82–85. doi: 10.1038/359082a0. [DOI] [PubMed] [Google Scholar]
- Sibley LD, S. H, Carruthers VB. Gliding motility: an efficient mechanism for cell penetration. Curr Biol. 1998;8:12–14. doi: 10.1016/s0960-9822(98)70008-9. [DOI] [PubMed] [Google Scholar]
- Soldati D, Boothroyd JC. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D, Lassen A, Dubremetz JF, Boothroyd JC. Processing of Toxoplasma ROP1 protein in nascent rhoptries. Mol Biochem Parasitol. 1998;96:37–48. doi: 10.1016/s0166-6851(98)00090-5. [DOI] [PubMed] [Google Scholar]
- Spudich JA. How molecular motors work. Nature. 1994;372:515–518. doi: 10.1038/372515a0. [DOI] [PubMed] [Google Scholar]
- Strom Morgan N, Skovronsky DM, Artavanis-Tsakonas S, Mooseker MS. The molecular cloning and characterization of Drosophila melanogaster myosin-IA and myosin-IB. J Mol Biol. 1994;239:347–356. doi: 10.1006/jmbi.1994.1376. [DOI] [PubMed] [Google Scholar]
- Sultan AA, Thathy V, Frevert U, Robson KJ, Crisanti A, Nussenzweig V, Nussenzweig RS, Menard R. TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell. 1997;90:511–522. doi: 10.1016/s0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- Vargas M, Voigt H, Sansonetti P, Guillen N. Molecular characterization of myosin IB from the lower eukaryote Entamoeba histolytica, a human parasite. Mol Biochem Parasitol. 1997;86:61–73. [PubMed] [Google Scholar]
- Wan KL, Carruthers VB, Sibley LD, Ajioka JW. Molecular characterization of an expressed sequence tag locus of Toxoplasma gondii encoding the micronemal protein MIC2. Mol Biochem Parasitol. 1997;84:203–214. doi: 10.1016/s0166-6851(96)02796-x. [DOI] [PubMed] [Google Scholar]
- Webb SE, Fowler RE, O'Shaughnessy C, Pinder JC, Dluzewski AR, Gratzer WB, Bannister LH, Mitchell GH. Contractile protein system in the asexual stages of the malaria parasite Plasmodium falciparum. Parasitology. 1996;112:451–457. doi: 10.1017/s0031182000076915. [DOI] [PubMed] [Google Scholar]
- Wes PD, Shawn Xu X-Z, Li H-S, Chien F, Doberstein SK, Montell C. termination of phototransduction requires binding of the NINAC myosin III and the PDZ protein INAD. Nat Neurosci. 1999;2:447–453. doi: 10.1038/8116. [DOI] [PubMed] [Google Scholar]