Abstract
Although homologues of the yeast checkpoint kinases Cds1 and Chk1 have been identified in various systems, the respective roles of these kinases in the responses to damaged and/or unreplicated DNA in vertebrates have not been delineated precisely. Likewise, it is largely unknown how damaged DNA and unreplicated DNA trigger the pathways that contain these effector kinases. We report that Xenopus Cds1 (Xcds1) is phosphorylated and activated by the presence of some simple DNA molecules with double-stranded ends in cell-free Xenopus egg extracts. Xcds1 is not affected by aphidicolin, an agent that induces DNA replication blocks. In contrast, Xenopus Chk1 (Xchk1) responds to DNA replication blocks but not to the presence of double-stranded DNA ends. Immunodepletion of Xcds1 (and/or Xchk1) from egg extracts did not attenuate the cell cycle delay induced by double-stranded DNA ends. These results imply that the cell cycle delay triggered by double-stranded DNA ends either does not involve Xcds1 or uses a factor(s) that can act redundantly with Xcds1.
INTRODUCTION
DNA damage, if not properly repaired, results in genomic instability, which is highly mutagenic and potentially lethal to the cell. During the cell cycle, the integrity of chromosomal DNA is ensured by surveillance systems that sense damaged DNA and arrest the cell cycle, giving the cell more time for repair processes. The surveillance systems that inhibit the entry into mitosis in the presence of damaged (or unreplicated) DNA consist of checkpoint signaling pathways that ultimately regulate the Cdc2-cyclin B complex, also known as maturation or M-phase promoting factor (Morgan, 1997; Rhind and Russell, 1998; Ohi and Gould, 1999). In various organisms, these pathways contain the phosphoinositide kinase relatives ATM, ATR, Rad3, and Mec1 and the effector kinases Chk2, Cds1, Rad53, and Chk1 (Elledge, 1996). The phosphatase Cdc25 and the kinases Wee1 and Myt1 directly control the inhibitory phosphorylation of Cdc2, which is maintained when upstream checkpoint regulators detect damaged or unreplicated DNA in the cell (Elledge, 1996; Morgan, 1997; Rhind and Russell, 1998; Ohi and Gould, 1999).
Although many types of DNA damage can activate a checkpoint, it is largely unknown what DNA structure is the ultimate damage signal. The radiomimetic agent methylmethane sulfonate, which elicits a strong Mec1-dependent checkpoint response in budding yeast, creates adducts and apurinic sites, which become single- and double-stranded breaks (Lindahl and Wood, 1999). Likewise, both UV and ionizing radiation induce DNA damage checkpoints in various organisms. UV causes the formation of pyrimidine dimers, which are repaired predominantly by nucleotide excision, whereas ionizing radiation generates mainly double-stranded breaks. Because DNA damage can arise by multiple mechanisms and the processing of primary DNA lesions can be complex in eukaryotic cells, it has been difficult to characterize at the molecular level the DNA structures that elicit checkpoint responses.
In addition to DNA damage, DNA replication blocks can also trigger cell cycle arrest. The signal(s) that elicits this arrest is unknown, but possibilities include replication intermediates, such as single-stranded DNA, which might accumulate when DNA synthesis is stalled (Li and Deshaies, 1993). In budding yeast, the length of cell cycle arrest in response to DNA damage correlates with the amount of single-stranded DNA that is generated by endonucleolytic processing (Lee et al., 1999). Furthermore, the addition of single-stranded M13 DNA to Xenopus egg extracts results in a strong cell cycle delay (Kornbluth et al., 1992).
Studies of the yeasts have revealed that DNA damage and replication blocks may be recognized by a group of proteins, each of which is required for a normal checkpoint. In the fission yeast Schizosaccharomyces pombe, for example, this group of gene products includes Rad1, Rad3, Rad9, Rad17, Rad26, Hus1, Cut5, and Crb2. Rad3 has substantial structural similarity to the human ATM and ATR proteins, each of which possesses protein kinase activity (Bentley et al., 1996; Banin et al., 1998; Canman et al., 1998; Tibbetts et al., 1999). A similar pathway exists in the budding yeast Saccharomyces cerevisiae (Elledge, 1996). The biochemical functions of the other proteins in this group are poorly understood, but they are currently thought to be involved in sensing damaged DNA or stalled replication complexes. Cds1 and Chk1 are two effector kinases with overlapping functions that receive signals from upstream checkpoint sensors. DNA damage and replication blocks activate Cds1 by a mechanism that requires these proteins (Lindsay et al., 1998). Chk1, however, is normally activated only in response to DNA damage (Walworth and Bernards, 1996; Lindsay et al., 1998). Both Cds1 and Chk1 phosphorylate and inhibit the function of Cdc25, the protein phosphatase that dephosphorylates tyrosine 15 of the cdk Cdc2 (Zeng et al., 1998; Furnari et al., 1999). Mammalian homologues of many of these checkpoint proteins have been isolated and have been shown to possess similar biochemical functions (Cimprich et al., 1996; Sanchez et al., 1997; Matsuoka et al., 1998; Parker et al. 1998; Blasina et al., 1999; Brown et al., 1999; Chaturvedi et al., 1999; Freire et al., 1999), indicating that many features of these checkpoint pathways have been conserved throughout evolution.
We have been using Xenopus egg extracts to study vertebrate checkpoint mechanisms. Previously, we demonstrated that immunodepletion of a Xenopus homologue of Chk1 (Xchk1) resulted in a substantial but not complete abrogation of the cell cycle delay in Xenopus egg extracts in response to replication blocks induced by aphidicolin or to UV-damaged DNA (Kumagai et al., 1998a). Evidence was presented that Xchk1 is involved in a caffeine-sensitive pathway but that a caffeine-insensitive pathway is also involved in the response to aphidicolin and UV radiation. In this report, we have cloned and characterized a Xenopus homologue of Cds1 (Xcds1). Using defined DNA templates, we present evidence that Xcds1 is phosphorylated and activated in response to the presence of DNA molecules with double-stranded ends. The response of Xcds1 to specific DNA templates is also abolished by caffeine. However, Xcds1 is not affected by aphidicolin or UV-damaged DNA. Conversely, Xchk1 does not respond to double-stranded DNA ends. Thus, Xcds1 and Xchk1 appear to respond to quite different signals from the genome. Significantly, immunodepletion of Xcds1 from Xenopus egg extracts did not compromise the cell cycle delay triggered by double-stranded DNA ends. This finding implies that the cell cycle delay induced by double-stranded DNA ends is mediated, at least in part, by an Xcds1-independent mechanism. Overall, our studies indicate that the organization and complexity of the DNA replication and damage checkpoint pathways in this vertebrate system bear some significant differences from analogous pathways in simpler eukaryotes.
MATERIALS AND METHODS
Isolation of a cDNA Encoding Xcds1
An internal fragment of a cDNA encoding Xcds1 was obtained by PCR with the use of the degenerate oligonucleotides AA(T/C)GGIACIT(T/G)(T/C/G)ITIAA and ATIA(G/A)IAT(A/G)TTITCIG-G(T/C)TTIAI(A/G)TCTC(T/G)(A/G)TG, which were designed according to conserved areas found in Cds1 homologues (Figure 1). The PCR reactions contained Xenopus oocyte cDNAs as template, 50 pmol of the degenerate oligonucleotides, 200 μM deoxynucleoside triphosphate, and 0.5 U of Taq polymerase in the buffer supplied by the manufacturer (GIBCO-BRL, Gaithersburg, MD). PCR reactions were heated at 94°C for 2 min followed by 30 cycles of amplification. Each cycle consisted of segments of 94°C for 1 min, 48°C for 2 min, and 72°C for 1 min. An extra 10 min was added to the 72°C extension step for the last cycle. The PCR products were separated on a 1% agarose gel. A 600-base pair (bp) DNA fragment was isolated from the gel and subsequently ligated to the vector pCR2.1 with the use of the TA cloning kit (Stratagene, La Jolla, CA). The 600-bp insert was recovered from the vector by digesting with EcoRI and was then labeled with 32P. The probe (3 × 105 cpm/μl) was denatured in boiling water for 5 min before being used to screen a Xenopus oocyte cDNA library (Mueller et al., 1995). Approximately 1 × 106 independent colonies were screened. After secondary screening, a full-length cDNA clone was obtained. Both strands of the cDNA were sequenced by primer walking with a dye terminator cycle sequencing kit and an ABI model 373 automated sequencer (Perkin Elmer-Cetus, Norwalk, CT). The cDNA sequence contains a large ORF, a 3′ poly(dA) stretch representing the poly(A) tail of the mRNA, and multiple in-frame translation termination codons upstream of the coding region.
Figure 1.
Sequence of Xcds1. Sequences of Cds1 homologues were aligned with the use of the PrettyPlot function of the Genetics Computer Group (Madison, WI) program. Identical residues are boxed. Sequences that were used to design degenerate PCR primers are underlined. The GenBank accession number for Xcds1 is AF174295.
Antibody Production
To clone the coding sequence of Xcds1 into the expression vector pET30(a)+ (Novagen, Madison, WI), a BamHI site upstream of the initiation codon and a XhoI site downstream of the termination codon were introduced by performing PCR with the primers GGACGTCGGATCCTCTCGTGATACTAAAACAGAG and GGA-CGTCCTCGAGTTATCTTTTTGCTCTCTTTTCGG. The resulting 1.5-kilobase PCR product was digested with BamHI and XhoI and ligated to pET30(a)+ that had been digested with BamHI and XhoI. The pET30(a)-Xcds1 construct was introduced into the Escherichia coli host strain BL21(DE3)pLysS (Novagen). Expression of His6-Xcds1 was induced by growing the cells at 37°C for 3 h in the presence of 0.4 mM isopropylthio-β-galactoside. His6-Xcds1 was purified by nickel agarose chromatography, subjected to SDS-PAGE, and subsequently excised from the gel. Polyclonal rabbit antibodies against gel-purified His6-Xcds1 were produced commercially (Covance Research Products, Richmond, CA). Antibodies against a peptide corresponding to the COOH-terminal end of Xcds1 (CSEILPTSAEKRAKR) were generated at another commercial facility (Zymed Laboratories, San Francisco, CA)
Preparation of Various DNA Templates and Egg Extracts
M13 single-stranded DNA was prepared according to a protocol included in a mutagenesis kit from Amersham (Arlington Heights, IL; oligonucleotide-directed in vitro mutagenesis system version 2). The pBS plasmids were prepared according to an alkaline lysis protocol (Sambrook et al., 1989). To generate linearized or open-circularized plasmids, 10 μg of plasmid pBS (Bluescript SK− phagemid, Stratagene) was either digested with restriction enzymes in the appropriate buffers under standard conditions or incubated at 56°C for 36 h in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The resulting linearized and open-circularized plasmids were then diluted with distilled water to a concentration of 0.2 μg/μl. Untreated pBS was also diluted accordingly. Oligonucleotides were synthesized at the Caltech DNA Synthesis Facility. Concentrated oligonucleotides were diluted with distilled water to a concentration of 2 μg/μl. The sequences for the two complementary “random”-sequence oligonucleotides (oligos) are GACTCCGAGAACCAATTGC (oligo 1) and GCAATTGGTTCTCGGAGTC (oligo 2). The sequence for the oligonucleotide capable of forming a hairpin structure is GCGGCACGTTCTCGTGCCGC (oligo 3). Oligo 1 and oligo 2 (10 μg each) were annealed in 20 μl of HBS buffer (10 mM HEPES, pH 7.5, 150 mM NaCl) in a water bath (500 ml of water) that was kept at 70°C for 5 min and then cooled at room temperature for 2.5 h. Oligo 3 (20 μg) was self-annealed by following the same procedure. To disrupt the duplex region, oligo 3 was boiled in water for 10 min and then cooled on ice immediately.
Xenopus egg extracts were prepared as described (Murray, 1991). Extracts supplemented with 100 μg/ml cycloheximide were incubated at 23°C for 90 min after the addition of DNA to a final concentration of either 10 ng/μl for plasmids and M13 DNA or 50 ng/μl for oligonucleotides.
Oligonucleotide Replication Assay
Oligonucleotides were labeled at the 5′ end with T4 polynucleotide kinase. Briefly, 4 μg of each oligonucleotide was incubated with 100 μCi of [32P]ATP and 20 U of T4 polynucleotide kinase (New England Biolabs, Beverly, MA) in a total volume of 40 μl of reaction buffer (50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 5 mM DTT). The reaction was performed at 37°C for 45 min. At the end of the reaction, 60 μl of distilled water was added to the reaction followed by a spin-column purification (Sephadex G-50) to remove the unincorporated [32P]ATP. Ten microliters of the 32P-labeled oligonucleotides (∼1 × 105 cpm/μl) was added to 100 μl of interphase egg extract at room temperature. The first sample (10 μl of extract) was taken and frozen in liquid nitrogen immediately after addition of the oligonucleotides. An aliquot (10 μl) of extract was then taken and frozen every 15 min. All samples were thawed on ice and diluted 20-fold with HBS buffer. The diluted extract was deproteinized by extractions with an equal volume of phenol:chloroform (1:1) once and with chloroform three times. The protein-free sample (5 μl) was loaded onto a 10% native polyacrylamide gel for electrophoresis. The radiolabeled oligonucleotides were visualized by autoradiography.
Production of His6-Xcds1 and His6-Xcds1-N324A Proteins from Bacteria
To introduce the N324A mutation into the Xcds1 gene, two pairs of primers were used to amplify the wild-type Xcds1 sequence: GGACGTCGGATCCTCTCGTGATACTAAAACAGAG and GGACTGGGTCGACGACAACAGCACAGCTTCAGGCTTCAG; GGACGTCCTCGAGTTATCTTTTTGCTCTCTTTTCGG and GGTTGTCGTCGA-CTAGTGAAGAATGTTGCAT. The resulting PCR products were digested with BamHI and SalI, and SalI and EcoRI, respectively, and ligated into the expression vector pET30(a)+ (Novagen) that had been digested with BamHI and EcoRI. A three-fragment ligation produced the construct pET30(a)-Xcds1-N324A. Both pET30(a)-Xcds1-N324A and pET30(a)-Xcds1 (constructed as described above for antibody production) were introduced into the E. coli strain BL21(DE3)pLysS (Novagen). Expression of the His6-Xcds1 and His6-Xcds1-N324A proteins was induced by growing the cells at 30°C for 3 h in the presence of 0.4 mM isopropylthio-β-galactoside in the medium. The proteins were purified by means of nickel agarose chromatography.
Kinase Assays
His6-Xcds1 or His6-Xcds1-N324A proteins were incubated with GST-Cdc25(254–316)-WT or GST-Cdc25(254–316)-S287A in 20 μl of kinase buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT) containing 2 μCi of [32P]ATP and 10 μM ATP (Kumagai et al., 1998a). The reaction was performed at 23°C for 15 min and terminated by adding 20 μl of gel loading buffer. The proteins were separated by SDS-PAGE, and the phosphorylated proteins were detected with the use of a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). To detect the kinase activity of endogenous Xcds1 protein, interphase egg extracts either lacking or containing poly(dT)40 were incubated at 23°C for 90 min. Next, the extracts were incubated with affinity-purified anti-Xcds1 antibodies bound to 10 μl of Affiprep protein A beads (Bio-Rad Laboratories, Richmond, CA) at 4°C for 60 min. After centrifugation, the extract supernatant was removed. The Affiprep protein A beads were washed three times with 1 ml of 10 mM HEPES, pH 7.5, 150 mM NaCl, 30 mM glycerophosphate, 0.1 mM Na3VO4, 0.5% NP-40, 0.1% SDS, 0.1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml chymostatin, and 10 μg/ml pepstatin and once with 1 ml of 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 0.1 mM PMSF. Kinase reactions were performed at 23°C for 15 min by incubating the immunoprecipitates in 20 μl of buffer containing 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 2 μCi of [32P]ATP, 10 μM ATP, and 2 μg of GST-Cdc25(254–316)-WT.
Immunodepletion of Xchk1 and Xcds1 from Egg Extracts
One hundred microliters of M-phase extract was incubated with 20 μg of affinity-purified anti-Xcds1 antibodies, 10 μg of affinity-purified anti-Xchk1 antibodies, or both bound to 10 μl of Affiprep protein A beads (Bio-Rad Laboratories) at 4°C for 50 min. The same amount of control rabbit immunoglobulin G (IgG) (Zymed Laboratories) was used for mock depletion. After incubation, the beads were removed by centrifugation. The supernatants were treated again under the same conditions to ensure that Xcds1 and/or Xchk1 were quantitatively removed from the extracts.
Miscellaneous
The assay for kinase activity toward Ser-287 of Xenopus Cdc25C in egg extracts was performed as described previously (Kumagai et al., 1998a). To block chromosomal DNA replication, aphidicolin (dissolved in DMSO at 10 mg/ml) was added to egg extracts to a final concentration of 100 μg/ml.
RESULTS
Isolation of a Xenopus Cds1 Homologue
To study the functional properties of Cds1 in Xenopus egg extracts, we isolated a cDNA encoding Xenopus Cds1 with the use of database analysis, PCR amplification, and library screening. The Xcds1 cDNA encodes a 58-kDa translation product of 517 amino acids (Figure 1). Xcds1 is most related to Chk2 (63% identical and 76% similar), the human homologue of Cds1 (Matsuoka et al., 1998; Blasina et al., 1999; Brown et al., 1999; Chaturvedi et al., 1999). The most conserved areas are the COOH-terminal kinase domain and the NH2-terminal forkhead-associated domain (Matsuoka et al., 1998). The extreme NH2-terminal region of Xcds1 is less conserved at the primary sequence level but is rich in SQ and TQ amino acid pairs, which are part of the consensus site for phosphorylation by the DNA-dependent protein kinase family (Anderson, 1993). Using histidine-tagged Xcds1 (His6-Xcds1) as antigen, we generated polyclonal antibodies against Xcds1. Affinity-purified anti-Xcds1 antibodies recognized a single protein of ∼58 kDa in Xenopus egg extracts (Figure 2A, lane 1). Antibodies against the COOH-terminal 15 amino acids of Xcds1 recognized a polypeptide of the same size (our unpublished observation).
Figure 2.
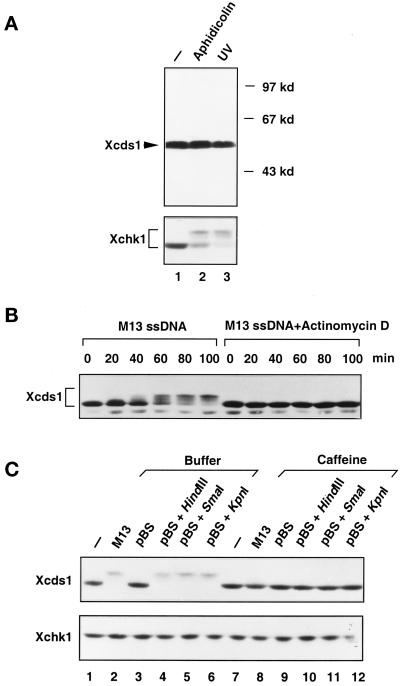
Modification of Xcds1 in response to M13 DNA and linearized plasmids. (A) Interphase extracts containing 2000 sperm nuclei/μl (lane1), 2000 sperm nuclei/μl and 100 μg/ml aphidicolin (lane 2), or 2000 UV-damaged sperm nuclei/μl (lane 3) were incubated at 23°C for 100 min. Seventy microliters of each extract was centrifuged through a sucrose cushion to isolate the nuclear fractions, which were subjected to SDS-PAGE and immunoblotting. After detection of Xcds1, the immunoblot was stripped and probed with anti-Xchk1 antibodies. (B) Interphase egg extracts containing 10 ng/μl M13 single-stranded (ss) DNA or the same amount of M13 DNA and 5 μg/ml actinomycin D were incubated at 23°C. An aliquot of extract (2 μl) was taken and frozen every 20 min after the addition of M13 DNA. Xcds1 protein in each aliquot was then detected by immunoblotting. (C) Extracts containing various DNA templates (lanes 1–6) or the same DNA templates and 5 mM caffeine (lanes 7–12) were incubated at 23°C for 90 min and then analyzed for Xcds1 protein by immunoblotting. The immunoblot was subsequently stripped and probed for Xchk1 protein.
Xcds1 Is Phosphorylated in Response to Double-stranded DNA Ends
In fission yeast and human cells, Cds1 is phosphorylated and activated in response to DNA damage and/or replication blocks (Murakami and Okayama, 1995; Sanchez et al., 1996; Boddy et al., 1998; Lindsay et al., 1998; Matsuoka et al., 1998; Blasina et al., 1999; Brondello et al., 1999; Brown et al., 1999; Chaturvedi et al., 1999). To test whether Xcds1 is similarly modified, the endogenous Xcds1 protein in the nuclear fraction of Xenopus egg extracts was examined by immunoblotting. As shown in Figure 2A, Xcds1 did not show a reduction in mobility during SDS-PAGE when chromosomal DNA replication was blocked by the DNA polymerase inhibitor aphidicolin or when the nuclear DNA was damaged with UV. In contrast, as we reported previously and confirmed here, Xchk1 was modified in response to both aphidicolin and UV (Figure 2A) (Kumagai et al., 1998a).
It has been reported that single-stranded M13 DNA also elicits a checkpoint response in Xenopus egg extracts (Kornbluth et al., 1992). Although Xchk1 mediates the response to aphidicolin and UV (Kumagai et al., 1998a), it is unclear which effector kinase is regulated by M13 DNA. To examine this issue, single-stranded M13 DNA was added to interphase extracts at a concentration (10 ng/μl) that delays mitosis substantially (Kornbluth et al., 1992). Incubation of the extracts with M13 DNA for 40 min or longer resulted in a dramatic decrease in the electrophoretic mobility of Xcds1 (Figure 2B). This modification of Xcds1 was blocked by caffeine, an agent that overrides checkpoint controls (Figure 2C, top, lane 8). Conversely, M13 DNA did not trigger the modification of Xchk1 (Figure 2C, bottom, lane 2).
Although M13 DNA was added to egg extracts in a single-stranded form, it is a very efficient template for DNA synthesis in such extracts. Within 40 min, it is quantitatively converted to double-stranded DNA, which mainly consists of three forms: closed circular, open circular, and linearized (Mechali and Harland, 1982). Significantly, the modification of Xcds1 did not occur until the M13 DNA had been incubated for 40–60 min in egg extracts (Figure 2B), by which time the replication of this template has reached completion. Furthermore, we found that inhibition of M13 DNA replication with actinomycin D (5 μg/ml) prevented the modification of Xcds1 (Figure 2B) (Mechali and Harland, 1982). Therefore, it is possible that a form of double-stranded DNA derived from M13, but not the single-stranded DNA itself, is the checkpoint signal. To test this hypothesis, closed-circular plasmid DNA, plasmids linearized by restriction enzymes, or open-circular plasmids generated by heating at 56°C were added separately to the extracts (Hofferer et al., 1995). Plasmids that had been digested with three different restriction enzymes, HindIII, SmaI, and KpnI, which produce 3′-protruding, blunt, and 5′-protruding ends, respectively, strongly induced the modification of Xcds1 (Figure 2C). In contrast, closed-circular or open-circular plasmids did not have any effect (Figure 2C, lane 3) (our unpublished observations). The modification of Xcds1 induced by linearized plasmids was also abolished by caffeine (Figure 2C, lanes 10–12).
Unlike single-stranded DNA, double-stranded plasmid DNA is not efficiently replicated in whole egg extracts. Its metabolism in the extracts is not quite clear, except that it has been reported that linearized plasmids are partially recircularized and multimerized by nonhomologous end joining (Labhart, 1999). Nonetheless, the linearized plasmids appeared to mimic the presence of damaged DNA with double-stranded breaks, which induced the modification of Xcds1 in the extracts. To evaluate further the nature of the checkpoint signal, we added some other defined DNA molecules, i.e., four types of DNA homopolymers, each 40 nucleotides long, to the extracts. Interestingly, modification of Xcds1 occurred only in the extracts containing poly(dT)40, and this modification was likewise sensitive to caffeine (Figure 3A). Significantly, as is the case for M13 DNA, linearized plasmids and poly(dT)40 did not induce the modification of Xchk1 under these conditions (Figures 2C and 3A).
Figure 3.
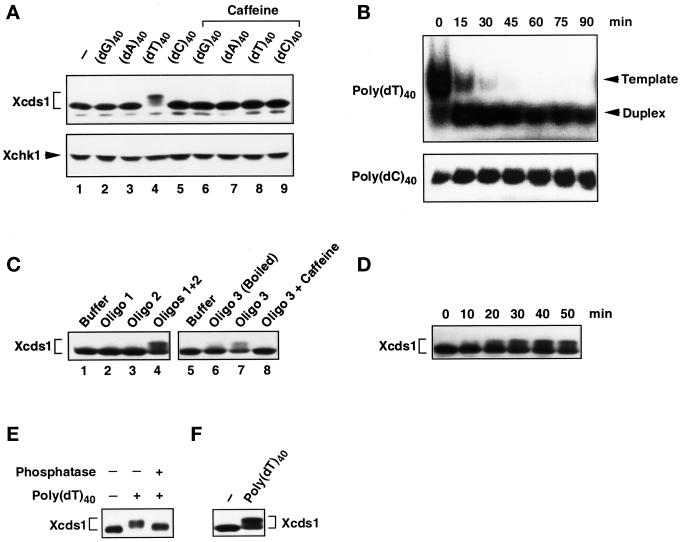
Modification of Xcds1 in response to various oligonucleotides. (A) Extracts containing DNA homopolymers (lanes 2–5) or the same DNA templates and 5 mM caffeine (lanes 6–9) were incubated at 23°C for 90 min and analyzed for Xcds1 protein by immunoblotting. The immunoblot was stripped and probed for Xchk1 protein. (B) Radiolabeled poly(dT)40 and poly(dC)40 were incubated with 100 μl of interphase extract. Just after the addition of the homopolymers (0 min) and every 15 min (15–90 min) afterward, a 10-μl sample was taken and deproteinized. The homopolymers were detected with autoradiography after native PAGE. (C) Same as A except that the indicated oligonucleotides were used. (D) Interphase egg extracts containing 50 ng/μl oligonucleotide duplex (oligo 1 + oligo 2) were incubated at 23°C. An aliquot of extract (2 μl) was taken and frozen every 10 min after the addition of oligonucleotides. Xcds1 protein in each aliquot was detected by immunoblotting. (E) Xcds1 protein immunoprecipitated from extracts containing 50 ng/μl poly(dT)40 was treated with either λ phosphatase or buffer for 60 min at 30°C. As a control, Xcds1 was also immunoprecipitated from untreated interphase extracts. Xcds1 protein in the immunoprecipitates was examined by immunoblotting. (F) Same as A except that interphase egg cytosol containing 50 ng/μl poly(dT)40 or no DNA was incubated for 100 min.
Why did poly(dT)40 but not the homopolymers poly(dC)40, poly(dA)40, and poly(dG)40 induce the modification of Xcds1? One plausible reason is that poly(dT)40 could be converted to double-stranded DNA by replication in the egg extract, whereas the others remained as single-stranded oligonucleotides. Indeed, the eukaryotic DNA primase-polymerase α complex initiates RNA primer and DNA synthesis efficiently on polypyrimidinic templates in vitro (Gronostajski et al., 1984; Grosse and Krauss, 1985; Yamaguchi et al., 1985a,b), whereas primer synthesis on the polypurines poly(dG) and poly(dA) is negligible. Xenopus egg extracts contain a high concentration of ATP (>1 mM), which may both promote primer synthesis on poly(dT) and suppress the formation of primers on poly(dC) (Yamaguchi et al., 1985b). One approach to test this hypothesis would be to examine the electrophoretic mobility of the homopolymers in a native polyacrylamide gel after incubation in egg extracts, because homopolymer duplexes usually migrate faster than the single-stranded duplexes. We confirmed that a poly(dT)40-poly(dA)40 duplex showed an increased mobility in a native gel compared with single-stranded poly(dT)40 (our unpublished observation). Poly(dT)40 and the other homopolymers were labeled at the 5′ end with 32P with the use of T4 polynucleotide kinase and subsequently incubated in interphase egg extracts for 90 min. As shown in Figure 3B, most of the poly(dT)40 was converted within 15 min to a form that migrated faster in the gel at the position of a duplex. This conversion did not occur in the case of poly(dC)40 (Figure 3B), poly(dG)40, or poly(dA)40 (our unpublished observations). In agreement with these observations, it was found that an oligonucleotide duplex (19 bp), preformed from two complementary oligonucleotides with random sequences, and an oligonucleotide containing a hairpin structure both elicited the modification of Xcds1 (Figure 3C). In these experiments, double-stranded DNA ends were required for the modification of Xcds1, because each of the two random-sequence oligonucleotides by themselves did not have this effect and because disruption of the hairpin structure by heating the oligonucleotide abolished its signaling capacity. The response of Xcds1 to the preformed oligonucleotide duplex occurred more quickly (within 20 min) than was the case for M13 DNA (Figure 3D), which would be consistent with the notion that the duplex does not need to undergo replication to trigger a response from Xcds1.
Collectively, these findings strongly suggest that double-stranded DNA ends are the signal that triggers the modification of Xcds1. This modification represents phosphorylation because treatment of the modified Xcds1 with λ phosphatase reversed the mobility alteration (Figure 3E). Interestingly, the signaling from double-stranded DNA to Xcds1 is a process that does not require membranes or an intact nuclear environment, because the phosphorylation of Xcds1 in response to poly(dT)40 occurred in membrane-free egg cytosol that was incubated under the same conditions as whole egg extracts (Figure 3F).
Xcds1 Is Activated by the Addition of Poly(dT)40 to Egg Extracts
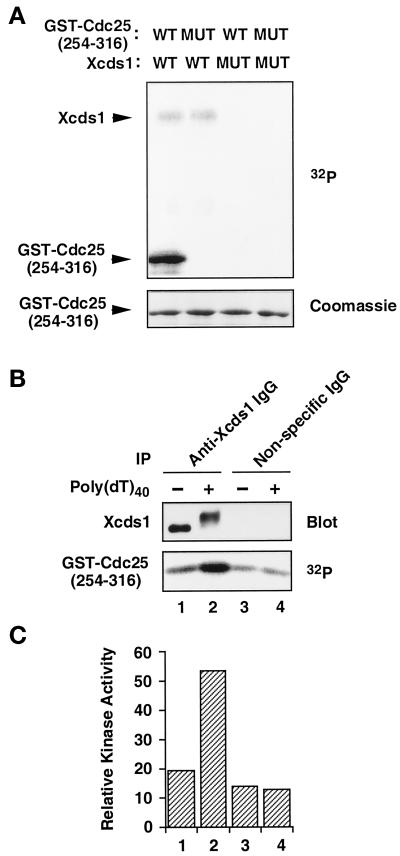
It has been shown that fission yeast Cds1 and human Chk2/Cds1 phosphorylate Cdc25 within a 14-3-3 binding site (Matsuoka et al., 1998; Zeng et al., 1998; Blasina et al., 1999; Brown et al., 1999). The binding of 14-3-3 proteins to Cdc25 is required for a normal checkpoint response in humans, Xenopus egg extracts, and fission yeast (Peng et al., 1997; Kumagai et al., 1998b; Zeng et al., 1998). Therefore, we tested whether Xcds1 would phosphorylate Xenopus Cdc25 in vitro. His6-Xcds1, but not a catalytically inactive mutant (His6-Xcds1-N324A), phosphorylated a 62-amino acid region of Xenopus Cdc25 fused to GST (GST-Cdc25[254–316]-WT) (Kumagai et al., 1998a). The phosphorylation occurs on Ser-287 in the 14-3-3 binding site of Xenopus Cdc25, because a serine-to-alanine mutation at this position abolished the phosphorylation (Figure 4A). Similarly, endogenous Xcds1 immunoprecipitated from egg extracts phosphorylated this substrate well (Figure 4B). Moreover, the hyperphosphorylated Xcds1 protein immunoprecipitated from extracts containing poly(dT)40 showed a five- to sixfold increase over background in its kinase activity toward GST-Cdc25[254–316]-WT (Figure 4, B and C), indicating that this simple template also triggers the activation of Xcds1.
Figure 4.
Activation of Xcds1 kinase in response to poly(dT)40. (A) His6-Xcds1 (WT) and His6-Xcds1-N324A (MUT) were purified from E. coli and incubated with GST-Cdc25[254–316]-WT (WT) or GST-Cdc25[254–316]-S287A (MUT) in kinase buffer containing [32P]ATP. The proteins were separated by SDS-PAGE and stained with Coomassie blue. The phosphorylated proteins were detected with the use of a PhosphorImager. (B) Immunoprecipitation (IP) was performed from interphase egg cytosol containing poly(dT)40 or no DNA with the use of anti-Xcds1 antibodies or nonspecific rabbit IgG. One-third of each immunoprecipitate was analyzed for the modification of Xcds1 by immunoblotting; two-thirds of each immunoprecipitate was incubated with GST-Cdc25[254–316]-WT to measure kinase activity as in A. (C) Quantitation of the data presented in B with the use of a PhosphorImager.
DNA Templates with Double-stranded DNA Ends Delay Mitosis
Single-stranded M13 DNA delays mitosis in cycling egg extracts, an effect that can be overridden by the base analogue caffeine (Kornbluth et al., 1992). In this report, we have demonstrated that M13 DNA, as well as other simple DNA molecules that either contain or generate double-stranded ends, promoted the phosphorylation of Xcds1. It is possible that M13 DNA delays mitosis by activating Xcds1. If this were the case, the other simple DNA molecules, such as poly(dT)40, linearized plasmids, and oligonucleotide duplexes, might also delay mitosis because of their capacity to activate Xcds1. Significantly, poly(dT)40 was found to greatly delay mitosis in egg extracts (Figure 5), as did M13 DNA and an oligonucleotide duplex (Figure 6). The delay of mitosis by poly(dT)40 was partially but not completely reversed by caffeine (Figure 5A). This observation is reminiscent of the fact that the aphidicolin-induced checkpoint in Xenopus egg extracts involves both caffeine-sensitive and caffeine-insensitive pathways (Kumagai et al., 1998a). In contrast, poly(dG)40, which is not converted to a double-stranded form, did not significantly delay the timing of mitosis (Figure 5A).
Figure 5.
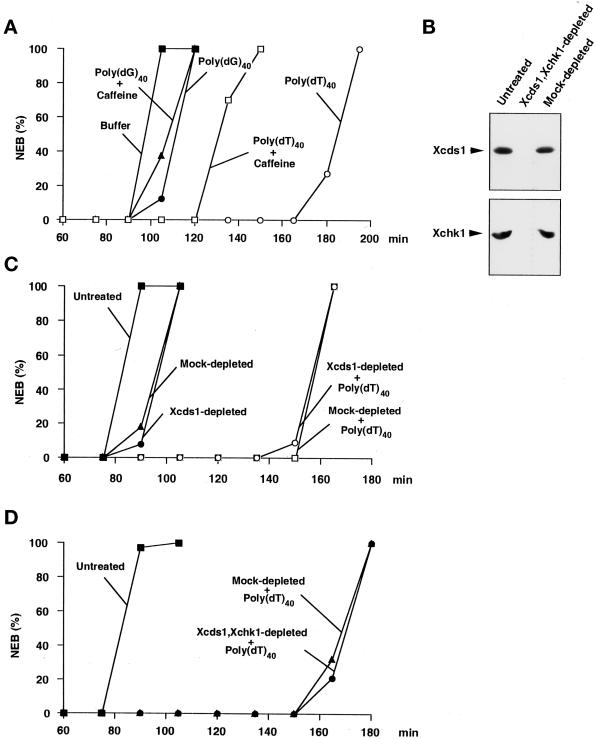
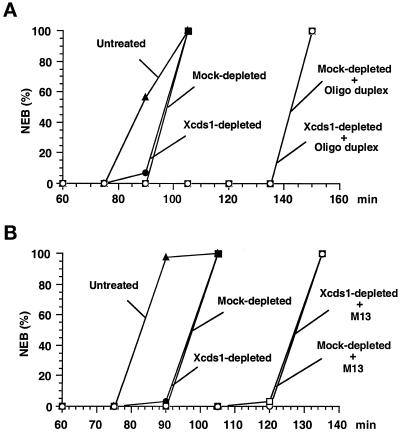
Response of Xenopus egg extracts to poly(dT)40 in the presence and absence of Xcds1. (A) A simple DNA homopolymer delays mitosis. Poly(dT)40 (○, □) or poly(dG)40 (●, ▴) was added to the extracts at a final concentration of 50 ng/μl in the presence (□, ▴) or absence (○, ●) of 5 mM caffeine. The extracts were activated with CaCl2 before the addition of DNA. Sperm nuclei (200 nuclei/μl) were added to the extracts to monitor the timing of nuclear envelope breakdown (NEB) by microscopy. (B) Removal of Xcds1 and Xchk1 proteins from egg extracts by immunodepletion. M-phase extract (100 μl) was incubated with a mixture of anti-Xcds1 antibodies (20 μg) and anti-Xchk1 antibodies (10 μg) bound to Affiprep protein A beads for 50 min at 4°C with constant rocking. Protein A beads were removed by centrifugation. A second round of depletion was then performed to ensure that both Xcds1 and Xchk1 were completely removed, which was assessed by immunoblotting. As a control, M-phase extracts underwent the same procedure with nonspecific rabbit IgG. For depletion of Xcds1 alone, anti-Xchk1 antibodies were omitted. (C) Depletion of Xcds1 does not diminish the mitotic delay caused by poly(dT)40. Sperm nuclei (500 nuclei/μl) (●, ▴) or both sperm nuclei (500 nuclei/μl) and poly(dT)40 (50 ng/μl) (○, □) were added to Xcds1-depleted (○, ●) or mock-depleted (□, ▴) extracts. (D) Same as C except that both Xcds1 and Xchk1 were depleted.
Figure 6.
Response of Xenopus egg extracts to an oligonucleotide duplex and M13 DNA in the presence and absence of Xcds1. (A) Depletion of Xcds1 does not diminish the mitotic delay caused by an oligonucleotide duplex. Sperm nuclei (500 nuclei/μl) (●, ▪) or both sperm nuclei (500 nuclei/μl) and oligonucleotide duplex (oligo 1 + oligo 2) (50 ng/μl) (○, □) was added to Xcds1-depleted (○, ●) or mock-depleted (□, ▪) extracts. Sperm nuclei (500 nuclei/μl) were also added to untreated extracts (▴). The timing of nuclear envelope breakdown (NEB) was monitored by microscopy. (B) Same as A except that the response to M13 DNA (10 ng/μl) was examined.
Immunodepletion of Xcds1 and/or Xchk1 Does Not Compromise the Mitotic Delay Induced by DNA Molecules with Double-stranded Ends
To test whether Xcds1 mediates the mitotic delay induced by poly(dT)40, we completely removed Xcds1 from egg extracts by immunodepletion (Figure 5B). Interestingly, egg extracts lacking Xcds1 still displayed an intact mitotic delay in response to poly(dT)40 (Figure 5C). Furthermore, depletion of Xcds1 did not compromise the delay of mitosis induced by M13 DNA or a double-stranded oligonucleotide (Figure 6). The fact that egg extracts containing double-stranded DNA ends arrest efficiently in the absence of Xcds1 could have several explanations. For example, it is possible that Xchk1 could compensate for the absence of Xcds1. Indeed, in fission yeast, Chk1 is required mainly for the damage checkpoint, but it can substitute for Cds1 in the replication checkpoint when Cds1 is missing (Murakami and Okayama, 1995; Boddy et al., 1998; Lindsay et al., 1998; Brondello et al., 1999). However, egg extracts lacking both Xcds1 and Xchk1 still underwent mitotic delay in the presence of poly(dT)40 (Figure 5D). Consistent with this observation, Xchk1 did not undergo checkpoint-associated phosphorylation in the presence of double-stranded DNA ends (Figure 3), even when Xcds1 had been removed (our unpublished observation).
Kinase Activity toward Ser-287 of Xcdc25 Remains in Egg Extracts Lacking Xcds1 and/or Xchk1
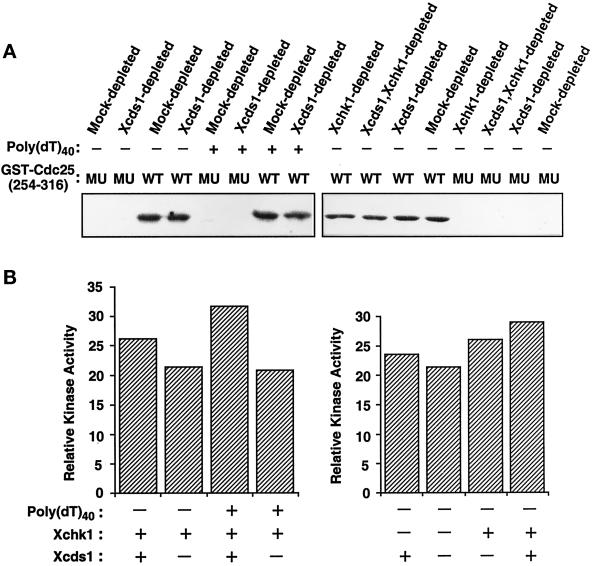
Another possibility is that there is a distinct checkpoint pathway(s) that acts independently of Xcds1 and Xchk1 (see “DISCUSSION”). This pathway could be either normally activated by the presence of double-stranded DNA ends or activated when Xcds1 is absent. It seems plausible that vertebrates such as Xenopus could have checkpoint systems that are more complex than those of lower eukaryotes. To address this possibility, we examined total kinase activity toward the substrate GST-Cdc25[254–316]-WT in egg extracts lacking Xchk1, Xcds1, or both (Figure 7, A and B). We observed that ∼70% of the kinase activity toward Ser-287 of Cdc25 remained in egg extracts when both Xcds1 and Xchk1 were removed (Figure 7B).
Figure 7.
Kinase activity toward Ser-287 of Xenopus Cdc25 in egg extracts lacking Xcds1 and/or Xchk1. (A) M-phase extracts were immunodepleted with anti-Xcds1 antibodies, anti-Xchk1 antibodies, both anti-Xcds1 and anti-Xchk1 antibodies, or nonspecific rabbit IgG. The treated extracts were activated with CaCl2 and incubated at 23°C for 90 min in the presence or absence of poly(dT)40. Finally, total kinase activity toward GST-Cdc25[254–316]-WT (WT) or GST-Cdc25[254–316]-S287A (MU) was assayed in each sample. (B) Quantitation of the data shown in A. Only kinase activity toward GST-Cdc25[254–316]-WT was quantitated, because phosphorylation of the S287A mutant was negligible.
DISCUSSION
In this report, we have analyzed the biochemical properties of Xcds1, a Xenopus homologue of the checkpoint kinase Cds1. Xcds1 becomes highly phosphorylated when M13 DNA or linearized, double-stranded plasmids are added to Xenopus egg extracts. Likewise, Xcds1 undergoes checkpoint-associated phosphorylation in response to poly(dT)40 and various double-stranded oligonucleotides. Although M13 and poly(dT)40 were added to the extracts in a single-stranded form, both of these templates are replicated very efficiently by the DNA synthetic machinery present in egg extracts. Indeed, inhibition of M13 DNA replication by treatment with actinomycin D abolished the modification of Xcds1. Furthermore, various single-stranded oligonucleotides that are not replicated by egg extracts did not bring about the phosphorylation of Xcds1. Collectively, we believe that these data make a strong case that templates containing double-stranded DNA ends trigger the phosphorylation of Xcds1. Nonetheless, the exact chemical nature of the DNA structure that is being recognized remains to be elucidated. Furthermore, poly(dT)40 also elicited an increased kinase activity of Xcds1 toward Ser-287 in the 14-3-3 binding site of Xenopus Cdc25. We suspect that the other templates would also activate Xcds1. Because double-stranded DNA ends would typically be formed upon exposure to DNA-damaging agents such as ionizing radiation, Xcds1 may normally be activated in response to this type of DNA damage in Xenopus cells.
Interestingly, the response of Xcds1 to DNA templates is quite distinct from that of Xchk1, a Xenopus homologue of Chk1 (Figure 8). As we described previously, Xchk1 undergoes a marked phosphorylation in egg extracts when replication is blocked by the DNA polymerase inhibitor aphidicolin or when UV-damaged sperm chromatin is added to the extracts (Kumagai et al., 1998a). In contrast, aphidicolin and UV damage do not elicit phosphorylation of Xcds1, as monitored by one-dimensional SDS-PAGE. Conversely, M13 DNA, linearized plasmids, double-stranded oligonucleotides, and poly(dT)40, each of which caused a strong phosphorylation of Xcds1, did not have detectable effects on Xchk1. It appears that Xcds1 and Xchk1 respond to quite different signals from DNA in the egg extract system. As shown here, Xcds1 responds to double-stranded DNA ends, but the signal that leads to modification of Xchk1 is not known at this time. In the presence of aphidicolin, the initial firing of replication origins can proceed but replication is blocked after the priming stage (Mahbubani et al., 1997), suggesting that some aspect of stalled DNA replication forks triggers the phosphorylation of Xchk1. UV radiation, which causes the formation of pyrimidine dimers, is also a very efficient signal for the modification of Xchk1. The recognition, excision, and/or repair of pyrimidine dimers by repair/replication factors in egg extracts could also lead to the modification of Xchk1. Furthermore, it is important to note that at the doses of UV that we have used, replication of UV-treated sperm chromatin in Xenopus egg extracts is strongly impaired. Thus, it is possible that aphidicolin and UV radiation both cause accumulation of a similar DNA structure, which may in turn lead to the modification of Xchk1.
Figure 8.
Model for the response of the kinases Xcds1 and Xchk1 to different DNA signals.
Although Cds1 and Chk1 have been well conserved throughout evolution, the respective roles of these kinases in various checkpoint responses appear to vary depending on the species. In budding yeast, the closest homologue of Cds1 (Rad53) responds to both DNA damage (induced by exposure to methylmethane sulfonate) and DNA replication blocks (induced by treatment with hydroxyurea), although the response to hydroxyurea was less pronounced (Sanchez et al., 1996; Sun et al., 1996). In wild-type fission yeast cells, Chk1 responds to DNA damage (induced by ionizing radiation, methylmethane sulfonate, or UV light) but not hydroxyurea treatment (Walworth and Bernards, 1996). However, in fission yeast mutants lacking Cds1, Chk1 does respond to hydroxyurea (Boddy et al., 1998; Lindsay et al., 1998; Brondello et al., 1999). Although this finding may suggest that fission yeast Chk1 can also respond to DNA replication blocks, it has been proposed that Cds1 is required to suppress DNA damage in hydroxyurea-treated cells (Lindsay et al., 1998; Brondello et al., 1999). Finally, fission yeast Cds1 responds strongly to treatment with hydroxyurea (Murakami and Okayama, 1995; Lindsay et al., 1998; Brondello et al., 1999). Fission yeast Cds1 can also be activated in response to DNA damage, but its responsiveness to damage is restricted to S-phase (Lindsay et al., 1998; Brondello et al., 1999). Collectively, the available data indicate that fission yeast Cds1 is activated by DNA replication blocks and exposure to damaging agents during S-phase, which may indirectly induce replication blocks. In contrast, Chk1 responds mainly to DNA damage and plays a more specialized role in the response to hydroxyurea. Thus, there appear to be significant differences between fission yeast and Xenopus egg extracts with respect to how Cds1 and Chk1 respond to the DNA structures that trigger checkpoints.
In human cells, Chk2, a Cds1 homologue, is activated upon exposure to ionizing radiation, UV light, and hydroxyurea, but the response to ionizing radiation is the strongest (Matsuoka et al., 1998; Brown et al., 1999). Significantly, the checkpoint kinase ATM, a member of the DNA-dependent protein kinase family that is defective in individuals with ataxia telangiectasia, has been implicated as an upstream regulator of human Chk2/Cds1 (Matsuoka et al., 1998; Brown et al., 1999). Ataxia telangiectasia cells are very sensitive to ionizing radiation but display a normal DNA replication checkpoint (Cliby et al., 1998). Thus, although human Chk2/Cds1 appears to respond to a wider variety of signals than Xcds1, human Chk2/Cds1 is similar to Xcds1 in that it is clearly involved in a pathway that responds to ionizing radiation/double-stranded DNA breaks. Relatively less has been reported about the signals to which human Chk1 responds. Sanchez et al. (1997) found that human Chk1 is modified in response to ionizing radiation, but the extent of modification was much less than what has been observed for human Chk2/Cds1 (Matsuoka et al., 1998). The effect of hydroxyurea or other agents that induce DNA replication blocks on human Chk1 has not been reported. Accordingly, the role of human Chk1 in the DNA replication checkpoint is unclear at this time. Significantly, the Drosophila homologue of Chk1 (Grapes) has also been implicated in the replication checkpoint in fly embryos (Fogarty et al., 1997; Sibon et al., 1997), suggesting that Xchk1 and at least one other metazoan Chk1 homologue may fulfill similar functions.
Previously, we demonstrated that the cell cycle delay in Xenopus egg extracts that is induced in response to treatment with aphidicolin involves multiple pathways: a caffeine-sensitive pathway containing Xchk1 and a caffeine-insensitive pathway. In view of the findings reported here, Xcds1 is not a viable candidate for an effector of the caffeine-insensitive component of the aphidicolin-induced checkpoint for a number of reasons. First, Xcds1 is not phosphorylated in response to aphidicolin. Second, the phosphorylation of Xcds1 by the presence of double-stranded DNA ends in egg extracts is completely abolished by caffeine. Finally, immunodepletion of both Xcds1 and Xchk1 from egg extracts did not further compromise the aphidicolin-induced checkpoint in relation to extracts from which Xchk1 alone had been removed (our unpublished observation).
The pathways that control the response of egg extracts to double-stranded DNA ends may be even more complex than those that are triggered by DNA replication blocks (Figure 8). As is the case for aphidicolin, the delay of the cell cycle induced by poly(dT)40 is only partially abrogated by treatment with caffeine. Thus, the cell cycle delay in response to poly(dT)40 also appears to involve both caffeine-insensitive and caffeine-sensitive pathways. The nature of the caffeine-insensitive pathway(s) that is triggered by poly(dT)40 is unknown. Likewise, it is unclear whether aphidicolin and poly(dT)40 trigger the same or distinct caffeine-insensitive pathways.
Because the phosphorylation/activation of Xcds1 in response to poly(dT)40 is abolished by caffeine, Xcds1 would be a candidate for a component of the caffeine-sensitive pathway(s). However, immunodepletion of Xcds1 does not compromise the cell cycle delay that is induced by poly(dT)40. Similar results were obtained when M13 DNA or a double-stranded oligonucleotide was used to delay the cell cycle. It is possible that Xcds1 becomes phosphorylated as a result of the presence of various DNA templates but does not play a role in the cell cycle delay that is triggered by them. This explanation would be somewhat unexpected in that Xcds1 does phosphorylate Xenopus Cdc25 well on Ser-287 in its 14-3-3–binding site. Cdc25 has been implicated as a target of the DNA replication/damage checkpoints in fission yeast, humans, and Xenopus egg extracts (Furnari et al., 1997; Peng et al., 1997; Sanchez et al., 1997; Kumagai et al., 1998b), although an additional target(s) such as Wee1 could also be involved (O'Connell et al., 1997). On the other hand, Lindsay et al. (1998) have proposed that fission yeast Cds1 may play a distinct role in regulating the formation or stabilization of replication structures. If the function of Xcds1 does not involve delaying the entry into mitosis, the implication would be that there is another factor(s) in egg extracts that carries out this role in response to double-stranded DNA ends.
Another explanation for the intact checkpoint delay in Xcds1-depleted extracts is that another factor in these extracts acts redundantly with Xcds1. This putative factor would not appear to be Xchk1, because immunodepletion of Xchk1 alone or in combination with Xcds1 did not compromise the cell cycle delay in response to poly(dT)40. However, ∼70% of the total kinase activity toward Ser-287 of Cdc25 remains behind in egg extracts lacking both Xcds1 and Xchk1. It will be necessary to identify this Ser-287–specific kinase to evaluate the possibility that it contributes to cell cycle regulation in response to double-stranded DNA ends.
The kinase(s) that lies upstream of Xcds1 in the pathways triggered by double-stranded DNA ends is currently unknown. In the human system, convincing evidence has been presented that ATM is an upstream regulator of human Chk2/Cds1 (Mutsuoka et al., 1998; Brown et al., 1999). Ataxia telangiectasia cells are hypersensitive to ionizing radiation (Meyn, 1995), which suggests that ATM is involved in mediating the response to double-stranded DNA ends. The recently described Xenopus homologue of ATM would be a good candidate for a regulator of Xcds1 (Robertson et al., 1999). Because the agents to which Xchk1 responds are quite different from those that activate Xcds1, the implication is that ATM may not be the principal upstream regulator of Xchk1 in the replication checkpoint response.
ACKNOWLEDGMENTS
We thank members of the Dunphy laboratory for suggestions and comments on the manuscript. We are grateful to Yuling Sheng for technical assistance. We thank A. Kumagai for providing anti-Xchk1 antibodies and K.H. Emami, S.X. Wang, and Y. Li for supplying reagents. This work was supported in part by a grant from the National Institutes of Health. Z.G. is an associate and W.G.D. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- Anderson CW. DNA damage and the DNA-activated protein kinase. Trends Biochem Sci. 1993;18:433–437. doi: 10.1016/0968-0004(93)90144-c. [DOI] [PubMed] [Google Scholar]
- Banin S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Blasina A, Van de Weyer I, Laus MC, Luyten WHML, Parker AE, McGowan CH. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- Boddy MD, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinase Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- Brondello J, Boddy MN, Furnari B, Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol Cell Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Lee CH, Schwarz JK, Mitiku N, Piwnica-Worms H, Chung JH. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, et al. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–168. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, Goldberg ML, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- Freire R, Murguia JR, Tarsounas M, Lowndes NF, Moens PB, Jackson SP. Human and mouse homologs of Schizosaccharomyces pombe rad1+ and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev. 1999;12:2560–2573. doi: 10.1101/gad.12.16.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari B, Blasina A, Boddy MN, McGowan CH, Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- Gronostajski RM, Field J, Hurwitz J. Purification of a primase activity associated with DNA polymerase α from HeLa cells. J Biol Chem. 1984;259:9479–9484. [PubMed] [Google Scholar]
- Grosse F, Krauss G. The primase activity of DNA polymerase alpha from calf thymus. J Biol Chem. 1985;260:1881–1888. [PubMed] [Google Scholar]
- Hofferer L, Winterhalter KH, Althaus FR. Xenopus egg lysates repair heat-generated DNA nicks with an average patch size of 36 nucleotides. Nucleic Acids Res. 1995;23:1396–1397. doi: 10.1093/nar/23.8.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S, Smythe C, Newport JW. In vitro cell cycle arrest induced by using artificial DNA templates. Mol Cell Biol. 1992;12:3216–3223. doi: 10.1128/mcb.12.7.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998a;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Yakowec PS, Dunphy WG. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol Biol Cell. 1998b;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P. Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol Cell Biol. 1999;19:2585–2593. doi: 10.1128/mcb.19.4.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1999;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Li JJ, Deshaies RJ. Exercising self-restraint: discouraging illicit acts of S and M in eukaryotes. Cell. 1993;74:223–226. doi: 10.1016/0092-8674(93)90413-k. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJF, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani HM, Chong JP, Chevalier S, Thommes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:893–897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Mechali M, Harland RM. DNA synthesis in a cell-free system from Xenopus eggs: priming and elongation on single-stranded DNA in vitro. Cell. 1982;30:93–101. doi: 10.1016/0092-8674(82)90015-0. [DOI] [PubMed] [Google Scholar]
- Meyn MS. Ataxia-telangiectasia and cellular responses to DNA damage. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Dunphy WG. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- O'Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Gould KL. Regulating the onset of mitosis. Curr Opin Cell Biol. 1999;11:267–273. doi: 10.1016/s0955-0674(99)80036-2. [DOI] [PubMed] [Google Scholar]
- Parker AE, Van de Weyer I, Laus MC, Verhasselt P, Luyten WH. Identification of a human homologue of the Schizosaccharomyces pombe rad17+ checkpoint gene. J Biol Chem. 1998;273:18340–18346. doi: 10.1074/jbc.273.29.18340. [DOI] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K, Hensey C, Gautier J. Isolation and characterization of Xenopus ATM (X-ATM): expression, localization, and complex formation during oogenesis and early development. Oncogene. 1999;18:7070–7079. doi: 10.1038/sj.onc.1203194. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Hendrickson EA, DePamphilis ML. DNA primase-DNA polymerase α from simian cells. J Biol Chem. 1985a;260:6254–6262. [PubMed] [Google Scholar]
- Yamaguchi M, Hendrickson EA, DePamphilis ML. DNA primase-DNA polymerase α from simian cells: sequence specificity of initiation sites on simian virus 40 DNA. Mol Cell Biol. 1985b;5:1170–1183. doi: 10.1128/mcb.5.5.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]