Abstract
The behavior of nuclear pre-mRNA-binding proteins after their nuclease and/or salt-induced release from RNA was investigated. After RNase digestion or salt extraction, two proteins that initially exist as tetramers (A2)3B1 in isolated heterogeneous nuclear ribonucleoprotein (hnRNP) complexes quantitatively reassociated to form regular helical filaments ranging in length from 100 nm to >10 μm. In highly magnified preparations prepared for scanning transmission electron microscopy, single filaments have diameters near 18 nm. In conventional negatively stained preparations viewed at low magnification, the diameters of the thinnest filaments range from 7 to 10 nm. At protein concentrations of >0.1 mg/ml, the filaments rapidly aggregated to form thicker filamentous networks that look like the fibrogranular structures termed the “nuclear matrix.” Like the residual material seen in nuclear matrix preparations, the hnRNP filaments were insoluble in 2 M NaCl. Filament formation is associated with, and may be dependent on, disulfide bridge formation between the hnRNP proteins. The reducing agent 2-mercaptoethanol significantly attenuates filament assembly, and the residual material that forms is ultrastructurally distinct from the 7- to 10-nm fibers. In addition to the protein rearrangement leading to filament formation, nearly one-third of the protein present in chromatin-clarified nuclear extracts was converted to salt-insoluble material within 1 min of digestion with RNase. These observations are consistent with the possibility that the residual material termed the nuclear matrix may be enriched in, if not formed by, denatured proteins that function in pre-mRNA packaging, processing, and transport.
INTRODUCTION
When nuclei are extensively digested with DNase and RNase and extracted with high-salt solutions, an insoluble residue remains that retains the gross architecture of the nucleus (reviewed by Pederson, 1998, 2000). The residue, termed the “nuclear matrix,” represents a small percentage of the total nuclear protein mass, but it is very heterogeneous in protein composition (Hodge et al., 1977; Staufenbiel and Deppert, 1983; Kuzmina et al., 1984). The major proteins present in the residue are the nuclear lamins, proteins that constitute elements of the metaphase chromosome and interphase nuclear scaffold, nuclear envelope and pore complex proteins, and numerous proteins associated with heterogeneous nuclear ribonucleoprotein (hnRNP) complexes (as discussed and referenced by Pederson, 1998).
Among the numerous other proteins present in the residual material are those that have been hypothesized to form a fibrous network or fibrogranular complex (Berezney and Coffey, 1977; Kaufmann et al., 1981; Gallinaro et al., 1983). On the basis of various experimental observations it has been suggested that the matrix functions in interphase nuclei to bind DNA and organize chromatin (Basler et al., 1981; Brasch and Peters, 1985; Nickerson et al., 1997), that actively transcribing genes and nascent transcripts are associated with the matrix (Mariman et al., 1982; Robinson et al., 1983; Jost and Seldran, 1984), that hnRNA processing is dependent on a transient association of nascent transcripts with the matrix (Herman et al., 1978; Ben-Ze'ev and Aloni, 1983), that RNA transport occurs via matrix-mediated events (Baglia and Maul, 1983), and that DNA replication occurs at sites where the DNA is associated with elements of the nuclear matrix (Dijkwel et al., 1979; Smith and Berezney, 1980). The fibrogranular appearance of the insoluble residue underlies the idea that a continuous salt-insoluble fibrous network (distinct from pre-mRNP fibrils and chromatin) exists in interphase nuclei. This network is thought to have functional similarities to intermediate filaments and the cytoskeleton but to possess a dynamic role in nuclear events.
The postulated properties of the nuclear matrix imply that it is both a regular structure formed by the self-assembly of one or more unique structural proteins but that it is composed mostly of nucleic acid-binding proteins. Only occasionally has attention has been paid to the solubility characteristics of the abundant nonchromatin proteins, or to the potential for protein aggregation and rearrangement during nuclear isolation, RNA removal, and exposure to salt at >150 mM and to detergents (Kaufmann and Shaper, 1984; Kaufmann et al., 1986; Mirkovitch et al., 1984; Small et al., 1985; Kirov and Tsanev, 1986). In this paper we report that the conditions typically used to generate the nuclear matrix induce specific hnRNP rearrangements leading to the formation of salt-insoluble filaments that ultrastructurally resemble the filaments observed in nuclear matrix preparations. In addition, nuclease digestion alone in low salt rapidly converts 20–30% of the soluble nuclear proteins into salt-insoluble filaments and fibrogranular networks. These observations do not preclude the existence of a salt-insoluble filamentous network in nuclei but suggest that its presence may not be indicative of the existence of similar structures in vivo. The acquired insolubility of hnRNP and other proteins after removal from their endogenous substrates may also result in the trapping of numerous enzymatic activities and nucleic acid fragments within the residual network. These results add to previously discussed methodological issues that have led to a degree of skepticism regarding the existence an interphase nuclear matrix in vivo (Zakian, 1985; Pederson, 1998, 2000).
MATERIALS AND METHODS
Nuclear Matrix Preparation, hnRNP Isolation, and hnRNP Filament Formation
For electrophoretic comparison of nuclear matrix fractions in the presence and absence of reducing agent, matrix was prepared from isolated HeLa S3 nuclei following the procedures of Gallinaro et al. (1983), from rat liver nuclei as described by Berezney and Coffey (1977), and from mouse erythroleukemia (MEL) nuclei as described by Long et al. (1979).
HeLa cell hnRNP complexes were isolated from exponentially growing cells in suspension culture as described previously (Huang and LeStourgeon, 1994). Fractions containing essentially pure nuclease-dissociated or salt-dissociated hnRNPs A2 and B1 were prepared as described by Lothstein et al. (1985). Initially, RNA-dissociated proteins were allowed to spontaneously form filaments either at 4°C in sucrose gradient fractions containing STM buffer (90 mM NaCl, 10 mM Tris-HCl, pH 8, and 1 mM MgCl) or in STM buffer (after dialysis to remove sucrose). More rapid and quantitative yields of filaments were obtained by overnight dialysis at 4°C against STMM buffer (90 mM NaCl, 10 mM Tris-HCl, pH 8, and 5 mM MgCl) or in 10-fold dilute STMM buffer (0.1× STMM) after an additional 2-h dialysis at 27°C. In routine preparations, gradient fractions containing proteins A2 and B1 (1–3 ml) were dialyzed against 1 l of the above STMM buffer. In some cases, 0.2 or 2.0% (vol/vol) 2-mercaptoethanol was included in the diluted STMM buffer during dialysis.
To determine whether high-salt insoluble RNP rearrangement and/or aggregation can occur instantaneously upon RNA digestion, isolated HeLa packed nuclei from 2.5 × 109 cells (1.8 ml) were brought to a volume of 3.6 ml with STM buffer and exposed at 0°C to four 10-s burst of ultrasound at 50 W. Chromatin was removed by centrifugation for 10 min at 9500 rpm in a Sorvall HB-4 rotor (Becton Dickenson, Palo Alto, CA). The Mg++ concentration of the chromatin-clarified nuclear sonicate (histone free) was brought to 5 mM by adding 1.0 M MgCl2. RNase A was added (to 65 μg/ml) to the concentrated nuclear sonicate, and this preparation was incubated at 37°C for 15 min. Turbidity was noted after 30 s, and samples were taken for electron microscopic examination. After an additional 30-s period, solid NaCl was added slowly with stirring to a final concentration of 2.0 M. After 30 min an aliquot was taken for ultrastructural analysis. Insoluble material was collected via centrifugation. The soluble protein was precipitated with alcohol, and both were prepared for SDS-PAGE.
For electron microscopic examination, either the grids were prepared by floating them on the surface of a drop of the sample and then subsequently stained with uranyl acetate as described previously (Lothstein et al., 1985), or the grids were allowed to float for 1 min on samples that had been fixed for 5 h in 0.1% glutaraldehyde and then allowed to float for 1 min on a 1% solution of ammonium molybdate or uranyl acetate and finally air dried.
Effect of Solvent and Temperature on Filament Stability
To observe the effects of high-salt, reducing agent, and additional nuclease activity on filament stability, filament preparations were treated with the conditions used to generate rat liver nuclear matrix (Berezney and Coffey, 1977). After dialysis of filaments overnight at 4°C against 10 mM Tris-HCl, pH 7.4, and 0.2 mM MgCl2, solid NaCl was added at room temperature with stirring to a concentration of 2.0 M. An aliquot was taken for ultrastructural analysis 3 h later. This preparation was then dialyzed overnight at 4°C against two changes of 10 mM Tris-HCl, pH 7.4, and 0.2 mM MgCl2, and an aliquot was taken for electron microscopy. Next, 1.0 M MgCl2 and 10% Triton X-100 were added to final concentrations of 5.0 mM and 1%, respectively. After overnight dialysis against two changes of 1 mM Tris-HCl, pH 7.4, and 5 mM MgCl2, DNase 1 and RNase A were each added to a final concentration of 200 μg/ml, and the sample was incubated for 1 h at 22°C. Any remaining filaments were then collected by centrifugation for 15 min at 10,000 rpm in a Sorvall HB-4 rotor. In separate experiments, filaments were equilibrated in 0.1× STMM and then exposed to 1.0 M salt by the addition of solid NaCl. After 3 h of stirring, samples were taken for ultrastructural examination, and filamentous and soluble protein species were separated and determined as above.
The effect of elevated temperature on filament composition and stability was investigated by placing suspensions of filaments, in 0.1× STM buffer, in a small plastic microfuge tube and incubating for 15 min each at successive 10° intervals between 30 and 70°. After each incubation, specimens were prepared for electron microscopy. Filamentous and soluble protein was determined as above.
To determine the effect of pH on filament stability, a suspension of filaments was split into eight equal aliquots and dialyzed separately against 10 mM NaCl, 10 mM Tris-acetate, and 5 mM MgCl2 buffered to various values over the range of pH 3–10. After overnight dialysis, aliquots were prepared for electron microscopy, and, after centrifugation, protein in the pellet and supernatant was prepared for electrophoresis. In an attempt to dissociate the filaments with urea, a filament preparation, previously equilibrated in 0.1× STMM, was made 6.2 M in urea. After 2.5 h of gentle stirring at room temperature, an aliquot was taken for ultrastructural analysis, and soluble and insoluble protein was determined as above.
Protein Reduction and RNA and Protein Electrophoresis
In the efforts described above to solubilize the RNP filaments, no reducing agent was present in the various buffers used. To investigate the effect of reducing agent on filament solubility, filaments were equilibrated in 0.1× STMM containing 2% (vol/vol) 2-mercaptoethanol. This preparation was allowed to stand 5 h at 23°C with occasional agitation. Filament preparations were also dialyzed against a high-salt EDTA buffer (1.0 M NaCl, 10 mM Tris-HCl, pH 8, and 5 mM EDTA). Ultrastructural analysis and determination of soluble and insoluble protein were preformed as described above.
When recovery of disulfide-linked proteins from gel slices was desired, bis-acrylylcylstamine was substituted for bis-acrylamide, and polymerization was catalyzed by illumination after riboflavin and N,N,N′,N′-tetramethyl-ethylenediamine were added to final concentrations of 18.5 μM and 0.17%, respectively. Coomassie blue-stained bands were excised from the bis-acrylylcylstamine cross-linked gels and placed in 40 μl of fresh 2-mercaptoethanol. Then, 160 μl of 0.15 M NaCl, 50 mM Tris-HCl, pH 7.5, and 5 mM EDTA were added. To dissolve the gel slices, these preparations were then titrated with 10 M NaOH to pH 8. After the gel slices were completely dissolved, 500 μl of 0.15 M NaCl, 50 mM Tris, and 5 mM EDTA were added, and the sample was centrifuged 10 min at 9000 rpm in a Sorvall HB-4 rotor to pellet the protein. Before electrophoresis, protein recovered from gels in this way and also some of the nuclear matrix preparations were aggressively reduced in 2% 2-mercaptoethanol in boiling water for 2 min and then loaded on standard SDS-PAGE as described elsewhere (Huang et al., 1994).
RESULTS
Our studies on hnRNP filament formation during isolation and manipulation arose from a set of observations on the effects of salt and nuclease on nuclear extracts enriched in crude hnRNP complexes and on purified 40S hnRNP monoparticles. Intact hnRNP monoparticles sediment broadly around a peak centered at 40S and are composed of multiple copies of six major proteins that appear to exist as three sets of tetramers, (A1)3B2, (A2)3B1, and (C1)3C2 (McAfee et al., 1997). A fragment of pre-mRNA near 700 nucleotides in length is packaged in each 40S monoparticle (Huang et al., 1994). hnRNP complexes, which are not exposed to nuclease, sediment as polyparticle complexes (from a dimer peak centered at ∼60S to complexes containing six to eight particles that sediment near 300S (McAfee et al., 1997). The basic A and B group proteins dissociate from RNA at 150 mM NaCl (Beyer et al., 1977). The spontaneous appearance of filaments in sucrose gradient fractions containing these salt-dissociated proteins prompted the studies described here.
hnRNP Filament Formation Is Associated with Disulfide Bridge Formation
In this study we report that hnRNP filament formation is more efficient at 25°C than on ice and that dialysis against STM buffer containing elevated magnesium ion contributes to filament assembly. At high protein concentrations, evidence of filament formation can be seen as turbidity within seconds after nuclease addition. If hnRNP complexes are not exposed to dissociating salt concentrations or to RNase, filaments are not observed.
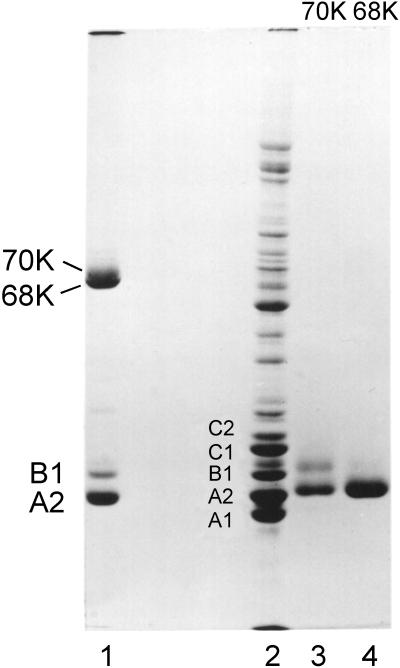
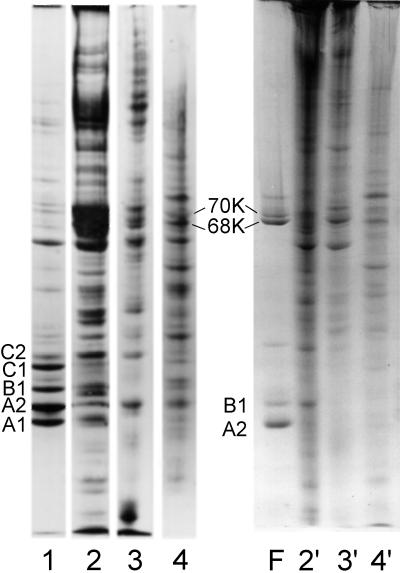
Gel electrophoresis of SDS-solubilized filaments collected by brief centrifugation reveal proteins A2 and B1 in the same 3:1 molar ratio as in intact hnRNP complexes (Figure 1). However, upon filament formation two additional bands appeared in gels at ∼68 and 70 kDa. As filament preparations aged in solution, the 68- and 70-kDa bands increased in relative concentration. To determine whether the 68- and 70-kDa bands result from sulfhydryl oxidation, mercaptoethanol was increased to 2%, and the samples were heated in boiling water for 2 min before electrophoresis. This led to the disappearance of the 68- and 70-kDa bands in SDS-PAGE (Figure 1).
Figure 1.
Coomassie blue-stained SDS-PAGE of freshly formed filaments prepared from sucrose gradient fractions containing essentially pure proteins A2 and B1 (lane 1) and excised and aggressively reduced 68- and 70-kDa bands (lanes 3 and 4, respectively). Lane 2 is a sample of 40S hnRNP particles included as an internal marker and to show the proteins typically present in isolated 40S monoparticles.
To determine the protein composition of the putative dimer bands, the latter were excised from gels and reduced as described in MATERIALS AND METHODS. The 68-kDa band yielded only A2, and the 70-kDa band yielded both A2 and B1 (Figure 1). Thus, disulfide-linked dimers of A2–A2 and A2–B1 form upon filament assembly. Sulfhydryl cross-linked protein is not detected in hnRNP preparations unless the proteins are dissociated from RNA via nuclease or by salt extraction. The 70- and 68-kDa disulfide-linked bands migrate in gels near the lamin bands A and B present in standard preparations of nuclear matrix.
Filament Insolubility
Proteins A2 and B1 dissociate from isolated hnRNP complexes at 0.15 M NaCl (Beyer et al., 1977), and the majority but not all of these proteins can be recovered from isolated nuclei by extensive extraction at this salt concentration (LeStourgeon et al., 1978; Peters and Commings, 1980). Upon filament formation, these proteins cannot be solubilized in 1.0 or 2.0 M salt over the pH range of 3–10 or at temperatures as high as 70°C (Figure 2). Filaments were also not solubilized after 4 h at 23°C in 2% 2-mercaptoethanol or by overnight dialysis against an EDTA-containing high-salt buffer (1.0 M NaCl, 10 mM Tris-HCl, pH 8, and 10 mM EDTA) (Figure 2). Thus, although magnesium ion and oxidation favor filament assembly, once formed, they cannot be dissociated by reducing agent or metal ion chelators.
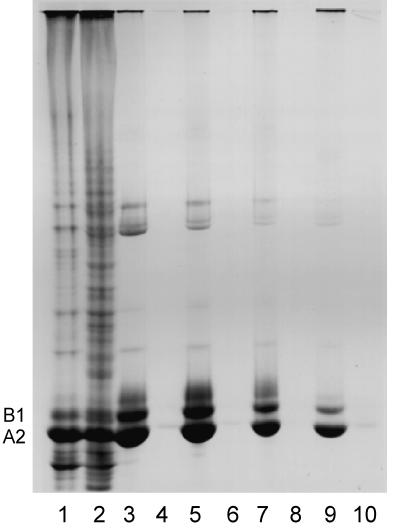
Figure 2.
SDS-PAGE of the insoluble (lane 1) and soluble (lane 2) protein after nuclease digestion of a chromatin-clarified nuclear sonicate. The insoluble (lane 3) and soluble (lane 4) protein formed in the presence of 0.2% mercaptoethanol. Insoluble filaments (lane 5) and soluble protein (lane 6) formed in the presence of 2% mercaptoethanol. Lane 7 shows filaments allowed to stand 5 h at room temperature in the presence of 2% mercaptoethanol. Lane 8 shows the protein solubilized by reducing agent. Insoluble (lane 9) and soluble (lane 10) protein after high-salt EDTA extraction is also shown. Essentially no protein is detected in lanes 4, 6, 8, and 10.
Ultrastructural Features of Matrix-like Complexes
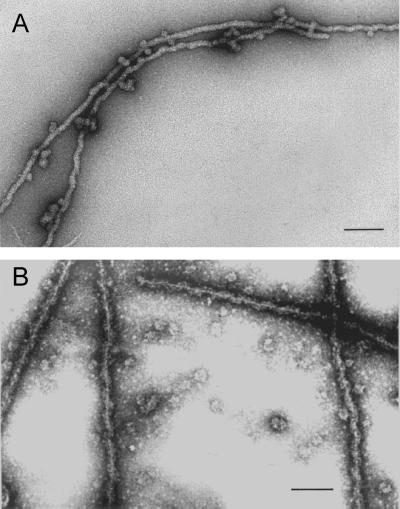
At high magnification, single filaments formed at low protein concentration from gradient-purified proteins A2 and B1 appeared highly regular and were of indeterminate length (Figure 3, A and B). Filaments prepared for scanning transmission electron microscopy (STEM) (Figure 3B) had a diameter of 18 nm and displayed a helical appearance with a pitch of ∼60 nm. A precise value for the pitch was complicated by filament flexibility and the appearance of filament stretching. In high-resolution STEM micrographs of filaments stained with ammonium molybdate, there were suggestions of a central fiber about which globular units are wrapped in a spiral manner (Figure 3B).
Figure 3.
(A) Electron micrograph of negatively stained filaments formed spontaneously in sucrose gradient fractions containing proteins A2 and B1 after salt-induced dissociation from 40S hnRNP complexes. (B) STEM of filaments dialyzed 10 h against STM buffer to remove sucrose. In both preparations a small amount of globular A2 and B1 aggregates can be seen often in contact with the filaments. Bar, 100 nm.
Filaments that formed from purified A2–B1 preparations or by brief dialysis against 0.1× STMM or by allowing sucrose gradient fractions to stand overnight at 4°C frequently revealed spherical complexes of A2–B1 associated with the filaments, often at regular intervals (Figure 3A). This phenomenon may contribute to the granules or globular structures seen associated with aggregates that formed at higher protein concentrations.
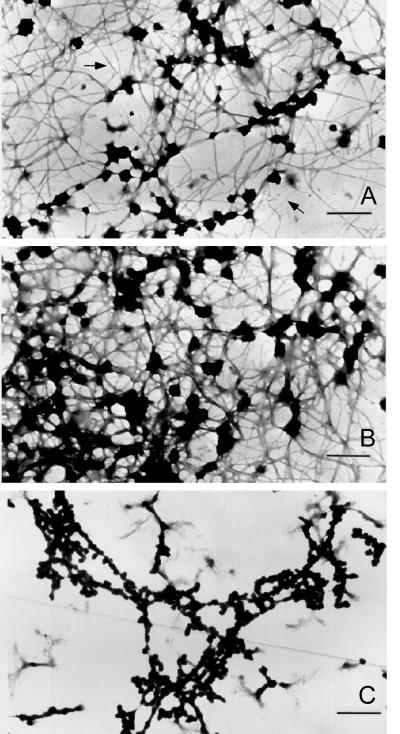
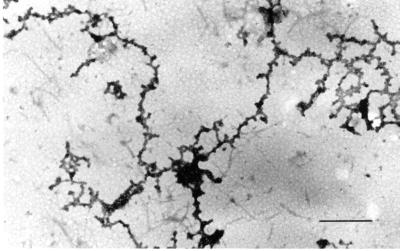
Filaments that formed at increasing protein concentration or by dialysis at 27°C showed extensive aggregation into thicker fibers and a more amorphous fibrogranular or matrix-like structure. Occasionally, in preparations at low protein concentration, the spiral nature of the filaments could be observed (Figure 5) with diameters of ∼7–10 nm (denoted by arrows). Presumably these filaments are synonymous with the single filaments shown in Figure 3. The difference in apparent filament diameter can only be attributed to differences in sample preparation, sucrose concentration, and staining procedures. Globular complexes ranging from 20 to 50 nm embedded within the matrix-like structure were more frequently observed at higher protein concentrations (Figure 4B). This was true whether the starting material was pure A2–B1 (Figure 4AB) or isolated 40S hnRNP monoparticles (Figure 4C). At higher protein concentrations the filaments are thicker, spiral filaments are not observed, and more extensive branching is apparent. This indicates that two or more filaments associate for various distances along their long axis.
Figure 5.
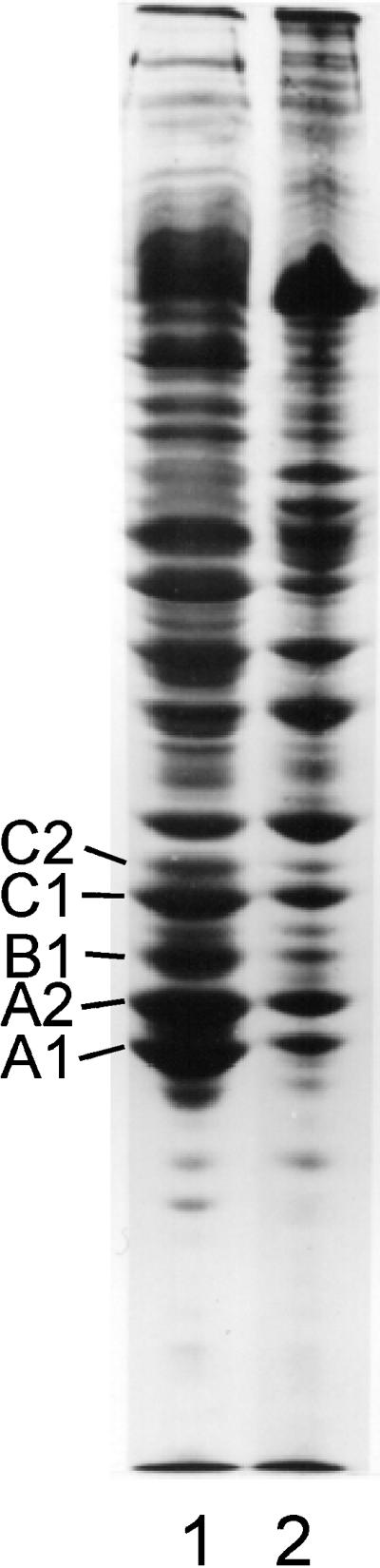
SDS-PAGE of the high-salt insoluble material (lane 2) generated in a chromatin-clarified crude nuclear sonicate 15 min after adding RNase at 30°C. Lane 1, insoluble protein.
Figure 4.
Electron micrographs (low magnification) of filaments formed by dialysis of A2 and B1 preparations against 0.1× STMM buffer. Initial protein concentrations were near 0.1 mg/ml (A) and 0.2 mg/ml (B). Bars, 500 nm. Note increasing filament anastomosis, aggregation, and globular particle formation at higher protein concentrations. (C) Salt-insoluble material formed upon RNase digestion of concentrated 40S hnRNP particle preparations in 0.1× STMM buffer. The thinner fibers in A are 7–10 nm in diameter. At higher protein concentration most fibers fall in the diameter range of 10–20 nm. Bar 100 nm.
Nuclease-induced Nuclear Protein Insolubility
When RNase was added to chromatin-clarified nuclear sonicates at 37°C, the preparation became turbid within 20–30 s. This phenomenon indicates the occurrence of protein rearrangement, denaturation, and aggregation. To determine whether salt-insoluble RNP complexes and filaments contribute to the turbidity observed, NaCl was added to 2.0 M, an aliquot was taken for ultrastructural examination, and the insoluble material was collected for protein analysis. Approximately 20–30% of the soluble protein present in nuclear sonicates was rendered insoluble in 2.0 M salt after nuclease digestion. The protein composition of the insoluble and soluble material was essentially the same (Figure 5). Electron micrographs of the salt-insoluble material revealed large amorphous networks of precipitated material in which individual filaments could not definitively be identified (Figure 6).
Figure 6.
Electron micrograph of the salt-insoluble material shown in Figure 5, lane 2. Bar, 100 nm.
In the above case in which ∼30% of the proteins present in a nuclear sonicate was rapidly converted to salt-insoluble material, no enrichment of A2 and B1 was observed in the residue (Figure 5). However, if crude nuclear sonicates were briefly digested with ribonuclease and then dialyzed overnight against dilute STM, the insoluble material remained heterogeneous in protein composition, but proteins A2 and B1 were the major proteins present. This indicates that filament formation can continue to occur from soluble hnRNP pools. In nuclear sonicates incubated at 30–37°C in the presence of high magnesium, significant levels of protease activity can be detected. This is the likely source of the bands positioned below A1 in Figure 2, lanes 1 and 2. This is also likely to be the explanation for the loss of A1, C1, and C2, because these proteins have been shown to be especially labile to protease activity (Lothstein et al., 1985).
The insoluble material formed from purified proteins A2 and B1 in the presence of 2% mercaptoethanol was an irregular thick branched or overlapping aggregate fibrillar network (Figure 7), and filaments with diameters near 10 nm could not definitively be identified. This may parallel the reduced levels of disulfide cross-linked A2–A2 and A2–B1 dimers formed in the presence of reducing agent (Figure 2, compare lanes 3, 5, and 7).
Figure 7.
Electron micrograph of the high-salt insoluble material generated when salt-dissociated and purified A2 and B1 tetramers are dialyzed into low salt (0.1× STMM buffer containing 2% mercaptoethanol). Bar, 500 nm.
HnRNP in Reduced Nuclear Matrix Preparations
Although the major hnRNP proteins are known to be significant components of nuclear matrix preparations (Comings and Okada, 1976; Peters and Commings, 1980; He et al., 1991; Pederson, 2000), it was of interest in this study to examine the extent of hnRNP oxidation and aggregation in various matrix preparations. In Figure 8, the Coomassie blue-stained bands in lane 1 are from a typical preparation of HeLa cell 40S hnRNP particles. Lanes 2–4 show the proteins present in partially reduced matrix preparations from rat liver, MEL cells, and HeLa cell nuclei. Lanes 2′, 3′, and 4′ show the same fractions solubilized and resolved in the absence of mercaptoethanol. The absence of bands corresponding to the major hnRNPs in nonreduced samples indicates that these proteins probably exist as oxidized and aggregated material in typical matrix preparations.
Figure 8.
Coomassie blue-stained SDS-PAGE showing the proteins present in a preparation of 40S hnRNP particles (lane 1) and rat liver, MEL cell, and HeLa cell nuclear matrix (lanes 2–4, respectively). The samples in lanes 1–4 were solubilized in standard SDS sample buffer containing mercaptoethanol. Matrix samples 2′, 3′, and 4′ are the same as those in lanes 2–4, but no reducing agent was present in the electrophoresis sample buffer. Lane F shows the proteins in an hnRNP filament preparation. The oxidized dimers of A2 and B1 are noted in lanes 4 and F. In the nonreduced samples (lanes 2′, 3′, and 4′) significant amounts of protein were trapped in the 3% stacking gel (our unpublished observations).
DISCUSSION
The salt-insoluble matrix-like filaments reported here do not form spontaneously in nuclear extracts containing hnRNP complexes or in preparations of purified hnRNP core particles. Insoluble filament formation is dependent on dissociation of the hnRNPs from their pre-mRNA substrate either by 150 mM salt or by nuclease activity. The disulfide-linked dimers of A2–A2 and A2–B1 recovered from filaments also do not exist before the formation of insoluble filaments. This phenomenon seems directly related to the finding that nuclear matrix could not be isolated if sulfhydryl blocking reagents were present during all steps of matrix preparation (Kaufmann and Shaper, 1984). As in nuclear matrix preparations, hnRNP filament formation is enhanced in the presence of magnesium. At hnRNP protein concentrations near or >0.1 mg/ml the filaments aggregate to form fibrogranular structures that very closely resemble published images of the nuclear matrix (He et al., 1990, 1991). Whether nuclear matrix is generated by nuclease digestion or by salt-only extraction, the ultrastructural appearance of the fibrogranular complexes appears similar (Capco et al., 1982; Fey et al., 1986). This is also true for the hnRNP filaments that formed in this study. In previous characterizations of nuclear matrix, spiral filaments have not been reported. However, as reported here, spiral filaments were only observed when prepared from purified A2 and B1 at low protein concentrations. Spiral filaments were not observed in the matrix-like complexes that rapidly formed at higher protein concentration or from more heterogeneous nuclear extracts. Filaments like those described here were previously observed in nuclease digests of RNP complexes from Triturus oocytes and were interpreted to be elements of a nuclear matrix (Kloetzel et al., 1982).
Although the above observations do not preclude the existence of an in vivo interphase nuclear matrix composed of salt-insoluble components, several related findings suggest that some of the fibrogranular material described as matrix is of hnRNP origin. For example, in metabolically active cells in tissues or in rapidly dividing cells in culture, the hnRNPs are major nuclear constituents (Dreyfuss et al., 1993). In rapidly dividing HeLa cells there is approximately as much of the individual A1, A2, and C1 proteins as there is individual histone species (LeStourgeon et al., 1990). Perhaps more significantly, nuclear matrix cannot be prepared from the nuclei of transcriptionally inactive cells (deficient in hnRNP) by conventional means (LaFond et al., 1983). These investigators also observed that the sulfhydryl cross-linking reagent sodium tetrathionate was only effective in stabilizing salt-resistant nuclear networks in metabolically active cells. The apparent absence of hnRNP observed here in nonreduced matrix preparations is consistent with this observation and with the findings of Kaufman and Shaper (1984) mentioned above. Although gold bead immunolabeling with a monoclonal antibody has been used to localize the core hnRNPs to the globular material in matrix preparations and not to the fibrous material (He et al., 1991), the highly specific rearrangement and oxidation leading to hnRNP filament assembly could mask the determinant for a monoclonal antibody.
High-salt insolubility is a defining property of nuclear matrix. As observed here, not only do the conditions used to generate nuclear matrix induce the formation of matrix-like filaments from hnRNP, but they also render 20–30% of the protein present in concentrated nuclear extracts insoluble in 2 M NaCl. Thus, high-salt insolubility after experimental manipulation may not adequately reflect the solubility properties of proteins in vivo, and, as shown by others (Kirov et al., 1984; Mirkovitch et al., 1984; Small et al., 1985), it is likely that a wide range of enzymatic activities become artifactually associated with the residual material. In the studies of Mirkovitch et al. (1984), glutaraldehyde prefixation was used to prevent protein rearrangement, with the result being an inability to isolate matrix complexes. This would be expected if the conditions for matrix isolation involve hnRNP rearrangement and the oxidation of thiol groups. In a more recent study, formaldehyde (a short-distance cross-linker) was used to stabilize matrix before nuclear extraction (Nickerson et al., 1997). Because chromatin could be removed via DNase without removal of the cross-linked material, these investigators argued for the preexistence of a separate fibrogranular nuclear matrix. This finding prompted the suggestion that nuclear matrix exists before salt extraction and that it makes few contacts with chromatin. However, this would be expected if hnRNP fibrils are a source of matrix, because each hnRNP fibril has but one noncovalent association with DNA in the transcription complex (Beyer et al., 1981; Beyer and Osheim, 1990).
ACKNOWLEDGMENTS
We thank Joseph Wall (Brookhaven National Laboratory) for assistance with the scanning transmission electron micrographs and Thoru Pederson for numerous helpful suggestions. This work was supported by National Science Foundation grant PFM-8209420 to W.M.L., American Cancer Society grant CD-15 to J.C.W., and National Institutes of Health grant RR-00715, which supported the STEM facility at the Brookhaven National Laboratory.
REFERENCES
- Baglia FA, Maul GG. Nuclear ribonucleoprotein release and nucleoside triphosphatase activity are inhibited by antibodies directed against one nuclear matrix glycoprotein. Proc Natl Acad Sci USA. 1983;80:2285–2289. doi: 10.1073/pnas.80.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler J, Hastie ND, Pietras D, Matsui SI, Sandberg AA, Berezney R. Hybridization of nuclear matrix attached deoxyribonucleic acid fragments. Biochemistry. 1981;20:6921–6929. doi: 10.1021/bi00527a027. [DOI] [PubMed] [Google Scholar]
- Ben-Ze'ev A, Aloni Y. Processing of SV40 RNA is associated with the nuclear matrix and is not followed by the accumulation of low-molecular-weight RNA products. Virology. 1983;125:475–479. doi: 10.1016/0042-6822(83)90218-0. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol. 1977;73:616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer AL, Bouton AH, Miller OL., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981;26:155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Ultrastructural analysis of the ribonucleoprotein substrate for pre-mRNA processing. In: Strauss PR, Wilson SH, editors. The Eukaryotic Nucleus: Molecular Biochemistry and Macromolecular Assemblies. Vol. 2. Caldwell, NJ: Telford Press; 1990. pp. 431–451. [Google Scholar]
- Brasch K, Peters KE. Nuclear protein, matrix and structural changes in rooster liver after estrogenic induction of vitellogenesis. Biol Cell. 1985;54:109–121. doi: 10.1111/j.1768-322x.1985.tb00386.x. [DOI] [PubMed] [Google Scholar]
- Capco DG, Wan KM, Penman S. The nuclear matrix: three-dimensional architecture and protein composition. Cell. 1982;29:847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- Comings DE, Okada TA. Nuclear proteins. III. The fibrillar nature of the nuclear matrix. Exp Cell Res. 1976;103:341–360. doi: 10.1016/0014-4827(76)90271-8. [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Mullenders LH, Wanka F. Analysis of the attachment of replicating DNA to a nuclear matrix in mammalian interphase nuclei. Nucleic Acids Res. 1979;6:219–230. doi: 10.1093/nar/6.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Fey EG, Krockmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinaro H, Puvion E, Kister L, Jacob M. Nuclear matrix and hnRNP share a common structural constituent associated with premessenger RNA. EMBO J. 1983;2:953–960. doi: 10.1002/j.1460-2075.1983.tb01527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He DC, Martin T, Penman S. Localization of heterogeneous nuclear ribonucleoprotein in the interphase nuclear matrix core filaments and on perichromosomal filaments at mitosis. Proc Natl Acad Sci USA. 1991;88:7469–7473. doi: 10.1073/pnas.88.17.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman R, Weymouth L, Penman S. Heterogeneous nuclear RNA-protein fibers in chromatin-depleted nuclei. J Cell Biol. 1978;78:663–674. doi: 10.1083/jcb.78.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge LD, Mancini P, Davis FM, Heywood P. Nuclear matrix of HeLa S3 cells. Polypeptide composition during adenovirus infection and in phases of the cell cycle. J Cell Biol. 1977;72:194–208. doi: 10.1083/jcb.72.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, LeStourgeon WM. The rapid preparation of the hnRNP core proteins and stepwise assembly of hnRNP particles in vitro. In: Celis JE, editor. Handbook of Cell Biology. I. New York: Academic Press; 1994. [Google Scholar]
- Huang M, Rech JE, Northington SJ, Flicker PF, Mayeda A, Krainer AR, LeStourgeon WM. The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1994;14:518–533. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost JP, Seldran M. Association of transcriptionally active vitellogenin II gene with the nuclear matrix of chicken liver. EMBO J. 1984;3:2005–2008. doi: 10.1002/j.1460-2075.1984.tb02082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Coffey DS, Shaper JH. Considerations in the isolation of rat liver nuclear matrix, nuclear envelope, and pore complex lamina. Exp Cell Res. 1981;132:105–123. doi: 10.1016/0014-4827(81)90088-4. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Okret S, Wikstrom AC, Gustafsson JA, Shaper JH. Binding of the glucocorticoid receptor to the rat liver nuclear matrix. The role of disulfide bond formation. J Biol Chem. 1986;261:11962–11967. [PubMed] [Google Scholar]
- Kaufmann SH, Shaper JH. A subset of non-histone nuclear proteins reversibly stabilized by the sulfhydryl cross-linking reagent tetrathionate. Polypeptides of the internal nuclear matrix. Exp Cell Res. 1984;155:477–495. doi: 10.1016/0014-4827(84)90208-8. [DOI] [PubMed] [Google Scholar]
- Kirov N, Djondjurov L, Tsanev R. Nuclear matrix and transcriptional activity of the mouse alpha-globin gene. J Mol Biol. 1984;180:601–614. doi: 10.1016/0022-2836(84)90029-9. [DOI] [PubMed] [Google Scholar]
- Kirov N, Tsanev R. Activated murine alpha-globin gene is not preferentially associated with the nuclear matrix. Int J Biochem. 1986;18:155–159. doi: 10.1016/0020-711x(86)90148-5. [DOI] [PubMed] [Google Scholar]
- Kloetzel P, Johnson M, Sommerville J. Interaction of the hnRNA of amphibian oocytes with fibril forming protein. Eur J Biochem. 1982;127:301–308. doi: 10.1111/j.1432-1033.1982.tb06870.x. [DOI] [PubMed] [Google Scholar]
- Kuzmina SN, Buldyaeva TV, Akopov SB, Zbarsky IB. Protein patterns of the nuclear matrix in differently proliferating and malignant cells. Mol Cell Biochem. 1984;58:183–186. doi: 10.1007/BF00240618. [DOI] [PubMed] [Google Scholar]
- LaFond RE, Woodcock H, Woodcock CL, Kundahl ER, Lucas JJ. Generation of an internal matrix in mature avian erythrocyte nuclei during reactivation in cytoplases. J Cell Biol. 1983;96:1815–1819. doi: 10.1083/jcb.96.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeStourgeon WM, Barnett SF, Northington SJ. Tetramers of the core proteins of 40S nuclear ribonucleoprotein particles assemble to package nascent transcripts into a repeating array of regular particles. In: Strauss PR, Wilson SH, editors. The Eukaryotic Nucleus: Molecular Biochemistry and Macromolecular Assemblies. Vol. 2. Caldwell, NJ: The Telford Press; 1990. [Google Scholar]
- LeStourgeon WM, Lothstein L, Walker BW, Beyer AL. The composition and general topology of RNA protein in monomer 40S ribonucleoprotein particles. In: Busch H, editor. The Cell Nucleus. Vol. 9. New York: Academic Press; 1981. pp. 49–86. [Google Scholar]
- Long BH, Huang CY, Pogo AO. Isolation and characterization of the nuclear matrix in Friend erythroleukemia cells: chromatin and hnRNA interactions with the nuclear matrix. Cell. 1979;18:1079–1090. doi: 10.1016/0092-8674(79)90221-6. [DOI] [PubMed] [Google Scholar]
- Lothstein L, Arenstorf HP, Chung S, Walker BW, Wooley JC, LeStourgeon WM. General organization of protein in HeLa 40S nuclear ribonucleoprotein particles. J Cell Biol. 1985;100:1570–1581. doi: 10.1083/jcb.100.5.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariman EC, van Eekelen CA, Reinders RJ, Berns AJ, van Venrooij WJ. Adenoviral heterogeneous nuclear RNA is associated with the host nuclear matrix during splicing. J Mol Biol. 1982;154:103–119. doi: 10.1016/0022-2836(82)90420-x. [DOI] [PubMed] [Google Scholar]
- McAfee JG, Huang M, Soltaninassab S, Rech JE, Iyengar S, LeStourgeon WM. The packaging of pre-mRNA. In: Krainer A, editor. Frontiers in Molecular Biology: Eukaryotic mRNA Processing. New York: Oxford University Press; 1997. pp. 68–102. [Google Scholar]
- Mirkovitch J, Mirault ME, Laemmli UK. Organization of the higher-order chromatin loop: specific ENA attachment sites on nuclear scaffold. Cell. 1984;1:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Nickerson JA, Krockmalnic G, Wan KM, Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc Natl Acad Sci USA. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Thinking about a nuclear matrix. J Mol Biol. 1998;277:147–159. doi: 10.1006/jmbi.1997.1618. [DOI] [PubMed] [Google Scholar]
- Pederson T. Half a century of “the nuclear matrix.”. Mol Biol Cell. 2000;11:799–805. doi: 10.1091/mbc.11.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters KE, Commings DE. Two-dimensional gel electrophoresis of rat liver nuclear washes, nuclear matrix, and hnRNA proteins. J Cell Biol. 1980;86:135–155. doi: 10.1083/jcb.86.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SI, Small D, Idzerda R, McKnight GS, Vogelstein B. The association of transcriptionally active genes with the nuclear matrix of the chicken oviduct. Nucleic Acids Res. 1983;11:5113–5130. doi: 10.1093/nar/11.15.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small D, Nelkin B, Vogelstein B. The association of transcribed genes with the nuclear matrix of Drosophila cells during heat shock. Nucleic Acids Res. 1985;13:2413–2431. doi: 10.1093/nar/13.7.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HC, Berezney R. DNA polymerase alpha is tightly bound to the nuclear matrix of actively replicating liver. Biochem Biophys Res Commun. 1980;97:1541–1547. doi: 10.1016/s0006-291x(80)80041-6. [DOI] [PubMed] [Google Scholar]
- Staufenbiel M, Deppert W. Nuclear matrix preparations from liver tissue and from cultured vertebrate cells: differences in major polypeptides. Eur J Cell Biol. 1983;31:341–348. [PubMed] [Google Scholar]
- Zakian VA. Taken with a grain of salt. Nature. 1985;314:223–224. doi: 10.1038/314223a0. [DOI] [PubMed] [Google Scholar]