Abstract
Glycosylphosphatidylinositols (GPIs) are critical for membrane anchoring and intracellular transport of certain secretory proteins. GPIs have a conserved trimannosyl core bearing a phosphoethanolamine (EthN-P) moiety on the third mannose (Man-3) through which the glycolipid is linked to protein, but diverse GPI precursors with EthN-Ps on Man-1 and Man-2 have also been described. We report on two essential yeast genes whose products are required late in GPI assembly. GPI11 (YDR302w) encodes a homologue of human Pig-Fp, a protein implicated in the addition of EthN-P to Man-3. PIG-F complements the gpi11 deletion, but the rescued haploids are temperature sensitive. Abolition of Gpi11p or Pig-Fp function in GPI11 disruptants blocks GPI anchoring and formation of complete GPI precursors and leads to accumulation of two GPIs whose glycan head groups contain four mannoses but differ in the positioning and number of side chains, probably EthN-Ps. The less polar GPI bears EthN-P on Man-2, whereas the more polar lipid has EthN-P on Man-3. The latter finding indicates that Gpi11p is not required for adding EthN-P to Man-3. Gpi13p (YLL031cp), a member of a family of phosphoryltransferases, is a candidate for the enzyme responsible for adding EthN-P to Man-3. Depletion of Gpi13p in a Gpi11p-defective strain prevents formation of the GPI bearing EthN-P on Man-3, and Gpi13p-deficient strains accumulate a Man4-GPI isoform that bears EthN-P on Man-1. We further show that the lipid accumulation phenotype of Gpi11p-deficient cells resembles that of cells lacking Gpi7p, a sequence homologue of Gpi13p known to add EthN-P to Man-2 of a late-stage GPI precursor. This result suggests that in yeast a Gpi11p-deficiency can affect EthN-P addition to Man-2 by Gpi7p, in contrast to the Pig-Fp defect in mammalian cells, which prevents EthN-P addition to Man-3. Because Gpi11p and Pig-Fp affect EthN-P transfer to Man-2 and Man-3, respectively, these proteins may act in partnership with the GPI-EthN-P transferases, although their involvement in a given EthN-P transfer reaction varies between species. Possible roles for Gpi11p in the supply of the EthN-P donor are discussed. Because Gpi11p- and Gpi13p-deficient cells accumulate isoforms of Man4-GPIs with EthN-P on Man-2 and on Man-1, respectively, and because the GPIs that accumulate in Gpi11p-defective strains are likely to have been generated independently of one another, we propose that the yeast GPI assembly pathway is branched.
INTRODUCTION
Glycosylphosphatidylinositols (GPIs) are made by all eukaryotes (McConville and Ferguson, 1993; Tiede et al., 1999), and their formation is essential for the growth of mammalian, yeast, and protozoal cells alike (Leidich et al., 1994; Kawagoe et al. 1996; Ilgoutz et al., 1999b). Many GPIs become covalently attached to the COOH terminus of secretory proteins and are critical for membrane anchoring, intracellular transport, and targeting of those proteins (Lisanti and Rodriguez-Boulan, 1990; Horvath et al., 1994; Doering and Schekman, 1996). In yeast, GPIs are essential for normal cellular morphogenesis and have an additional role in mediating cross-linking of glycoproteins to cell wall β-glucan (Orlean, 1997; Lipke and Ovalle, 1998).
GPIs have the conserved core structure NH2-CH2-CH2-PO4-6Manα1,2Manα1,6-Man-α1,4GlcNα1,6Ins-PO4-lipid, which can be modified by the addition of an acyl chain to the inositol and of phosphoethanolamine (EthN-P) to the first and possibly second mannoses (Homans et al., 1988; Roberts et al., 1988; Hirose et al., 1992; Kamitani et al., 1992; Puoti and Conzelmann, 1992, 1993). In yeast, an α1,2-linked Man becomes attached to the third Man of the GPI precursor (Sipos et al., 1994).
GPIs are preassembled and transferred to the COOH terminus of certain proteins in the lumen of the endoplasmic reticulum (ER). Synthesis of the GPI precursor proceeds in a stepwise manner. Reactions common to all eukaryotes are the transfer of GlcNAc to phosphatidylinositol (PI), the de-N-acetylation of GlcNAc-PI, the successive addition of three α-linked mannoses, and the attachment of EthN-P to the 6′ position of the third Man (reviewed by Stevens, 1995; Takeda and Kinoshita, 1995; Tiede et al., 1999). It is through this EthN-P moiety that the GPI becomes attached to protein in a transamidation reaction (Udenfriend and Kodukula, 1995; Sharma et al., 1999). Addition of side branches can occur concomitantly with the extension of the GPI “backbone.” In yeast and mammals, the 2′-OH of inositol is acylated before addition of the first Man, and the first and second mannoses of mammalian GPIs can bear EthN-P side chains (Puoti et al., 1991; Deeg et al., 1992; Hirose et al., 1992; Kamitani et al.; 1992, Puoti and Conzelmann, 1992, 1993). EthN-P has also been detected on Man-1 of yeast GPIs (Canivenc-Gansel et al., 1998; Sütterlin et al., 1998).
Genes involved in GPI biosynthesis in mammalian and yeast cells have been identified after isolation of GPI anchoring-deficient mammalian cell lines and temperature-sensitive yeast mutants (reviewed by Takeda and Kinoshita, 1995; Orlean, 1997; Tiede et al., 1999). Sequence comparisons of the proteins involved in the synthesis and de-N-acetylation of GlcNAc-PI, in the addition of the third Man, and in anchor transfer to protein, together with the demonstration that heterologous GPI synthesis genes can complement a deficiency in another organism's counterpart of that gene, indicate that the machinery for the formation of the GPI core has been highly conserved (Leidich et al., 1995; Schönbächler et al., 1995; Vossen et al., 1995; Hamburger et al., 1995; Benghezal et al., 1996; Inoue et al., 1996; Hiroi et al., 1998; Sütterlin et al., 1998; Tiede et al., 1998; Watanabe et al., 1998, 1999).
Not only are the proteins that assemble the GPI backbone conserved, but proteins involved in EthN-P side branching may be, too. A family of three phosphoryltransferase-related proteins is conserved in yeast and mammals, and two of its members have been implicated in EthN-P side chain addition (Benachour et al., 1999; Gaynor et al., 1999; Hong et al., 1999). Yeast Mcd4p is essential for growth and GPI anchoring, and a deficiency in Mcd4p, as well as in its nonessential homologue Gpi7p, leads to defects in cell wall morphogenesis. Addition of at least one EthN-P side chain may therefore be a critical event in GPI assembly in yeast and mammals.
Despite the conservation of the steps and proteins involved in core GPI assembly and modification, GPI biosynthesis is not as fully understood as other conserved ER pathways for glycosylation and export of secretory proteins. In particular, the pattern of addition of EthN-P side chains to mannosylated GPIs has made the later stages in the GPI synthetic pathway difficult to chart. In mammalian cells, GPIs with one or two mannoses can sometimes also bear EthN-P on the Man linked to GlcN, and Man3-GPIs bearing between zero and three EthN-Ps can also be radiolabeled with [3H]Man. Indeed, in mammalian cells, three Man3-containing GPIs can be detected that are, in principle, competent for transfer to protein by virtue of having EthN-P on their third Man (the “bridging” EthN-P) (Hirose et al., 1992; Kamitani et al., 1992). In yeast, two “complete precursors” with four mannoses and the bridging EthN-P have been characterized (Sipos et al., 1994). It is not known which of these GPIs are transferred protein, whether these species are all obligatory intermediates in the formation of protein-linked GPIs, or whether some represent a separate pool of non–protein-linked GPIs (Kamitani et al., 1992).
An intriguing feature of GPI assembly is that GPI synthetic activities in both mammalian cells and in Leishmania are localized in distinct subcompartments of the ER. This raises the possibility that a GPI assembly pathway can be split between two or more successive subcompartments, or that different species of GPIs can be generated independently in different compartments (Ralton and McConville, 1998; Ilgoutz et al., 1999a; Vidugiriene et al., 1999).
Among the proteins required for GPI assembly, Pig-Fp has an enigmatic role at a late stage in formation of the mammalian GPI precursor. The largest and most polar GPI accumulated in mammalian class F mutants contains three mannoses but lacks EthN-P on its third Man, suggesting that the mutants are defective in addition of the bridging EthN-P to the 6′-position of the third, α1,2-linked Man (Sugiyama et al., 1991; Hirose et al., 1992; Kamitani et al., 1992; Puoti and Conzelmann, 1993). The PIG-F gene, cloned by complementation of the GPI anchoring defect of the class F mutant, restores the ability of the cells to make later-stage GPIs (Inoue et al., 1993). However, class F mutants accumulate multiple GPIs with one, two or three mannoses as well as side chain EthN-Ps (Lemansky et al., 1991; Sugiyama et al., 1991; Kamitani et al., 1992; Puoti and Conzelmann, 1993), indicating that the mutation is pleiotropic. Interestingly, Thy-1 class F mutants are also defective in ether lipid synthesis and make PI that contains exclusively base-labile diacylglycerols (Stevens and Raetz, 1990). The relationship of this defect to the cells' GPI anchoring deficiency is unclear, although it has been speculated that EthN-P addition may require the mammalian acceptor GPI to have an alkyl-acyl-PI (Inoue et al., 1993).
To understand the basis of the complexity of the late stages in GPI assembly, we are generating yeast strains deficient in the proteins involved, characterizing the structures of the GPIs that accumulate in them, and defining the order in which the proteins act. We report that GPI11, which encodes the yeast Pig-Fp homologue, is essential for viability and required for GPI anchoring. Gpi11p-deficient mutants accumulate two Man4-containing GPI precursors, one of which bears an HF-labile substituent (HFLS), probably EthN-P, on Man-2, whereas the other is substituted on Man-3, the latter finding suggesting that Gpi11p is not solely responsible for adding the bridging EthN-P. We show that, instead, YLL031cp (Gpi13p) is a better candidate for the protein that adds EthN-P to the third GPI Man. The likeliest models for the structures of the lipids that accumulate on Gpi11p and Gpi13p deficiency require that these GPIs are generated in different arms of a branched GPI synthetic pathway.
MATERIALS AND METHODS
Materials
[2-3H]myo-Inositol (specific activity, 15–20 Ci/mmol), [2,6-3H]mannose (specific activity, 20 Ci/mmol), [1-3H]ethanolamine (specific activity, 10–30 Ci/mmol), and NaB[3H]4 (specific activity, 5–15 Ci/mmol) were obtained from American Radiolabeled Chemicals (St. Louis, MO), and [α-32P]dCTP (specific activity, >3000 Ci/mol) was from ICN Biochemicals (Costa Mesa, CA). Jack Bean and Aspergillus satoi α-mannosidases were from Oxford GlycoSciences (Oxford, United Kingdom), and phosphatidylinositol-specific phospholipase C was from ICN Biochemicals. Calcofluor White was from Sigma (St. Louis, MO).
Yeast Strains and Growth Media
The yeast strains used in this work are listed in Table 1, and the construction of those strains made specifically for this study is detailed below. The majority of the strains used were derived from YMW3, a diploid obtained by crossing strains YMW1 and YMW2 (Zieler et al., 1995). Details about the S-40 (pmi40), FBY11 (gpi8), and RYY51 (Δpsd1/Δpsd2) strains are given by Payton et al., (1991), Benghezal et al., (1995), and Trotter and Voelker (1995), respectively. Heterozygous YLL031c deletion strain BY4743 MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 lys2Δ0 met15Δ0 YLL031c::KANR/YLL031c was generated in the Saccharomyces Gene Deletion Project and obtained from Research Genetics (Huntsville, AL). The gaa1 strain, defective in GPI transfer to protein (Hamburger et al., 1995), was isolated in a screen for mutants synthetically lethal with Δgpi1 (B.A. Westfall, L. St. Louis, and P. Orlean, unpublished data). “Double mutants” between gpi11::LEU2-pPIG-F and pmi40, gpi8 or YLL031c::KANR/pGAL-YLL031c and the gpi11/Δpsd1/Δpsd2 “triple mutant” were created by introducing pPIG-F into the gpi8, pmi40, YLL031c::KANR/pGAL-YLL031c, or Δpsd1/Δpsd2 strains, respectively, and then disrupting the GPI11 locus in these strains with LEU2. The gpi1/gpi11 double mutant was made by disrupting GPI1 with URA3 in the gpi11::LEU2-pPIG-F strain.
Table 1.
Saccharomyces cerevisiae strains
| Strain | Plasmid | Genotype |
|---|---|---|
| YMW3 | None | ade2-1, ade3Δ22, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100, MATa/α |
| GPI11/gpi11∷LEU2 | None | GPI11/gpi11∷LEU2, ade2-1, ade3Δ22, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100, MATa/α |
| gpi11∷LEU2-pPIG-F | pPIG-F | gpi11∷LEU2, ade2-1, ade3Δ22, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100, MATa |
| gpi1∷URA3/gpi11∷LEU2-pPIG-F | pPIG-F | gpi1∷URA3, gpi11∷LEU2, ade2-1, ade3Δ22, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100, MATa |
| gpi11∷LEU2-pGAL-GPI11 | pGAL-GPI11 | gpi11∷LEU2, ade2-1, ade3Δ22, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100, MATa |
| FBY11 | None | gpi8-1, ade2-1, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, MATa |
| gpi8/gpi11∷LEU2pPIG-F | pPIG-F | gpi8-1, gpi11∷LEU2, ade2-1, ura3-1, leu2-3, 112, trp1-1, his3-11, 15, MATa |
| S-40 | None | pmi40-1, leu2-3, 112, ura3-1, his3-11, trp1-1, MATa |
| pmi40/gpi11∷LEU2-pPIG-F | pPIG-F | pmi40-1, gpi11∷LEU2, leu2-3, 112, ura3-1, his3-11, trp1-1, MATa |
| RYY51 | None | psd1∷TRP1, psd2∷HIS3, trp1-1, ura3-1, leu2-3, 112, his3-11, suc2, rho+, lys2, MATα |
| psd1∷TRP1/psd2∷HIS3/gpi11∷LEU2-pPIG-F | pPIG-F | psd1∷TRP1, psd2∷HIS3, gpi11∷LEU2, trp1-1, ura3-1, leu2-3, 112, his3-11, suc2, rho+, lys2, MATα |
| YLL031c/ YLL031c∷KANR | None | YLL031c/YLL031c∷KANR, his3Δ1, leu2Δ0, met15/met15Δ0, lys2/lys2Δ0, ura3Δ0, ade2-1, ade3Δ22, trp1-1, MATa/α |
| YLL031c∷KANR-pGAL-YLL031c | pGAL-YLL031c | YLL031c∷KANR, his3Δ1, leu2Δ0, met15, ura3Δ0, lys2 MATa |
| YLL031c∷KANR-pGAL-YLL031c/gpi11∷LEU2-pPIG-F | pPIG-F pGAL-YLL031c | YLL031c∷KANR, gpi11∷LEU2, his3Δ1, leu2Δ0, met15, ura3Δ0, lys2 MATa |
| gpi7∷LEU2 | None | gpi7∷LEU2, ade2-1, ade3Δ22, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100, MATα |
| gpi7∷URA3/gpi11∷LEU2-pPIG-F | pPIG-F | gpi7∷URA3, gpi11∷LEU2, ade2-1, ade3Δ22, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100, MATa |
SD and YPD media were as described (Sherman, 1991). Inositol-free synthetic medium was prepared from the ingredients listed for Difco (Detroit, MI) vitamin-free yeast nitrogen base in the Difco Manual. SGlyYE, SGlcYE, and SGalYE media contain 0.67% (wt/vol) Difco yeast nitrogen base, 0.2% (wt/vol) Difco yeast extract, and 3% (vol/vol) glycerol, 5% (wt/vol) glucose, or 2% (wt/vol) galactose, respectively, as carbon sources, as well as the necessary supplements to complement strain auxotrophies (Sütterlin et al., 1998). [3H]Mannose labeling was carried out in 0.67% (wt/vol) Difco yeast nitrogen base containing 2% (wt/vol) sodium pyruvate and 0.1% (wt/vol) glucose (Sipos et al., 1994). Calcofluor White sensitivity was tested using YPD medium supplemented with 0.032 mg/ml Calcofluor White (Sigma).
Database Searching and Sequence Analyses
The deduced amino acid sequence of human Pig-Fp (Inoue et al., 1993) was used as a probe to search the GenBank Sequence Database using the BLASTP and TBLASTN algorithms (Altschul et al., 1990). Sequences were analyzed using the programs DNA Strider 1.1 (Marck, 1988) and Clustal V (Higgins et al., 1991). Searches were also conducted using the PSI-BLAST program (Altschul et al., 1997).
PCR Amplification
Amplification of DNA fragments by the PCR was carried out using Vent DNA polymerase (New England Biolabs, Beverly, MA), Taq DNA polymerase (Life Technologies, Gaithersburg, MD), or Pwo DNA polymerase (Boehringer Mannheim, Indianapolis, IN) in 50-μl reaction mixtures containing 1 μM oligonucleotide primers, 200 μM dNTPs, and 60 ng of plasmid DNA, 1 μg of genomic DNA, or 1–2 μg of a cDNA library as template. Amplification reactions using Vent DNA polymerase contained 5 μl of 10× Vent polymerase buffer and 4 mM MgSO4; reactions using Taq DNA polymerase contained 5 μl of 10× Taq polymerase buffer and 6 mM MgCl2; and reactions using Pwo DNA polymerase contained 5 μl of 10× Pwo polymerase buffer and 25 mM MgSO4. Amplification reactions were started using a “hot start” procedure consisting of a 2-min incubation period at 99°C and a 1-min incubation period at 80°C at which time 0.5 U of Taq, 2.5 U of Pwo, or 2 U of Vent DNA polymerase were added.
Disruption of GPI11 and GPI7
The first 60 bp of the GPI11 open reading frame (ORF) and 1 kb of DNA upstream of the gene were amplified from Saccaromyces cerevisiae genomic DNA by PCR using the forward oligonucleotide primer 5′-CGGGATCCGTGGCATTTTTGGATATTCCTGTTT-3′ and the reverse primer 5′-CCTTAATTAATGTCATCGGAGAATGATACGGTTTT-3′, and the resulting DNA fragment was cloned into the BamHI–PacI sites of pNEB193 (New England Biolabs) to create pCTGPI11-1-1. The last 60 bp of the GPI11 ORF and 1 kb of flanking downstream DNA were PCR amplified from genomic DNA using the oligonucleotide primers 5′-CCTTAATTAACCTATTGTTGTTGGAGGTTATTTGG-3′ and 5′-GCGCGAAGCTTTCTAATCTACAGGAATTC-3′ as forward and reverse primers, respectively. The amplified DNA fragment was cloned into the PacI and HindIII sites of pNEB193 to form pCTGPI11-1-3. This PacI–HindIII fragment was then excised from pCTGPI11-1-3 and cloned into the PacI and HindIII sites of pCTGPI11-1-1 to create the plasmid pCTGPI11-1-4. The selectable marker gene LEU2 was amplified from pIRT2 (Hindley et al., 1987) by PCR using primers 5′-CCTTAATTAACTCGAGGACTTCTAGTATAT-3′ and 5′-CCTTAATTAACCGTTTCTGACAGAGTAAAATTC-3′ and cloned into the PacI site of pCTGPI11-1-4 to produce pCTGPI11-1-6, a plasmid containing the complete gpi11::LEU2 disruption fragment. Approximately 1 μg of a HindIII fragment containing gpi11::LEU2 was excised from pCTGPI11-1-6 and used to transform the wild-type diploid YMW3. Leucine prototrophs were selected on solid SD medium lacking leucine at 25°C for 3–5 d, after which integration of the gpi11::LEU2 fragment at the GPI11 chromosomal locus was confirmed by Southern blotting (Maniatis et al., 1982), using a 629-bp HindIII–EcoRV fragment isolated from the GPI11 promoter region as hybridization probe.
To disrupt GPI7 (YJL062w), ∼80% of the gene was replaced with LEU2 using a strategy similar to that used for GPI11. DNA fragments consisting of approximately the first 200 bp of GPI7 and some 850 bp 5′ to the gene and of some 220 bp of the 3′ end of GPI7 and 800 bp of 3′-flanking DNA were amplified from chromosomal DNA using the primer pair 5′-CTTGTATCTAGAGAGTTTCCTAGCAATACCCACTG-3′ (forward primer) and 5′-GAATCATTAATTAAGTCTGATCTGAGAGCATCAATG-3′ (reverse primer) for the 5′ fragment and the primer pair 5′-GCACAAGTTAATTAACAAAC-GTTCATCAGAAGTAAG-3′ (forward) and 5′-CTTACTGAATTCTAAATCTGTGATGTCGTTAACACG-3′ (reverse) for the 3′ fragment. The two PCR fragments were successively ligated into pNEB193 using XbaI and PacI sites engineered respectively at the 5′ and 3′ ends of the 5′ fragment, and PacI and EcoRI sites incorporated at the 5′ and 3′ ends of the 3′ fragment. A PacI cassette consisting of the LEU2 gene was then inserted in the PacI site joining the residual GPI7 coding region in the upstream and downstream fragments. The entire fragment consisting of gpi7::LEU2 and 5′ and 3′ flanking chromosomal DNA was excised and used to transform diploid YMW3 to leucine prototrophy. An analogous strategy was used to make a gpi7::URA3 fragment to disrupt GPI7 in the gpi11::LEU2-pPIG-F strain.
In the case of the gpi7::LEU2 and gpi7::URA3 disruptants and of the double or triple mutants generated by introduction of the gpi11::LEU fragment into a mutant strain, the presence of the disrupting allele at the correct genomic locus was verified by whole-cell PCR using a primer complementary to DNA specific to the disrupting marker gene and a primer complementary to genomic DNA outside the flanking DNA present in the disrupting DNA fragment. Introduction of gpi1::URA3 into gpi11::LEU2-pPIG-F was confirmed by demonstrating that in vitro GlcNAc-PI synthetic activity had been abolished in Ura+ haploids.
Plasmids for Expression of GPI11, PIG-F, and YLL031c in Yeast
To place GPI11 and under control of the galactose inducible/glucose repressible GAL10-1 promoter (Johnson and Davis, 1984), GPI11 was amplified by PCR with Vent polymerase using the oligonucleotides 5′-CCGGGATCCAATATGCCAGCTAAAAA-AAG-G-3′ and 5′-GGGGATCCGGACCAGTTTATGTTACCTCTA-3′ as forward and reverse primers, respectively. Thermocycling consisted of 20 successive cycles of 94°C for 30 s, 52°C for 60 s, and 72°C for 60 s and then a single final extension of 72°C for 5 min. The resulting product was digested with BamHI and ligated into the BamHI site of the GAL10-1 expression vector pMW29 (Zieler et al., 1995) to create pGAL-GPI11.
Similarly, YLL031c was cloned behind the GAL10-1 promoter by amplifying the ORF from yeast genomic DNA by PCR with Pwo DNA polymerase with the forward primer 5′-GCGGATCCAATATGGATGAAAAGACAATTAAAAAGTCG-3′ and the reverse primer 5′-CGGCGTCTAGATTTGTAAGTAAAGAGTGGAAATGAAGTTCG-3′. Amplification consisted of 25 successive cycles of incubations at 95°C for 30 s, 58°C for 30 s, and 72°C for 3.5 min and then a final incubation for 7 min at 72°C. The amplified fragment was digested with BamHI and XbaI and cloned into the BamHI and XbaI sites of pMW20 (Zieler et al., 1995) to produce pGAL-YLL031c.
A cDNA of human PIG-F was cloned from a SuperScript human juvenile female (9 y old) liver cDNA library (Life Technologies) by PCR with Taq DNA polymerase, the primers 5′-CGGGATCCCCCCGCTTCCCTTCCGCGGGAGGG-3′ and 5′-CGGGATCC-GCACAAAGAAATATCTCCCTTTGC-3′ (PIG-F primer 2), and 30 cycles of consecutive incubations at 95°C for 30 s, 62°C for 60 s, and 72°C for 60 s, followed by one cycle of incubation at 72°C for 3 min. The 758-bp product was digested with BamHI and ligated into the BamHI site of pNEB193 to create pCTPIGF-4-1. A DNA construct allowing expression of human PIG-F to be driven by the native yeast GPI11 promoter was assembled by recombinant PCR (Higuchi, 1990) using the primer sets 5′-CGGGATCCGCGCATTTTGAAGCATGGAGAG-3′ and 5′-TCTCTTGATATCGTTATCTTTCATATTTAAATTGGACCTTCTTTAGTG-3′ to amplify the GPI11 promoter region from genomic DNA and the primer 5′-CACTAAAGAAGGTCCAATTTAAATATGAAAGATAACGATATCAAGAGA-3′ and PIG-F primer 2 (see above) to amplify the PIG-F coding region from pCTPIGF-4-1. Amplification of each fragment and recombinant PCR with Vent DNA polymerase consisted of 30 cycles of successive incubations at 95°C for 30 s, 52°C for 60 s, and 72°C for 60 s, followed by a final cycle of 72°C for 3 min. The 1082-bp chimeric fragment was digested with BamHI and cloned into the BamHI site of pRS414 (TRP1, CEN), pRS426 (URA3, 2μ; Christianson et al., 1992), to form pCTPIGF-7-1 and pCTPIGF-7-2, respectively.
Radiolabeling of S. cerevisiae Lipids and Proteins
For [3H]inositol labeling of lipids in temperature-sensitive strains, cultures of each strain were grown at 25°C in inositol-free medium to an OD600 of ∼1, and cells from 10 ml of culture were then harvested by centrifugation and resuspended in 1 ml of fresh inositol-free medium. These cultures were then either shifted to 37°C for 20 min or maintained at 25°C before 15 μCi of [3H]inositol were added to them, and radiolabeling was continued for 2 h at the same temperature. For [3H]inositol labeling of lipids in the gpi11::LEU2- pGAL-GPI11, YLL031c::KANR-pGAL-YLL031c, and YLL031c::KANR- pGAL-YLL031c/gpi11::LEU2-pPIG-F strains, the procedure of Sütterlin et al. (1998) was adapted. Cells were maintained on SGlyYE medium and then shifted to SGalYE or SGlcYE medium for 16h. In the case of gpi11::LEU2-pGAL-GPI11, 10 OD600 units of cells were then resuspended in 1 ml of inositol-free medium containing galactose or glucose, respectively, and 15 μCi of [3H]inositol were added, and radiolabeling was continued for 60 min at 30°C. For the YLL031c::KANR-pGAL-YLL031c strain, 10 OD600 units of cells from a 16-h culture in SGlcYE medium were resuspended in 1 ml of inositol-free, glucose-containing medium and then radiolabeled with 15 μCi of [3H]inositol for 2 h at 25°C. In the case of the YLL031c::KANR/pGAL-YLL031c/gpi11::LEU2/pPIG-F strain, batches of 10 OD600 units of SGlcYE-repressed cells were resuspended and incubated in inositol-free medium containing glucose for 30 min at 25°C, after which one batch was shifted to 37°C for 20 min, and the other was maintained at 25°C. Fifteen microcuries of [3H]inositol were then added, and radiolabeling continued for 2 h at 37 and 25°C, respectively. [3H]Inositol labeling of proteins in the gpi11::LEU2-pGAL-GPI11 strain was carried out similarly, with the exception that 100 μCi of [3H]inositol were used to label ∼10 OD600 units of cells for 60 min at 30°C in 1 ml of galactose- or glucose-containing medium (Colussi and Orlean, 1997). Radiolabeled proteins were separated by SDS-PAGE through a 10% acrylamide gel and then made visible by fluorography.
[2-3H]Mannose labeling of lipids in cells harboring the pmi40 mutation was performed essentially as described previously (Sipos et al., 1994), using a cell density of 10 OD600 units/ml in synthetic medium containing 2% sodium pyruvate and 0.1% glucose, 100 μCi of [2-3H]mannose, and a labeling period of 60 min. For [3H]ethanolamine labeling of lipids in the gpi11::LEU2-pPIG-F/psd1::TRP1/psd2::HIS3 strain, cells were grown to midlog phase in SD medium supplemented with 2 mM EthN and 2 mM choline. Ten OD600 units of cells were then washed three times with 5 ml of SD medium lacking EthN and choline and resuspended in 1 ml of SD containing 50 μCi of [3H]EthN. Radiolabeling was continued for 14–20 h.
Extraction, Mild Base and Phospholipase C Treatment, and Chromatography of Candidate GPI Precursors
After radiolabeling, NaN3 was added to cultures to give a concentration of 10 mM, and yeast cells were then washed twice with cold water. Cell pellets were resuspended in 30 μl of a 10 mg/ml solution of lyticase in 1 M sorbitol and incubated for 30 min at 37°C, after which 100 μl of methanol and 100 μl of chloroform were added. Cell debris was removed by centrifugation, and the lipid extract was evaporated to dryness, then extracted into 200 μl of 1-butanol saturated with 10 mM Tris-HCl, pH 7.5, containing 0.1 mM EDTA, and then back-extracted with 100 μl of water. For phosphatidylinositol-specific phospholipase C (PI-PLC) treatment, lipid samples were evaporated to dryness, then resuspended in 10 mM Tris-HCl, pH 7.5, containing 0.2 mM EDTA, 20% (vol/vol) 1-propanol, and 0.05 U of bacterial PI-PLC (ICN Biochemicals), and incubated for 16–24 h at 37°C (Puoti and Conzelmann, 1993). Control incubations lacking PI-PLC were performed in parallel. For complete mild base hydrolysis, lipids were resuspended in a mixture of 100 μl of methanol and 100 μl of 30% aqueous ammonia and incubated at 37°C for 16–24 h (Mayor et al., 1990). Control incubations in methanol alone were carried out in parallel. After PI-PLC or mild base treatment, lipids were again extracted with buffer-saturated 1-butanol, and the organic phase was analyzed by TLC.
TLC of radiolabeled lipids was performed on 20-cm Silica Gel 60 plates (Altech, Deerfield, IL). Plates were prerun in solvent A (chloroform:methanol:water, 65:25:4, vol/vol), after which the samples were applied and separated in solvent B (chloroform:methanol:water, 10:10:2.5, vol/vol) or solvent C (chloroform:methanol:water, 10:10:3, vol/vol). After separation, TLC plates were exposed to BioMax MS film (Eastman Kodak, Rochester, NY) for 4–7 d using a TranScreen-LE (Kodak) intensifying screen. TLC plates of [3H]inositol-labeled lipids were exposed for 4–7 d, those of [3H]EthN-labeled lipids were exposed for 10 d, and high-performance TLC (HPTLC) plates were exposed from 6 to 30 d as indicated in the legends to Figures 7 and 9.
Figure 7.
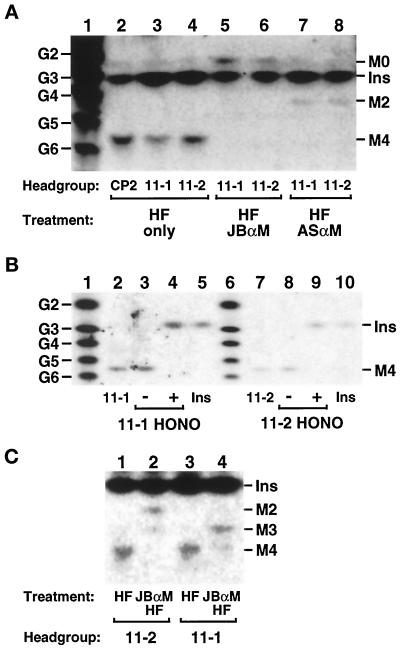
Analysis of the glycan head groups of lipids 11-1 and 11-2. Lipids 11-1 and 11-2 were radiolabeled at 37°C with [3H]inositol in the gpi11:: LEU2-pPIG-F strain, and CP2 was radiolabeled at 37°C in gpi8 cells. The three lipids were isolated by preparative TLC and deacylated by treatment with mild base. (A) Size analysis and α-mannosidase sensitivity. Deacylated head groups were re-N-acetylated with acetic anhydride, then dephosphorylated with HF, and samples of them were treated with JBαM (lanes 5 and 6), with ASαM (lanes 7 and 8), or not treated (lanes 2–4). Glycans were then separated by HPTLC and detected by fluorography. Lane 1 displays a series of NaB[3H]4-reduced glucose2 (G2)-glucose6 (G6) oligomers, and the known mobilities of [3H]inositol-labeled GPI glycans and [3H]inositol are indicated. (B) Nitrous acid deamination. Deacylated, N-acetylated, and dephosphorylated glycans were submitted to nitrous acid (HONO) deamination (lanes 4 and 9) or mock treated (lanes 3 and 8). Lanes 2 and 7 of the HPTLC contain [3H]inositol-labeled material released from lipids 11-1 and 11-2 after mild base, acetic anhydride, and HF treatment; lanes 5 and 10 contain [3H]inositol; and lanes 1 and 6 contain NaB[3H]4-reduced glucose oligomers. (C) Positioning of HF-labile substituents. Deacylated, N-acetylated head groups were dephosphorylated directly with HF or treated first with JBαM and then with HF before separation by HPTLC. Traces of Man3 and Man4 glycans in lanes 2 and 4, respectively, most likely originate from incomplete JBαM digestion. The [3H]inositol present in glycan samples in A and C is a contaminant from the preparative TLC. Ins, M0, M2, M3, and M4, [3H]Ins, GlcNAc-[3H]Ins, Man2-GlcNAc-[3H]Ins, Man3-GlcNAc-[3H]Ins, and Man4-GlcNAc-[3H]Ins, respectively. HPTLC plates were exposed to x-ray film for 30 d (A), 10 d (B), and 21 d (C).
Figure 9.
Depletion of YLL031cp leads to accumulation of a Man4-GPI bearing an HF-labile substituent on Man-1. (A) Accumulation of a [3H]inositol-labeled lipid. The YLL031c:: KANR-pGAL-YLL031c strain was maintained on SGlyYE medium, shifted to SGlcYE medium for 16 h, then resuspended in inositol-free medium containing glucose, and radiolabeled with [3H]inositol for 2 h at 25°C (lane 1). The gpi11::LEU2-pPIG-F (lane 2, Δgpi11) and gaa1 (lane 3) strains were radiolabeled with [3H]inositol at 37°C. Lipids were extracted and separated by TLC in solvent B. (B) Size analysis of the glycan head group of lipid 13-1 and positioning of HF-labile substituents on it. The procedures followed are detailed in the legend to Figure 7. [3H]Inositol-labeled lipid 13-1 was isolated by preparative TLC, deacylated, re-N-acetylated, and dephosphorylated with HF, and the resulting neutral glycan head group was separated by HPTLC without further treatment (lanes 2 and 4) or after treatment with JBαM (lane 3) or with ASαM (lane 5). A sample of deacylated, re-N-acetylated 13-1 was first digested with JBαM and then dephosphorylated with HF before separation by HPTLC (lane 6). Lane 1 displays NaB[3H]4-reduced glucose oligomers. The abbreviations used are defined in the legend to Figure 7. The HPTLC plate was exposed to x-ray film for 6 d.
Glycan Head Group Analysis
[3H]Inositol-labeled 11-1, 11-2, 13-1, and complete precursor 2 (CP2) from ∼500 OD600 units of pulse-labeled gpi11::LEU2-pPIG-F, YLL031c::KANR/pGAL-YLL031c, or gpi8 cells were each purified by two rounds of preparative TLC using 1000-μm-thick Silica-G plates (Altech). Preparative plates were prerun in solvent A (chloroform:methanol:water, 65:25:4, vol/vol), after which [3H]inositol-labeled lipids were applied and separated in solvent B (chloroform:methanol:water, 10:10:2.5, vol/vol). Silica was scraped from the region of the plate containing [3H]inositol-labeled lipids 11-1, 11-2, 13-1, and CP2 into the barrel of a 10-ml syringe (plugged with a small wad of glass wool) to create a minicolumn, from which the lipid was eluted by passing 25 ml of solvent E (chloroform:methanol:water, 10:20:7, vol/vol) through the silica, after which the eluate was dried under nitrogen. Soluble head groups were released from the purified lipids by complete mild base hydrolysis for 16–24 h at 37°C with a mixture of 5 ml of 30% aqueous ammonia and 5 ml of methanol and then butanol extracted and dried. Head groups were treated with jack bean α-mannosidase (JBαM), or A. satoi α-mannosidase (ASαM) in 0.1 M sodium acetate, pH 5.0, containing either 0.5 U JBαM or 5 μU ASαM for 16–24 h at 37°C, and the mixture was then desalted by passage through AG 501-X8 resin (Bio-Rad, Hercules, CA) as described (Schneider and Ferguson, 1995). Re-N-acetylation and HF dephosphorylation (for 4 d at 4°C) of head groups were performed as described by Schneider and Ferguson (1995). Nitrous acid deamination of head group glycans was performed in 440 μl of 0.1 M sodium acetate, pH 3.5, to which 50 μl of freshly made 1 M NaNO2 were then added, and reactions were allowed to proceed at 50°C for 4 h (Puoti and Conzelmann, 1992). Neutral head groups or head group fragments were separated on high-performance silica TLC plates (Altech). Radiolabeled neutral glycans were separated by developing the chromatogram four consecutive times in solvent D (1-butanol:ethanol:water, 4:3:3, vol/vol; Schneider and Ferguson, 1995) and then detected by fluorography for 6–30 d. Mobilities of head group glycan fragments were compared with those of standards of radiolabeled glucose polymers generated by partial acid hydrolysis of dextran followed by reductive labeling with NaB[3H]4 as described (Schneider and Ferguson, 1995).
RESULTS
Identification of the S. cerevisiae Homologue of PIG-F
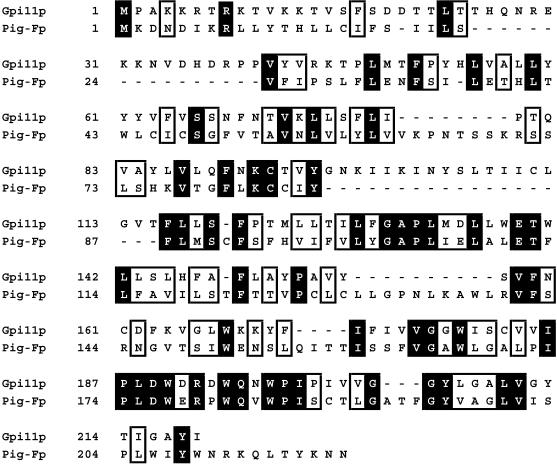
Searches of the GenBank sequence database using BLAST (Altschul et al., 1990) for S. cerevisiae counterparts of the mammalian Pig-F protein yielded an unidentified open reading frame, YDR302w, encoding a hydrophobic 219-amino-acid protein with 26% identity and 49% similarity to Pig-Fp. We designate this gene GPI11. The alignment of the deduced amino acid sequences of the human Pig-F and Gpi11 proteins is presented in Figure 1.
Figure 1.
Alignment of deduced amino acid sequences of Pig-Fp and Gpi11p. The alignment was generated using the Clustal V program (Higgins, 1994). Identical amino acids are represented in black boxes, and amino acids defined as similar are in open boxes. Numbers correspond to sequence position of the first amino acid in each line.
A database search using the PSI-BLAST program (Altschul et al., 1997) revealed that larger proteins that contain sequences homologous to the same ∼110-amino-acid segment of Pig-Fp and Gpi11p (corresponding to amino acids 90–200 of Pig-Fp) are encoded in the Schizosaccharomyces pombe and Caenorhabditis elegans genomes. The S. pombe protein (accession number CAB40182) contains 503 amino acids, of which residues 390–503 are similar to Pig-Fp and Gpi11p, but amino acids 20–200 resemble short-chain dehydrogenases and reductases. In contrast, amino acids 10–120 of the 571-residue C. elegans protein (accession number AAB03156) resemble Pig-Fp/Gpi11p, whereas amino acids 130–520 are similar to nitrogen permease regulators. We have not confirmed that these S. pombe and C. elegans proteins are each encoded by a single gene, and it is possible that these apparently larger open reading frames are the result of sequencing errors. We have not identified any other proteins that show sequence similarity over their entire lengths to these putative fission yeast and nematode proteins.
Lethal Disruption of GPI11
To test whether GPI11 is an essential gene and to create Gpi11p-deficient strains, the chromosomal GPI11 locus was disrupted by deleting 82% of its coding region and replacing that DNA with the LEU2 gene. A linear gpi11::LEU2 DNA fragment was used to transform a GPI11/GPI11 leu2/leu2 diploid to leucine prototrophy. Correct integration of gpi11::LEU2 at the GPI11 chromosomal locus was confirmed by Southern blot analysis. This diploid was sporulated, and tetrad analysis of the meiotic segregants from 10 asci revealed that in each case, only the two leucine auxotrophic, GPI11 haploids grew into colonies. The remaining two spores of a tetrad germinated and completed one or two rounds of cell division before ceasing further growth. We conclude that the gpi11::LEU2 haploids were inviable, and that GPI11 is essential for vegetative growth.
Complementation of the Lethal gpi11::LEU2 Mutation by PIG-F
To establish whether Gpi11p and human Pig-Fp have equivalent functions in vivo, we tested whether human PIG-F can complement the lethal null mutation in GPI11. The human PIG-F coding region was amplified from a human liver cDNA library and cloned in frame behind the native yeast GPI11 promoter. GPI11 promoter-PIG-F fusions were cloned into both centromeric and 2μ plasmids, which were in turn introduced into the heterozygous gpi11::LEU2 diploids. Tetrad analysis of the meiotic segregants obtained upon sporulation of the transformed diploids revealed that PIG-F expressed on a 2μ plasmid restored viability to the gpi11::LEU2 haploids at 25°C, but that PIG-F expressed from the low-copy, centromeric plasmid did not. In control experiments, native GPI11 complemented gpi11::LEU2 when expressed on both centromeric and 2μ plasmids. Human Pig-Fp can therefore substitute for Gpi11p at high copy in vivo. The simplest interpretation of this is that these two proteins have the same function in vivo. A number of other mammalian GPI biosynthetic genes can also complement null mutations in their S. cerevisiae homologues (Benghezal et al., 1996; Sütterlin et al., 1998; Tiede et al., 1998).
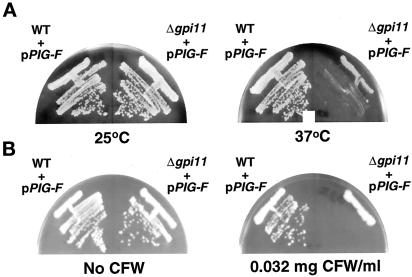
Interestingly, the gpi11::LEU2 haploids complemented by PIG-F (henceforth, “gpi11::LEU2-pPIG-F“) grew very slowly at 37°C (Figure 2A) and also showed hypersensitivity to Calcofluor White, a phenotype common to strains defective in GPI anchoring as well as cell wall synthesis mutants (Ram et al., 1994; Benghezal et al., 1995) (Figure 2B). The partial temperature-sensitivity of gpi11::LEU2-pPIG-F could be exploited to test for GPI anchoring defects.
Figure 2.
Temperature-sensitive growth and Calcofluor White sensitivity of gpi11:: LEU2-pPIG-F cells. Wild-type (GPI11) and gpi11:: LEU2 strains harboring pPIG-F were streaked on YPD agar and incubated for 4 d at 25 or 37°C (A) or on YPD agar with or without 0.032 mg of Calcofluor White/ml and incubated for 4 d at 30°C (B).
GPI Anchoring Defects of Gpi11p-deficient Strains
If Gpi11p participates in GPI assembly, then Gpi11p-deficient strains should be blocked in GPI attachment to protein and accumulate a GPI biosynthetic precursor. A Gpi11p deficiency was created in two different ways. In the first approach, the GPI11 gene was placed under the control of the glucose-repressible GAL10 promoter to allow GPI11 expression to be halted and Gpi11p to become depleted upon shift of the culture to the repressing carbon source. In the second approach, the partially temperature-sensitive gpi11::LEU2-pPIG-F strain was shifted to 37°C.
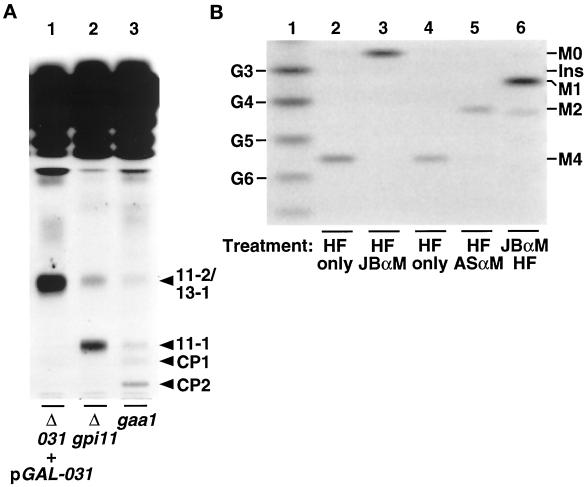
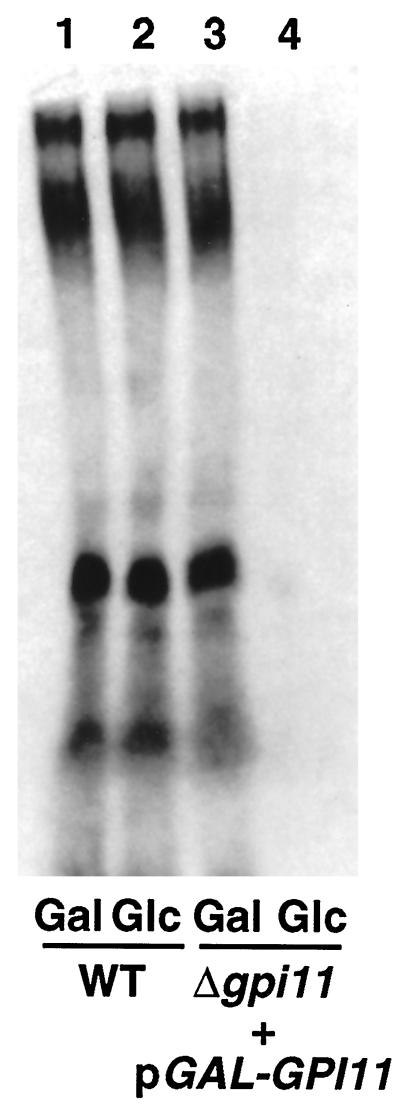
gpi11::LEU2-pGAL-GPI11 cells were tested for a GPI anchoring defect by pulse labeling them with [3H]inositol and examining whether [3H]inositol incorporation into protein is blocked when GPI11 expression is repressed. Because all detectable protein-linked inositol in yeast is present in GPI anchors, radiolabeling with [3H]inositol provides a test of the ability of mutants to carry out GPI anchoring (Conzelmann et al., 1990). For these experiments, gpi11::LEU2-pGAL-GPI11 cells were maintained on the nonrepressing carbon source glycerol, shifted either to galactose- or to glucose-containing medium for 16 h, and then pulse labeled with [3H]inositol for 1 h. Cultures shifted to glucose are blocked in [3H]inositol incorporation into protein, whereas the control culture in galactose remains capable of incorporating this precursor (Figure 3). Depletion of Gpi11p therefore leads to a GPI anchoring defect.
Figure 3.
Depletion of Gpi11p results in a block in [3H]inositol incorporation into proteins. Wild-type (WT) and gpi11:: LEU2-pGAL-GPI11 (Δgpi11 + pGAL-GPI11) cells were maintained on SGlyYE medium, shifted to SGalYE or SGlcYE medium for 16 h, then resuspended in 1 ml of inositol-free medium containing Gal or Glc, respectively, and radiolabeled with 100 μCi of [3H]inositol for 60 min at 30°C. Proteins were then extracted and separated by SDS-PAGE, and radiolabeled GPI-anchored proteins were visualized by fluorography.
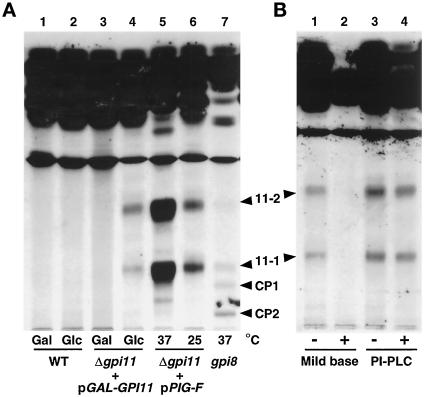
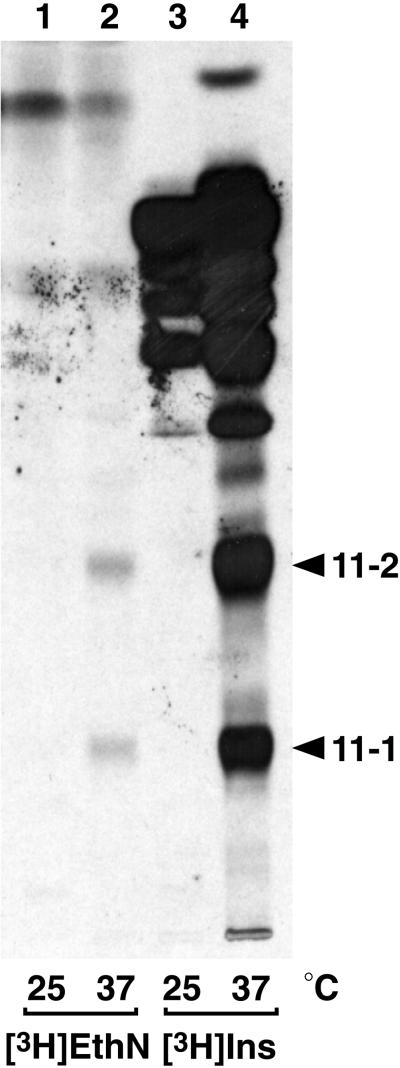
We next tested whether any [3H]inositol-labeled GPI biosynthetic intermediates accumulate in Gpi11p-depleted strains, as is the case with mammalian class F mutants. Shift of gpi11::LEU2-pGAL-GPI11 cells to glucose leads to the accumulation of two [3H]inositol-labeled lipids, designated 11-1 and 11-2 (Figure 4A, lane 4), and these are absent from cells shifted to galactose (Figure 4A, lane 3). These lipids are sensitive to mild base hydrolysis but resistant to PI-PLC, indicating that they bear ester-linked fatty acyl chains and suggesting they are acylated on their inositol moiety (Figure 4B). Both are properties of known late-stage yeast GPI precursors (Sipos et al., 1994). Lipids 11-1 and 11-2 have mobilities distinct from those of CP1 and CP2 (Sipos et al., 1994), which accumulate in the gpi8 strain (Figure 4A, lane 7), a mutant blocked in GPI transfer to protein (Benghezal et al., 1995).
Figure 4.
Accumulation of mild base-sensitive and PI-PLC-resistant [3H]inositol-labeled lipids in gpi11:: LEU2-pGAL-GPI11 and gpi11:: LEU2-pPIG-F cells. (A) Wild-type (WT) and gpi11:: LEU2-pGAL-GPI11 (Δgpi11 + pGAL-GPI11) cells were maintained on SGlyYE medium, shifted to SGalYE or SGlcYE medium for 16 h, then resuspended in 1 ml of inositol-free medium containing Gal (lanes 1 and 3) or Glc (lanes 2 and 4), respectively, and radiolabeled with 15 μCi of [3H]inositol for 60 min at 30°C. gpi11:: LEU2-pPIG-F cultures were grown at 25°C in inositol-free medium and then divided into two equal portions, one of which was shifted to 37°C for 20 min and the other maintained at 25°C. Shifted and control cultures were then radiolabeled with 15 μCi of [3H]inositol for 2 h at 37 and 25°C, respectively (lanes 5 and 6). The gpi8 strain was grown in inositol-free medium, shifted to 37°C, and pulse labeled at 37°C (lane 7). Radiolabeled lipids were then extracted as detailed in MATERIALS AND METHODS. (B) Lipids from gpi11::LEU2-pGAL-GPI11 cells that had been shifted to glucose and [3H]inositol-labeled in glucose-containing medium were incubated with methanol/water or methanolic ammonia (mild base) (lanes 1 and 2) or without or with PI-PLC (lanes 3 and 4). After either treatment, incubation mixtures were extracted with buffer-saturated 1-butanol and submitted to phase partitioning, after which the butanol phase was separated by TLC using solvent B, and radiolabeled lipids were detected by fluorography. Arrowheads indicate the positions of lipids 11-1, 11-2, and complete precursors CP1 and CP2 (Sipos et al., 1994).
Lipids with the same mobility as 11-1 and 11-2 also accumulate in gpi11::LEU2-pPIG-F cells, accumulation being most pronounced at 37°C (Figure 4A, lanes 5 and 6). The absence, from extracts of gpi11::LEU2-pPIG-F cells, of any lipids whose mobility differs from those of lipids 11-1 and 11-2 indicates that when Pig-Fp is expressed in yeast, it does not generate aberrant lipids by acting on a yeast GPI precursor that Gpi11p itself does not recognize. The fact that comigrating lipids accumulate in gpi11::LEU2 strains depleted of native yeast Gpi11p or partially complemented by PIG-F is consistent with the notion that the comigrating lipids have the same structure, and that the two strains have the same GPI biosynthetic defect. However, we cannot exclude the possibility that the comigrating lipids that accumulate in the gpi11::LEU2-pGAL-GPI11 and gpi11::LEU2-pPIG-F strains are different structural isoforms with the same size and charge (see DISCUSSION). With this caveat, though, we exploited the gpi11::LEU2-pPIG-F strain as a bona fide temperature-sensitive “gpi11 “ mutant.
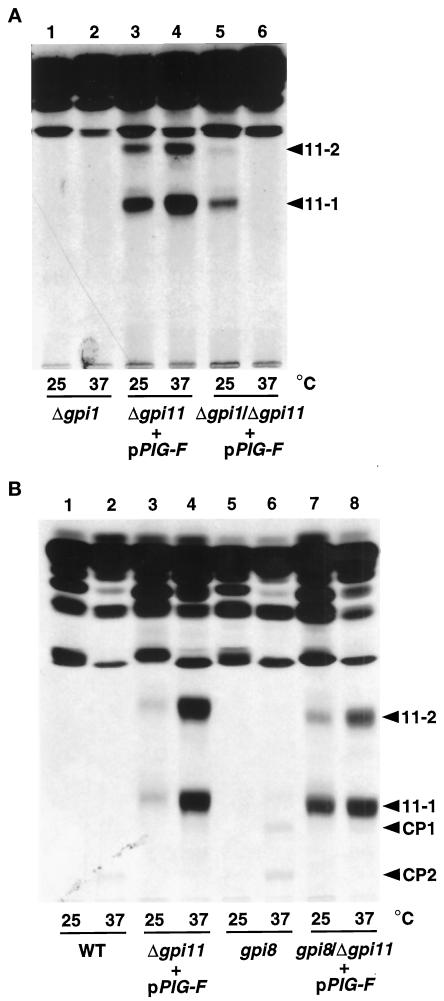
The finding that Gpi11p-deficient strains accumulate potential GPI precursors allowed us to obtain genetic evidence that GPI11 is involved in the GPI biosynthetic pathway. We tested the epistasis relationships of the temperature-sensitive gpi11::LEU2-pPIG-F “mutation” with known GPI anchoring mutations by introducing the gpi11::LEU2-pPIG-F background into the gpi1 and gpi8 mutants and testing the double mutants for the accumulation of [3H]inositol-labeled GPI precursors 11-1, 11-2, CP1, and CP2. The gpi1 mutation, which blocks formation of GlcNAc-PI, the first step in GPI assembly (Leidich et al., 1994; Leidich and Orlean, 1996), is epistatic to gpi11: low levels of both lipids 11-1 and 11-2 accumulate in gpi1::URA3/gpi11::LEU2-pPIG-F strains labeled at 25°C, at which temperature gpi1 strains exhibit only a partial GPI synthetic defect, but shift to 37°C completely abolishes accumulation of these two GPIs (Figure 5A, lanes 5 and 6). Formation of lipids 11-1 and 11-2 is therefore dependent on GlcNAc-PI synthesis.
Figure 5.
Epistasis relationships of a Gpi11p-deficient strain determined from lipid accumulation phenotypes of gpi11:: LEU2-pPIG-F harboring additional mutations in the gpi1 or gpi8 genes. Strains were pulse labeled with [3H]inositol at 25 and 37°C as described in the legend of Figure 4A. (A) Lipids from the gpi1:: URA3 (lanes 1 and 2), gpi11:: LEU2-pPIG-F (lanes 3 and 4), and gpi1:: URA3/gpi11:: LEU2-pPIG-F (lanes 5 and 6) strains, separated by TLC using solvent C. (B) Lipids from wild-type (WT; lanes 1 and 2), gpi11:: LEU2-pPIG-F (lanes 3 and 4), gpi8 (lanes 5 and 6), and gpi8/gpi11:: LEU2-pPIG-F (lanes 7 and 8) strains, separated in solvent B. The positions of lipids 11-1, 11-2, CP1, and CP2 are indicated.
Gpi11p is required for the formation of complete GPI precursors CP1 and CP2. A gpi8/gpi11::LEU2-pPIG-F double mutant was made by disrupting GPI11 in a gpi8 strain that had been transformed with pPIG-F. The resulting strain was likely to be a double mutant, because it failed to grow at 37°C, whereas the gpi11::LEU2-pPIG-F and gpi8 strains are but partially temperature sensitive at 37°C (our unpublished data). Pulse labeling of the gpi8/gpi11::LEU2-pPIG-F mutant with [3H]inositol, followed by TLC display of GPI precursors, reveals that neither CP1 nor CP2 accumulated (Figure 5B, lanes 6–8). Furthermore, no new polar [3H]inositol-labeled lipids accumulated in the gpi8/gpi11::LEU2-pPIG-F strain. This makes it unlikely that Pig-Fp rescues a Gpi11p-depleted strain by transferring the bridging EthN-P to a Man3-containing GPI: such an aberrant lipid would be expected to accumulate because of the gpi8 block.
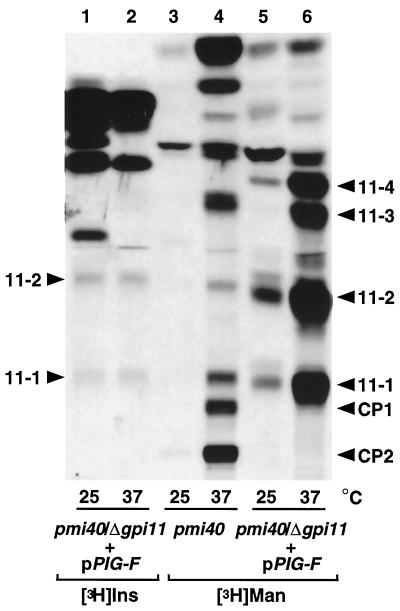
This finding was corroborated by the results of pulse-labeling experiments with [2-3H]mannose. These were carried out with strains harboring the pmi40 mutation, which causes conditional mannose auxotrophy and enhances [2-3H]mannose labeling of late-stage GPI precursors (Sipos et al., 1994). The pmi40/gpi11::LEU2-pPIG-F double mutant was created by disrupting GPI11 in a pmi40 strain into which pPIG-F had previously been introduced. In the pmi40 strain (Figure 6, lane 4), [3H]mannose-labeled lipids with mobilities corresponding to those of CP1 and CP2 (Sipos et al., 1994) are radiolabeled at 37°C. In addition, [3H]mannose-labeled species with the same mobilities as lipids 11-1 and 11-2 are present in this sample, a finding that suggests that these two lipids are normally made in GPI11 cells.
Figure 6.
Abolition of [2-3H]mannose labeling of CP1 and CP2 in the pmi40/gpi11:: LEU2-pPIG-F strain and accumulation of four [2-3H]mannose-labeled lipids. The pmi40/gpi11:: LEU2-pPIG-F strain was radiolabeled at 25 or 37°C with [3H]inositol (lanes 1 and 2) or with [2-3H]mannose according to Sipos et al. (1994) (lanes 5 and 6), and pmi40 was radiolabeled with [2-3H]mannose (lanes 3 and 4). Lipids were separated by TLC using solvent B. The positions of lipids 11-1, 11-2, 11-3, 11-4, CP1, and CP2 are indicated.
pmi40/gpi11::LEU2-pPIG-F cells radiolabeled with [2,6-3H]mannose at 25°C accumulate two lipids with mobilities corresponding to [3H]inositol-labeled lipids 11-1 and 11-2 (Figure 6, lanes 1 and 5). At 37°C, pmi40/gpi11::LEU2-pPIG-F cells show pronounced accumulation of four [3H]mannose-labeled lipids, the two most polar of which comigrate with [3H]inositol-labeled lipids 11-1 and 11-2 (Figure 6, lane 6). [3H]mannose labeling of 11-1 and 11-2 corroborates the notion that they are mannosylated GPIs. We do not yet know whether lipids 11-3 and 11-4 are also GPIs, but the accumulation of multiple [3H]mannose-labeled lipids in a Gpi11p-depleted S. cerevisiae strain has its parallel in mammalian Thy-1 class F mutants (Lemansky et al., 1991; Hirose et al., 1992; Kamitani et al., 1992; Puoti and Conzelmann, 1993). Strikingly, this [3H]mannose-labeling experiment shows that CP1 and CP2 do not become radiolabeled in pmi40/gpi11::LEU2-pPIG-F cells, although both lipids are present in extracts of pmi40 cells (Figure 6, lanes 4 and 6). The formation of CP1 and CP2 is therefore dependent on Gpi11p.
Taken together, the results of these genetic analyses indicate that Gpi11p acts in the GPI biosynthetic pathway between GlcNAc-PI synthesis and GPI transfer to protein. To investigate what step in GPI biosynthesis is blocked in the gpi11::LEU2-pPIG-F strain and to determine the difference between lipids 11-1 and 11-2, we analyzed the structure of the glycan head group of these two lipids.
Structural Analysis of the Glycan Head Groups of the Lipids 11-1 and 11-2
Lipids 11-1 and 11-2 could differ in the number of mannose residues present, as is the case in mammalian Thy-1 class F mutants, in which Man1-, Man2-, and Man3-containing GPIs accumulate (Sugiyama et al., 1991; Hirose et al., 1992; Kamitani et al., 1992; Puoti and Conzelmann, 1993), by the presence of one or more polar substituents (most likely EthN-Ps; Hirose et al., 1992; Kamitani et al., 1992; Puoti and Conzelmann, 1993) or in their lipid composition. We tested the first two possibilities with the [3H]inositol-labeled lipids 11-1 and 11-2 isolated from the gpi11::LEU2-pPIG-F strain. Because of the lipids that accumulate in this strain, only these two can be radiolabeled in sufficient quantity for analysis and separated from other radiolabeled, non-GPI species. We are assuming that these lipids are identical to those that accumulate in cells depleted of native Gpi11p: [3H]inositol-labeled lipids 11-1 and 11-2 accumulate in gpi11::LEU2-pGAL-GPI11 cells at levels that are too low for glycan head group analysis (see caveat above).
Lipids 11-1 and 11-2 were isolated after two rounds of preparative TLC, and full-size neutral glycan head groups were isolated from them after deacylation, re-N-acetylation, and treatment with HF to remove any phosphodiester-linked substituents. The sizes of the glycans were determined by comparing their TLC mobilities with those of a series of NaB[3H]4-reduced dextran standards, with the mobility of the neutral glycan derived from CP2 from the control gpi8 mutant, and with the published mobilities of GPI standards. The full-size neutral glycans were also tested for their susceptibility to jack bean α-mannosidase and to an α1,2-specific mannosidase from A. satoi.
The neutral glycan head groups from both lipids 11-1 and 11-2 migrated with the Man4-GlcNAc-[3H]Ins standard prepared from CP2 to a position on the HPTLC between those of the reduced penta- and hexasaccharide standards, consistent with the published mobility of Man4-GlcNAc-Ins (Benghezal et al., 1995) (Figure 7A, lanes 2–4). Treatment of the neutral glycan head groups with JBαM converted each to a [3H]inositol-labeled species with the mobility of GlcNAc-Ins, consistent with the removal of four α-linked mannoses (Figure 7A, lanes 5 and 6), whereas α1,2-mannosidase treatment converted the three glycans to species migrating to the positions expected for Man2-GlcNAc-Ins, consistent with the removal of two α1,2-linked mannoses (Figure 7A, lanes 7 and 8). The neutral glycan head groups of lipids 11-1 and 11-2 are therefore identical to that of the complete GPI precusor. These findings rule out the possibility that lipids 11-1 and 11-2 differ in their number of mannose residues and that 11-2 is a Man3-containing species observed, because earlier intermediates in the GPI assembly pathway become “backed up.” Also ruled out is the possibility that lipid 11-2 is an abnormal species that accumulates, because heterologously expressed Pig-Fp aberrantly transfers EthN-P to a Man3-containing yeast GPI that resembles the Pig-Fp natural acceptor, creating a GPI that cannot be transferred to protein.
To obtain additional evidence that 11-1 and 11-2 are GPIs and contain nonacetylated GlcN adjacent to inositol, [3H]inositol-labeled glycan head groups that had been dephosphorylated with HF but not re-N-acetylated were submitted to nitrous acid deamination. The head groups from 11-1 and 11-2 both released [3H]inositol, as expected for a GPI (Figure 7B, lanes 4 and 9). These findings, together with the results of size analyses and α-mannosidase digestion, indicate that the dephosphorylated head groups of 11-1 and 11-2 are both Man4-GlcNAc-Ins.
We next tested whether lipids 11-1 and 11-2 differ in the extent to which they are decorated with phosphodiester-linked side branches that would have been removed upon HF treatment to generate the neutral glycan. Both mammalian and yeast GPI precursors have been shown to bear HFLSs (presumed to be EthN-P) on one or more of the first three GPI mannoses (Hirose et al., 1992; Kamitani et al., 1992; Puoti and Conzelmann, 1993; Canivenc-Gansel et al., 1998; Sütterlin et al., 1998; Benachour et al., 1999). The occurrence of EthN-P side branches on GPI mannoses can be inferred from the presence of HF-labile substituents that block removal by JBαM of the first substituted Man that the glycosidase encounters and of any Man residues internal to it. In these experiments, deacylated and re-N-acetylated lipids 11-1 and 11-2 were first treated with JBαM and then with HF, and the resulting glycans were submitted to size analysis by HPTLC. The results reveal a clear difference between 11-1 and 11-2: JBαM digestion followed by HF treatment results in conversion of 11-1 to Man3-GlcNAc-Ins (Figure 7C, lanes 3 and 4) and of 11-2 to Man2-GlcNAc-Ins (Figure 7C, lanes 1 and 2). Lipid 11-1 therefore bears an HFLS on its third Man, whereas 11-2 has one on its second. The neutral glycan obtained after JBαM treatment, and then HF dephosphorylation, of deacylated 11-2 is essentially homogeneous Man2-GlcNAc-Ins: although the sample in Figure 7C, lane 4, might contain Man-GlcNAc-[3H]Ins that is obscured by contaminating [3H]inositol, independent analysis of a sample free of [3H]Ins confirmed that Man2-GlcNAc-Ins is the major component, because only a trace of material migrating at the position predicted for Man-GlcNAc-Ins was detected (our unpublished results). Therefore, if lipid 11-2 is a mixture of Man4 species bearing a single HFLS on either Man-1 or Man-2, then the former is at best a very minor component. Similarly, the fact that lipid 11-1 yields no detectable Man2-GlcNAc-[3H]Ins after JBαM digestion followed by HF treatment indicates that this species does not contain significant amounts of a Man4-GPI that bears an HFLS on its second mannose but that is unsubstituted on Man-3. However, we cannot rule out the possibility that 11-1 is a mixture of Man4 species, both with EthN-P on Man-3, but with a second EthN-P on either Man-1 or Man-2. If the HFLSs are indeed EthN-Ps, lipids 11-1 and 11-2 should become radiolabeled with [3H]EthN. To enhance [3H]EthN incorporation into lipids, the gpi11::LEU2-pPIG-F background was introduced into an EthN-auxotrophic Δpsd1/Δpsd2 (Trotter and Voelker, 1995) strain. Both 11-1 and 11-2 are radiolabeled with [3H]EthN (Figure 8); therefore, each contains at least one EthN-P moiety.
Figure 8.
Lipids 11-1 and 11-2 are both radiolabeled with [3H]ethanolamine. The ethanolamine-auxotrophic psd1::TRP1/psd2:: HIS3/gpi11::LEU2-pPIG-F strain was suspended in SD medium lacking ethanolamine and choline and radiolabeled at 25 or 37°C with either [3H]ethanolamine (50 μCi) or [3H]inositol. Radiolabeled lipids were extracted, separated by TLC in solvent B, and detected by fluorography. Lipids 11-1 and 11-2 are marked with arrows.
A New Yeast Gene Required for Addition of EthN-P to the Third GPI Man
The possibility that the HFLS linked to the third Man in lipid 11-1 is the bridging EthN-P implies either that Gpi11p is not solely responsible for adding EthN-P to Man-3 or that in Gpi11p's absence, another protein can do so. Candidates for potential GPI EthN-P transferases have recently been identified. These are the three members of the conserved Mcd4/Gpi7 family of proteins, which contain a domain showing similarity to phosphoryl and sulfuryltransferases (Galperin et al., 1998; Benachour et al., 1999; Gaynor et al., 1999). Of these, Mcd4p is an essential protein required for GPI anchoring, and mcd4 mutants weakly accumulate a number of candidate GPIs that have been speculated to be a series of aberrant GPIs that lack an EthN-P side branch (Gaynor et al., 1999). Gpi7p, a nonessential protein required for growth at high temperature, is not required for GPI transfer to protein but has been proposed to be involved in the addition of EthN-P to the second Man of the GPI precursor (Benachour et al., 1999). The role of the third Mcd4p-related protein, YLL031cp, was unknown, but the existence of three Mcd4-like proteins, and the likelihood that the yeast GPI can bear three EthN-P side branches, suggested to us that YLL031cp might be involved in EthN-P transfer to the third GPI Man, a possibility we tested.
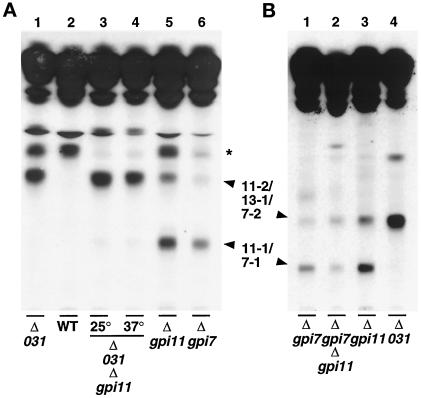
Deletion of YLL031c had been established by the Saccharomyces Deletion Project to be lethal. We constructed a YLL031c disruptant complemented by YLL031c under the control of the GAL10 promoter and used this strain to test whether depletion of YLL031cp leads to the accumulation of [3H]inositol-labeled GPIs. The YLL031c::KANR-pGAL-YLL031c strain was maintained on SGlyYE medium and then shifted to glucose-containing SGlcYE medium for 16 h before radiolabeling: a prominent [3H]inositol-labeled lipid (“13-1”) was accumulated that had the same mobility as lipid 11-2 of the gpi11::LEU2-pPIG-F strain (Figure 9A, lanes 1 and 2). In contrast to the latter mutant, a lipid with the mobility of 11-1 was not observed in the YLL031c::KANR-pGAL-YLL031c strain. The lipids that accumulate in YLL031cp- and Gpi11p-deficient strains are all less polar than CP2, which accumulates in the anchor attachment-defective gaa1 mutant (Hamburger et al., 1995) (Figure 9A, lane 3). Because CP2 has been reported to bear three EthN-Ps (Benachour et al., 1999), the decreased polarity of lipids 11-1, 11-2, and 13-1 is consistent with the notion that the latter three lipids bear fewer EthN-Ps.
The fact that lipid 13-1 comigrates with lipid 11-2 suggested that it too is a Man4-GPI bearing the same number of EthN-Ps as lipid 11-2. To confirm this, and to test whether lipid 13-1 indeed lacks EthN-P on Man-3, we determined the size of its neutral glycan head group and the position of its EthN-P side branches following the procedures we used to analyze lipids 11-1 and 11-2. The head group from lipid 13-1 had the mobility of Man4-GlcNAc-Ins (Figure 9B, lanes 2 and 4) and was converted to GlcNAc-Ins upon incubation with JBαM and to Man2-GlcNAc-Ins when treated with ASαM (Figure 9B, lanes 3 and 5). JBαM treatment followed by HF dephosphorylation yielded Man1-GlcNAc-Ins and only traces of Man2-GlcNAc-Ins (Figure 9B, lane 6), in contrast to the lipid 11-2′ head group, which generated Man2-GlcNAc-Ins. The results of these analyses therefore reveal that lipid 13-1 is predominantly a Man4-GPI that bears an EthN-P side branch on Man-1, although we cannot rule out the presence of a minor amount of a Man4-GPI with EthN-P on Man-2.
This finding has three important implications. First, the absence of EthN-P on Man-3 is consistent with the notion that YLL031cp is indeed involved in adding the bridging EthN-P. Second, this result indicates that 13-1 bears only a single EthN-P moiety, which in turn allows us to conclude that lipid 11-2 also has only one EthN-P side branch. Third, the fact that lipids 11-2 and 13-1 are different isoforms of Man4-GPI with one EthN-P is inconsistent with the notion that GPI synthesis proceeds along linear biosynthetic pathway. Because our results clearly implicate YLL031cp in GPI assembly, we designate this yeast open reading frame GPI13.
To obtain genetic corroboration of the notion that Gpi13p is responsible for adding EthN-P to Man-3, we created a YLL031c::KANR-pGAL-GPI13/gpi11::LEU2-pPIG-F strain and tested whether depletion of Gpi13p also prevents formation of lipid 11-1. The double mutant was shifted to glucose for 16 h to allow the cells to become depleted of Gpi13p and then shifted to 37°C or maintained at 25°C before pulse labeling with [3H]inositol. The strain accumulated a lipid migrating at the position of 11-2 and 13-1, but only traces of lipid 11-1 could be detected (Figure 10A, lanes 3–5). Because lipid 11-1 differs from 11-2 and 13-1 by the presence of an HFLS on the third GPI Man, these results strongly implicate Gpi13p in the addition of EthN-P to the third mannose of the GPI. The simplest explanation for the formation of traces of lipid 11-1 is that this material was generated by small amounts of Gpi13p remaining in the Gpi13p-depleted cells, although it is formally possible that the Pig-Fp present in these cells was capable of very low levels of EthN-P transfer.
Figure 10.
Formation of lipid 11-1 is dependent on YLL031c but can occur in the absence of GPI7. (A) The YLL031c:: KANR- pGAL-YLL031c/gpi11::LEU2-pPIG-F strain was maintained on SGlyYE medium, then shifted to SGlcYE medium for 16 h at 25°C. Cells were then resuspended and incubated in inositol-free medium containing glucose for 30 min at 25°C, after which equal portions were shifted to 37°C for 20 min or maintained at 25°C and then labeled for 2 h with [3H]inositol at 37 or 25°C respectively (lanes 3 and 4, Δ031Δgpi11), as detailed in MATERIALS AND METHODS. The gpi11::LEU2-pPIG-F (lane 5) and gpi7::LEU2 (lane 6) strains were pulse labeled with [3H]inositol at 37°C. A YLL031c::KANR- pGAL-YLL031c strain (lane 1) and a YLL031c+ sibling of that strain harboring pGAL-YLL031c (lane 2, WT) were grown and labeled as described for Figure 9A, lane1. Radiolabeled lipids were separated by TLC using solvent B. The species indicated by an asterisk is not seen reproducibly, nor is its presence correlated with a deficiency in any single Gpi protein. (B) The gpi7::URA3/gpi11::LEU2-pPIG-F strain was pulse labeled at 37°C with [3H]inositol, and radiolabeled lipids were separated by TLC using solvent B (lane 2). Samples of [3H]inositol-labeled lipids that accumulate in the gpi7::LEU2, gpi11::LEU2-pPIG-F and YLL031c::KANR-pGAL-YLL031c strains were loaded in lanes 1, 3, and 4 respectively.
We next exploited strains harboring a GPI7 disruption to obtain an estimate of the number of phosphodiester-linked substituents on lipid 11-1 and to determine whether lipid 11-1 bears EthN-P on its second mannose. GPI7 disruptants have been reported to accumulate Man-(EthN-P)Man-Man-(EthN-P)Man-GlcN-(acyl-Ins)PI and therefore to be defective in addition of EthN-P to Man-2 of that GPI (Benachour et al., 1999). We compared the TLC mobility of the GPIs that accumulate in Gpi11- and Gpi13p-depleted strains with the mobility of the GPI that accumulates in the gpi7 disruptant. The major [3H]inositol-labeled lipid that accumulates in gpi7::LEU2 (“7-1”) comigrates with lipid 11-1 (Figure 10A, lanes 5 and 6), suggesting that these Man4-GPIs have the same number of phosphodiester-linked substituents, namely two. Interestingly, the gpi7::LEU2 strain also accumulates traces of a lipid with the same mobility as lipid 11-2 (“7-2”). Although the amounts of this material are too small for the position of the HFLS it bears to be determined, it is possible that this lipid is a Man4-GPI with EthN-P on Man-1, but we cannot exclude the possibility that lipid 7-2 is or contains a Man4-GPI species bearing EthN-P on Man-2. If the latter holds, then the gpi7 deletion may only affect EthN-P addition to Man-2 of Man-(EthN-P)Man-Man-(EthN-P)Man-GlcN-(acyl-Ins)PI and not abolish modification of Man-2 on all GPIs.
If lipid 11-1 bears EthN-P on its second mannose, then this GPI should not be formed if the GPI7 deletion mutation is present in the gpi11::LEU2-pPIG-F strain. However, formation of 11-1 was not abolished in the gpi7::URA3/gpi11:: LEU2-pPIG-F double mutant, indicating that at least some of this species does not bear EthN-P on its second mannose (Figure 10B, lane 2). The gpi7::URA3/gpi11::LEU2-pPIG-F strain also accumulates a GPI comigrating with lipids 7-2, 11-2, and 13-1 (Figure 10B, lane 2).
DISCUSSION
We show that the essential Gpi11 and Gpi13 proteins are involved in late stages in the formation of the yeast GPIs, and we identify and characterize three new candidate GPI precursors. Gpi11p is the sequence and functional homologue of human Pig-Fp, a protein implicated in the addition of EthN-P to the third Man of the mammalian GPI precursor. Depletion of Gpi11p prevents the formation of complete GPI precursors and results in the accumulation of two new Man4-GPIs. Of these, lipid 11-2, a Man4-GPI lacking EthN-P on its third Man, is a species that might be predicted to accumulate if Gpi11p were responsible for adding EthN-P to Man-3; however, the other, lipid 11-1, is a Man4-GPI that bears an HFLS on its third Man. One interpretation of the presence of this substituent on Man-3 is that Gpi11p has no role, or at best a very minor one, in the addition of the bridging EthN-P to the yeast GPI precursor. Indeed, we show that Gpi13p is a better candidate for the enzyme responsible for adding EthN-P to the third GPI mannose: depletion of Gpi13p prevents formation of GPIs bearing EthN-P on Man-3 and results in accumulation of a Man4-GPI with EthN-P on Man-1.
Possible Structures of Lipids 11-1 and 11-2
The results of our analyses of the head groups of lipids 11-1 and 11-2 from the gpi11::LEU2-pPIG-F strain, together with the structure we determined for lipid 13-1 and that published for the lipid that accumulates in gpi7, allow us to propose a single structure for lipid 11-2 and two alternative structures for lipid 11-1. The composition of the GPIs that migrate at the positions of lipids 11-1 and 11-2 from the heterologously complemented gpi11::LEU2-pPIG-F strain can be accounted for in three different models. In addition, because we cannot rule out the formal possibility that the GPIs from gpi11::LEU2-pGAL-GPI11 that comigrate with lipids 11-1 and 11-2 from gpi11::LEU2-pPIG-F are different structural isoforms of 11-1 and 11-2, we consider an alternative model for the structures of 11-1 and 11-2 from the gpi11::LEU2-pGAL-GPI11 strain. Our models are based on the following assumptions.
First, we are assuming that the difference in TLC mobilities of lipids 11-1 and 11-2 can be accounted for by differences in their head groups, but we note that our data showing differences in the positioning of HFLSs do not exclude the possibility that the two GPIs also differ in their lipid moieties. Second, we assume that the HF-labile substituents are all EthN-Ps. So far, the only HFLSs that have been detected or proposed to occur on yeast and mammalian GPIs are EthN-Ps (Homans et al., 1988; Roberts et al., 1988; Hirose et al., 1992; Kamitani et al., 1992; Puoti and Conzelmann, 1993; Ueda et al., 1993). In addition, lipids 11-1 and 11-2 can be radiolabeled with [3H]EthN. Third, we assume that no GPI precursor can bear more than three EthN-Ps, which is consistent with the fact that a maximum of three EthN-P substituents has been detected on a mammalian GPI (Ueda et al., 1993). Fourth, we make the assumption that only a single EthN-P can be present on any one of mannoses 1–3. To date, EthN-Ps have been positioned at the 2′-OH of Man1 and the 6′-OHs of Man2 and Man3 (Homans et al., 1988; Ueda et al., 1993). Although it is possible that more than one EthN-P may be linked to a single GPI mannose, this has not been documented. Fifth, we are assuming that the only difference between the 11-1 and 11-2 head groups is the presence of one additional EthN-P on 11-1. This is based on the fact that 11-2 comigrates with 13-1, a Man4-GPI with one EthN-P, and the fact that 11-1 comigrates with a Man4-GPI that bears two EthN-Ps (Benachour et al., 1999). In our models, therefore, lipids 11-2 and 11-1 bear one and two EthN-Ps, respectively. Sixth, we assume that the HFLS attached to Man-3 of lipid 11-1 is the EthN-P moiety that is normally added at the 6′ position of that sugar. Although Man-3 has not been reported to bear EthN-P at any other position, it is possible that the HFLS on Man-3 of lipid 11-1 is not esterified to the 6′-OH, or, even if it is, that it was transferred there by the aberrant action of an enzyme on a GPI that accumulates in Gpi11p-depleted cells. We cannot yet exclude the former possibility, but the fact that lipids 11-1 and 11-2 are radiolabeled with [3H]Man in pmi40 GPI11+ cells (Figure 6, lane 4) suggests that these species are normally formed. Last, we recognize the possibility that GPIs bearing EthN-P substituents could be generated in more complex pathways involving removal and reattachment of EthN-Ps. However, removal of EthN-P substituents from GPI precursors has yet to be documented, and we will not invoke additional steps as these in our present discussion.
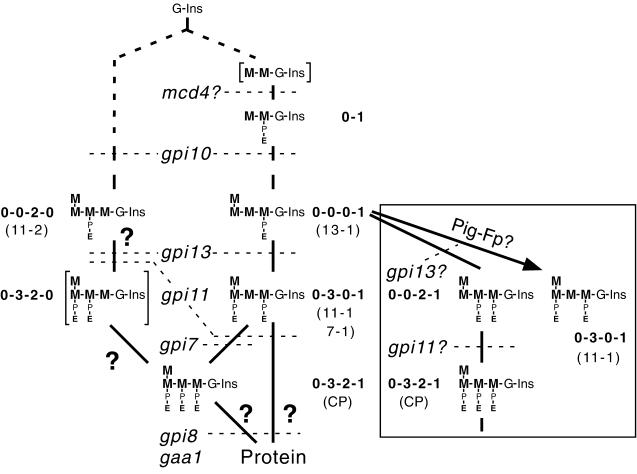
In the following discussion, we will use a shorthand for GPI head group configurations in which the number of zeros and numerals corresponds to the number of mannoses in the glycan, the first zero or numeral corresponds to the outermost mannose, a zero indicates that the mannose at that position is unsubstituted, and the numbers 1–3 indicate that mannose residues 1–3, respectively, bear EthN-P (Figure 11).
Figure 11.
Structural models for the head groups of lipids 11-1, 11-2, and 13-1 and proposed branched pathway for yeast GPI biosynthesis. Shown are the structures discussed in the text and the shorthand notation used for each of them. Lipids 11-1, 11-2, 13-1, and CP are identified. The structures in square brackets have not been shown unambiguously to be formed in yeast mutants. The steps known or proposed to be blocked in yeast GPI assembly mutants on the basis of their lipid accumulation phenotype, are indicated with dotted lines. The possibility that gpi13 affects EthN-P addition to Man-3 of lipid 11-2 is indicated, although gpi13 accumulates predominantly a 0-0-0-1 lipid. Alternative routes to formation of complete precursors and protein-bound anchors are marked by ?. Inset, alternative lipid structures formed from the 0-0-0-1 GPI precursor according to model 4 (see DISCUSSION), the steps proposed to be blocked in Gpi11p- and Gpi13p-deficient strains, and the EthN-P addition step proposed to be carried out by heterologously expressed Pig-Fp. M, mannose; G, glucosamine; P, phosphate; E, ethanolamine.
In model 1, the 11-1′ head group is Man-(EthN-P-)Man-(EthN-P-)Man-Man-GlcNAc-Ins (“0-3-2-0” configuration), and that of 11-2′ is Man-Man-(EthN-P-)Man-Man-GlcNAc-Ins (“0-0-2-0”). However, the formation of two GPIs that lack an HFLS on Man-1 is inconsistent with the phenotypes of Gpi10p and Gpi13p-depleted strains: the former accumulates Man-(EthN-P)Man (Canivenc-Gansel et al., 1998; Sütterlin et al., 1998), and the latter an “0-0-0-1” GPI. Moreover, the gpi7::URA3/gpi11::LEU2-pPIG-F strain still forms a lipid with the mobility of 11-1, indicating that at least some of 11-1 bears two EthN-Ps but is unsubstituted on Man-2 (Figure 10B, lane 2). The head group configuration of lipid 11-1 is therefore unlikely to be exclusively 0-3-2-0.
In model 2, the 11-1′ head group is Man-(EthN-P-)Man-Man-(EthN-P-)Man-GlcNAc-Ins (“0-3-0-1”), and that of 11-2′ is 0-0-2-0. These structures are consistent with schemes for the mammalian GPI synthetic pathway in which Man-1 and Man-3 are first modified, and then Man-2 (Hirose et al., 1992). Model 3 suggests that the lipid 11-1′ head group is a mixture of 0-3-0-1 and 0-3-2-0, but that the 11-2′ head group is predominantly, if not exclusively 0-0-2-0, although a 0-0-0-1 species could be present. Traces of the latter may accumulate in gpi7::LEU2.
Model 4 (Figure 11, inset) is based on the possibility that the GPIs accumulated in gpi11::LEU2-pGAL-GPI11 cells have the same number of EthN-P side branches as lipids 11-1 and 11-2 from the gpi11::LEU2-pPIG-F strain, but that these lipids are structural isoforms of 11-1 and 11-2 that bear their EthN-P side branches on different mannoses on the GPI glycan. In model 4, 11-1 has the “0-0-2-1” configuration, and 11-2 can be a mixture of 0-0-2-0 and 0-0-0-1. Models 2 and 3, on the one hand, and model 4, on the other, lead to alternative interpretations of the function of Gpi11p and are discussed next in this context.
Possible Functions for Gpi11p
Previous studies of the GPI anchoring-defective mammalian class F mutants showed that the most polar GPI that accumulates in these cells is a Man3 species lacking EthN-P on its third mannose (Sugiyama et al., 1991; Puoti and Conzelmann, 1993), suggesting that Pig-Fp is involved in adding the bridging EthN-P during GPI assembly. Our finding that lipid 11-1 from gpi11::LEU2-pPIG-F cells bears EthN-P on Man-3 can be explained in two ways, depending on whether depletion of native Gpi11p results in accumulation of a GPI that is unsubstituted on its third mannose (model 4) or one that bears EthN-P on Man-3 (models 2 and 3). In the first case, Gpi11p and Pig-Fp are responsible for adding EthN-P to Man-3, whereas in the second, they are not or at best have a very minor role relative to Gpi13ps.
If Gpi11p-depletion prevents EthN-P addition to Man-3 and normally leads to accumulation of a 0-0-2-1 GPI, then the 0-3-0-1 GPI that accumulates in gpi11::LEU2-pPIG-F cells could be formed when heterologously expressed Pig-Fp transfers EthN-P to a 0-0-0-1 lipid that is generated in gpi11::LEU2-pGAL-GPI11 cells according to model 4. Accumulation of the 0-3-0-1 species presumably occurs because this is an aberrant structure that cannot be processed further. Depletion of Gpi11p in any GPI11-disrupted strain should lead to accumulation of a 0-0-2-1 GPI, and this species might be present alongside the 0-3-0-1 lipid that accumulates in gpi11::LEU2-pPIG-F cells. Our inability to detect a 0-0-2-1 GPI in the latter strain can be explained by its conversion to the complete “0-3-2-1” precursor by Pig-Fp. This proposal for Gpi11p and Pig-Fp function also implies that Pig-Fp does not act on a 0-0-2-0 GPI, either because this species is not recognized as an acceptor or because it is never exposed to Pig-Fp.
An exclusive role for Gpi11p and Pig-Fp in transferring EthN-P to Man-3 seems unlikely for at least three reasons. First, if Pig-Fp is an EthN-P transferase that acts on 0-0-0-1, then we would predict that YLL031c::KANR/pGAL- YLL031c/gpi11::LEU2-pPIG-F cells should accumulate large amounts of 0-3-0-1, because a Gpi13p-deficiency normally leads to accumulation of 0-0-0-1, the very substrate that Pig-Fp is proposed to recognize. However, the YLL031c:: KANR-pGAL-YLL031c/gpi11::LEU2-pPIG-F strain accumulates but traces of 0-3-0-1. Second, the predictions this proposal makes for the structures of the GPIs that accumulate in gpi11::LEU2-pGAL-GPI11 and gpi11::LEU2-pPIG-F cells in turn require that Gpi13p converts the 0-0-0-1 GPI (which accumulates in YLL031c::KANR-pGAL-YLL031c) to the 0-0-2-1 form of lipid 11-1 proposed to accumulate in gpi11::LEU2-pGAL-GPI11. This again conflicts with our results with the YLL031c::KANR-pGAL-YLL031c/gpi11::LEU2- pPIG-F strain, which accumulates virtually no GPI with EthN-P on Man-3, clearly implicating Gpi13p in EthN-P transfer to Man-3. Moreover, if Gpi13p can add EthN-P to Man-2, it might be expected to compensate for the GPI7 null mutation and prevent formation of the 0-3-0-1 species whose accumulation is a characteristic phenotype of GPI7 disruptants (Benachour et al., 1999). Gpi13p is therefore most likely to add EthN-P to Man-3, not Man-2. Third, Pig-Fp failed to exhibit EthN-P transferring activity in two strain backgrounds in which this heterologously expressed protein might be expected to have encountered its natural mammalian substrate. Thus, Pig-Fp would be predicted to transfer EthN-P to a Man3-GPI, and a Man3-GPI with EthN-P on Man-3 might be “trapped” in the gpi8/gpi11::LEU2-pPIG-F strain; however, no new aberrant lipid accumulated in that strain (Figure 5B, lane 8). Furthermore, when PIG-F or GPI11 was expressed in the yeast smp3 mutant, which is defective in the addition of Man-4 and accumulates Man3-GPIs bearing EthN-P on either Man-1 or Man-2 (S. Grimme, B. Westfall, C. Taron, and P. Orlean, unpublished results), they neither suppressed the temperature sensitivity of smp3 nor caused the accumulation of an aberrant, “3-0-1” or “3-2-1” GPI. Because of these arguments, we believe that model 4 and the role it predicts for heterologously expressed Pig-Fp provide a less satisfactory explanation for the structures of lipids 11-1 and 11-2 than models 2 and 3 do.
If Gpi11p depletion does not prevent EthN-P addition to Man-3 and normally leads to accumulation of Man4-GPIs bearing EthN-P on Man-3, then models 2 and 3 apply. We include model 3, a variant of model 2, because we cannot exclude the possibility that 11-1, in addition to a 0-3-0-1 lipid, might contain a 0-3-2-0 component. Both models, however, have the same implications for the function of Gpi11p. The simplest interpretation for the accumulation of 0-3-0-1 and 0-3-2-0 GPIs in Gpi11p-deficient cells is that another protein can add the EthN-P moiety to the 6′ position of Man-3, and the following discussion of Gpi11p function will be based on this notion. However, we note that we cannot yet exclude the possibility that the EthN-P in lipid 11-1 is not at the 6′-position of Man-3, or that it has been added by another EthN-P transferase in a pathway or compartment that does not generate protein-linked GPIs. Likewise, we cannot completely exclude a role for Gpi11p/Pig-Fp in EthN-P transfer to Man-3, but the contribution these proteins can make to this reaction in yeast is at best a very small one. Furthermore, because a Gpi11p deficiency prevents the formation of complete precursors, Gpi11p may be directly involved in EthN-P addition to the unsubstituted mannoses in 0-3-0-1, 0-3-2-0, and 0-0-2-0 GPIs to convert these species to complete precursors with the 0-3-2-1 configuration (Benachour et al., 1999). However, if Gpi11p were an EthN-P transferase itself, it would have to recognize up to three different substrates and transfer EthN-P to two different mannoses. Our results with the gpi11::LEU2-pPIG-F strain indicate that Pig-Fp does not fulfill this role.
Although any role for Gpi11p/Pig-Fp in Gpi13p-dependent transfer of EthN-P to Man-3 can only be a very minor one in yeast, Gpi11p may be more closely involved in the function of Gpi7p. Thus, the lipid accumulation phenotypes of gpi7::LEU2 and gpi11::LEU2-pPIG-F cells are superficially similar: both accumulate a 0-3-0-1 GPI and a Man4-GPI with a single EthN-P, as does the gpi7::URA3/gpi11::LEU2-pPIG-F double mutant. Although we do not know whether lipid 7-2, like 11-2, bears EthN-P on Man-2, the similarity of the biochemical phenotypes of Gpi7p- and Gpi11p-deficient strains raises the possibility that Gpi11p cooperates with Gpi7p. Gpi11p, however, must have additional functions, because it is required for anchor transfer to protein, whereas Gpi7p is not.
The possibility that Gpi11p cooperates with Gpi7p, together with the finding that in mammalian cells, a Pig-Fp deficiency prevents EthN-P addition to Man-3, implicates the functional homologues Gpi11p and Pig-Fp in EthN-P transfer to both Man-2 and Man-3, albeit in two different species. One explanation is that in yeast and mammalian cells, respectively, Gpi11p and Pig-Fp function in concert with each member of the Mcd4p/Gpi7p/Gpi13p family of homologous EthN-P transferases. The fact that Gpi11p has little or no role in EthN-P transfer to Man-3 in yeast, whereas the role of Pig-F in this reaction in mammalian cells is a major one, may reflect a difference in the extent to which yeast Gpi13p and its mammalian homologue depend on a partnership with Gpi11p and Pig-Fp, respectively.
There are striking parallels between the GPI accumulation phenotypes of Gpi11p-depleted yeast cells and Thy-1 class F mutants in that both cell types accumulate at least four prominent [3H]Man-labeled lipids (Figure 6, lane 6; Lemansky et al., 1991; Sugiyama et al., 1991; Hirose et al., 1992, Kamitani et al., 1992; Puoti and Conzelmann 1993). However, it is not clear from the available structural information whether the array of lipids of each cell type could have been generated in the same way in the two species. Thus, the GPIs that accumulate in Thy-1 class F mutants contain one, two, or three mannoses, whereas the two most polar GPIs that accumulate in Gpi11p-deficient strains are both Man4 species, suggesting that Gpi11p acts after addition of the fourth Man. However, because we do not yet know whether the less polar lipids 11-3 and 11-4 have fewer than four mannoses, we cannot exclude the possibility that Gpi11p can affect earlier stages in GPI synthesis. Regardless, the conservation of function of Pig-Fp and Gpi11p suggests that the lipids that accumulate in yeast and mammalian cells deficient in these proteins do so for the same underlying biochemical reasons.
We do not know what lipids 11-1 (0-3-0-1) and 11-2 (0-0-2-0) are next converted to. Because formation of complete precursors 1 and 2 is dependent on Gpi11p, it is possible that these species are formed directly or indirectly from lipid 11-1, 11-2, or both. Proposals for the function of Gpi11p must be consistent with the likelihood that the 0-0-2-0 structure of lipid 11-2 is not a precursor of the 0-3-0-1 head group of lipid 11-1. The accumulation of these two structures can be explained by a model for a branched GPI synthetic pathway (discussed later) and by the ability of Gpi11p to act in different pathway branches.
The intimate involvement of Gpi11p/Pig-Fp in EthN-P transfer at two or more stages in GPI assembly can be explained by the direct participation of these proteins as components of different EthN-P transferases (although their role can nonetheless be dispensable depending on the EthN-P transferase and species) or by an indirect role of these proteins in facilitating side branch addition. If Gpi11p is not directly involved in EthN-P transfer, then its possible roles fall into two categories. In the first, Gpi11p either generates the phosphatidylethanolamine (PE) species that donates (Menon and Stevens, 1992) the EthN-Ps for conversion of lipids 11-1 and 11-2 to complete precursors or supplies PE to a compartment(s) containing lipids 11-1 and 11-2 and specific EthN-P transferases. The latter possibility is in accord with the notion that addition of EthN-P to GPIs may be dependent on PE export from the mitochondria to one or more separate GPI biosynthetic compartments (Vidugiriene et al., 1999). Indeed, Gpi11p could be both a “supplier” of EthN-P and a transferase subunit if it functioned to translocate PE across a bilayer to the phosphoryltransferase domain of an Mcd4 family protein. The second category of functions for Gpi11p includes a role for the protein in the transport of GPIs to the site where further EthN-Ps are added, or one in making a modification to GPIs—for example, by remodeling their lipid—that is necessary for their transport or for further EthN-P addition to them. The conjectured roles for Gpi11p in GPI remodeling or transport recall the observation that Thy-1 class F mutants make PI and GPIs containing diacylglycerols rather than base-resistant ether lipids (Stevens and Raetz, 1990; Puoti and Conzelmann, 1993). This phenotype does not have a parallel in yeast, because the diacylglycerol moieties of yeast GPI precursors are not replaced with base-resistant lipids. However, the Thy-1 class F mutation could instead affect the formation of a specialized form of PE that is required as EthN-P donor in GPI assembly.
Although Gpi11p and Pig-Fp clearly affect EthN-P addition to GPIs, we note that a Pig-Fp deficiency may have further, pleiotropic effects that are not explained by an EthN-P supply or transfer defect. Thus, Lemansky et al. (1991) found that in class F mutants, [3H]Man labeling of dolichyl phosphate mannose is much decreased, and indeed, in pmi40/gpi11::LEU2-pPIG-F yeast cells, [3H]Man labeling of two very nonpolar, but unidentified species is much reduced (Figure 6, lanes 4 and 6).
Why does Pig-Fp partially rescue the GPI11 deletion? If Pig-Fp is an EthN-P transferase, then it might rescue by transferring EthN-P to a Man3-containing yeast GPI to generate a GPI that is competent for transfer to protein. This seems unlikely because the gpi8/gpi11::LEU2-pPIG-F mutant does not accumulate a trimannosylated GPI bearing the bridging EthN-P. The possibility that Pig-Fp inefficiently transfers EthN-P to Man-3 of a 0-0-2-0 GPI to generate a species that can subsequently be transferred to protein might explain an accumulation of lipid 11-2 but not of lipid 11-1. If Pig-Fp has a function other than EthN-P transfer or has an auxiliary role in one or more of these reactions, then the temperature sensitivity of the gpi11::LEU2-pPIG-F strain may reflect the fact that this hydrophobic protein is functioning in a non-native membrane environment. Furthermore, if Pig-Fp interacts with other proteins in mammalian membranes, then its associations with possible yeast counterparts of its normal partners may be weak at high temperature.
Function of YLL031cp and Its Homologues in GPI Synthesis
Our results indicate that Gpi13p is a better candidate for the enzyme that adds EthN-P to the third GPI mannose than Gpi11p is, because the structure of the lipid that accumulates upon Gpi13p-depletion—a Man4-GPI that bears EthN-P on its first mannose—is consistent with the role of Gpi13p in EthN-P addition to Man-3. Furthermore, although a GPI bearing EthN-P on Man-3 is still formed in the absence of Gpi11p function, only traces of this lipid are made in a strain doubly deficient in Gpi11p and Gpi13p.
The features of the amino acid sequence of Gpi13p are also consistent with a role for the protein in EthN-P transfer. This protein and its homologues Mcd4p and Gpi7p contain a domain resembling phosphoryltransferases (Benachour et al., 1999; Gaynor et al., 1999), whereas Gpi11p and Pig-Fp show no similarities to proteins of known function. Strains deficient in Mcd4p and Gpi7p also have phenotypes consistent with defects in EthN-P transfer to GPIs, in the case of the latter, EthN-P transfer to Man-2 of a Man4-GPI (Benachour et al., 1999). Because MCD4 and GPI13 are essential genes, no other gene's product can substitute for theirs. In the case of nonessential Gpi7p, either another protein can substitute for it in some cases, or the EthN-P it transfers is not necessary for growth under some conditions.
The evidence so far from mammalian and yeast cells is consistent with the notion that their GPI precursors can receive up to three EthN-Ps, one on each of mannoses 1–3, with the EthN-P at the 6 position of the third, α1,2-Man becoming amide linked to protein. The transfer of three EtN-Ps to three different mannoses correlates with the presence in eukaryotes of three Mcd4/Gpi7 family proteins. Each of these proteins might transfer EthN-P to a different mannose, and the available evidence leads us to speculate that Mcd4p, Gpi7p, and Gpi13p add EthN-P to mannoses 1–3, respectively, in yeast, although they may not act on every potential acceptor GPI. Consistent with this, it has recently been reported that mammalian cells deficient in the mammalian homologue of Mcd4p are defective in the addition of EthN-P to Man-1 (Hong et al., 1999).
Implications of the Structures of Lipids 11-1, 11-2, and 13-1 for GPI Biosynthesis in Yeast
The four models proposed for the structures of lipids 11-1 and 11-2 that occur upon depletion of native Gpi11p have different implications for the biosynthetic origin of the GPIs that can be radiolabeled in yeast. The structures in models 1 and 4 above are consistent with these two species being successive intermediates in a linear biosynthetic pathway in which lipid 11-2 is a precursor of lipid 11-1. Lipid 11-1 would be the primary species accumulating behind the block created by Gpi11p deficiency, and 11-2 (and possibly 11-3 and 11-4) would in turn accumulate as a result of backing up of earlier intermediates. The possibility that the “0-0-2-0→0-3-2-0” sequence implied by model 1 represents the sole route to completion of GPIs seems unlikely, because this excludes the formation of any GPIs bearing EthN-P on Man-1. The sequence suggested in model 4 (0-0-0-1 → 0-0-2-1) and evaluated above was also judged less likely to occur. In principle, if lipid 11-2 were a precursor of 11-1, it should be chased into the latter species. However, demonstration of a precursor product relationship between 11-2 and 11-1 in pulse–chase experiments is precluded by the fact that any 11-1 that had accumulated as a result of Gpi11p depletion would obscure the subsequent conversion of 11-2 to material comigrating with 11-1.
If lipids 11-1 and 11-2 have the structures proposed in models 2 and 3, then the 0-3-0-1 structure of lipid 11-1 is most likely generated independently of the 0-0-2-0 structure of 11-2 (Figure 11). For this to occur, 1) the 0-3-0-1 species or its Man2- or Man3-containing precursors must either not be substrates for or not become exposed to an enzyme that adds EthN-P to Man-2; and 2) Man1–4-containing precursors of 11-2 must escape attachment of EthN-P to Man-1, and the Man3 and Man4 precursors of 11-2 must either not be recognized by or not be presented to an enzyme that transfers EthN-P to their third Man. Lipids 11-1 and 11-2 would therefore be generated in alternative pathways that differ in their complement of EthN-P transferases but both of which are dependent on Gpi11p. The notion that the yeast GPI synthetic pathway is branched is strongly supported by the fact that strains depleted of Gpi11p and Gpi13p accumulate two different Man4-EthN-P isoforms, 0-0-0-1 and 0-0-2-0, respectively, each of which could have been generated in a different arm of the pathway. Evidence for pathway branching also comes from the smp3 mutant, which is defective in the addition of the fourth Man to the yeast GPI precursor: this strain accumulates two Man3-GPI isoforms, one bearing EthN-P on Man-1 and the other with EthN-P on Man-2 (Grimme, Westfall, Taron, and Orlean, unpublished results).
The stage in GPI assembly at which these pathways separate and start generating distinguishable head groups cannot yet be pinpointed. Although this could occur as soon as Man-GlcN-(acyl-Ins)PI is formed, the earliest well-characterized mannosylated GPI intermediate labeled in vivo is the Man2 species that accumulates in the gpi10 mutant. This strain accumulates Man-(EthN-P-)Man-GlcN-(acyl-Ins)PI (Canivenc-Gansel et al., 1998; Sütterlin et al., 1998), indicating that Man-GlcN-(acyl-Ins)PI or Man2-GlcN-(acyl-Ins)PI could be substrates for the enzyme that transfers EthN-P to Man-1. Interestingly, gpi10 does not accumulate (EthN-P)Man-Man-GlcN-(acyl-Ins)PI, suggesting that Man2-GlcN-(acyl-Ins)PI either is not a substrate for or is not exposed to the EthN-P transferase that modifies Man-2.
The notion that there are alternative pathways for extension and side chain modification of GPI precursors has been invoked to explain the diversity of GPIs that can be radiolabeled in mammalian cells (Hirose et al., 1992; Kamitani et al., 1992). One way in which alternative pathways could be maintained is for them to be physically separated, and indeed, there is evidence for this from three different organisms. In mammalian cells, steps from GlcNAc-PI de-N-acetylation to generation of (EthN-P)Man-GlcN-(acyl-Ins)PI are enriched in a subcompartment of the ER associated with mitochondria, but it is possible that this structure is elongated and modified in another membrane (Vidugiriene et al., 1999). In Leishmania mexicana, the biosynthetic activities that generate (EthN-P)Man-Man-Man-GlcN-PI are also localized in a subcompartment of the ER that may be the counterpart of the mitochondria-associated GPI biosynthetic domain in mammalian cells (Ilgoutz et al., 1999a). In yeast, at least one protein involved in GPI modification, Gpi7p, is found predominantly in a plasma membrane-associated fraction (Benachour et al., 1999).
The GPIs generated in a given compartment may not be transferred to protein but, instead, may remain as “free GPIs.” Such glycolipids may be among the multiple GPIs made in mammalian and yeast cells that bear EthN-P on Man-3 and that could, in principle, be transferred to protein (Hirose et al., 1992; Kamitani et al., 1992; Ueda et al., 1993; Sipos et al., 1994; Benachour et al., 1999). Indeed, in neither yeast nor mammalian cells is it yet clear which apparently complete GPI precursor is transferred to protein (Kamitani et al., 1992; Benachour et al., 1999). The alternative GPI synthetic pathways whose existence in yeast is implied by models 2 and 3 may represent separate routes for the synthesis of the GPIs that are transferred to protein and those that remain free. This is consistent with the suggestion that in mammalian cells, non–protein-linked GPIs may leave their site of synthesis in the ER and receive side chain modifications in other, post-ER compartments (Kamitani et al., 1992).
In summary, we have made two sets of findings about GPI assembly in yeast. First, we have identified two new yeast genes required for GPI assembly. Of these, GPI13 encodes a candidate for the EthN-P transferase that adds the bridging EthN-P, whereas GPI11 encodes a protein that may be a “generic” EthN-P transferase subunit whose importance varies from transferase to transferase and from organism to organism, or which may serve to supply the EthN-P donor to a given transferase, or which may play both roles. Second, we have shown that in yeast, the GPI biosynthetic pathway branches to generate at least two series of structurally distinct GPIs, and we suggest these may be elaborated in separate intracellular membranes. The existence of separate GPI assembly pathways raises the possibility that the GPIs they generate may have distinct fates and functions in the cell.
ACKNOWLEDGMENTS
We thank M. Walberg, A. Conzelmann, M. DeTiani, and D.R. Voelker for the YMW3, FBY11, S-40, and RYY51 strains, respectively, and we thank B. Westfall for helpful discussions. This work was supported by grant GM-46220 from the National Institutes of Health and by a Helen Corley Petit Professorship from the College of Liberal Arts and Sciences of the University of Illinois at Urbana–Champaign. P.O. is a recipient of a Burroughs Wellcome Fund Scholar Award in Pathogenic Mycology.
Abbreviations used:
- ASαM
Aspergillus satoi α-1,2 mannosidase
- ER
endoplasmic reticulum
- EthN-P
phosphoethanolamine
- GPI
glycosylphosphatidylinositol
- HFLS
HF-labile substituent
- HPTLC
high-performance TLC
- JBαM
jack bean α-mannosidase
- ORF
open reading frame
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PI-PLC
phosphatidylinositol-specific phospholipase C
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benachour A, Sipos G, Flury I, Reggiori F, Canivenc-Gansel E, Vionnet C, Conzelmann A, Benghezal M. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J Biol Chem. 1999;274:15251–15261. doi: 10.1074/jbc.274.21.15251. [DOI] [PubMed] [Google Scholar]
- Benghezal M, Benachour A, Rusconi S, Aebi M, Conzelmann A. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 1996;15:6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Benghezal M, Lipke PN, Conzelmann A. Identification of six complementation classes involved in the biosynthesis of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae. J Cell Biol. 1995;130:1333–1344. doi: 10.1083/jcb.130.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canivenc-Gansel E, Imhof I, Reggiori F, Burda P, Conzelmann A, Benachour A. GPI anchor biosynthesis in yeast: phosphoethanolamine is attached to the alpha1,4-linked mannose of the complete precursor glycophospholipid. Glycobiology. 1998;8:761–770. doi: 10.1093/glycob/8.8.761. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Colussi PA, Orlean P. The essential Schizosaccharomyces pombe gpi1+ gene complements a bakers yeast GPI anchoring mutant and is required for efficient cell separation. Yeast. 1997;13:139–150. doi: 10.1002/(SICI)1097-0061(199702)13:2<139::AID-YEA69>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Conzelmann A, Fankhauser C, Desponds C. Myoinositol gets incorporated into numerous membrane glycoproteins of Saccharomyces cerevisiae: incorporation is dependent on phosphomannomutase (SEC53) EMBO J. 1990;9:653–661. doi: 10.1002/j.1460-2075.1990.tb08157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeg MA, Humphrey DR, Yang SH, Ferguson TR, Reinhold VN, Rosenberry TL. Glycan components in the glycoinositol phospholipid anchor of human erythrocyte acetylcholinesterase. Novel fragments produced by trifluoroacetic acid. J Biol Chem. 1992;267:18573–18580. [PubMed] [Google Scholar]
- Doering TL, Schekman R. GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Bairoch A, Koonin EV. A superfamily of metalloenzymes unifies phosphopentomutase and cofactor-independent phosphoglycerate mutase with alkaline phosphatases and sulfatases. Protein Sci. 1998;7:1829–1835. doi: 10.1002/pro.5560070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Mondesert G, Grimme SJ, Reed SI, Orlean P, Emr SD. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol Biol Cell. 1999;10:627–648. doi: 10.1091/mbc.10.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D, Egerton M, Riezman H. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J Cell Biol. 1995;129:629–639. doi: 10.1083/jcb.129.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1991;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Higuchi R. Recombinant PCR. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. pp. 177–183. [Google Scholar]
- Hindley J, Phear G, Stein M, Beach D. Sucl+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol Cell Biol. 1987;1:504–511. doi: 10.1128/mcb.7.1.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi Y, Komuro I, Chen R, Hosoda T, Mizuno T, Kudoh S, Georgescu SP, Medof ME, Yazaki Y. Molecular cloning of human homologue of yeast GAA1 which is required for attachment of glycosylphosphatidylinositols to proteins. FEBS Lett. 1998;421:252–258. doi: 10.1016/s0014-5793(97)01576-7. [DOI] [PubMed] [Google Scholar]
- Hirose S, Prince GM, Sevlever D, Ravi L, Rosenberry TL, Ueda E, Medof ME. Characterization of putative glycoinositol phospholipid anchor precursors in mammalian cells. Localization of phosphoethanolamine. J Biol Chem. 1992;267:16968–16974. [PubMed] [Google Scholar]
- Homans S, Ferguson M, Dwek R, Rademacher T, Anand R, Williams A. Complete structure of the glycosylphosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature. 1988;333:269–272. doi: 10.1038/333269a0. [DOI] [PubMed] [Google Scholar]
- Hong Y, Maeda Y, Watanabe R, Ohoshi K, Mishkind M, Riezman H, Kinoshita T. Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J Biol Chem. 1999;274:35099–35106. doi: 10.1074/jbc.274.49.35099. [DOI] [PubMed] [Google Scholar]
- Horvath A, Sütterlin C, Manning-Krieg U, Movva NR, Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgoutz SC, Mullin KA, Southwell B, McConville MJ. Glycosylphosphatidylinositol. biosynthetic enzymes are localized to a stable tubular subcompartment of the endoplasmic reticulum in Leishmania mexicana. EMBO J. 1999a;18:3643–3654. doi: 10.1093/emboj/18.13.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilgoutz SC, Zawadzki JL, Ralton JE, McConville MJ. Evidence that free GPI. glycolipids are essential for growth of Leishmania mexicana. EMBO J. 1999b;18:2746–2755. doi: 10.1093/emboj/18.10.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Kinoshita T, Orii T, Takeda J. Cloning of a human gene, PIG-F, a component of glycosyl-phosphatidylinositol anchor biosynthesis, by a novel expression cloning strategy. J Biol Chem. 1993;268:6882–6885. [PubMed] [Google Scholar]
- Inoue N, Watanabe R, Takeda J, Kinoshita T. PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis is a homologue of Saccharomyces cerevisiae GPI2. Biochem Biophys Res Commun. 1996;226:193–199. doi: 10.1006/bbrc.1996.1332. [DOI] [PubMed] [Google Scholar]
- Johnson M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T, Menon AK, Hallaq Y, Warren CD, Yeh ETH. Complexity of ethanolamine phosphate addition in the biosynthesis of glycosylphosphatidylinositol anchors in mammalian cells. J Biol Chem. 1992;267:24611–24619. [PubMed] [Google Scholar]
- Kawagoe K, Kitamura D, Okabe M, Taniuchi I, Ikawa M, Watanabe T, Kinoshita T, Takeda J. Glycosylphosphatidylinositol-anchor-deficient mice: implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria. Blood. 1996;87:3600–3606. [PubMed] [Google Scholar]
- Leidich SD, Drapp DA, Orlean P. A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J Biol Chem. 1994;269:10193–10196. [PubMed] [Google Scholar]
- Leidich SD, Kostova Z, Latek RR, Costello LC, Drapp DA, Gray W, Fassler JS, Orlean P. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. Cloning of the GPI2 gene. J Biol Chem. 1995;270:13029–13035. doi: 10.1074/jbc.270.22.13029. [DOI] [PubMed] [Google Scholar]
- Leidich SD, Orlean P. Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycosylphosphatidylinositol anchor synthesis. J Biol Chem. 1996;271:27829–27837. doi: 10.1074/jbc.271.44.27829. [DOI] [PubMed] [Google Scholar]
- Lemansky P, Gupta DK, Meyale S, Tucker G, Tartakoff AM. Atypical mannolipids characterize Thy-1-negative lymphoma mutants. Mol Cell Biol. 1991;11:3879–3885. doi: 10.1128/mcb.11.8.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke PN, Ovalle R. Cell wall architecture in yeast: new structure and new challenges. J Bacteriol. 1998;180:3735–3740. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti MP, Rodriguez-Boulan E. Glycophospholipid membrane anchoring provides clues to the mechanism of protein sorting in polarized epithelial cells. Trends Biochem Sci. 1990;15:113–118. doi: 10.1016/0968-0004(90)90195-h. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch E, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1982. [Google Scholar]
- Marck C. “DNA Strider”: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Menon AK, Cross GAM. Glycolipid precursors for the membrane anchor of Trypanosoma brucei variant surface glycoproteins. II. Lipid structures of phosphatidylinositol-specific phospholipase C sensitive and resistant glycolipids. J Biol Chem. 1990;265:6174–6181. [PubMed] [Google Scholar]
- McConville MJ, Ferguson MAJ. The structure, biosynthesis, and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon AK, Stevens VL. Phosphatidylethanolamine is the donor of the ethanolamine residue linking a glycosylphosphatidylinositol anchor to protein. J Biol Chem. 1992;267:15277–15280. [PubMed] [Google Scholar]
- Orlean P. Biogenesis of yeast wall and surface components. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cell Cycle and Cell Biology. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 229–362. [Google Scholar]
- Payton MA, Rheinnecker M, Klig LS, DeTiani M, Bowden E. A novel Saccharomyces cerevisiae secretory mutant posses a thermolabile phosphomannose isomerase. J Bacteriol. 1991;173:2006–2010. doi: 10.1128/jb.173.6.2006-2010.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A, Conzelmann A. Structural characterization of free glycolipids which are potential precursors for glycophosphatidylinositol anchors in mouse thymoma cell lines. J Biol Chem. 1992;267:22673–22680. [PubMed] [Google Scholar]
- Puoti A, Conzelmann A. Characterization of abnormal free glycophosphatidylinositols accumulating in mutant lymphoma cells of classes B, E, F, and H. J Biol Chem. 1993;268:7215–7224. [PubMed] [Google Scholar]
- Puoti A, Desponds C, Fankhauser C, Conzelmann A. Characterization of a glycophospholipid intermediate in the biosynthesis of glycosylphosphatidylinositol anchors accumulating in the Thy-1-negative lymphoma line SIAb. J Biol Chem. 1991;266:21051–21059. [PubMed] [Google Scholar]
- Ralton JA, McConville MJ. Delineation of three pathways of glycosylphosphatidylinositol biosynthesis in Leishmania mexicana. Precursors from different pathways are assembled on distinct pools of phosphatidylinositol and undergo fatty acid remodeling. J Biol Chem. 1998;273:4245–4257. doi: 10.1074/jbc.273.7.4245. [DOI] [PubMed] [Google Scholar]
- Ram AFJ, Wolters A, Hoopen RT, Klis FM. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to Calcofluor White. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- Roberts WL, Santikarn S, Reinhold VN, Rosenberry TL. Structural characterization of the glycoinositol phospholipid membrane anchor of human erythrocyte acetylcholinesterase by fast atom bombardment mass spectrometry. J Biol Chem. 1988;263:18776–18784. [PubMed] [Google Scholar]
- Schneider P, Ferguson MAJ. Microscale analysis of glycosylphosphatidylinositol structures. Methods Enzymol. 1995;250:614–630. doi: 10.1016/0076-6879(95)50100-2. [DOI] [PubMed] [Google Scholar]
- Schönbächler M, Horvath A, Fassler J, Riezman H. The yeast spt14 gene is homologous to the human PIG-A gene and is required for GPI anchor synthesis. EMBO J. 1995;14:1637–1645. doi: 10.1002/j.1460-2075.1995.tb07152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma DK, Vidugiriene J, Bangs JD, Menon AK. A cell-free assay for glycosylphosphatidylinositol anchoring in African trypanosomes. Demonstration of a transamidation reaction mechanism. J Biol Chem. 1999;274:16479–16486. doi: 10.1074/jbc.274.23.16479. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sipos G, Puoti A, Conzelmann A. Glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae: absence of ceramides from complete precursor glycolipids. EMBO J. 1994;13:2789–2796. doi: 10.1002/j.1460-2075.1994.tb06572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens VL. Biosynthesis of glycosylphosphatidylinositol membrane anchors. Biochem J. 1995;310:361–370. doi: 10.1042/bj3100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens VL, Raetz CRH. Class F Thy-1-negative murine lymphoma cells are deficient in ether lipid biosynthesis. J Biol Chem. 1990;265:15653–15658. [PubMed] [Google Scholar]
- Sugiyama E, DeGasperi R, Urakaze M, Chang HM, Thomas LJ, Hyman R, Warren CD, Yeh ETH. Identification of defects in glycosylphosphatidylinositol anchor biosynthesis in the Thy-1 expression mutants. J Biol Chem. 1991;266:12119–12122. [PubMed] [Google Scholar]
- Sütterlin C, Escribano MV, Gerold P, Maeda Y, Mazon MJ, Kinoshita T, Schwarz RT, Riezman H. Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem J. 1998;332:153–159. doi: 10.1042/bj3320153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda J, Kinoshita T. GPI-anchor biosynthesis. Trends Biochem Sci. 1995;20:367–371. doi: 10.1016/s0968-0004(00)89078-7. [DOI] [PubMed] [Google Scholar]
- Tiede A, Bastisch I, Schubert J, Orlean P, Schmidt RE. Biosynthesis of glycosylphosphatidylinositols in mammals and unicellular microbes. Biol Chem. 1999;380:503–523. doi: 10.1515/BC.1999.066. [DOI] [PubMed] [Google Scholar]
- Tiede A, Schubert J, Nischan C, Jensen I, Westfall B, Taron CH, Orlean P, Schmidt RE. Human and mouse Gpi1p homologues restore glycosylphosphatidylinositol membrane anchor biosynthesis in yeast mutants. Biochem J. 1998;334:609–616. doi: 10.1042/bj3340609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter PJ, Voelker DR. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- Ueda E, Sevlever D, Prince GM, Rosenberry TL, Hirose S, Medof ME. A candidate mammalian glycoinositol phospholipid precursor containing three phosphoethanolamines. J Biol Chem. 1993;268:9998–10002. [PubMed] [Google Scholar]
- Vidugiriene J, Sharma DK, Smith TK, Baumann NA, Menon AK. Segregation of glycosylphosphatidylinositol biosynthetic reactions in a subcompartment of the endoplasmic reticulum. J Biol Chem. 1999;274:15203–15212. doi: 10.1074/jbc.274.21.15203. [DOI] [PubMed] [Google Scholar]
- Vossen JH, Ram AF, Klis FM. Identification of SPT14/CWH6 as the yeast homologue of hPIG-A, a gene involved in the biosynthesis of GPI anchors. Biochim Biophys Acta. 1995;1243:549–551. doi: 10.1016/0304-4165(95)00002-s. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Inoue N, Westfall B, Taron CH, Orlean P, Takeda J, Kinoshita T. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 1998;17:877–885. doi: 10.1093/emboj/17.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R, Ohishi K, Maeda Y, Nakamura N, Kinoshita T. Mammalian PIG-L. and its yeast homologue Gpi12p are N-acetylglucosaminyl-phosphatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem J. 1999;339:185–192. [PMC free article] [PubMed] [Google Scholar]
- Zieler HA, Walberg M, Berg P. Suppression of mutations in two Saccharomyces cerevisiae genes by the adenovirus E1A protein. Mol Cell Biol. 1995;15:3227–3237. doi: 10.1128/mcb.15.6.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]