Abstract
The molecular nature of determinants that mediate degradation of unassembled, polytopic subunits of oligomeric membrane proteins and their stabilization after partner subunit assembly is largely unknown. Expressing truncated Na,K-ATPase α subunits alone or together with β subunits, we find that in unassembled α subunits neither the four N-terminal transmembrane segments acting as efficient alternating signal anchor–stop transfer sequences nor the large, central cytoplasmic loop exposes any degradation signal, whereas poor membrane insertion efficiency of C-terminal membrane domains M5, M7, and M9 coincides with the transient exposure of degradation signals to the cytoplasmic side. β assembly with an α domain comprising at least D902 up to Y910 in the extracytoplasmic M7/M8 loop is necessary to stabilize Na,K-ATPase α subunits by favoring M7/M8 membrane pair formation and by protecting a degradation signal recognized from the endoplasmic reticulum (ER) lumenal side. Thus our results suggest that ER degradation of Na,K-ATPase α subunits is 1) mainly mediated by folding defects caused by inefficient membrane insertion of certain membrane domains, 2) a multistep process, which involves proteolytic and/or chaperone components acting from the ER lumenal side in addition to cytosolic, proteasome-related factors, and 3) prevented by partner subunit assembly because of direct protection and retrieval of degradation signals from the cytoplasm to the ER lumenal side. These results likely represent a paradigm for the ER quality control of unassembled, polytopic subunits of oligomeric membrane proteins.
INTRODUCTION
In eukaryotic cells, membrane and secretory proteins are translocated into the endoplasmic reticulum (ER) during synthesis through a channel (translocon) formed by the Sec61 complex. Secretory proteins are completely transferred into the ER lumen, whereas membrane proteins integrate into the lipid bilayer by lateral exit of hydrophobic sequences from the translocon (High, 1995). During translocation, α-helical packing (Lemmon et al., 1997), interaction with molecular chaperones and cotranslational modifications (Ruddon and Bedows, 1997) and, in the case of oligomeric proteins, assembly with partner subunits (Geering, 1997) favor the correct folding into a tertiary protein structure. For many proteins, it is well documented that this maturation process in the ER is necessary for intracellular trafficking and function. The ER exerts an efficient quality control on misfolded forms of proteins, which can be produced because of a mutation, because a partner subunit of an oligomeric protein is missing, or because a protein naturally folds slowly. Misfolded proteins are recognized, their exit from the ER is prevented, and their final fate is degradation. This ER quality control is an important process because it prevents structurally and functionally altered proteins from accumulating in the cell but also because it directly contributes to the pathophysiology of several genetic diseases (Brooks, 1997).
The process called ER degradation was assumed to occur in the ER lumen or a pre-Golgi compartment until recent studies provided evidence that soluble as well as membrane proteins, in particular misfolded mutant proteins, were ultimately degraded by the cytosolic proteasome after retrograde transport back to the cytoplasm via the Sec61 translocon (Sommer and Wolf, 1997; Cresswell and Hughes, 1997; Kopito, 1997). However, ER degradation is a multistep pathway, and several concerted processes may precede proteasomal degradation. Initial steps of the degradation pathway such as recognition of the misfolded protein and targeting to the translocon may indeed occur in the ER lumen. The mechanisms involved in these processes may differ among soluble and membrane proteins. For instance, interaction with the molecular ER chaperone BiP (binding protein) appears to be necessary for the retrograde transport and proteasomal degradation of soluble proteins such as mutant yeast carboxypeptidase ysc Y (Plemper et al., 1997) but not for that of polytopic membrane proteins such as the mutant ATP-binding cassette transporter Pdr5* (Plemper et al., 1998). In the same line, interaction with calnexin, another ER chaperone, facilitates the degradation of the soluble mutant α1-anti-trypsin Z (Qu et al., 1996) and prepro-α factor (McCracken and Brodsky, 1996) but is not important for the degradation of the polytopic cystic fibrosis conductance regulator (CFTR) (Loo et al., 1998). On the other hand, soluble and membrane proteins may share common mechanisms for recognition and targeting to proteasomal degradation. Deletion of the lumenal RING-H2 finger domain of the ER membrane Hrd1/Der3 protein, which is necessary for degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase (Hampton et al., 1996), was shown to impede proteasomal degradation of soluble carboxypeptidase ysc Y (Bordallo et al., 1998), of the mutated membrane protein Sec61 (Bordallo et al., 1998), and of Pdr5* (Plemper et al., 1998).
The degradation of large, polytopic membrane proteins raises some particular questions, which so far have not been resolved. Retrotranslocation of lipid-inserted membrane domains to the translocon, which is necessary for proteasomal degradation, appears energetically very costly, and therefore it was speculated (Sommer and Wolf, 1997; Lord, 1996) that it may be sufficient that the proteasome only shaves off the large cytoplasmic domains present in polytopic membrane proteins such as CFTR. Recent evidence, however, suggests that cleavage of extracytoplasmic loops of polytopic membrane proteins by proteolytic enzymes in the ER lumen may facilitate the extraction of individual transmembrane segments by retrograde transport. For instance, HMG-CoA reductase is cleaved near membrane span 8 by a membrane-bound cysteine protease (Moriyama et al., 1998), whereas mutant P-glycoprotein is accessible to an unidentified protease in the first extracellular loop before proteasome degradation (Loo and Clarke, 1998).
The recognition by the various components of a multistep degradation pathway requires the existence and the exposure of specific signals in the misfolded proteins. Little is known on the molecular nature of these signals and on the conformational changes a membrane protein undergoes to expose these signals and permit protein degradation or, on the contrary, to mask these signals and protect the protein from degradation. These questions are relevant not only for mutated proteins, which are prone to degradation because of significant misfolding, but also in fact for any nascent protein. During synthesis and maturation, proteins pass through several partially unfolded states (Ashkenas and Byers, 1997), which may permit transient exposure of degradation signals. This situation is particularly striking in the case of subunits of oligomeric proteins, which in the absence of an appropriate partner subunit may be significantly misfolded and are rapidly degraded (Geering, 1997). To guarantee efficient expression, proteins normally assemble and/or fold rapidly, avoiding significant degradation. However, in some cases of polytopic membrane proteins, significant ER degradation occurs even of monomeric, wild-type proteins, as demonstrated for CFTR (Ward et al., 1995), or of assembled, oligomeric proteins as in the case of the epithelial Na channel (eNac) α-β-γ complexes (Valentijn et al., 1998), a phenomenon that can be explained by a particularly slow folding of these proteins.
To identify the existence and the molecular nature of potential degradation signals in a polytopic membrane protein and to determine the structural and molecular requirements that govern the exposure and the protection of these signals during protein maturation, we have studied the ER degradation of the α subunit of the hetero-oligomeric Na,K-ATPase. Na,K-ATPase belongs to the P-type ATPase superfamily of cation transporters and is a ubiquitous plasma membrane enzyme responsible for intra- and extracellular Na and K homeostasis (for review, see Horisberger, 1994). Among the P-type ATPases, only the Na,K- and H,K-ATPase α subunits require assembly with a partner β subunit in the ER to become stably expressed, functionally active, and competent for intracellular transport (Geering et al., 1996; Beggah et al., 1999). Similar to most P-type ATPases, the α subunits of Na,K- and H,K-ATPases have 10 transmembrane segments and a large central, cytoplasmic loop and expose the N and the C termini to the cytoplasmic side. To get insight into the structural and molecular mechanisms and the sequence of events that lead to degradation or protection from degradation of these α-proteins, we produced a series of truncated Na,K-ATPase α mutants, expressed them in Xenopus oocytes in the absence or presence of β subunits, and followed in parallel the stability and the topological features of the α variants.
Our results show that degradation of unassembled Na,K-ATPase α subunits is a multistep process and is favored by the poor membrane insertion efficiency of certain membrane domains. Indeed, neither N-terminal membrane segments, which act as efficient signal anchor–stop transfer sequences, nor the large, cytosolic loop exposes any degradation signals during synthesis. On the other hand, several degradation signals that initiate degradation are transiently exposed during synthesis in the C-terminal membrane domain because of inefficient membrane insertion. These degradation signals are specifically recognized either from the lumenal or the cytoplasmic side and mediate degradation by proteasome-dependent or -independent mechanisms. Interaction of the β subunit with a defined stretch of residues in the extracytoplasmic domain between transmembrane segments M7 and M8 permits the correct folding of the α subunit and protects it from degradation. Most likely our data are examples for a general mechanism involved in the ER quality control of polytopic subunits of oligomeric proteins.
MATERIALS AND METHODS
Truncated Constructs, Chimera, and Site-directed Mutagenesis of the α Subunit of Na,K-ATPase
Truncated constructs of the α1 subunit of Xenopus laevis (Verrey et al., 1989) were prepared by introducing a stop codon at different points in the α cDNA cloned into the pSD5 vector by using the PCR method (Nelson and Long, 1989). Single, double, or triple point mutations were introduced into the cDNA of truncated or wild-type constructs by the PCR method. For description of mutants see Table 1.
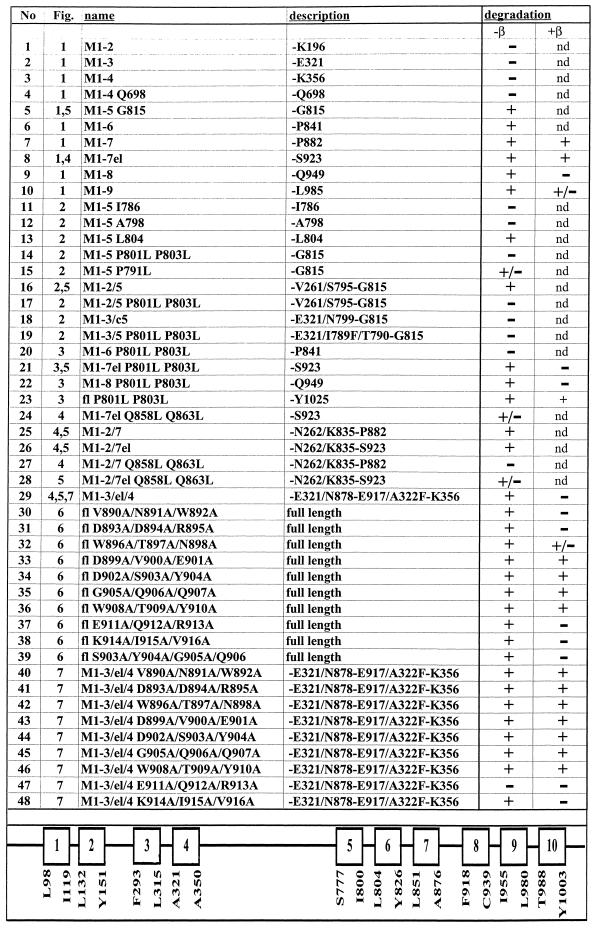
Table 1.
Description of α mutants and summary results
Indicated is the figure in which the mutants appear with the indicated name, which specifies the number of membrane segments contained in the mutant and the point mutations introduced. All mutants start at Met-1. The description indicates the last amino acid of the α mutants or the composition of chimeric constructs. fl, full length α proteins, which contain the indicated point mutations. In the degradation column, the results on the degradation of α variants synthesized in the oocyte in the absence or presence of β subunits are summarized. +, α-proteins degraded after a 48-h chase; −, α-proteins not degraded after a 48-h chase; M, membrane segment; nd, not determined. At the bottom, a putative, linear model of the α subunit of Xenopus Na,K-ATPase is shown, which indicates the amino acids that define the putative transmembrane segments according to a 10-transmembrane segment model (Moller et al., 1996).
The constructs M1–2/5, M1–2/5 P801L/P803L, M1–2/7, M1–2/7el, M1–2/7 Q858L/Q869L, M1–2/7el Q858L/Q869L M1–2/7el G860L/G867L, M1–3/C5, and M1–3/el/4 were prepared as described (Béguin et al., 1998).
To check the membrane topology of the α variants, we used a reporter glycosylation scanning (RGS) assay (Bayle et al., 1997; Béguin et al., 1998; Beggah et al., 1999). For this purpose, we generated chimera between the constructs containing N-terminal domains of the α subunit and the 223 amino acids (M81–L303) of the ectodomain of the β1 subunit of Bufo marinus Na,K-ATPase (Jaisser et al., 1992) containing four glycosylation sites. The chimera were produced as described (Béguin et al., 1998).
All constructs generated by PCR amplification were sequenced by dideoxy sequencing. In vitro–synthesized RNA (cRNA) was prepared according to the method of Melton et al. (1984).
Expression of the Na,K-ATPase in Xenopus Oocytes and Immunoprecipitation of α and β Subunits
Stage V–VI oocytes were obtained from X. laevis females (African Xenopus Facilities, Noordhoek, Republic of South Africa) as previously described (Geering et al., 1989). Routinely, 8–10 ng of α cRNA were injected into oocytes in either the absence or presence of 0.5–1 ng cRNA coding for Xenopus Na,K-ATPase β1 subunits (Verrey et al., 1989). To study the degradation of α variants, oocytes were metabolically labeled at 19°C for 6 or 24 h in modified Barth's medium containing 0.6 mCi/ml [35S]methionine (New England Nuclear, Boston, MA) and subjected to a chase period of 24 and/or 48 h in modified Barth's medium containing 10 mM unlabeled methionine. Digitonin extracts were prepared after the pulse and chase period and subjected to immunoprecipitation under denaturing or nondenaturing conditions as previously described (Jaunin et al., 1993) by using polyclonal anti-α or anti-β antibodies (Ackermann and Geering, 1990). To distinguish glycosylated from nonglycosylated species of α-β chimera, immunoprecipitated samples were subjected to endoglycosidase H (Endo H; Calbiochem-Novabiochem, La Jolla, CA) treatment as described (Jaunin et al., 1993). Immunoprecipitated proteins were subjected to SDS-PAGE, revealed by fluorography, and quantified by densitometry with an LKB (Piscataway, NJ) 2202 Ultrascan.
Inhibition of Proteasomal Degradation
To study the importance of the proteasome in the degradation of Na,K-ATPase α subunits, Xenopus oocytes were preincubated overnight in the absence or presence of 50 μM lactacystin (provided by E.J. Corey, Harvard University, Cambridge, MA) before injection of wild-type or mutant Na,K-ATPase α subunit cRNA. Oocytes were then metabolically labeled for 6 h in the absence or presence of 100 μM lactacystin and subjected to a 24-h chase period in the presence or absence of 25 μM lactacystin before preparation of digitonin extracts and immunoprecipitation. As a control protein, we expressed the α subunit of the renal epithelial Na channel (α rENaC) (Canessa et al., 1993), which was shown to be degraded by the proteasome (Staub et al., 1997).
RESULTS
To identify structural determinants that mediate the cellular degradation of individual Na,K-ATPase α subunits and to determine the protective role of β subunit assembly, we expressed truncated, wild-type, or mutant α subunits in the presence or absence of β subunits in Xenopus oocytes and followed the fate of the newly synthesized α-proteins by immunoprecipitation after pulse–chase labeling with [35S]methionine.
Cellular Degradation of Truncated and Full-Length α Subunits of Na,K-ATPase and Protection by β Subunit Assembly
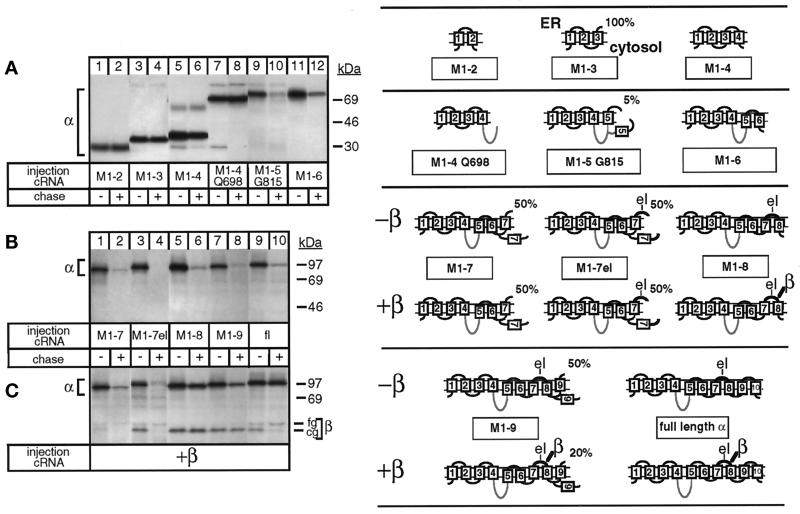
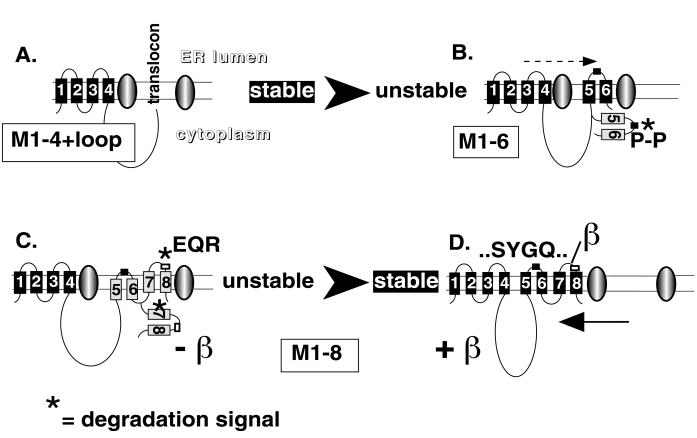
In contrast to the full-length α subunit (Figure 1B, lanes 9 and 10), truncated α-proteins (for description see Table 1) containing the transmembrane segments M1 and M2 (M1–2; Figure 1A, lanes 1 and 2), M1 up to M3 (M1–3; lanes 3 and 4), and M1 up to M4 (M1–4; lanes 5 and 6) were stable during a 48-h chase period. Significantly, an M1–4 α-protein containing 348 of 426 amino acids of the second, cytoplasmic loop of the α subunit (M1–4 Q698; Figure 1A, lanes 7 and 8) was also stably expressed. However, elongation of the protein up to Gly-815 including the first C-terminal membrane domain M5 was completely degraded during a 48-h chase, without production of proteolytic fragments (Figure 1A, lanes 9 and 10). Similarly, M1–6 (Figure 1A, lanes 11 and 12) up to M1–9 α-proteins (Figure 1B, lanes 1–8) were degraded as the full-length α subunit (Figure 1B, lanes 9 and 10).
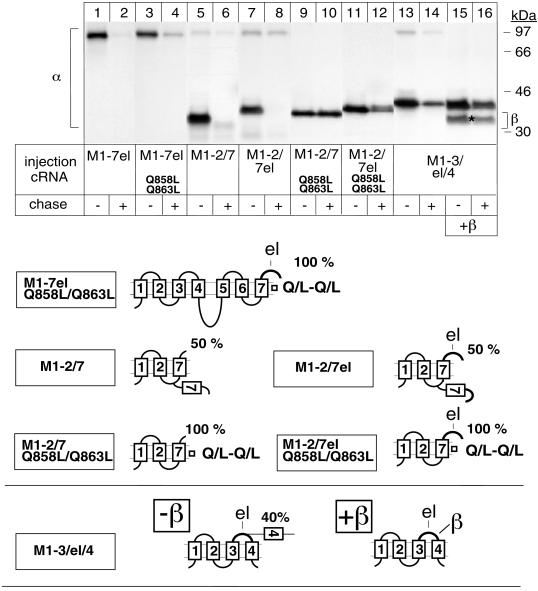
Figure 1.
Degradation of the unassembled Na,K-ATPase α subunit and its protection by β subunit assembly. Oocytes were injected with cRNA coding for truncated Na,K-ATPase α subunits (for description see Table 1) in the absence (A and B) or presence (C) of cRNA coding for Na,K-ATPase β subunits, as described in MATERIALS AND METHODS, metabolically labeled for 24 h and subjected to a 48-h chase period before preparation of digitonin extracts and immunoprecipitation with an α antibody under denaturing (A and B) or nondenaturing (C) conditions. Shown are fluorograms of immunoprecipitates resolved by SDS-PAGE. (A) Truncated α-proteins containing the 2 N-terminal membrane pairs (lanes 1–6) and, in addition, the large cytosolic loop (lanes 7 and 8) of Na,K-ATPase α subunits are not degraded during a 48-h chase period, but a truncated α-protein containing transmembrane segment M5 (lanes 9 and 10) or M6 (lanes 11 and 12) is degraded. (B) M1–7 up to M1–9 truncated α-proteins are degraded (lanes 1–8) during the chase period similar to full-length (fl), wild-type α subunits (lanes 9 and 10). (C) Association of β subunits with truncated α-proteins. Indicated are the positions of truncated α-proteins, coimmunoprecipitated β subunits, and proteins of known molecular masses. The weak band visible in lanes 3 and 4 and migrating at the level of the fully glycosylated β subunit seen in lane 10 represents an artifact occasionally produced at the migration front of the antibodies (Jaunin et al., 1993). cg, core glycosylated β subunits; fg, fully glycosylated β subunits. One of three or four representative experiments is shown. Also summarized at right are results obtained by Béguin et al. (1998) on the putative membrane topology of truncated α-proteins determined by an RGS assay. Indicated is the percentage of truncated α-proteins containing the ectodomain of a β subunit that became glycosylated in an RGS assay performed in Xenopus oocytes in the absence or presence of β subunits. The loops M5/M6 and M7/M8 are only partially inserted in the membrane until complete synthesis and β association have occurred. For further description, see RESULTS. el, extracellular loop between M7 and M8 containing a β-assembly domain. The cytoplasmic loop is drawn as a gray line, indicating that it is not on the scale.
Previously, we have determined the efficiency of membrane insertion and the topology of truncated α-proteins by an RGS assay of proteins expressed in Xenopus oocytes (Béguin et al., 1998). For these topology studies, the ectodomain of a β subunit containing four glycosylation sites was added to the C-terminal end of each truncated α-protein. The presence or absence of glycosylation could be used to determine whether a translated sequence ends on the lumenal or the cytoplasmic side of the ER membrane and, consequently, whether a C-terminal membrane domain acts as a signal anchor (SA) or a stop transfer (ST) sequence. In this assay, M1 and M1–3 α-proteins are 100% glycosylated, whereas M1–2 and M1–4 α-proteins are not glycosylated, suggesting that the formation of the first two N-terminal membrane pairs of the α subunit is mediated by membrane insertion of alternating SA and ST sequences. On the other hand, M5, M7, and M9 in M1–5, M1–7, and M1–9 α-proteins are poor SA sequences, as reflected by the partial glycosylation of these α-proteins in an RGS assay. For clarity, the results of these topology studies (Béguin et al., 1998) are summarized in Figure 1. If we assume that the topology studies previously performed with the RGS assay reflect the topology of the molecules devoid of a glycosylation reporter, then the stability of the M1–2, M1–3, and M1–4 α-proteins coincides with the efficient SA or ST properties of M1 and M3 or M2 and M4, respectively. On the other hand, the high susceptibility to degradation of M1–5, M1–7, and M1–9 α-proteins correlates with the poor SA function of M5, M7, and M9, suggesting that the poor membrane insertion efficiency may be related to the degradation process.
By using the two-hybrid approach, Colonna et al. (1997) have recently identified an essential β-association site consisting of an SYGQ motif located in the C-terminal half of the extracytoplasmic loop between M7 and M8 of the α subunit of Na,K-ATPase. Coexpression of β subunits with M1–7 or with M1–7el α-proteins (containing the M7/M8 extracytoplasmic loop “el”) did not change the membrane topology, e.g., the glycosylation pattern of these α-proteins (see models in Figure 1), and did not protect them from degradation (Figure 1C, lanes 1–4). Although β assembly was observed with M1–7el α-proteins during the pulse period (lane 3), stable interaction with these α variants may not be possible because of some steric hindrance. On the other hand, β subunits permanently associated with and stabilized M1–8 α-proteins (lanes 5 and 6) similar to full-length α subunits (lanes 9 and 10). β subunits also interacted with M1–9 α-proteins, but the resulting stabilization of this α-protein was less efficient (lanes 7 and 8) than that of M1–8 or full-length α-proteins. Interaction of β subunits with M1–9 α-proteins reduces the proportion of glycosylated, membrane-inserted M1–9 α-proteins (see models in Figure 1), which may be related to its higher sensitivity to degradation. In contrast to full-length α-β complexes, which could leave the ER, as reflected by the full glycosylation of the β subunit (Figure 1C, lanes 9 and 10), α-β complexes containing M1–8 or M1–9 α-proteins remained in the ER in their core glycosylated form (lanes 5–8). The results of Figure 1 are summarized in Table 1.
These data reflect the complex mechanism that governs the correct packing and the stable membrane insertion of a polytopic, oligomeric protein. The N-terminal membrane domain of the α subunit of Na,K-ATPase has intrinsic molecular characteristics that permit efficient membrane integration and, consequently, protection against degradation. Furthermore, the large cytoplasmic loop does not expose any degradation signals during synthesis. On the other hand, our results show that the inefficient membrane insertion properties of C-terminal membrane domains correlate with the degradation of unassembled α subunits and that β assembly with the extracytoplasmic loop between M7 and M8 favors membrane insertion and packing of the C-terminal membrane domain and, consequently, the stabilization of the entire α subunit. In the following, we analyzed 1) the existence and the molecular nature of putative degradation signals in individual α subunits, 2) the implication of the proteasome in the degradation of individual α subunits, and 3) the molecular and topogenic basis of the protection against degradation of the α subunit by β assembly.
Proline Residues in the M5/M6 Connecting Loop of the α Subunit Are Part of a Putative Degradation Signal, Which Is Recognized from the Cytoplasmic Side
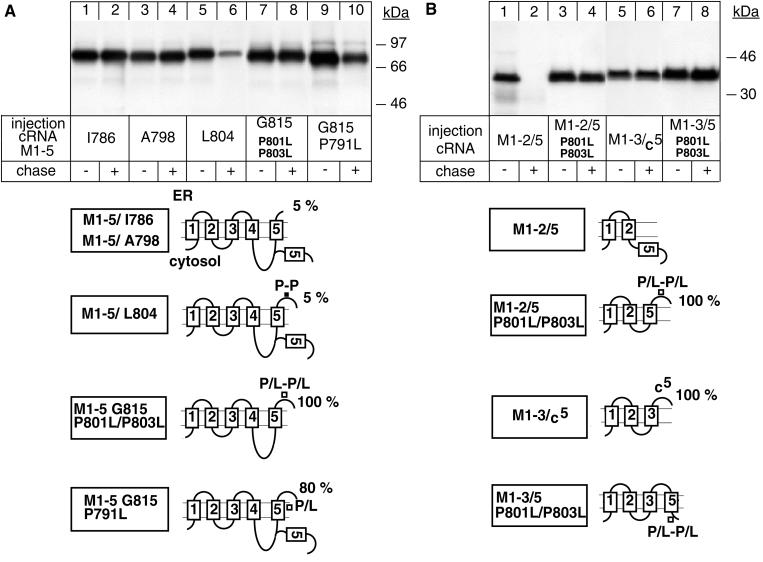
The observation that a truncated α-protein ending at Gln-698 (M1–4 Q698) was not degraded, whereas one ending at Gly-815 (M1–5 G815) was degraded (Figure 1A) suggested that a degradation signal exists within the domain encompassed by Gln-698 and Gly-815. To identify the critical amino acids involved, we tested the degradation of truncated M1–5 α-proteins of different length. According to a recently proposed topology model (Moller et al., 1996; Béguin et al., 1998), M5 of the α subunit of Na,K-ATPase starts at Ser-777, ends at Ile-800, and is connected to M6 by a three-amino-acid–long extracellular loop containing two proline residues (see Table 1). M1–5 α-proteins ending at Ile-786 (Figure 2A, lanes 1 and 2) or at Ala-798 (lanes 3 and 4) were stable during a 48-h chase similar to a M1–4 Q698 α-protein (Figure 1A, lanes 7 and 8), whereas an M1–5 α-protein ending at Leu-804 was degraded (Figure 2A, lanes 5 and 6) similar to the M1–5 G815 α-protein (Figure 1A, lanes 9 and 10). The M5 sequences included in these proteins had a similar, poor SA function as reflected by the low percentage of glycosylated forms revealed by the RGS assay (see models in Figure 2). These data suggested that the six amino acids within the Ala-798–Leu-804 domain mediate degradation of the M1–5 G815 α-protein. We have previously shown that Pro-801 and Pro-803 present within this domain are partly responsible for the poor SA function of M5 because their mutation permits complete membrane insertion of M5 in an M1–5 α-protein (Béguin et al., 1998). Mutation of these proline residues also prevented degradation of M1–5 α-proteins (Figure 2A, lanes 7 and 8), whereas mutation of Pro-791, which permits more efficient but not complete membrane insertion of M5, only partially protected wild-type M1–5 α-proteins from degradation (lanes 9 and 10). These results suggest that Pro-801 and Pro-803 may be part of a putative degradation signal, which becomes protected after membrane integration of M5. To verify this hypothesis, we added M5 to an M1–2 α-protein, which is intrinsically stable (Figure 1A, lanes 1 and 2), and followed the degradation of the resulting M1–2/5 α-protein. The RGS assay showed that M5 in this protein was not glycosylated and thus cannot act as an SA sequence (see models in Figure 2). M5 in the M1–2/5 α-protein was entirely exposed to the cytoplasm, and it mediated the degradation of M1–2 α-proteins (Figure 2B, lanes 1 and 2). Mutation of Pro-801 and Pro-803 induced complete membrane integration of M5 and stabilization of the M1–2/5 α-protein (lanes 3 and 4). These data suggest that a putative degradation signal containing P801/P803 is recognized by a component of a cytosolic degradation system. This is supported by the observation that addition of the C-terminal domain of M5 from Thr-790 to Gly-815 to an M1–3 α-protein did not induce degradation of the resulting M1–3/C5 α-protein (lanes 5 and 6). In this α-protein the C-terminal domain of M5 is entirely exposed to the ER lumen, as reflected by its full glycosylation in the RGS assay (Figure 2). Furthermore, a proline-mutated M1–3/5 P801L/P803L α-protein, which, according to the RGS assay, is not glycosylated and in which M5 acts as a stop transfer sequence, was stable after a chase period (Figure 2B, lanes 7 and 8).
Figure 2.
The extracytoplasmic loop between M5 and M6 contains a putative P-P degradation motif. Oocytes were injected with cRNA coding for truncated α-proteins, treated as described in Figure 1, and digitonin extracts were immunoprecipitated with an α antibody under denaturing conditions. (A) P801L/P803L mutations favor membrane insertion of M5 and impede degradation of an M1–5 G815 α-protein. (B) Transposition of M5 mediates degradation of the intrinsically stable M1–2 α-protein from the cytoplasmic side. One of two to four representative experiments is shown. At the bottom, putative topology models determined by an RGS assay are shown, indicating the percentage of glycosylated ER forms of truncated α-proteins. Also indicated are the positions of the mutated proline residues.
M7 and the Extracellular Loop between M7 and M8 Can Mediate Degradation of the α Subunit
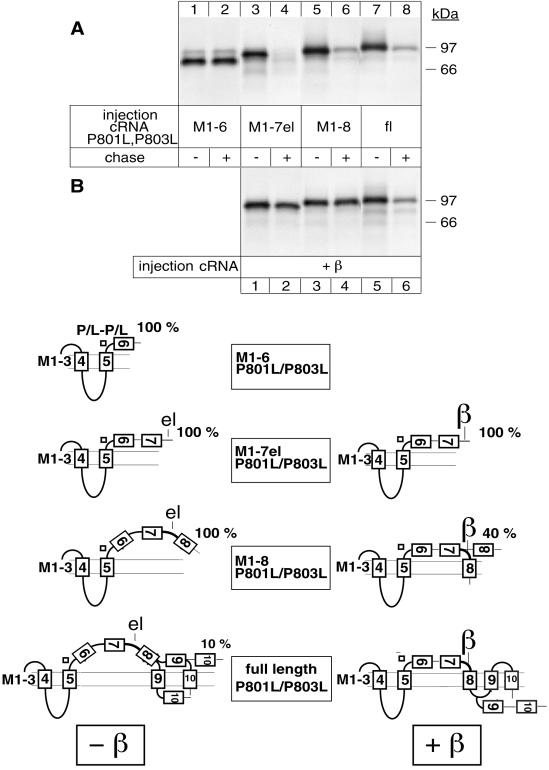
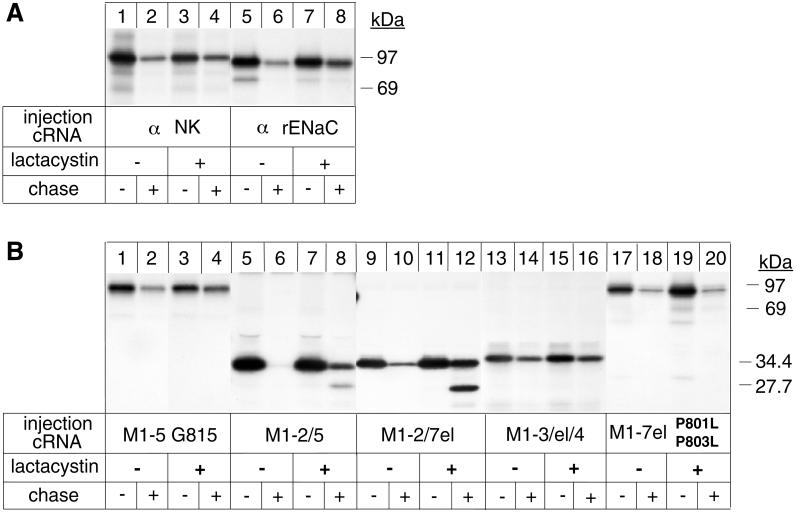
To identify additional degradation signals in the α subunit, we followed the stability of P801L/P803L variants of truncated α-proteins. P801L/P803L mutations not only permit complete membrane insertion of M5 and stabilization of M1–5 α-proteins, but they also impede the formation of the M5/M6 pair in M1–6 α-proteins (Béguin et al., 1998). As a consequence, in P801L/P803L variants of M1–6, M1–7el, M1–8, M1–9, and M1–10 α-proteins, M6, M7, M8, and most of M9 are released to the ER lumen. Only M10 in a P801L/P803L variant of M1–10 α-proteins acts as an efficient ST sequence (see models in Figure 3).
Figure 3.
A region encompassing M7 up to M8 contains a putative degradation signal that is recognized from the ER lumenal side. Oocytes were injected with cRNA coding for truncated α-proteins in the absence (A) or presence (B) of cRNA coding for β subunits, treated as described in Figure 1 and immunoprecipitated with an α antibody under denaturing conditions. (A) Truncated M1–7el α-proteins containing P801L/P803L mutations are degraded from the ER lumenal side. (B) β subunits can efficiently stabilize topologically aberrant M1–7el P801L/P803L and M1–8 P801L/P803L α-proteins but not full-length P801L/P803L α subunits. One of three or four representative experiments is shown. Also indicated are putative topology models determined by an RGS assay of α-proteins expressed in the absence or presence of β subunits. The model of full-length P801L/P803L α subunits expressed in the presence of β subunits is not definitively established (Béguin et al., 1998).
Of these P801L/P803L variants, M1–6 α-proteins were stable (Figure 3A, lanes 1 and 2), whereas M1–7el (lanes 3 and 4), M1–8 (lanes 5 and 6), and full-length α-proteins (lanes 7 and 8) were degraded. Coexpression of β subunits nearly completely protected the P801L/P803L variant of M1–7el (Figure 3B, lanes 1 and 2) and of M1–8 (lanes 3 and 4) α-proteins from degradation. These results suggest that M7 and/or the M7/M8 extracellular loop contain degradation signals and that at least one of these signals is recognized from the ER lumenal side. Furthermore, the results show that, despite severe misfolding of the P801L/P803L variant of M1–7el and of M1–8 α-proteins, β assembly can mask this degradation signal either by direct protection or by a conformational change of the proteins. In contrast to the P801L/P803L variant of M1–7el and M1–8, the P801L/P803L variant of the full-length α subunit could not be stabilized by coexpression with β subunits (lanes 5 and 6) although β assembly occurred and was shown to produce a topological change in this protein (Béguin et al., 1998). It is possible that in this protein, putative degradation signals located further downstream in M9 and/or M10 become available and abrogate the protective effect of β assembly.
To further characterize the role in the α subunit degradation of M7 and/or the β assembly domain, we followed the degradation of α-proteins with Q858L/Q863L mutations in M7, which, according to results from RGS assays, permit complete membrane insertion of M7 in M1–7el or M1–2/7el α-proteins (Béguin et al., 1998) (see models in Figure 4). The M1–7el Q858L/Q863L (Figure 4, lanes 3 and 4) α-proteins were significantly degraded during a 48-h chase period, although to a lesser extent than M1–7el proteins (lanes 1 and 2), indicating that complete membrane insertion of M7 cannot entirely stabilize M1–7el α-proteins and suggesting that perhaps both the M7 and the β assembly domain may be implicated in degradation.
Figure 4.
M7 as well as the extracytoplasmic loop between M7 and M8 of the Na,K-ATPase α subunit contain putative degradation signals. Oocytes were injected with cRNA coding for truncated α-proteins in the absence (lanes 1–14) or presence (lanes 15 and 16) of cRNA coding for β subunits, treated as described in Figure 1 and immunoprecipitated with an α antibody under denaturing conditions (lanes 1–14) or nondenaturing conditions (lanes 15 and 16). Complete membrane insertion of M7 does not prevent degradation of M1–7 α-proteins, and transposition of the extracytoplasmic loop between M7 and M8 (el) mediates degradation of intrinsically stable, truncated M1–4 α-proteins. Also, β subunits can associate with and stabilize M1–3/el/4 α-proteins. Indicated are the positions of α-proteins. The β subunit coimmunoprecipitated with M1–3/el/4 α-proteins is indicated by an asterisk. Immunoprecipitated samples shown in lanes 15 and 16 were treated with Endo H to permit separation of the α and β bands. One of two or three representative experiments is shown. The putative membrane topology of truncated α-proteins expressed in the absence or presence of β subunits is shown.
To test this hypothesis, we compared the degradation of two α variants in which M7, lacking or containing the β assembly domain, was added to the intrinsically stable M1–2 α-protein. Both M7 and M7el in the M1–2/7 (Figure 4, lanes 5 and 6) and in the M1–2/7el (lanes 7 and 8) α-proteins, respectively, showed only partial SA function and induced degradation of M1–2 α-proteins. On the other hand, Q858L/Q863L mutations, which fix M7 in the membrane, completely stabilized the M1–2/7 (lanes 9 and 10) and to a lesser extent the M1–2/7el (lanes 11 and 12) α-proteins. The greater lability of M1–7el Q858L/Q863L compared with that of M1–2/7el Q858L/Q863L can be explained by the existence of additional, distal degradation signals in M1–7el Q858L/Q863L (e.g., in M5), which may cooperate in its degradation.
Thus, these results support the hypothesis that M7 as well as the β assembly domain may contain degradation signals. The putative degradation signal in M7 is likely to be recognized from the cytoplasmic side, similar to that in M5, and is masked by membrane insertion, whereas that in the β assembly domain is accessible from the ER lumenal side. To further characterize the existence of a putative degradation signal in the β assembly domain, we also followed the stability of an M1–3/el/4 α variant in which the extracellular loop between M3 and M4 in an intrinsically stable M1–4 α-protein (Figure 1A, lanes 5 and 6) was replaced by the M7/M8 loop. In an RGS assay, ∼40% of the M1–3/el/4 α-protein was glycosylated (see models in Figure 4), indicating that in this α variant, the ST function of M4 is partially impeded. Nevertheless, the M7/M8 el loop is entirely exposed to the ER lumenal side and mediates the degradation of the M1–4 α-protein (Figure 4, lanes 13 and 14). Coexpressed β subunits assembled with the M1–3/el/4 α variant (Figure 4, lanes 15 and 16), reestablished efficient ST function of M4 and stabilized the M1–3/el/4 α-protein (Figure 4, lanes 15 and 16). This result is consistent with the protection of a degradation signal in the M7/M8 loop, although it cannot definitively be excluded that M4 contains a degradation signal that becomes protected after correct membrane retention of M4 in β-associated M1–3/el/4 α-proteins.
Implication of the Proteasome in the Degradation of Individual α Subunits
To study the implication of the cytoplasmic proteasomal system in the degradation of individual α subunits, we tested the effect of lactacystin, a specific inhibitor of the proteasome, on the stability of wild-type and several α variants expressed in oocytes in the absence of β subunits. Although lactacystin is not a very efficient inhibitor of the proteasome of Xenopus oocytes, we consistently observed a partial inhibition of the degradation of α subunits (Figure 5A, lanes 1–4), which was comparable with the degree of inhibition of the degradation of α subunits of the rat rENaC (Figure 5A, lanes 5–8), a protein that was shown to depend on the proteasome for its degradation (Staub et al., 1997). Partial inhibition of α subunit degradation was also observed with the proteasome inhibitor MG-132 (our unpublished results). Inhibition of degradation by lactacystin was also tested for some topology-representative α variants. M1–5 G815 (Figure 5B, lanes 1–4) and M1–2/5 α-proteins (lanes 5–8) were also partially protected from degradation in the presence of lactacystin, suggesting that the putative P-P degradation motif in the M5/M6 extracellular loop, which in these α variants is mainly exposed to the cytoplasm (see models in Figure 1), may be a direct or indirect target for proteasomal degradation. Similarly, degradation of M1–2/7el (Figure 5B, lanes 9–12) and M1–2/7 (our unpublished results) α-proteins was partially impeded by lactacystin. In these proteins, as in M1–2/5 proteins, a proteolytic fragment accumulated in the presence of lactacystin, which according to its molecular mass could correspond to the N-terminal domain including M1 and M2 of the α subunit. Significantly, the degradation of M1–3/el/4 was also partially inhibited by lactacystin (lanes 13–16) despite the premise that the putative degradation signal in the el domain is entirely exposed to the ER lumenal side in this α variant (see models in Figure 4). This result indicates that the el domain may be necessary as a mediator in an early step of proteasomal degradation, which is initiated by factors acting from the ER lumenal side. Finally, lactacystin did not influence the degradation of M1–7el P801L/P803L α proteins (Figure 5B, lanes 17–20). Because in these α variants, the C-terminal domain starting with M6 is entirely exposed to the ER lumen (see models in Figure 3), this result demonstrates that proteases acting from the ER lumenal side may participate in the degradation of misfolded proteins.
Figure 5.
Proteasome-dependent and -independent degradation of Na,K-ATPase α variants. Oocytes were preincubated or not with 50 μM lactacystin for 16 h before injection with cRNA coding for Na,K-ATPase α subunits (α NK), for truncated α variants, or for α subunits of the renal, epithelial Na channel (α rENaC). After a 6-h pulse with [35S]methionine in the absence or presence of 100 μM lactacystin and a 24-h chase in the absence or presence of 25 μM lactacystin, digitonin extracts were prepared, and immunoprecipitations under denaturing conditions were performed with an Na,K-ATPase α antibody (A, lanes 1–4, and B) or with an α rENaC antibody (A, lanes 5–8). (A) Degradation of unassembled Na,K-ATPase α subunits is partially inhibited by the proteasomal inhibitor lactacystin, similar to that of α rENaC. (B) Partial inhibition of degradation by lactacystin of topology-representative α variants. Indicated is the molecular mass of the uncleaved M1–2/7el α-protein and that of a proteolytic fragment accumulating in the presence of lactacystin. One of two or three representative experiments is shown.
Analysis of the Role of the M7/M8 Extracellular Loop in the Stabilization of the α Subunit after β Association
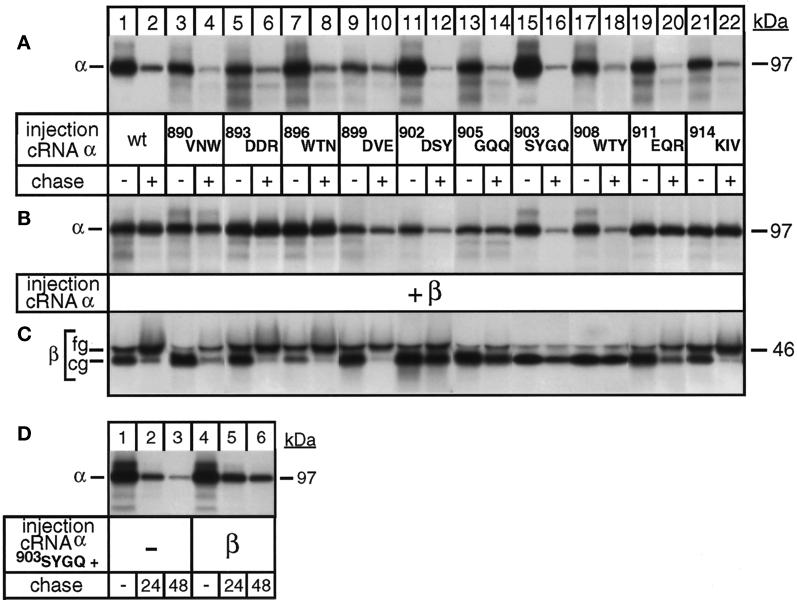
To characterize the protective effect of β assembly with the extracytoplasmic loop between M7 and M8, we produced alanine scanning variants of full-length α subunits by replacing three by three amino acids of the domain encompassing Val-890 and Val-916 (see Table 1) and followed the degradation of these mutant α-proteins after a 24-h pulse and a 48-h chase period in the absence or presence of coexpressed β subunits. In the absence of β subunits, all α mutants were degraded during the chase period (Figure 6A, lanes 3–22) similar to the wild-type α subunit (lanes 1 and 2). Coexpressed β subunits could associate with and stabilize α mutants with alanine replacements at the borders of the Val-890–Val-916 domain, e.g., the alanine variants 890VNW, 893DDR, 896WTN, 899DVE, 911EQR, and 914KIV (Figure 6B, lanes 3–10 and 19–22). Stabilization of α mutants by β subunits went in parallel with the acquisition of complex-type sugars by the β subunit (Figure 6C, lanes 3–10 and 19–22), indicating that these mutant α-β complexes can leave the ER and are routed to the plasma membrane similar to wild-type α-β complexes (lanes 1 and 2). In contrast, alanine mutants affected in the central region of the Val-890–Val-916 domain encompassing Asp-902 up to Tyr-910 were degraded during a 48-h chase period even in the presence of β subunits (Figure 6B, lanes 11–18). In agreement, β subunits that were coexpressed with these assembly-incompetent α-mutants remained in their core glycosylated ER form and were slowly degraded (Figure 6C, lanes 11–18), as expected for unassembled β subunits (Geering et al., 1996). These results indicate that the previously reported 903SYGQ906 motif of α-β interaction (Colonna et al., 1997) is important but not sufficient for complete stabilization of α subunits by the β subunit. A region that at least encompasses Asp-902 up to Tyr-910 is essential for the association of the β subunit and the stabilization of the α subunit. This result was further supported by a more detailed analysis of the degradation process. To avoid the significant degradation of the 903SYGQ variant, which occurs during a 24-h pulse period (Figure 6, compare A and B, lane 15) and which impedes to resolve subtle differences in the degradation after a single 48-h chase period, we labeled the newly synthesized proteins only during a 6-h pulse and followed the stability of the α variant after 24- and 48-h chase periods. This experiment revealed that 903SYGQ alanine mutants were partially protected from degradation in the presence of β subunits (Figure 6D, lanes 4–6) compared with 903SYGQ alanine mutants expressed without β subunits (lanes 1–3).
Figure 6.
The SYGQ motif in the M7/M8 extracellular loop of Na,K-ATPase α subunits is necessary but not sufficient for the correct folding of Na,K-ATPase α subunits after β assembly. Oocytes were injected with α cRNA coding for full-length α subunits with alanine replacements of the amino acids indicated in the absence (A) or presence (B–D) of cRNA coding for β subunits and subjected to a 24-h pulse and a 48-h chase period (A–C) or a 6-h pulse and 24- and 48-h chase periods (D) before preparation of digitonin extracts and immunoprecipitation with an α antibody (A, B, and D) or a β antibody (C) under denaturing conditions. (A) Alanine variants expressed in the absence of β subunits are degraded during a 48-h chase period (lanes 3–22) similar to wild-type (wt) α subunits (lanes 1 and 2). (B) A region encompassing D902 up to Y910 is essential for β interaction and stabilization. (C) β subunits coexpressed with alanine variants affected in the region encompassing D902 up to Y910 remain in their core glycosylated (cg) ER form (lanes 11–18), indicating that they cannot efficiently associate with α variants, in agreement with the lack of α stabilization. β subunits coexpressed with alanine variants affected in the region V890 up to E901 or E911 up to V916 become fully glycosylated (fg), indicating that the α-β complexes have left the ER. One of three representative experiments is shown. (D) Partial stabilization of 893SYGQ alanine variants by β subunits. One of two representative experiments is shown.
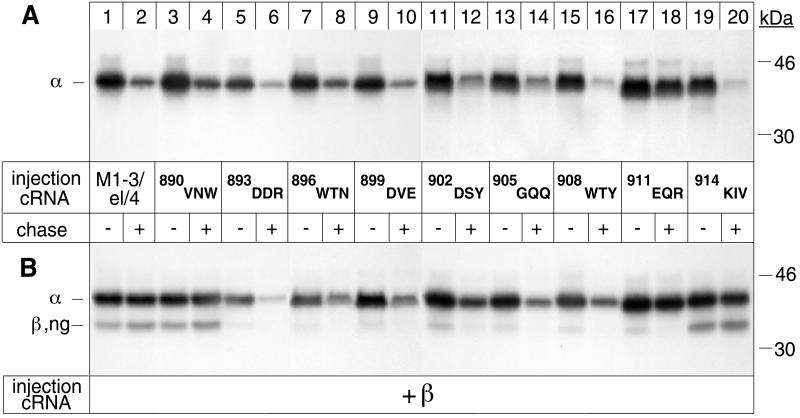
Alanine scanning in the extracellular el domain did not abolish degradation of individual α subunits. This result may indicate that the β assembly domain lacks a specific degradation signal, but it could as well be explained by the existence of other degradation signals in other regions (e.g., in M5 and/or M7) of the α subunit, which prevail over the abolition of a degradadtion signal in the Val-890–Val-916 domain. To distinguish between these two possibilities, we produced alanine scanning variants of the Val-890–Val-916 domain in the M7/M8 loop contained in the M1–3/el/4 α-protein and followed the stability of these mutants expressed in the presence and absence of β subunits. As shown before (Figure 4), the wild-type M7/M8 el loop transposed between M3 and M4 rendered the stable M1–4 α-protein susceptible to degradation (Figure 7A, lanes 1 and 2). In the absence of β subunits, all alanine variants of the M1–3/el/4 α-proteins were also degraded during a chase period (lanes 3–16, 19, and 20), with the exception of the 911EQR alanine variant (lanes 17 and 18), which was stable. This result suggests that the mutation had abolished a putative degradation signal. Coexpressed β subunits associated with and stabilized M1–3/el/4 α-proteins (Figure 7B, lanes 1 and 2), and the 890VNW (lanes 3 and 4) and 914KIV (lanes 19 and 20) alanine variants mutated in the N- and C-terminal borders, respectively, of the Val-890–Val-916 domain. In contrast, all other alanine scanning variants (lanes 5–18) could not permanently associate with β subunits and, with the exception of the intrinsically stable 911EQR variant, were not stabilized.
Figure 7.
The β assembly domain contains a putative degradation signal. Oocytes were injected with α cRNA coding for M1–3/el/4 α-proteins (lanes 1 and 2) or M1–3/el/4 α variants with alanine replacements of the amino acids indicated in the absence (A) or presence (B) of cRNA coding for β subunits and subjected to a 24-h pulse and a 48-h chase period before preparation of digitonin extracts and immunoprecipitation with an α antibody. (A) Alanine replacement of 901EQR impedes degradation of M1–3/el/4 α-proteins. (B) β assembly with M1–3/el/4 α-proteins depends on the integrity of a region encompassing D893 up to R913. Immunoprecipitated samples shown in B were treated with Endo H to permit separation of the α and β bands. One of two or three representative experiments is shown.
The results obtained with alanine variants of the M1–3/el/4 α-protein confirm that the Val-890–Val-916 domain may indeed contain a degradation signal that is recognized from the ER lumenal side. Because the Val-890–Val-916 domain encompasses the β assembly domain, it is likely that this degradation signal is directly masked by association of the β subunit. Furthermore, the analysis of the alanine variants in an unusual topographic context confirm that the integrity of a domain exceeding the 903SYGQ906 motif is necessary for efficient β association and correct folding of the α subunit.
DISCUSSION
In this study, we have used the α subunit of Na,K-ATPase as a model protein to analyze the mechanisms that are implicated in the degradation of polytopic membrane proteins and in their protection from degradation by assembly with a partner subunit. The parallel analysis of the topological features and the stability of a series of mutant Na,K-ATPase α subunits show that the degradation of unassembled polytopic subunits of oligomeric proteins is a highly complex event, which is tightly linked to the inefficient membrane insertion properties of certain membrane domains and which depends on the existence of several putative degradation signals, which during protein synthesis may transiently be exposed to cytosolic or ER lumenal components of the degradation machinery. Partner subunit interaction in specific regions mediates protection from degradation of polytopic membrane proteins by favoring the correct membrane insertion of sequences with insufficient hydrophobicity and by shielding potential degradation signals.
The Four N-terminal Membrane Sequences of Na,K-ATPase α Subunits Function as a Stable Membrane Anchor for Nascent α Subunits
Results obtained with an RGS assay show that the four N-terminal hydrophobic segments of α subunits of Na,K-ATPase (Béguin et al., 1998) as well as of α subunits of H,K-ATPase (Beggah et al., 1999) behave as efficient, alternating signal anchor–stop transfer sequences and permit the formation of the first two membrane pairs (Figure 8). In this study, we show that M1–2, M1–3, and M1–4 α-proteins are stably expressed in oocytes. This result indicates that the mechanism of sequential translocation of alternating SA and ST sequences proposed for the generation of polytopic membrane topology (Lipp et al., 1989) produces proteins that indeed are retained in the ER but that are not recognized by the ER quality control system as misfolded proteins to become degraded. At present, the question remains open of at which time point during synthesis of a polytopic membrane protein are transmembrane segments transferred from the translocon to the lipid bilayer (for review, see Hegde and Lingappa, 1997). The complete protection against degradation of the truncated α-proteins including M1 up to M4 suggests that the two N-terminal membrane pairs of α subunits are translocated into the lipid bilayer during or soon after synthesis and provide a stable membrane anchor for the nascent α-proteins.
Figure 8.
Model of the membrane insertion and degradation of unassembled Na,K-ATPase α subunits and their protection from degradation by β assembly. (A) The first four membrane segments, M1–4, of the Na,K-ATPase α subunit insert into the ER membrane by alternating signal anchor and stop transfer sequences and produce two membrane pairs, which adopt a stable conformation possibly by lateral exit from the translocon into the lipid bilayer. The large cytoplasmic loop does not expose any degradation signals during synthesis, indicating that it rapidly folds into a correct conformation or does not contain primary degradation signals. (B) Degradation of unassembled α subunits coincides with inefficient membrane insertion properties of C-terminal membrane segments, which permit transient exposure of various degradation signals (asterisk) to the cytoplasmic side. The first putative degradation signal, which is exposed in the nascent α subunit, is located in the loop connecting M5 and M6 and contains two proline residues, which potentially can be recognized by the degradation machinery from the cytoplasmic side. Because degradation of α subunits is an all-or-none process and proceeds without production of intermediate, proteolytic fragments, the potentially lipid-inserted, N-terminal membrane pairs must be retranslocated from the lipid bilayer into the translocon for degradation. (C) A further degradation signal was identified in M7, which is recognized from the cytoplasmic side. Finally, the β assembly domain located in the extracytoplasmic loop between M7 and M8 contains a putative degradation signal, which is recognized from the ER lumenal side. Degradation of unassembled α subunits is ultimately mediated by the cytosolic, proteasomal system, but our results provide evidence for a multistep mechanism, which involves proteolytic and/or chaperone components acting from the ER lumenal side. (D) β assembly with a short stretch of amino acids comprising Asp-902 up to Tyr-910 is necessary to stabilize Na,K-ATPase α subunits before completion of synthesis. A larger domain comprising Asp-893 up to Arg-913 may be involved in additional folding events after β interaction. β interaction initially favors the formation of the M7/M8 membrane pair and as a consequence may allow secondary intramolecular interactions, which permit correct packing of the C-terminal membrane domain and possibly its integration into the lipid bilayer.
The Large Cytoplasmic Loop Per Se Is Not a Target for Degradation of Unassembled Na,K-ATPase α Subunits
The premise that not only misfolded secretory but also multimembrane-spanning proteins are ultimately degraded by the cytosolic proteasome (Kopito, 1997) requires that membrane domains that may be integrated into the lipid bilayer during synthesis must be retranslocated to the translocon for retrotransport to the cytoplasm. From an energetic point of view, this process appears very costly, and it was envisaged (Lord, 1996; Sommer and Wolf, 1997) that stripping off the large cytoplasmic loops existing in polytopic proteins such as CFTR or the Na,K-ATPase α subunits may be sufficient for degradation. Significantly, our results on truncated Na,K-ATPase α subunits show that the second cytoplasmic loop, which comprises approximately half of the amino acids of the α subunit, is not susceptible to degradation. Rather than being targets for direct proteasomal degradation, our results indicate that large cytoplasmic loops may rapidly fold into a correct conformation, which may be necessary to avoid proteolytic attack from the cytosol during protein synthesis. Without the stable membrane anchor provided by the first two N-terminal pairs and the proper folding of the large cytoplasmic loop, Na,K-ATPase α subunits might never efficiently accumulate before interaction with the β subunit in the M7/M8 loop. This hypothesis does not exclude, however, the possibility that the large cytoplasmic loop may contain signals for proteasomal degradation but that their recognition may depend on the exposure of primary signals in the C-terminal domain, which are responsible for the initiation of the degradation process of unassembled α subunits.
Inefficient Membrane Insertion of C-terminal Membrane Segments and Transient Exposure of Potential Degradation Signals Are Determinants for the Degradation of Unassembled Na,K-ATPase α Subunits by the Cytosolic, Proteasomal System
In analogy to the efficient membrane insertion of N-terminal membrane segments M1-M4, which correlates with the stability of N-terminal α-proteins, inefficient membrane insertion of C-terminal membrane segments M5, M7, and M9, as determined by the glycosylation reporter scanning assay, coincides with the susceptibility to degradation of unassembled α-proteins containing C-terminal domains. Inefficient membrane insertion of C-terminal membrane domains is mainly due to specific sequence information within or near the hydrophobic sequences that reduces their interaction with nonpolar surfaces and prevents correct intramolecular interactions for proper packing to occur until β subunits associate (Béguin et al., 1998). Our results indeed indicate that certain α domains, which in the mature, β-assembled α subunit are located on the extracytoplasmic side, are transiently exposed to the cytoplasmic side during synthesis of the α protein because of inefficient membrane insertion of C-terminal membrane domains and potentially become targets for cytosolic, proteasomal attack (Figure 8). One of the candidate domains that could be implicated in such a degradation process comprises M5 and the M5/M6 extracytoplasmic loop, which contains of a Pro-Leu-Pro motif highly conserved in Na,K- and H,K-ATPases (Moller et al., 1996). According to mutational analysis, the two proline residues strongly impede membrane insertion of M5 (Béguin et al., 1998) and at the same time may be involved in the degradation of α proteins from the cytosolic side (Figure 2) most likely by the proteasomal system (Figure 5). In addition to this putative proline degradation motif in the M5/M6 loop, we have evidence that degradation signals exist in M7 (Figure 4) and possibly in M9 (Figure 1), which may also be transiently exposed to the cytoplasm during synthesis because of inefficient membrane insertion of these domains.
Knowledge about different signals in proteins that mediate proteasomal degradation is still limited and mainly concerns naturally short-lived, cytosolic proteins, which, with one exception, are targeted for degradation by covalent modifications (for review, see Pickart, 1997). Ubiquitin is conjugated onto a lysine residue of a target protein by an ubiquitin-conjugating enzyme and forms a polyubiquinated chain, which is recognized by the proteasome. Obviously, a crucial step for degradation is the initial recognition of a target protein by the ubiquitin-conjugating enzyme. Furthermore, phosphorylation of PEST elements, regions rich in Pro, Glu, Ser, and Thr residues, the nature of the N-terminal amino acid, or the presence of a so-called destruction box was found to be important for ubiquitinylation and degradation of certain cytosolic, short-lived proteins (for review, see Hershko and Ciechanover, 1998). So far, nothing is known about the molecular nature of the signals that are implicated in degradation of polytopic membrane proteins. Ubiquitinylation is necessary for proteasomal targeting of some membrane proteins such as CFTR (Ward et al., 1995) but not for others such as HMG-CoA reductase (McGee et al., 1996). Coppi and Guidotti (1997) have reported that Na,K-ATPase α subunits expressed in COS cells become ubiquitinylated. In light of the scant information available, the question remains open of whether the putative proline degradation signal in the M5/M6 loop or that in M7 and M9 of the Na,K-ATPase α subunit represents 1) specific signals that can be directly recognized by the proteasome or 2) targets for ancillary proteins such as molecular chaperones that assist the retrograde transport to the cytosol (Sommer and Wolf, 1997), ubiquitin ligases that permit ubiquitinylation of lysine residues, or protein kinases that, as in PEST sequences, mediate phosphorylation necessary for ubiquitinylation.
Degradation Signals Recognized from the ER Lumenal Side
In addition to degradation signals that may be directly exposed to the cytoplasmic side because of the inefficient membrane insertion of C-terminal segments, our combined analysis of the topology and the stability of α variants revealed potential degradation signals that are recognized from the ER lumenal side (Figure 8). One possible signal may be located in the β assembly domain in the extracellular loop between M7 and M8 of the Na,K-ATPase α subunit. This signal is transposable and can mediate degradation of an intrinsically stable membrane domain, and its mutation abolishes degradation (Figure 7). Because this signal was shown to be exclusively exposed to the ER lumenal side in the α variant tested, it can be concluded that it is recognized by a component of the degradation machinery that acts from the inside of the ER. The ultimate degradation mediated by this signal is at least partially proteasome dependent, suggesting that this signal may have a primary role in the recognition as a misfolded protein and as a mediator for retrograde transport through the translocon. Alternatively, this signal may be the target for ER lumenal proteases. Our analysis of artificial, misfolded α-proteins indeed revealed that ER lumenal proteases may exist that can degrade polytopic membrane proteins apparently independent of the proteasome system (Figure 3).
Protection of Degradation of Na,K-ATPase α Subunits by β Subunit Assembly
In this study, we show that association of the β subunit with an interaction site in the C-terminal end of the M7/M8 extracellular loop of Na,K-ATPase α subunits is the key event for the correct packing of the C-terminal membrane domain and, consequently, for an overall stabilization of the α subunit (Figure 8). As previously reported (Béguin et al., 1998), β interaction initially favors the formation of the M7/M8 membrane pair and as a consequence may support intramolecular interactions between other membrane segments to complete the proper folding of the α subunit. The present study also suggests that β interaction with the region in the M7/M8 loop may shield an intrinsic degradation signal that could be important for the recognition of the misfolded state of unassembled α subunits by a component acting from the ER lumenal side (Figure 7).
By using the two-hybrid approach, recent studies have identified a common SYGQ motif in the M7/M8 loop, which is the minimal sequence that permits assembly of β subunits with both Na,K-ATPase (Colonna et al., 1997) and H,K-ATPase (Melle-Milovanovic et al., 1998) α subunits. In this study, we provide evidence that the SYGQ motif is necessary but not sufficient for the correct packing of the α subunits of oligomeric P-type ATPases. Indeed, abolition of the SYGQ motif by mutagenesis does not completely impede interaction with β subunits and stabilization of the Na,K-ATPase α subunit (Figure 6D). Furthermore, mutational analysis of a region encompassing Val-890 and Val-916 in the M7/M8 extracellular loop, which is highly conserved among Na,K-ATPase α subunits of different species and among α isoforms, reveals a significant influence of amino acid residues surrounding the 903SYGQ906 motif on the efficiency of the β subunit to stabilize α subunits (Figures 6 and 7). According to our results, it is possible that the SYGQ motif represents a primary interaction site for β subunits that allows secondary interaction events to occur, which are necessary for the complete maturation of α subunits.
In conclusion, our study reveals the multistep nature and the sequence of events that govern the degradation of unassembled Na,K-ATPase α subunits and their protection from degradation by β assembly. Although mechanistic details of degradation may differ among different misfolded polytopic membrane proteins, it is likely that our data reflect several characteristics that are common to the degradation of such proteins. In particular, poor efficiency of membrane insertion, which is determined by specific, natural sequence information, or of mutations of certain membrane segments may be a general feature that is responsible for the transient exposure of degradation signals during synthesis. Furthermore, because our studies reveal degradation signals that function either from the ER lumenal or the cytoplasmic side in a proteasome-dependent or -independent manner, we may conclude that degradation of polytopic, misfolded membrane proteins involves ER lumen-exposed components, including proteases and molecular chaperones, as well as cytosolic components of the proteasomal system.
Finally, the example of the Na,K-ATPase α subunit permits some speculation on the necessity of partner subunit assembly in the maturation of oligomeric transport proteins. In the case of P-type ATPases, transport function is achieved through conformational changes that involve the C-terminal part of the α subunit (for review, see Moller et al., 1996). In contrast to other P-type ATPases, which do not need assembly with β subunits for maturation, in Na,K-and H,K-ATPase α subunits, the specific sequence information in the C-terminal membrane domains, which provide the conformational flexibility, may be so stringent that it requires, during evolution, the association of a helper protein to permit correct membrane insertion. Interaction with the β subunit is expected to increase the overall stability of the α subunit but not its conformational flexibility.
ACKNOWLEDGMENTS
We thank S. Girardet, S. Roy, and Danièle Schaer for technical assistance. This work was supported by Swiss National Fund for Scientific research grants 31-42954.95 and 31-53721.98 (to K.G.) and 3100-052178.97 (to O.S.).
REFERENCES
- Ackermann U, Geering K. Mutual dependence of Na,K-ATPase α-subunits and β-subunits for correct posttranslational processing and intracellular transport. FEBS Lett. 1990;269:105–108. doi: 10.1016/0014-5793(90)81130-g. [DOI] [PubMed] [Google Scholar]
- Ashkenas J, Byers PH. The final stage of gene expression: chaperones and the regulation of protein fate. Am J Hum Genet. 1997;61:267–272. doi: 10.1086/514865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle D, Weeks D, Hallen S, Melchers K, Bamberg K, Sachs G. In vitro translation analysis of integral membrane proteins. J Recept Signal Transduct Res. 1997;17:29–56. doi: 10.3109/10799899709036593. [DOI] [PubMed] [Google Scholar]
- Beggah AT, Béguin P, Bamberg K, Sachs G, Geering K. β-Subunit assembly is essential for the correct packing and the stable membrane insertion of the H,K-ATPase α-subunit. J Biol Chem. 1999;274:8217–8223. doi: 10.1074/jbc.274.12.8217. [DOI] [PubMed] [Google Scholar]
- Béguin P, Hasler U, Beggah A, Horisberger JD, Geering K. Membrane integration of Na,K-ATPase α-subunits and β-subunit assembly. J Biol Chem. 1998;273:24921–24931. doi: 10.1074/jbc.273.38.24921. [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DA. Protein processing: a role in the pathophysiology of genetic disease. FEBS Lett. 1997;409:115–120. doi: 10.1016/s0014-5793(97)00423-7. [DOI] [PubMed] [Google Scholar]
- Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- Colonna TE, Huynh L, Fambrough DM. Subunit interactions in the Na,K-ATPase explored with the yeast two-hybrid system. J Biol Chem. 1997;272:12366–12372. doi: 10.1074/jbc.272.19.12366. [DOI] [PubMed] [Google Scholar]
- Coppi MV, Guidotti G. Ubiquitination of Na,K-ATPase α-1 and α-2 subunits. FEBS Lett. 1997;405:281–284. doi: 10.1016/s0014-5793(97)00182-8. [DOI] [PubMed] [Google Scholar]
- Cresswell P, Hughes EA. Protein degradation: the ins and outs of the matter. Curr Biol. 1997;7:R552–R555. doi: 10.1016/s0960-9822(06)00279-x. [DOI] [PubMed] [Google Scholar]
- Geering K. Oligomerization and maturation of eukaryotic membrane proteins. In: Heijne G, editor. Membrane Protein Assembly. New York: Springer; 1997. pp. 173–188. [Google Scholar]
- Geering K, Beggah A, Good P, Girardet S, Roy S, Schaer D, Jaunin P. Oligomerization and maturation of Na,K-ATPase—functional interaction of the cytoplasmic NH2 terminus of the β subunit with the α subunit. J Cell Biol. 1996;133:1193–1204. doi: 10.1083/jcb.133.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geering K, Theulaz I, Verrey F, Häuptle MT, Rossier BC. A role for the β-subunit in the expression of functional Na+-K+-ATPase in Xenopus oocytes. Am J Physiol. 1989;257:C851–C858. doi: 10.1152/ajpcell.1989.257.5.C851. [DOI] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-coA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Lingappa VR. Membrane protein biogenesis: regulated complexity at the endoplasmic reticulum. Cell. 1997;91:575–582. doi: 10.1016/s0092-8674(00)80445-6. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- High S. Protein translocation at the membrane of the endoplasmic reticulum. Prog Biophys Mol Biol. 1995;63:233–250. doi: 10.1016/0079-6107(95)00005-8. [DOI] [PubMed] [Google Scholar]
- Horisberger J-D. The Na,K-ATPase: structure-function relationship. Austin, TX: R.G. Landes; 1994. [Google Scholar]
- Jaisser F, Canessa CM, Horisberger J-D, Rossier BC. Primary sequence and functional expression of a novel ouabain-resistant Na,K-ATPase. The β subunit modulates the potassium activation of the Na,K-pump. J Biol Chem. 1992;25:16895–168903. [PubMed] [Google Scholar]
- Jaunin P, Jaisser F, Beggah AT, Takeyasu K, Mangeat P, Rossier BC, Horisberger JD, Geering K. Role of the transmembrane and extracytoplasmic domain of β-subunits in subunit assembly, intracellular transport, and functional expression of Na,K-pumps. J Cell Biol. 1993;123:1751–1759. doi: 10.1083/jcb.123.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, MacKenzie KR, Arkin IT, Engelman DM. Transmembrane α-helix interactions in folding and oligomerization of integral membrane proteins. In: Heijne G, editor. Membrane Protein Assembly. New York: Springer; 1997. pp. 3–24. [Google Scholar]
- Lipp J, Flint N, Haeuptle M-T, Dobberstein B. Structural requirements for membrane assembly of proteins spanning the membrane several times. J Cell Biol. 1989;109:2013–2022. doi: 10.1083/jcb.109.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo MA, Jensen TJ, Cui L, Hou Y, Chang XB, Riordan JR. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Quality control by proteases in the endoplasmic reticulum. Removal of a protease-sensitive site enhances expression of human P-glycoprotein. J Biol Chem. 1998;273:32373–32376. doi: 10.1074/jbc.273.49.32373. [DOI] [PubMed] [Google Scholar]
- Lord JM. Protein degradation: go outside and see the proteasome. Curr Biol. 1996;6:1067–1069. doi: 10.1016/s0960-9822(02)70666-0. [DOI] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Assembly of ERr-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee TP, Cheng RH, Kumagai H, Omura S, Simoni RD. Degradation of 3-hydroxy-3-methylglutaryl-CoA reductase in endoplasmic reticulum membranes is accelerated as a result of increased susceptibility to proteolysis. J Biol Chem. 1996;271:25630–25638. doi: 10.1074/jbc.271.41.25630. [DOI] [PubMed] [Google Scholar]
- Melle-Milovanovic D, Milovanovic M, Nagpal S, Sachs G, Shin JM. Regions of association between the α and the β subunit of the gastric H,K-ATPase. J Biol Chem. 1998;273:11075–11081. doi: 10.1074/jbc.273.18.11075. [DOI] [PubMed] [Google Scholar]
- Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller JV, Juul B, Lemaire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- Moriyama T, Sather SK, McGee TP, Simoni RD. Degradation of HMG-CoA reductase in vitro. Cleavage in the membrane domain by a membrane-bound cysteine protease. J Biol Chem. 1998;273:22037–22043. doi: 10.1074/jbc.273.34.22037. [DOI] [PubMed] [Google Scholar]
- Nelson RM, Long GL. A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal Biochem. 1989;180:147–151. doi: 10.1016/0003-2697(89)90103-6. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and Bip to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Egner R, Kuchler K, Wolf DH. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem. 1998;273:32848–32856. doi: 10.1074/jbc.273.49.32848. [DOI] [PubMed] [Google Scholar]
- Qu DF, Teckman JH, Omura S, Perlmutter DH. Degradation of a mutant secretory protein, α(1)-antitrypsin Z, in the endoplasmic reticulum requires proteasome activity. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- Ruddon RW, Bedows E. Assisted protein folding. J Biol Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125. [DOI] [PubMed] [Google Scholar]
- Sommer T, Wolf DH. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1227–1233. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn JA, Fyfe GK, Canessa CM. Biosynthesis and processing of epithelial sodium channels in Xenopus oocytes. J Biol Chem. 1998;273:30344–30351. doi: 10.1074/jbc.273.46.30344. [DOI] [PubMed] [Google Scholar]
- Verrey F, Kairouz P, Schaerer E, Fuentes P, Geering K, Rossier BC, Kraehenbuhl J-P. Primary sequence of Xenopus laevis Na+-K+-ATPase and its localization in A6 kidney cells. Am J Physiol. 1989;256:F1034–F1043. doi: 10.1152/ajprenal.1989.256.6.F1034. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]