Abstract
Recent studies have shown that Cdc6 is an essential regulator in the formation of DNA replication complexes. However, the biochemical nature of the Cdc6 molecule is still largely unknown. In this report, we present evidence that the Saccharomyces cerevisiae Cdc6 protein is a double-stranded DNA-binding protein. First, we have demonstrated that the purified yeast Cdc6 can bind to double-stranded DNA (dissociation constant ∼ 1 × 10−7 M), not to single-stranded DNA, and that the Cdc6 molecule is a homodimer in its native form. Second, we show that GST-Cdc6 fusion proteins expressed in Escherichia coli bind DNA in an electrophoretic mobility shift assay. Cdc6 antibodies and GST antibodies, but not preimmune serum, induce supershifts of GST-Cdc6 and DNA complexes in these assays, which also showed that GST-Cdc6 binds to various DNA probes without apparent sequence specificity. Third, the minimal requirement for the binding of Cdc6 to DNA has been mapped within its N-terminal 47-amino acid sequence (the NP6 region). This minimal binding domain shows identical DNA-binding properties to those possessed by full-length Cdc6. Fourth, the GST-NP6 protein competes for DNA binding with distamycin A, an antibiotic that chelates DNA within the minor groove of the A+T-rich region. Finally, site-direct mutagenesis studies revealed that the 29KRKK region of Cdc6 is essential for Cdc6 DNA-binding activity. To further elucidate the function of Cdc6 DNA binding in vivo, we demonstrated that a binding mutant of Cdc6 fails to complement either cdc6-1 temperature-sensitive mutant cells or Δcdc6 null mutant cells at the nonpermissive temperature. The mutant gene also conferred growth impairments and increased the plasmid loss in its host, indicative of defects in DNA synthesis. Because the mutant defective in DNA binding also fails to stimulate Abf1 ARS1 DNA-binding activity, our results suggest that Cdc6 DNA-binding activity may play a pivotal role in the initiation of DNA replication.
INTRODUCTION
In eukaryotes, DNA synthesis initiates at multiple chromosomal sites called origins of DNA replication. In the budding yeast Saccharomyces cerevisiae, such origins are known as autonomously replicating sequences (ARSs). These sequences can act in plasmids to confer a high frequency of yeast transformation and in the chromosomal locations to initiate bidirectional DNA replication (Broach et al., 1983; Campbell and Newlon, 1991). Ample evidence supports ARS1 as a yeast chromosomal origin of DNA replication. It is organized into three functional domains: A, B, and C (Celniker et al., 1984). Domain A contains the 11-base pair (bp) core consensus sequences, called ARS consensus sequence (ACS). The ACS is found in all ARSs. Point mutations and linker-scanning mutagenesis of ARS1 show that the A domain is an essential element, although it is not alone sufficient for full origin activity in DNA replication (Bell and Stillman, 1992; Marahrens and Stillman, 1992). Linker-scanning mutagenesis also revealed that the B domain can be subdivided into three distinct elements, B1, B2, and B3, and that they are collectively critical for the origin function of ARS1 (Marahrens and Stillman, 1992). Domain C may be less important, because domains A and B are sufficient for ARS1 function (Marahrens and Stillman, 1992).
The trans binding factors that bind to cis ARS1 DNA fragments have also been studied intensively. Previous DNA-binding studies with crude yeast nuclear extracts detected an abundant nuclear protein called ARS-binding factor I (Abf1) that interacted specifically with the B3 element of ARS1 (Buchman et al., 1988; Diffley and Stillman, 1988; Sweder et al., 1988). This protein is essential for both DNA replication and transcription and is required for the viability of yeast (Rhode et al., 1989, 1992). Linker-scanning analysis of ARS1 indicates that binding of the Abf1 protein to the intact B3 element is required for full ARS function (Marahrens and Stillman, 1992). More recently, a multiprotein complex that contains six different polypeptides has been identified as the origin recognition complex (ORC) (Bell and Stillman, 1992). Upon specific recognition of ACS, ORC interacts with both the A and B1 elements of ARS1, as demonstrated in DNase I footprinting (Bell and Stillman, 1992; Diffley et al., 1994) and gel mobility shift assays (Rao and Stillman, 1995). Data obtained from two-dimensional gel electrophoresis reveal that orc mutants form few active replication origins, implying that the mutants are defective in the initiation of chromosomal DNA replication (Liang et al., 1995). Thus, ORC acts as an initiator protein complex at the A-B1 domain of ARS1 for yeast DNA replication (Bell and Stillman, 1992; Diffley et al., 1994).
How function of the replication origins is coupled to cell cycle progression has recently come under intensive investigation (Küntzel et al., 1996; Piatti et al., 1996; Stillman, 1996; Newlon, 1997). In vivo DNase I footprinting shows that yeast replication origins exist in two states during the cell cycle: a postreplicative state (postRC) in G2 and M phases, and a prereplicative state (preRC) in G1 phase (Diffley et al., 1994). Both ORC and Abf1 appear to be bound at ARS1 after DNA replication and before cell division. The in vivo postRC footprint closely resembles the footprint generated in vitro by purified ORC and Abf1. The footprints of DNA from G1 cells, however, differ significantly from those obtained in vitro with the purified ORC and Abf1 proteins. The data suggest that the binding of ORC and Abf1 to the origins does not control the initiation of DNA replication. Rather, modification of origins has to occur with the involvement of some additional factors. A number of genes have been suggested to associate with ORC. The Cdc6 protein is one such candidate.
The CDC6 gene may be involved in DNA replication directly. Exposing temperature-sensitive cdc6 mutant cells to restrictive conditions blocks S-phase entry (Hartwell, 1976). The mutant-expressing cells also have high plasmid and chromosome loss rates. The high rates of plasmid loss can be suppressed by supplying multiple copies of the ARS unit on the plasmid (Hogan and Koshland, 1992). Overexpression of ORC6 in cdc6-1 mutant cells reduces the restrictive temperature requirement (Li and Herskowitz, 1993), suggesting that these two proteins interact genetically with each other. The sequence of the cloned CDC6 gene predicts a 58-kDa protein with consensus elements of purine nucleotide-binding sites (Zhou et al., 1989; Kelly et al., 1993). Cdc6 ATP binding is essential for the loading of mini-chromosome maintenance proteins and the association with Orc1 (Wang et al., 1999; Weinreich et al., 1999). It has also been shown that Cdc6 expressed in insect cells can be coprecipitated with affinity-purified ORC, suggesting that Cdc6 may associate with ORC to form a complex in the initiation of DNA replication (Liang et al., 1995). Moreover, genomic footprinting and primer extension reactions have shown that Cdc6 is essential for the establishment and maintenance of preRC (Cocker et al., 1996). In a yeast strain with the CDC6 gene under the control of the regulatory MET3 promoter, preRC complexes formed only upon CDC6 expression (Cocker et al., 1996). The results suggest that Cdc6 is a component of preRC complexes and interacts with ORC subunits. Cdc6 homologues have been found in Schizosaccharomyces pombe (Lopez-Girona et al., 1998), Xenopus (Coleman et al., 1996; Tugal et al., 1998), and Homo sapiens (Jiang et al., 1999), which is indicative of conservation of this important pathway.
Despite the exciting possibility that Cdc6 and ORC orchestrate the assembly of the replication complexes, the role of Cdc6 in the initiation of replication is still obscure. Little information is available that would allow elucidation of this process at the biochemical level. We have demonstrated previously that there is a direct physical interaction between Orc1 and Cdc6 (Wang et al., 1999) and that yeast Cdc6 protein can stimulate Abf1 binding to the B3 domain of the ARS1 DNA fragment (Feng et al., 1998). In this report, we present evidence that Cdc6 binds double-stranded (ds) DNA nonspecifically and that the 47-amino acid N-terminal region of Cdc6 is sufficient for DNA binding. We have further performed site-direct mutagenesis studies, altering the 29KRKK sequence, which has also been found in several other A+T DNA-binding proteins. This mutant, which is defective in DNA binding in vitro, also exhibits compromised function in vivo. Our results suggest the following sequential events at replication origins at the intermolecular level: Orc1 (and other ORC components) recruit Cdc6, which allows Cdc6 binding to DNA to facilitate subsequent assembly of the replication complex, thus changing Abf1 affinity to the ARS1 fragment. Therefore, our results indicate that Cdc6 may initiate DNA replication by interacting with DNA at replication origins.
MATERIALS AND METHODS
Materials
Cdc6 antibodies have been described previously (Elasser et al., 1996; Jong et al., 1996). Anti-GST antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Distamycin A, actinomycin D, and echinomycin were purchased from Sigma Chemical (St. Louis, MO).
Purification of Cdc6 Protein
Cdc6 protein was purified from the protease-deficient yeast strain BJ2168 (a prc1-407 prb1-1122 pep4-3 leu2 trpl ura3-52), which was transformed with a high-copy-number plasmid carrying the GAL1,10 promoter driving the expression of the CDC6 gene. All procedures were carried out at 4°C. Ten liters of fermenter culture was grown in SD-ura to 1.5∼3.0 × 107 cells/ml with the use of raffinose as a carbon source. Galactose (final concentration, 2%) was added for induction for 4 h. Cells were harvested and washed with double-distilled water twice (∼25 g wet weight). The washed cells were resuspended into buffer A (50 mM Tris, pH 7.2, 1 mM DTT, 1 mM EDTA, 5% glycerol) with 100 mM NaCl, supplemented with protease inhibitors (3 mM benzamidine, 0.5 mM PMSF), and then disrupted with a Bead-beater (Bio-spec, Bartlesville, OK) eight times for 20 s each time. The supernatant was collected by centrifugation (18,000 × g, 30 min) and labeled as fraction I. This fraction contained 1500 mg of total protein in a volume of 200 ml. Fraction I was loaded onto a 300-ml DE-52 column (Whatman, Clifton, NJ), which was preequilibrated with buffer A. The Cdc6 protein passed through the column, whereas ∼75% of the total protein was retained in the column. Protein blotting was used to monitor Cdc6 recovery. The eluted fractions containing Cdc6 proteins were combined (300 ml, ∼400 mg) and designated as fraction II. Fraction II was diluted with buffer A 1:1 to reduce salt to 50 mM NaCl and then loaded onto a prepacked Mono-Q 16/10 column (20 ml; Amersham Pharmacia Biotech, Piscataway, NJ), which was preequilibrated with buffer A- 50 mM NaCl. The proteins were eluted with buffer A in a gradient from 50 to 120 mM NaCl. Cdc6 from the Mono-Q column was pooled (∼5 mg of total protein in 15 ml) and designated as fraction III. Fraction III was concentrated to ∼0.5 ml with a Centricon-30 (Amicon, Beverly, MA) and loaded onto a fast-protein liquid chromatography (FPLC) Superose 12 column (Amersham Pharmacia Biotech) in buffer A with 30 mM NaCl. The flow rate was 0.3 ml/min, and 0.33 ml of each fraction was collected. Marker proteins were run separately under the same conditions. The majority of Cdc6 protein was collected in fraction 32, corresponding to a size of ∼116 kDa. Fractions 31–34 were pooled (0.08∼0.1 mg in 1.2 ml) and designated as fraction V. Fraction V was loaded onto a 2-ml dsDNA or single-stranded (ss) DNA column (Pharmacia, Piscataway, NJ). Cdc6 protein was eluted with the use of buffer A with a linear salt gradient from 50 to 500 mM NaCl in buffer A. Cdc6 protein was eluted from the dsDNA column in the fractions containing 0.28∼0.3 M NaCl. The protein passed through the ssDNA column. After elution from the dsDNA column, Cdc6 protein was >98% pure.
GST Fusion Protein Expression and Purification
pGEX-KT (Pharmacia) was used to create various GST-Cdc6 expression constructs. GST-Cdc6 and its truncated fusion proteins were expressed in Escherichia coli and purified as described (Elasser et al., 1996; Feng et al., 1998). To construct GST-ΔPH6, PCR was performed with the use of pBlueScript-CDC6 and a 5′ primer (5′-GATGGTGCATTGCCAGC-3′) plus the M13/pUC reverse amplification primer (Life Technologies-BRL, Grand Island, NY). The PCR product of CDC6 deletion from 141 to 225 (Zhou et al., 1989) was digested with PstI and EcoRI and cloned into the pGEX-KT vector. The fusion proteins were stored in buffer H/0.1 (50 mM HEPES, pH 7.5, 0.1 M KCl, 1 mM EDTA, 50 mM Mg acetate, 10% glycerol, 0.02% NP40). Fusion protein GST-NP6 was overproduced in E. coli and purified with the use of glutathione–agarose beads. To separate GST-NP6 from GST or other contaminants, the protein preparation was further purified over a FPLC Mono-S column (Pharmacia) in 20 mM Tris-HCl, pH 8.0, with the use of a 25∼500 mM NaCl gradient. GST passed cleanly through the Mono-S column, whereas GST-NP6 was eluted out in the buffer containing ∼150 mM NaCl. SDS-PAGE, gel staining, and protein blot analysis were performed with the use of the Phast system (Pharmacia).
Nitrocellulose Filter DNA-binding Assay
The binding assay measures the amount of heat-denatured DNA or native DNA bound to nitrocellulose filters in the presence of the purified yeast Cdc6, as described previously (Jong et al., 1985). Briefly, YEp352 was nick translated and used to assess binding strength. ssDNA was prepared by heat denaturation. Labeled DNA (0.2 fg) was incubated with various concentrations of Cdc6 in 20 mM Tris-HCl, pH 8, 1 mM EDTA, 2 mM β-mercaptoethanol, 50 mM NaCl, 50% glycerol in a 0.05-ml reaction volume at 30°C for 5 min. The DNA/protein complexes retained by nitrocellulose filters were counted in a scintillation counter.
Electrophoretic Mobility Shift Assay
5′ primer 5′-GGTGTTGATGTAAGCGGAGGTTGGAGAC-3′ (691–719) and 3′ primer 5′-GCGGTGAAATGGTAAAAGTCAACCCCCTGCG-3′ (943–973) were used to amplify a 283-bp ARS1 DNA fragment from S. cerevisiae genomic DNA. 5′ primer 5′-AAAATAGCAAATTTCGTCAAAAATGC-3′ (750–767) and 3′ primer 5′-ACAATCAATCAAAAAGCCAAATG-3′ (904–922) were used to amplify a 173-bp ARS1 DNA fragment. The PCR products were subcloned into pBlueScript and verified by DNA sequencing. Plasmid pBS-ARS1/200 containing the 173-bp ARS1 fragment was cut with BamHI and digested with DNA exonuclease III. Plasmid pARS1-B3 was identified as a 45-bp deletion containing only the B3 subunit of ARS1. 32P-labeled ARS1 probes were prepared with the use of a Klenow fill-in reaction. DNA and protein interactions for electrophoretic mobility shift assays (EMSAs) were performed in a 20-μl mixture containing 25 mM HEPES, pH 7.6, 5 mM MgCl2, 50 mM KCl, 1 fg of BSA, various amounts of the GST-Cdc6 fusion proteins (as indicated in figure legends), and 5–10 fmol of the probe (∼15 × 103 cpm). DNA-binding reactions were incubated for 10 min at 25°C and then were loaded immediately onto a 6% nondenaturing polyacrylamide gel (acrylamide:bisacrylamide, 80:1) that had been prerun for 1 h at 25°C in 0.5× Tris-borate-EDTA solution. The EMSA gel was run for 4 h at 120 V and 25°C, dried, and then autoradiographed.
Other Methods
Site-directed mutagenesis to generate mutated cdc6 gene was described previously (Wang et al., 1999). In the following description, the mutated nucleotides are underlined. The primer for cdc6(P21S) is GAT GCT AGC GCA ACG CCT CCA CGA; the primer for cdc6(K46A) is a complementary strand, i.e., TGA GCC AAA CTG CAG AGC TTC TGG GGA. A pair of mutant primers were used to generate the cdc6(AG6) mutation, i.e., RK6: CAA GCT AGC TCC TGC CAA AGG TCG TGG AGG CGT TGC; RK3: TTT GCT AGC TTG CAG TTC ACA GAT GTT ACA. The mutated primer pairs create a NheI site to facilitate screening. The mutated cdc6 genes were subcloned into a pGST-KT vector. The mutated cdc6(AG6) gene was also subcloned into the single-copy plasmid YCplac33 (URA3 selection) to generate YCp33-AG6, which contains −226 bp upstream of the CDC6 promoter region. Plasmid stability assays were performed as follows. A colony grown on selective medium was suspended in 0.2 ml of water. A 0.1-ml aliquot was used to inoculate 5 ml of nonselective medium (SD + Leu), and cultures were grown with aeration for ∼10 generations. Dilutions of the initial suspension were plated on YPD plates, and colonies were counted to determine the initial concentration of viable cells. These plates were then replica plated to SD-leu to determine the initial percentage of plasmid-bearing cells. Final samples were treated similarly. The loss rate per cell per generation was calculated as 1 − (final percentage of plasmid-bearing cells/initial percentage of plasmid-bearing cells)1/n, where n is generation number.
RESULTS
Cdc6 Protein Purified from Yeast Can Bind to dsDNA
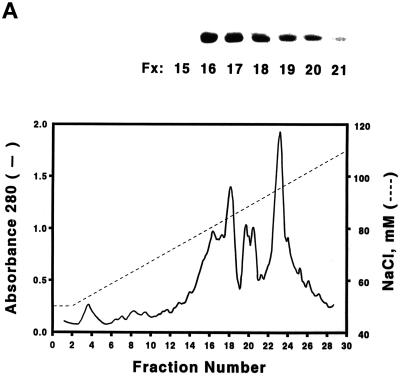
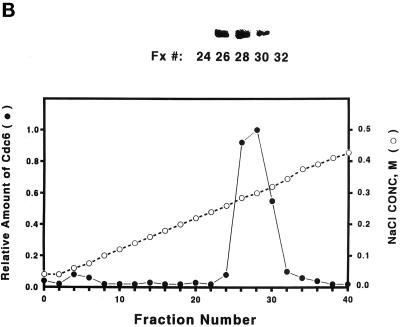
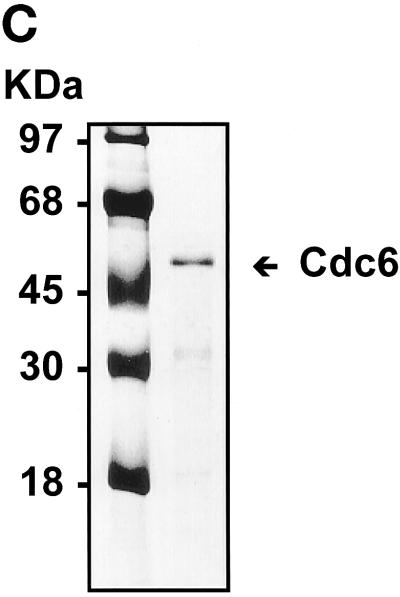
To investigate the biochemical nature of the yeast Cdc6 protein, we have used chromatography to purify Cdc6 protein from yeast extracts. Protein blots were used to monitor each fraction throughout the chromatographic process. Yeast crude extracts were first passed through a DE-52 anion exchanger column, and the clear pass-through supernatant was applied to a FPLC Mono-Q 16/10 column. The Cdc6 peak was eluted at around 80∼85 mM NaCl (Figure 1A), and the Cdc6 fractions were pooled and further purified over a FPLC Superose 12 gel filtration column (Figure 2A). Cdc6 eluted from the gel filtration column at the fractions corresponding to ∼116 kDa, suggesting that Cdc6 exists as a dimer in its native form. The pooled Cdc6 protein fractions were applied to either a dsDNA affinity column or a ssDNA affinity column. The protein can be retained by the dsDNA column up to a concentration of 280∼300 mM NaCl (Figure 1B). However, the protein did not bind to ssDNA but passed cleanly through the column. This result suggests that Cdc6 can bind dsDNA with moderate to high affinity. After elution from the dsDNA column, the protein is ∼98% pure and is available for biochemical studies (Figure 1C).
Figure 1.
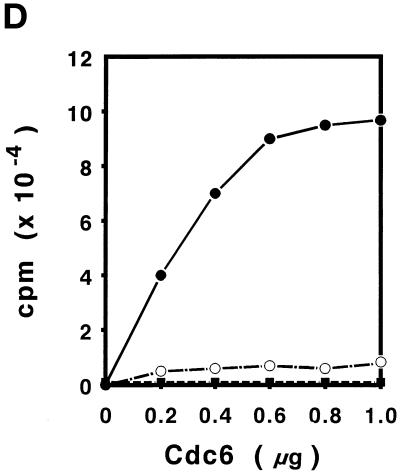
Purification of S. cerevisiae Cdc6. (A) Yeast extracts were passed through a DE-52 column and reloaded onto a FPLC Mono-Q 16/10 column. The location of Cdc6 was determined by protein blotting (top). The profile of Mono-Q chromatography is shown. Plotted is the relative position of Cdc6 (top), protein concentration (OD280), and NaCl concentration (mM) by FPLC fraction number. (B) dsDNA affinity column chromatography. The elution of Cdc6 was monitored by protein blotting (top). Plotted is the relative position of Cdc6 (●) and NaCl concentration (M, ○) by FPLC fraction number. (C) SDS-PAGE analysis of purification. (Lane 1) Prestained protein molecular mass standards from Life Technologies/BRL. Numbers correspond to the molecular masses of standard proteins. (Lane 2) Five nanograms of the purified yeast Cdc6 protein. Proteins were fractionated with the use of PhastGel SDS-PAGE (Pharmacia). The gel was silver stained. (D) The binding of purified yeast Cdc6 protein to DNA. The nitrocellulose DNA-binding assay was performed as described in MATERIALS AND METHODS. The effect of increasing concentrations of yeast Cdc6 protein on dsDNA (●) and ssDNA (○) is shown. Reaction without Cdc6 is also indicated (▪).
Figure 2.
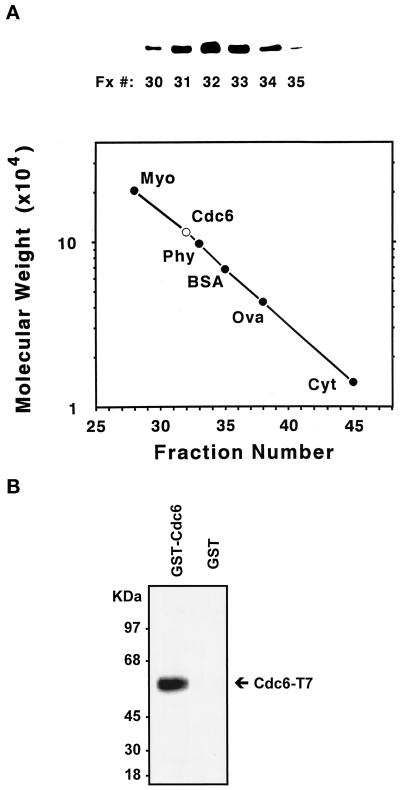
Cdc6 is a dimer in its native form. (A) FPLC Superose 12 gel filtration. Concentrated fraction III (0.2 ml) was applied to a FPLC Superose 12 column with a flow rate of 0.3 ml/min and 0.3 ml per fraction collection. Standard protein markers are myosin (Myo; 205,000 Da), phosphorylase b (Phy; 97,400 Da), BSA (68,000 Da), ovalbumin (Ova; 43,000 Da), and cytochrome C (Cyt; 12,300 Da). The peak of Cdc6 elution was found in fraction 32, as determined by protein blotting (top). Plotted is the molecular mass (kDa) by FPLC fraction number. The extrapolated size of Cdc6 is ∼116 kDa. (B) Protein blots made with the use of an anti-T7 mAb to detect T7-tagged Cdc6 copurified with GST-Cdc6. A yeast GST-Cdc6 fusion and a T7-tagged Cdc6 protein were coexpressed in yeast strain BJ2168. GST-Cdc6 was purified by glutathione affinity precipitation. The purified GST-Cdc6 fraction was resolved by 10% SDS-PAGE for detection of the associated T7-tagged Cdc6 (lane 1). Under the same experimental conditions, GST only used as the control (lane 2) did not react with anti-T7 antibody.
An aliquot of the purified protein was used for an in vitro nitrocellulose filter DNA-binding assay (Figure 1D). As was observed during dsDNA and ssDNA chromatography, the purified Cdc6 protein binds dsDNA preferentially. The apparent dissociation constant is ∼1 × 10−7 M, where the value is the concentration of protein that binds 50% of input DNA. However, in this in vitro assay, one cannot rule out the possibility that a trace amount of contaminant DNA-binding protein(s) contributes to the DNA-binding activity. Alternative approaches are required to substantiate this observation.
Cdc6 Is a Homodimer in Its Native Form
DNA sequence data revealed that the ORF of CDC6 encoded a protein of 58 kDa (Zhou et al., 1989). Gel filtration studies obtained during the Cdc6 purification show that the Cdc6 protein isolated from yeast eluted at ∼116 kDa (Figure 2A). This result suggests that the Cdc6 protein is either a homodimer in its native form or a heterodimer associated with another protein of similar size. To distinguish between these two possibilities, we have expressed yeast Cdc6 from a baculovirus construct in Sf9 cells. A similar dimeric Cdc6 molecule was observed by gel filtration purification of the expressed protein. Because of the conserved nature of the protein, it is unlikely that yeast Cdc6 interacts with protein components different from those expressed by Sf9 cells. To further confirm the dimeric nature of Cdc6, we have constructed a GST-Cdc6 fusion protein and a T7-tagged Cdc6 protein, both under the control of the galactose-inducible promoter. Upon induction, both proteins were coexpressed in yeast. The GST-Cdc6 protein was first isolated with the use of glutathione–agarose, and the resulting precipitate was blotted and probed with an anti-T7 mAb. GST alone was used as a negative control (Figure 2B). The result showed that GST-Cdc6 can associate with T7-tagged Cdc6, bolstering the hypothesis that Cdc6 exists as a homodimer.
E. coli–expressed GST-Cdc6 Can Also Bind to DNA
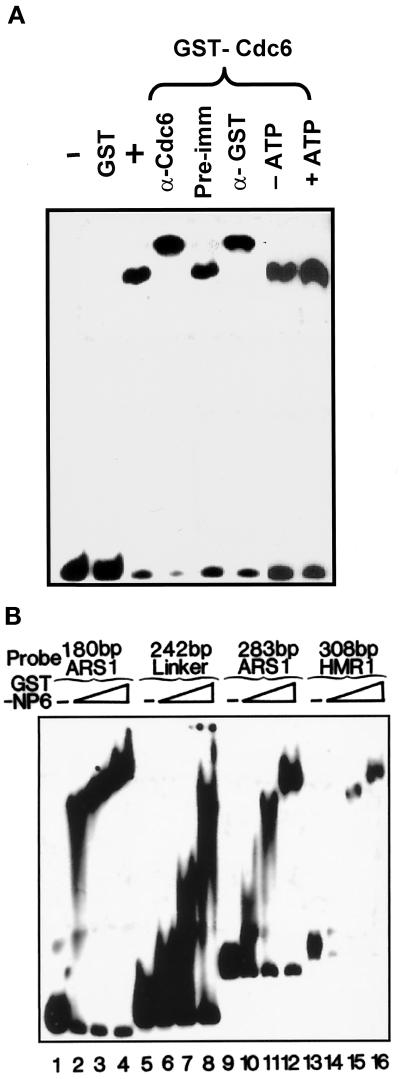
GST fusion proteins can be easily expressed and purified, thus facilitating deletion mapping analysis. Therefore, we used both full-length GST-Cdc6 and truncated mutant fusion proteins for DNA-binding studies. The GST-Cdc6 protein was incubated with the 283-bp ARS1 probe for EMSA. Several control experiments were performed to support the observation that the DNA binding is due to the activity of GST-Cdc6. First, GST protein alone showed no DNA-binding activity, even when 1000 ng of the GST protein was used (Figure 3A, lane 2). Second, incubation of an anti-Cdc6 antibody with the GST-Cdc6 protein resulted in the supershift of DNA/protein complexes (Figure 3A, lane 4), but incubation with rabbit preimmune serum showed no effect (Figure 3A, lane 5). The supershift of the DNA/GST-Cdc6 complexes was also observed upon incubation with an anti-GST antibody (Figure 3A, lane 6). The DNA-binding activity of GST-Cdc6 was not affected by the presence of ATP (1 mM) (Figure 3A, lane 7). Together, these results demonstrate that E. coli–expressed GST-Cdc6, like yeast- and baculovirus-expressed versions, bind DNA. We have further tested Cdc6 DNA binding with the use of a variety of DNA fragments as probes. Under similar conditions, GST-Cdc6 bound to the 180- and 283-bp ARS1, to a 242-bp polylinker, and to a 308-bp HMR silencer (Brand et al., 1987) (Figure 3B). As protein concentration increased, GST-Cdc6 formed nondistinct complexes with DNA. Smear bands were more significant in the absence of poly(dA-dG):poly(dC-dT). Together, these results demonstrate that GST-Cdc6 binds to DNA without any apparent sequence specificity.
Figure 3.
EMSA of GST-Cdc6. (A) Buffer only (lane 1) and GST only (1000 ng, lane 2) were used as controls. Thirty nanograms of the GST-Cdc6 fusion was used in lanes 3–8. One microgram of the anti-Cdc6 antiserum (lane 4), 3 μg of rabbit preimmune serum (lane 5), and 0.2 μg of a monoclonal anti-GST antibody (lane 6) were incubated with the GST-Cdc6 on ice for 60 min before the addition of 10 fmol of the 283-bp ARS1 probe. Lane 8 contains 1 mM ATP in the reaction mixture. Lane 7 is identical to lane 3 and is a no-ATP control. The arrow on the right indicates the supershifts. (B) Buffer only (lanes 1, 5, 9, and 13) was used as a control; 30, 60, and 90 ng of GST-Cdc6 were used with various DNA probes: 180 bp of ARS1, 242 bp of the polylinker DNA fragment from the plasmid pBlueScript, 283 bp of ARS1, and 308 bp of HMR DNA.
Determination of the Cdc6-binding Domain
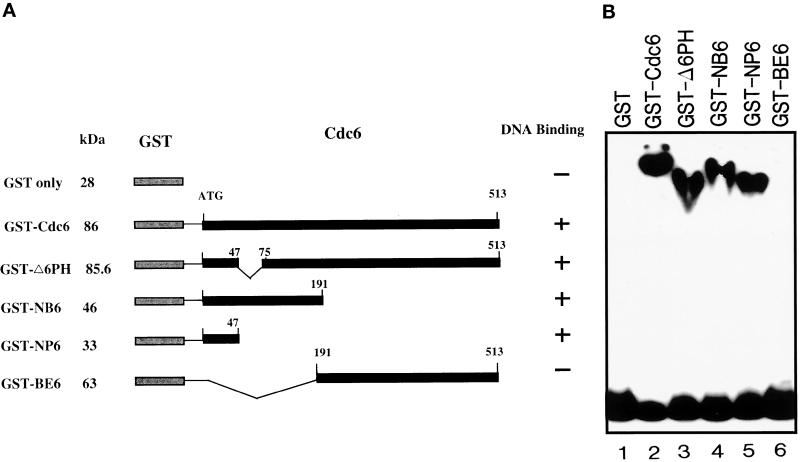
Several truncated GST-Cdc6 fusion proteins were generated to identify the specific region responsible for DNA binding. Expression and purification of the truncated GST-Cdc6 fusion proteins have been described (Elasser et al., 1996; Feng et al., 1998). Approximately 30 ng of the various deletion mutants was incubated with the 283-bp ARS1 probe. EMSAs showed that DNA-binding activity was retained by the 85.6-kDa GST-ΔPH6, which had a deletion from amino acids 47 to 74, by the 46-kDa GST-NB6, a C-terminal deletion mutant containing 191 amino acid residues, and by the 33-kDa GST-NP6, the smallest deletion mutant, containing only the first 47 N-terminal amino acids of the Cdc6 protein (Figure 4, A and B). However, GST-BE6, an N-terminal deletion containing amino acid residues from 191 to the stop codon, showed no DNA binding (Figure 4, A and B). According to these results, GST-NP6 is the smallest construct that can bind to the DNA fragment used in this study.
Figure 4.
Mapping of the DNA-binding domain of Cdc6. (A) A scheme of full-length and truncated GST-Cdc6 fusion proteins is shown. GST is indicated as a lightly shaded box, whereas Cdc6 and its deletions are indicated as dark solid boxes. The molecular mass (kDa) of each fusion protein is listed at the left. The numbers above the boxes indicate locations of amino acid residues. DNA-binding activity of each fusion protein is indicated as + or − on the right. (B) Approximately 30 ng of GST-Cdc6 fusions was added to each reaction containing 10 fmol of the 283-bp ARS1 probe to determine the DNA-binding activity of each mutant with the use of EMSA.
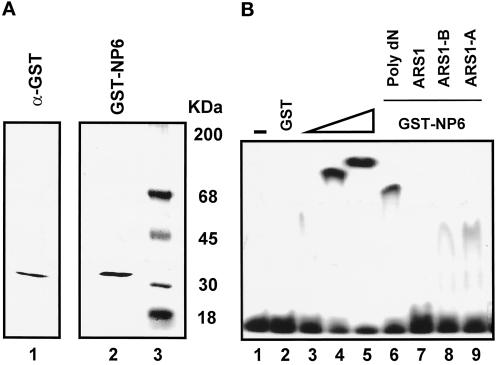
It was important to determine whether the GST-NP6 DNA-binding properties were similar to those of full-length Cdc6 protein. Therefore, we purified the E. coli–expressed GST-NP6 first via a glutathione affinity column and then over a FPLC Mono-Q column. GST proteins did not bind to the Mono-Q column, but GST-NP6 bound and was eluted in a buffer containing 150 mM NaCl. This process produced the fusion protein GST-NP6, which purified to homogeneity, as shown by a single band of 33 kDa in a silver-stained gel (Figure 5A, lane 2). The identity of the purified protein was confirmed by Western blotting with the use of an anti-GST antibody (Figure 5A, lane 1). The DNA-binding properties of the purified GST-NP6 were characterized with the use of this highly purified preparation.
Figure 5.
DNA-binding properties of GST-NP6. (A) The purity of 50 ng of GST-NP6 eluted from a FPLC Mono-S column was determined by PhastGel SDS-PAGE (Pharmacia) and silver staining (lane 2). A Western blot of a similar aliquot of the same Mono-S fraction, with the use of anti-GST antibody, is shown in lane 1. Protein molecular mass markers are in lane 3. (B) EMSA of GST-NP6 with 10 fmol of ARS1 as the probe. Buffer only (lane 1) and GST only (1000 ng, lane 2) were used as controls. GST-NP6 was added at 5 ng (lane 3), 10 ng (lane 4), and 30 ng (lanes 5–9). For a competition assay, 0.2 μg of poly(dA-dG):poly(dC-dT) (Poly dN; lane 6), 100 ng of the 283-bp ARS1 (lane 7), 30 ng of the 210-bp ARS1-B (lane 8), and 10 ng of the 70-bp ARS1-A (lane 9) were also added.
We determined the purified GST-NP6 binding activity by titrating the protein concentration and adding various competitors in gel retardation assays. Protein/DNA complexes with a distinct migration pattern could be seen in titration of the purified GST-NP6 from 5 to 10 to 30 ng per reaction with the180-bp ARS1 probe (Figure 5B, lanes 3–5). At lower concentrations (<10 ng per reaction), however, no predominant shift was observed (Figure 5B, lane 3). Thirty nanograms of the purified GST-NP6 was then used in a reaction containing 10 fmol of the 45-bp ARS1-B3 probe and various competitors. DNA binding was reduced by the addition of a 150-fold molar excess of poly(dA-dG):poly(dC-dT) (Figure 5B, lane 6). Compared with the reaction in which no competitor DNA was included (lane 5), <50% DNA binding could be observed with the addition of poly(dA-dG):poly(dC-dT) (lane 6). When a 500-fold molar excess of the competitor was included, the binding of the purified GST-NP6 to the 45-bp ARS1-B3 was completely suppressed. In a reaction in which a 40-fold molar excess of 283-bp ARS1 was added, DNA binding was also abolished completely (Figure 5B, lane 7). In addition, >50% of DNA binding could be competed by a 20-fold molar excess of either the 210-bp ARS1-B fragment or the 70-bp ARS1-A fragment (Figure 5B, lanes 8 and 9). Anti-GST antibody incubated with GST-NP6 induced a supershift of the DNA/protein complexes, but a rabbit preimmune serum had no effect. Thus, the data demonstrate unambiguously that the first 47 amino acids of the N terminus of Cdc6 contribute to DNA binding. An attempt to determine if a specific DNA-binding site(s) was recognized by the GST-Cdc6 protein with the use of DNase I footprinting failed to identify any defined region occupied by the protein in the 283-bp ARS1 probe. Consistent with the results shown in Figure 3B, GST-NP6 also binds to DNA nonspecifically in vitro.
Competition between DNA-binding Reagents and Cdc6 for DNA Binding
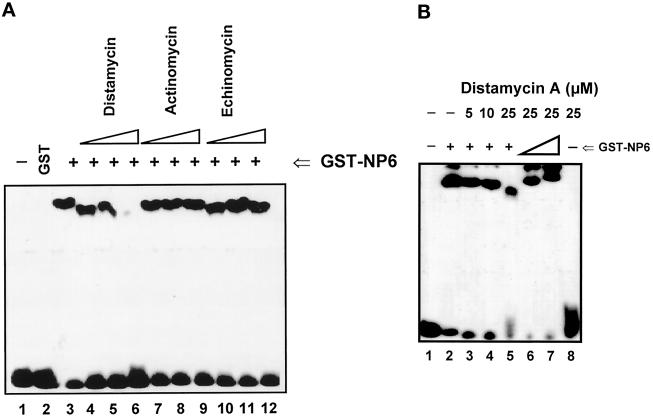
The DNA-binding properties of GST-Cdc6 described above revealed that the Cdc6 protein binds to DNA without recognition of individual bases or base pairs. The results prompted us to examine more closely the GST-Cdc6–DNA interaction. Certain DNA-binding reagents are potent inhibitors of complexes formed between proteins and their targeted DNA sequences (Kornberg and Baker, 1992). We addressed whether various DNA-binding drugs could inhibit the formation GST-Cdc6/DNA complexes. Distamycin A is an A+T-directed nonintercalating DNA-binding reagent that interacts with the minor groove of B-type DNA. Actinomycin D binds DNA by intercalating between alternating G+C base pairs in the minor groove. Echinomycin intercalates at two DNA sites simultaneously and binds preferentially to G+C-rich regions. EMSAs were performed with coincubation of 30 ng of the purified GST-NP6 and 10 fmol of the 45-bp ARS1-B3 probe and varying concentrations of distamycin A (5, 10, or 25 μM; Figure 6A, lanes 4–6), actinomycin D (5, 10, or 25 μM; lanes 7–9), or echinomycin (5, 10, or 25 μM; lanes 10–12). Cdc6 DNA binding was inhibited only by distamycin A (Figure 6A, lane 5 and 6). Neither actinomycin D nor echinomycin had any effect. As much as 25 μM distamycin A was required for disruption of the binding of purified GST-NP6 to DNA (Figure 6, A, lane 6, and B, lane 5). Therefore, we addressed whether the distamycin A inhibition of Cdc6 DNA binding could be restored by increasing the quantity of purified GST-NP6 in the reaction. As the amount of the GST-NP6 protein was increased from 30 to 100 ng, increased DNA binding was observed in EMSA complexes to which 25 μM distamycin A was added (Figure 6B, lanes 6 and 7). The incubation of 25 μM distamycin A alone with the probe did not result in any shifted band but caused smearing of the probe on the native polyacrylamide gel (Figure 6B, lane 8, bottom), indicating the association of distamycin A with the ARS-B3 DNA fragment, which is A+T rich (79% A+T). Thus, competition between the GST-NP6 protein and distamycin A in DNA binding was demonstrated. One possible interpretation of these data is that, like distamycin A, Cdc6 protein interacts with the minor groove at A+T-rich regions of DNA.
Figure 6.
DNA-binding competition between purified GST-NP6 and DNA-binding reagents. (A) Analysis of DNA-binding competition by various reagents. Buffer only (lane 1) and 1 μg of the GST protein (lane 2) were used as controls; 30 ng of GST-NP6 (+ at top of lanes) was incubated with 10 fmol of the 45-bp ARS1-B3 probe (lanes 3–12). The DNA-binding reagent distamycin A, actinomycin D, or echinomycin was added simultaneously with the protein to a final concentration of 5 μM (lanes 4, 7, and 10), 10 μM (lanes 5, 8, and 11), or 25 μM (lanes 6, 9, and 12). (B) Analysis of distamycin A competition. Buffer only was in lane 1; + at the top of lanes indicates 30 ng of GST-NP6. Increasing amounts of purified GST-NP6 (lanes 6 and 7) were incubated simultaneously with various concentrations of distamycin A (lanes 3–6) or without distamycin A (lane 2) and the 45-bp ARS1-B3 probe. In lane 8, no protein was added. Distamycin A is much smaller than GST-NP6; therefore, the shifted bands migrate faster when distamycin A concentration increases. The positions of the wells are indicated at the top of the panel. The band above the shifted complex in lanes 2, 6, and 7 indicates material retained within the well.
Physiological Consequences of the Mutation of cdc6 within Its DNA-binding Domain
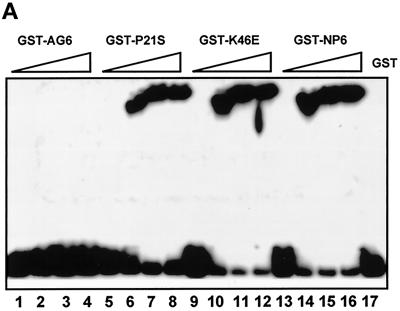
We have performed site-direct mutagenesis experiments to alter selected amino acid residues within the NP6 DNA-binding region and examine the physiological consequences. Single-point mutants cdc6(T7E), cdc6(P21S), cdc6(S34D), and cdc6(K46A) were constructed for these experiments. However, no alternation of DNA binding was observed with the use of these mutants. This result was not surprising, because the binding affinity of a non-sequence-specific DNA-binding protein toward DNA is commonly robust. In many cases, the mutation of one or even a few amino acid residues may not drastically alter binding activity. On the other hand, some A+T-rich DNA-binding proteins contain a mutation-sensitive proline–basic amino acid motif (Travers, 1993, Dutnall et al., 1996). A similar motif was found in Cdc6, 27RPLKRKK. Therefore, we mutated the four basic amino acid residues, 29KRKK, to the neutral amino acids, AGAS, to examine the effect on DNA-binding activity. The DNA-binding ability of the GST fusion protein prepared from this construct, GST-AG6, was completely lost (Figure 7A).
Figure 7.
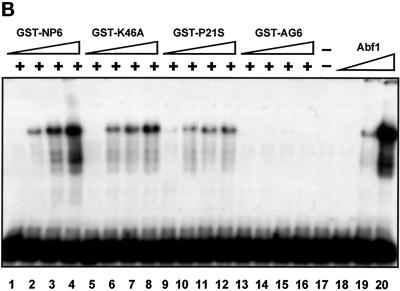
Mutagenesis analysis of Cdc6 DNA-binding and Abf1-stimulation activities. (A) EMSA was performed to examine the DNA-binding activity of wild-type and mutated GST fusions. Ten, 20, 30, and 40 ng of GST-AG6 (lanes 1–4), GST-P21S (lanes 5–8), GST-K46A (lanes 9–12), and GST-NP6 (lanes 12–16) were used. In lane 17, 100 ng of GST was used as a negative control. (B) The stimulation of Abf1 DNA-binding activities was described previously (Feng et al., 1998). EMSA was performed as in the DNA-binding assay, except that 10 ng of purified Abf1 protein was added in lanes 1–16. Lane 17 shows a no-protein control, and 10, 20, and 50 ng of Abf1 protein alone (lanes 18–20) were used as positive controls. The 283-bp ARS1 was used as the probe. Various amount of GST fusions (0.5, 1, 2, and 3 ng) were added in the reaction mixture: GST-NP6, lanes 1–4; GST-cdc6(K46A), lanes 5–8; GST-cdc6(P21S), lanes 9–12; and GST-AG6, lanes 13–16.
We have reported previously that the Cdc6 protein can stimulate Abf1 DNA-binding activity to the B3 domain of ARS1 (Feng et al., 1998). To investigate the relationship between Abf1 and Cdc6 and its DNA-binding activity, we performed an Abf1 DNA-binding stimulation assay with the use of wild-type and mutated cdc6 constructs. One caveat for these experiments is that suboptimal concentrations of Abf1 and Cdc6 proteins must be used to avoid a DNA band shift induced by Cdc6 or Abf1 alone. For our control studies, 10, 20, and 50 ng of purified Abf1 protein was tested for band shift (Figure 7B, lanes 18–20). Under these conditions, the suboptimal concentration of Abf1 (10 ng) cannot shift the DNA fragment, and this quantity of Abf1 protein was selected as the standard for stimulation studies with the use of various amounts of GST fusions [Figure 7B, GST-NP6, lanes 1–4; GST-cdc6(K46A), lanes 5–8; GST-cdc6(P21S), lanes 9–12; and GST-AG6, lanes 13–16]. Similar to the results obtained by analyzing the DNA-binding activity of the cdc6 mutants alone, we found that stimulation of Abf1 DNA binding was a feature of all mutants except GST-AG6. Thus, the DNA-binding activity and Abf1 DNA-stimulation activity of Cdc6 are closely linked.
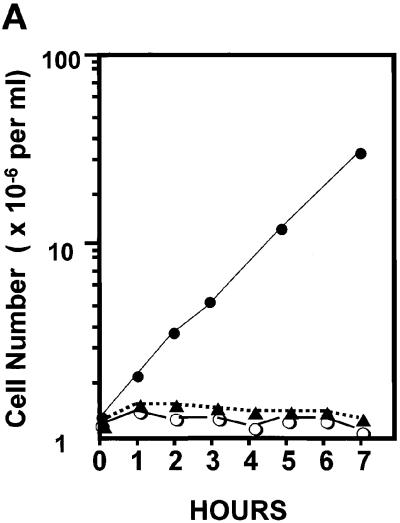
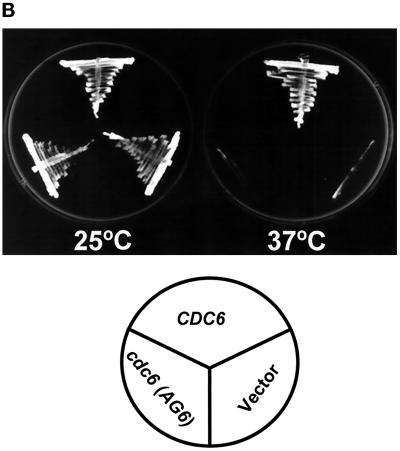
To further evaluate the effect of the presence of a mutant cdc6(AG6) allele in the temperature-sensitive mutant cdc6-1 background, cdc6-1 cells were transformed with the cdc6(AG6) mutant gene on a single-copy vector. A plasmid containing the wild-type allele, and vector alone, were used as controls. The resulting transformants were selected at room temperature with the use of the selection marker Leu2. Transformed cells were grown to log phase at the permissive temperature (25°C) and then transferred to the nonpermissive temperature (37°C) for another 7 h, during which growth rates were determined (Figure 8B). Cells harboring the wild-type CDC6 plasmid grew at a rate similar to that of wild-type strains under these conditions (∼100 min), but cdc6(AG6) growth essentially stopped immediately upon the shift to 37°C, similar to that of the vector control. Thus, the AG6 mutant showed marked impairment in growth, indicating that this motif, already shown to be crucial for DNA-binding activity and Abf1 stimulation, may also be critical for CDC6 function in vivo. Similar results were observed with inoculated plates incubated at 25°C (permissive temperature) and 37°C (nonpermissive temperature). No colonies formed from the AG6 mutant–transformed cdc6-1 cells (Figure 8B) or Δcdc6 null cells. It is clear that the cdc6(AG6) mutant represents a loss-of-function protein that cannot rescue the cdc6-1 mutation.
Figure 8.
Growth properties of transformants carrying wild-type CDC6 and mutant cdc6(AG6) genes. (A) cdc6-1 cells were transformed with the mutated plasmid YCp111-cdc6(AG6) carrying the cdc6 DNA-binding mutant. The plasmids YCp111-CDC6 (wild-type gene) and YCp111 (vector only) were used as positive and negative controls, respectively. Yeast transformants carrying the indicated plasmids were grown at SD-leuΔ at room temperature until early log phase. The cultures were then shifted to 37°C (nonpermissive temperature) and harvested at various times, and cells were counted with the use of a hemacytometer. The value given represents an average of three or four independent determinations. (●) YCp111-CDC6; (▴) YCp111-cdc6(AG6); (○) YCp111 only. (B) SD-leu plates were incubated with wild-type CDC6 and cdc6(AG6) mutant–transformed cells. Plates were then incubated at 25 and 37°C, respectively. Cells transformed with vector YCp111 only were used as the negative control. Plates were photographed after 30 h of incubation.
We further examined plasmid stability in cdc6-1 cells expressing mutant cdc6(AG6) genes. Because cdc6-1 shows enhanced plasmid loss at the nonpermissive temperature and plasmids may be stabilized by the inclusion of a functional copy of CDC6 on the plasmid, the mutated allele of cdc6 was tested for its ability to confer stability on plasmids. As shown in Table 1, cdc6-1 cells bearing a plasmid with the wild-type CDC6 gene had a plasmid loss rate of ∼0.02 per generation during a 10-generation period under nonselective conditions, a loss rate similar to that of wild-type cells. Inclusion of the cdc6(AG6) mutant gene, however, resulted in dramatic plasmids loss (0.2∼0.24 by the 10th generation), similar to that of vector-transformed cells. Loss of plasmid stability has been used as an indicator of defect(s) in DNA synthesis. In this case, it further illustrated the impact that the cdc6(AG6) mutation has on CDC6 function and the importance of CDC6 function to DNA replication.
Table 1.
Plasmid stability in cdc6-1 and transformants
| Strain | Loss rate of plasmid |
|---|---|
| cdc6-1/YCp111-CDC6 | 0.03 ± 0.005 |
| cdc6-1/YCp111-cdc6(AG6) | 0.22 ± 0.01 |
| cdc6-1/YCp111 only | 0.20 ± 0.02 |
The cdc6-1 mutant was transformed by wild-type and mutant plasmids as described in Figure 8. Transformants were grown in nonselective medium for 10 generations and plated in supplemented minimal medium or on YPD. A detailed protocol was described in MATERIALS AND METHODS. The loss rate per cell per generation was calculated as 1 − (final percentage of plasmid-bearing cells/initial percentage of plasmid-bearing cells)1/n, where n is generation number.
DISCUSSION
Yeast Cdc6 Is a DNA-binding Protein
In this report, we present evidence that Cdc6 isolated from the budding yeast S. cerevisiae binds DNA. Initially, we purified yeast Cdc6 protein via conventional and FPLC chromatography. The Cdc6 protein obtained was purified to >98% homogeneity by passage over DE-52, FPLC Mono-Q, Superose 12, and dsDNA affinity columns. The fact that Cdc6 protein bound the dsDNA column provided the first clue regarding its DNA-binding property. Gel filtration purification also revealed that Cdc6 apparently exists as a dimer in its native form. Currently, the significance of the Cdc6 dimerization is unknown. However, its importance to the DNA-binding activity of Cdc6 seems minimal, because various GST-truncated Cdc6 fusions still retain DNA-binding activities. Our studies also revealed that Cdc6 DNA-binding activity could be observed routinely with the use of yeast-purified Cdc6 protein, in vitro–translated Cdc6 protein, or GST-Cdc6 fusion proteins expressed in E. coli.
DNA-binding Properties of the Cdc6 Protein
Our studies revealed that Cdc6 DNA binding has some unique features. First, Cdc6 is a highly sequence-tolerant DNA-binding protein. Cdc6 binds to DNA fragments of varying lengths and nucleotide sequences. No DNA consensus sequence was identified by DNase I footprinting. Unlike sequence-specific transcription factors, which interact with DNA in a direct, “digital” style, GST-Cdc6 may interact with DNA by analogue recognition of the local conformation and configuration of DNA target sequences. Second, when DNA binding occurred in the presence of higher concentrations of the minimal Cdc6 DNA-binding region, NP6, nondistinct high-molecular-weight complexes were formed. This indicates that Cdc6 might bind to DNA in both a monomeric form and a multimeric form in a concentration-dependent manner. Third, this fully functional N terminus of Cdc6, GST-NP6, competed with distamycin A for DNA binding. Inhibition of DNA binding by distamycin A, and restoration of DNA binding with increasing amounts of the purified GST-NP6, indicate that the NP6 domain can effectively displace distamycin A bound in the minor groove of A+T-rich tracts. Distamycin A can also induce local structural distortions of DNA by bending the DNA helix and by inducing conformational changes in DNA A+T-rich tracts (Dorn et al., 1992). The fact that GST-NP6 competes with distamycin A for DNA binding suggests that Cdc6 may recognize some distinct structure of DNA, rather than a specific DNA sequence, and reverse the conformational changes induced by distamycin A.
Cdc6 DNA-binding Domain
We have determined that the 47-amino acid N terminus of Cdc6 possesses the primary DNA-binding properties of the molecule. We presented evidence that a purified GST fusion of NP6 competes with distamycin A for DNA binding. The DNA-binding property of Cdc6 resembles that of high-mobility-group (HMG) box proteins (Lilley, 1992). We have used the NIH computer program MACAW to analyze the Cdc6 amino acid sequences and search for sequence homology with HMG box or other DNA-binding proteins. Interestingly, the alignment of the first 77 amino acids of the N terminus of Cdc6, HMG1, and several other HMG box proteins (Diffley and Stillman, 1991) revealed a weak but readable similarity to the HMG box proteins, consistent with our data that the DNA-binding domain is located within the 47-amino acid N terminus. HMG box domains are found in many nonspecific DNA-binding proteins as well as in some sequence-specific transcription factors. This group of proteins is characterized by a number of highly conserved aromatic, basic, and proline residues with the lack of a clearly defined consensus sequence within an 80-amino acid sequence (Diffley and Stillman, 1991; Lilley, 1992). The sequence of the first 47 amino acids of the Cdc6 polypeptide contains 8 proline (>17%) and 10 basic amino acid (>21%) residues. However, DNA-binding activities may not simply be due to the prevalence of positive charges, because the Cdc6 protein does not bind ssDNA. Some HMG box DNA-binding proteins have been shown to stimulate complex formation by bending or wrapping DNA strands into specific configurations. This observation is also consistent with our previous observation that Cdc6 can stimulate Abf1 binding to the B3 domain of ARS1 (Feng et al., 1998). Our mutagenesis studies are consistent with these observations (Figure 7, A and B). It is clear that the cdc6(AG6) mutant is a loss-of-function protein that cannot bind to DNA and fails to rescue the cdc6-1 mutation (Figure 8, A and B). However, we cannot rule out the possibility that the phenotype observed with the use of the cdc6(AG6) mutant could be produced by defects in some other Cdc6 function(s), such as interactions with other components of the replication machinery, in addition to DNA.
Biological Significance of Cdc6 DNA-binding Activity
How does Cdc6 DNA-binding activity reconcile with its role in DNA replication? Recent reports indicate that Cdc6 is involved in the formation of so-called prereplicative complexes, preRC (Diffley et al., 1994; Rowley et al., 1995; Cocker et al., 1996), in which the Cdc6 associates with ORC (Liang et al., 1995). We have recently demonstrated that there is direct physical interaction between Cdc6 and Orc1 proteins (Wang et al., 1999). A proper molecular interaction between Orc1 and Cdc6 may be a prerequisite for assembly of the operational replicative complex at the G1/S transition. It is tempting to speculate that Cdc6 is recruited by ORC components, directly and/or indirectly, and then interacts with DNA in vivo at replication origins. Presumably, Cdc6 triggers DNA replication by interacting with DNA and other initiation components to form the functional preRC (Diffley et al., 1994; Liang et al., 1995; Rowley et al., 1995; Cocker et al., 1996). In the present study, an in vitro DNA-binding assay revealed that Cdc6 binds to DNA nonspecifically. However, in vivo, Cdc6 DNA binding could be restricted to certain DNA regions, particularly if Cdc6 must be recruited by ORC complexes to execute its function in DNA replication. It is intriguing that the Cdc6 protein can effectively displace distamycin A bound in the minor groove of A+T-rich tracts (Figure 6) and that the proline–basic amino acid residues (found in several preferentially A+T-binding proteins) of Cdc6 are essential for DNA binding (Figure 7). These observations would be consistent with a role for Cdc6 in initiation, because a previous study suggested that the ease of DNA unwinding (dependent on A+T concentration) determined replication origins in the yeast genome (Broach et al., 1983; Umek and Kowalski, 1988). A+T-rich sequences can usually be found in many ARS sequences. If Cdc6 proteins preferentially bind to A+T-rich sequences in vivo, yeast replication origins would certainly be reasonable targets for Cdc6.
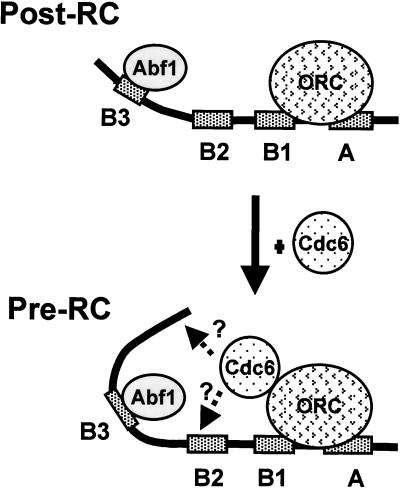
Recently, we also found that Cdc6 can stimulate Abf1 DNA-binding activity (Feng et al., 1998). One attractive model that can be drawn from these data is that Cdc6 may serve as an architectural factor involved in the formation of multiple protein/DNA complexes. The binding of Cdc6 to DNA may change the local conformation of the DNA, leading to increased affinity for sequence-specific DNA-binding proteins. The findings presented here, coupled with those produced by our previous work, suggest a model for the involvement of Cdc6 in DNA replication at the molecular level (Figure 9). ORC in its postRC form recruits Cdc6 (Wang et al., 1999). Cdc6 may distort the dsDNA juxtaposed to ACS and induce conformational changes favorable for the assembly of a higher order of initiation complex (preRC), as exemplified by the stimulation of Abf1 binding to the B3 domain of ARS1 (Feng et al., 1998). These dynamic conformational changes may invite the loading of members of mini-chromosome maintenance proteins and other initiation factors. The identification of yeast Cdc6 as a DNA-binding protein provides additional evidence for the sequence of events from the postRC to the preRC transition at the molecular level.
Figure 9.
A model illustrates CDC6 gene function in the post-RC-to-preRC transition. ORC and Abf1 associate with the A-B1 and B3 domains of ARS1, respectively, throughout the cell cycle. Sequential events involving Cdc6 at the intermolecular level are proposed. At the G1 phase, Cdc6 is recruited by Orc1 (Wang et al., 1999) and possibly by other ORC components. Cdc6 may bind to DNA in juxtaposition to the ORC (dotted arrows) and stimulate Abf1 DNA-binding affinity (Feng et al., 1998), thus triggering assembly of a functional preRC, which would include loading of mini-chromosome maintenance and other initiation factors. If the ARS element does not contain an Abf1-binding site, Cdc6 may stimulate other sequence-specific binding factors via its DNA-binding activity near ORC-binding sites.
ACKNOWLEDGMENTS
We thank Dr. Robert Chiu for critical reading. This research was supported by National Institutes of Health grant GM 48492 and by a grant to the Neil Bogart Memorial Laboratories by the T.J. Martell Foundation.
REFERENCES
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Brand AH, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Broach JR, Li YY, Feldman J, Jayaram M, Nasmyth K, Hicks JB. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harbor Symp Quant Biol. 1983;47:1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Buchman AR, Kimmerly WJ, Rine J, Kornberg RD. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Newlon CS. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. In: Broach JR, Jones EE, Pringle J, editors. Chromosomal DNA Replication. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1991. pp. 41–46. [Google Scholar]
- Celniker SE, Sweder K, Srienc F, Bailey JE, Campbell JL. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JFX. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci USA. 1988;85:2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A, Affolter M, Muller M, Gehring WJ, Leupin W. Distamycin-induced inhibition of homeodomain-DNA complexes. EMBO J. 1992;11:279–286. doi: 10.1002/j.1460-2075.1992.tb05050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutnall RN, Neuhaus D, Rhodes D. The solution structure of the first zinc finger domain of SWI5: a novel structural extension to a common fold. Structure. 1996;4:599–611. doi: 10.1016/s0969-2126(96)00064-0. [DOI] [PubMed] [Google Scholar]
- Elasser S, Feng L, Wang B, Campbell JL, Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Wang B, Jong AY. Saccharomyces cerevisiae Cdc6 stimulates Abf1 DNA binding activity. J Biol Chem. 1998;273:1298–1302. doi: 10.1074/jbc.273.3.1298. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hogan E, Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wells NJ, Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc Natl Acad Sci USA. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong A, Aebersold R, Campbell JL. Multiple species of single-stranded nucleic acid–binding proteins in Saccharomyces cerevisiae. J Biol Chem. 1985;260:16367–16374. [PubMed] [Google Scholar]
- Jong A, Yong M, Chen G-C, Zhang S-Q, Chen C. Intracellular location of the Saccharomyces cerevisiae CDC6 gene product. DNA Cell Biol. 1996;15:883–895. doi: 10.1089/dna.1996.15.883. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA Replication. 2nd ed. New York: WH Freeman; 1992. pp. 451–456. [Google Scholar]
- Küntzel H, Schulz A, Ehbrecht I-M. Cell cycle control and initiation of DNA replication in Saccharomyces cerevisiae. Biol Chem. 1996;377:481–487. [PubMed] [Google Scholar]
- Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Lilley DM. HMG has DNA wrapped up. Nature. 1992;357:282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol Biol Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Newlon CS. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- Piatti S, Böhm T, Cocker JH, Diffley JFX, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode PR, Elsasser S, Campbell JL. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1064–1077. doi: 10.1128/mcb.12.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode PR, Sweder KS, Oegema KF, Campbell JL. The gene encoding ARS-binding factor 1 is essential for the viability of yeast. Genes Dev. 1989;3:1926–1939. doi: 10.1101/gad.3.12a.1926. [DOI] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JFX. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Sweder KS, Rhode PR, Campbell JL. Purification and characterization of proteins that bind to yeast ARSs. J Biol Chem. 1988;263:17270–17277. [PubMed] [Google Scholar]
- Travers A. The three-dimensional architecture of DNA-protein complexes. In: Travers A, editor. DNA–Protein Interactions. London: Chapman & Hall; 1993. pp. 45–50. [Google Scholar]
- Tugal T, Zou-Yang XH, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J Biol Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- Umek RM, Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988;52:559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Wang B, Feng L, Hu Y, Huang SH, Reynolds CP, Jong AY. The essential role of Saccharomyces cerevisiae Cdc6 nucleotide-binding site in cell growth, DNA synthesis and association with Orc1. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang SH, Jong A. Molecular cloning of Saccharomyces cerevisiae CDC6 gene. J Biol Chem. 1989;264:9022–9029. [PubMed] [Google Scholar]