Abstract
In vitro DNA-binding assays demonstrate that the heat shock transcription factor (HSF) from the yeast Saccharomyces cerevisiae can adopt an altered conformation when stressed. This conformation, reflected in a change in electrophoretic mobility, requires that two HSF trimers be bound to DNA. Single trimers do not show this change, which appears to represent an alteration in the cooperative interactions between trimers. HSF isolated from stressed cells displays a higher propensity to adopt this altered conformation. Purified HSF can be stimulated in vitro to undergo the conformational change by elevating the temperature or by exposing HSF to superoxide anion. Mutational analysis maps a region critical for this conformational change to the flexible loop between the minimal DNA-binding domain and the flexible linker that joins the DNA-binding domain to the trimerization domain. The significance of these findings is discussed in the context of the induction of the heat shock response by ischemic stroke, hypoxia, and recovery from anoxia, all known to stimulate the production of superoxide.
INTRODUCTION
Since its discovery in 1962 (Ritossa, 1962), the heat shock response has been the focus of intensive investigation, leading to significant insights into protein folding and global gene regulation. In virtually all organisms, the heat shock response is manifested as the stress-induced, rapid, and dramatic increase in synthesis rates of a small number of protein chaperones. The chaperones bind to partially unfolded proteins and act to prevent their aggregation and to facilitate their refolding. Thus, this highly conserved system serves as an intricate means to protect cells against damage resulting from environmental stress.
The most common stresses that induce the heat shock response are elevated temperature and oxidative stress. The latter is particularly important medically, because it typically results from the production of superoxide anion that occurs during partial oxygen deprivation or during the recovery from anoxia that occurs upon reperfusion after ischemia. Induction of the heat shock system upon recovery from anoxia may be universal; it has been described not only in mammalian systems but in Drosophila (Ritossa, 1964) and yeast (Brazzell and Ingolia, 1984). It has been unclear, however, how reperfusion triggers the heat shock response. Superoxide is rapidly converted to hydrogen peroxide, which can stimulate the production of the highly reactive hydroxyl radical, which, in turn, causes considerable “reperfusion damage.” Thus, the heat shock response could be induced directly via one or more of these reactive oxygen species, or it could be induced by the protein damage caused by the hydroxyl radical.
In eukaryotes, the heat shock response depends on modulating the activity (rather than the concentration) of a transcription factor. The heat shock transcription factor (HSF) is synthesized constitutively; its activity is regulated posttranslationally. Despite this commonality, different species show remarkably dissimilar HSF proteins. Furthermore, there is diversity in the mechanisms of regulation of HSF activity. For example, under nonstressed conditions, metazoans sequester the majority of HSF as chaperone-bound monomers within the cytoplasm; upon heat-shock, HSF monomers undergo an intramolecular rearrangement that allows HSF subunits to trimerize (Mosser et al., 1990; Abravaya et al., 1992; Baler et al., 1992, 1993, 1996; Rabindran et al., 1993; Westwood and Wu, 1993; Zuo et al., 1994; Shi et al., 1998; Zou et al., 1998). Trimers bind DNA and facilitate transcription. Despite the apparent simplicity, transcriptional activation is not a necessary consequence of DNA binding. Treatment of human cells with salicylate induces HSF to trimerize and bind DNA but not to activate transcription (Jurivich et al., 1992). This suggests that there are at least two activation steps for metazoan HSF, one of which occurs subsequent to DNA binding.
HSF from Saccharomyces cerevisiae and Kluyveromyces cerevisiae (but not HSF from Schizosaccharomyces pombe) behaves differently from metazoan HSFs. Budding yeast HSF trimerizes and binds to promoters before stress (Jakobsen and Pelham, 1988; Gross et al., 1990). Nonetheless, upon heat shock, the level of HSF-dependent transcription is enhanced. This suggests that yeast HSF can exist in two conformations, an unstressed “low-activity” conformation and a stressed “high-activity” conformation. The analysis of deletions and gene fusions suggests that these two conformations differ in that the transcriptional activation domains are buried, or masked, in the low-activity form (Nieto-Sotelo et al., 1990; Sorger, 1990; Bonner et al., 1992).
To date, most of the analyses of conformational differences between HSF from nonshocked versus heat-shocked cells have focused on the metazoan monomer-to-trimer transition (Rabindran et al., 1993; Westwood and Wu, 1993; Zuo et al., 1994). Yet, the effects of salicylate on human HSF (Jurivich et al., 1992) and the natural DNA-bound state of yeast HSF (Gross et al., 1990; Giardina and Lis, 1995) suggest that functionally significant conformational differences may occur subsequent to DNA binding. In this study, we have sought to address the issue of a heat shock–dependent conformational change in DNA-bound HSF. Specifically, we have examined the DNA-binding patterns of single or multiple HSF trimers to synthetic heat shock elements (HSEs) in vitro and sought to determine the nature of the signals that trigger the HSF conformational change.
MATERIALS AND METHODS
β-Galactosidase assays were performed as described previously (Breeden and Nasmyth, 1985; Bonner et al., 1994). Reagents and human and Escherichia coli superoxide dismutases were purchased from Sigma (St. Louis, MO). Molecular and yeast manipulations were performed as described by Sambrook et al. (1989) and Sherman et al. (1986).
Strains and Plasmids
Yeast strain YJB341 (Torres and Bonner, 1995) was used for β-galactosidase assays after transformation with plasmids carrying various alleles of the HSF1 gene and expulsion of the resident plasmid by selection on 5-fluoro-orotic acid. The protease-deficient strain YJB270 (Torres and Bonner, 1995) was used similarly for the preparation of protein extracts.
The plasmid pJB398 was constructed as the wild-type HSF1 plasmid to which other alleles were compared; it carries the EcoRI–XhoI genomic fragment cloned into YEplac181 (Gietz and Sugino, 1988) and has been modified to carry additional BglII and KpnI sites at codons 268 and 440, respectively. This plasmid was modified as follows. To construct HSF1–583, we inserted an oligonucleotide carrying stop codons in all three reading frames into the StuI site at codon 583. To construct HSF1–424, we used oligonucleotide-directed mutagenesis to insert a stop codon at codon 425 and XhoI sites at codons 426 and 833; the material between 426 and 833 was deleted. To construct HSF161–833, we constructed PCR primers to amplify HSF from codons 161 to 833, with the stop codon intact, with the addition of BamHI sites for insertion into the BglII site of YCpGAL3 (Bonner, 1991). Mutant L269P was generated by PCR mutagenesis (Torres and Bonner, 1995), mutant F261L, I272L was generated by mutagenesis with an oligonucleotide carrying 1% random bases, and other mutants were generated by oligo-directed mutagenesis as described previously (Torres and Bonner, 1995). For HSF purification, a 6-histidine tag was inserted at codon 832 by oligonucleotide-directed mutagenesis. All mutations were verified by sequencing.
Gel Mobility Shift DNA-Binding Assays
Extracts were prepared and used in gel mobility shift DNA-binding assays as described previously (Bonner et al., 1994). The DNA probes used in the assays were BS4T (GGGATCCTGTGAAGCTTCCTGAAGCTTCCTAGAGTCGACCTGCAG) and BS6T (CATGACGTCGACCCTGAAGCTTCTAGAAGCTTCTAGAAGCTTCCTAGAGTC-GACCTGCAG). These were rendered double-stranded by annealing to BSb (CTGCAGGTCGACTCTAG) and extending with Sequenase (United States Biochemical, Cleveland, OH), with [α-32P]dCTP (Amersham, Arlington Heights, IL) to label the probe. For use as an unlabeled competitor, BS6T was annealed to an oligonucleotide complementary to its entire length. DNA-binding incubations were 15-μl reactions containing 15 μg of whole-cell extract (or an appropriate amount of purified HSF), 1.3 ng of labeled probe, 15 ng of bovine serum albumin, 1.5 ng of sonicated E. coli DNA, and 1 mM ATP in the binding buffer described by Sorger et al. (1987): 20 mM HEPES, pH 7.9, 1 mM EDTA, 60 mM KCl, 1 mM dithiothreitol, and 12% glycerol. The addition of ATP was necessary to dissociate bound hsp70 (Bonner et al., 2000). For kinetic analyses, samples were loaded onto the gel at appropriate time points; thus earlier samples were electrophoresed somewhat longer than later samples.
Purification of HSF
The HSF1 gene in pJB398, modified to include a 6-histidine tag at the C terminus, was introduced into BJJ5457 (α ura3-52 trp1 lys2-801 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL [Jones, 1991]) for extract preparation. HSF was purified by chromatography on immobilized nickel (Novagen, Madison, WI) according to the protocol supplied by Novagen.
RESULTS
Adjacent DNA-bound HSF Trimers Form a Complex That Correlates with In Vivo Activity
Our laboratory has isolated previously a mutation in yeast HSF that shows altered regulation of activity (Bonner et al., 1992). The mutation M232V affects a highly conserved position within the DNA-binding domain of HSF. We have sought to understand how this substitution causes this effect; our analysis led us to examine the interaction between DNA-bound trimers.
The most significant aspect of this mutation is that it dramatically elevates basal-level HSF activity. By the use of an HSE–lacZ reporter gene to assess HSF activity levels, wild-type HSF displays low basal-level activity before heat shock and ∼20-fold higher activity after a brief heat shock (Figure 1A). The mutation increases basal-level activity so that it is equivalent to the level of activity in heat-shocked wild-type cells. The mutant allele is not completely deregulated, however, because its activity is further induced approximately fivefold with heat shock.
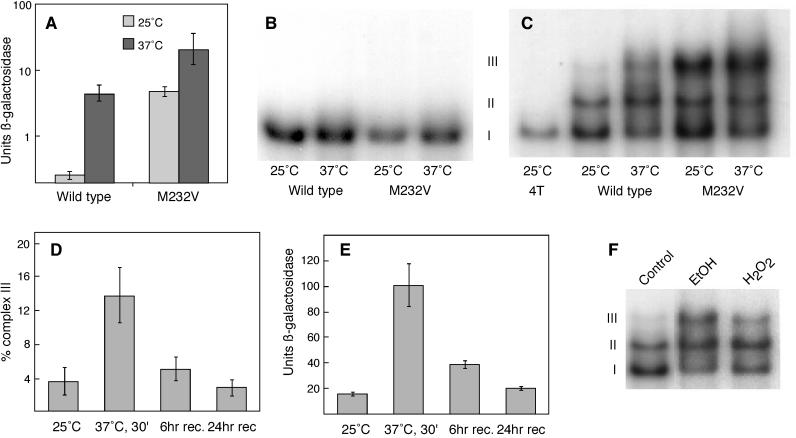
Figure 1.
Complex III correlates with HSF activity. (A) A yeast strain carrying wild-type HSF or HSFM232V and carrying an HSE–lacZ reporter gene was grown at 25°C and subjected to 1-h heat shock at 37°C, and β-galactosidase activity was measured. (B) DNA-binding assays of extracts from cells carrying either wild-type HSF or HSFM232V are shown. The cells were grown at 25°C or heat shocked at 37°C for 1 h. The probe BS4T accommodates a single HSF trimer. (C) The same extracts that were used in B were examined using probe BS6T, to which two trimers can bind. The first lane (on the left) used probe BS4T (4T) to mark the position of the HSF trimer (complex I). (D) Extracts from cells grown at 25°C, heat shocked for 30 min at 37°C, or heat shocked and returned to 25°C for 6 or 24 h were used in DNA-binding assays with the DNA probe BS6T, as in E. The percentage of counts in complex III was determined by a phosphorimager. (E) A yeast strain carrying the HSE–lacZ reporter gene was treated similarly to the cells used in B, and β-galactosidase activity was measured. (F) DNA-binding assays of extracts from cells grown at 25°C and treated with 6% ethanol (EtOH) or 0.125 mM H2O2 are shown. rec, recovery.
To assess whether the mutant might differ from wild-type HSF in DNA-binding ability, we examined HSF binding to a synthetic HSE that accommodates only a single trimer (Bonner et al., 1994). As shown in Figure 1B, there is no detectable difference in complex formation between wild-type and mutant HSF. Both form a single complex under nonshocked conditions. With heat shock the complexes become somewhat more heterodisperse because of phosphorylation (Sorger and Pelham, 1988). We extended our analysis to include the kinetics of binding; no difference was detected between mutant and wild type (our unpublished observations). From these observations, we conclude that the mutation does not affect the inherent DNA-binding ability of HSF.
Although the inherent DNA-binding ability of HSFM232V appears normal, we thought it might be possible that the lesion affects the ability of two bound trimers to interact. To test this possibility, we repeated the DNA-binding assays using a synthetic HSE that binds two HSF trimers (HSE6T; Figure 1C). On this HSE, HSF forms three distinct complexes (I, II, and III), whose abundances vary depending on the source of HSF. Complex I is of identical mobility to a complex that forms on an HSE that can accommodate a single HSF trimer (Figure 1C, compare with lane 1) and therefore represents the binding of a single trimer to this HSE. Complex II is of identical mobility to a complex comprised of six HSF monomers (Bonner et al., 1994) and thus represents the binding of two trimers to HSE6T. Wild-type HSF from nonshocked cells forms almost exclusively complexes I and II, whereas HSF from heat-shocked cells forms a modest amount of complex III in addition to complexes I and II. In previous work (using different DNA probes and exclusively wild-type HSF [see Bonner et al., 1994]), complex III was either undetectable or so minor as to be overlooked. However, complex III became very abundant when we examined HSFM232V. Comparison of the four samples shown in Figure 1C reveals that the amount of complex III increases with HSF activity, showing quite clearly that the DNA-binding pattern of HSF is affected by its in vivo activity state. These observations, in combination with results presented below, suggest that complex III might represent an active species of HSF.
If complex III does, indeed, represent a high-activity HSF species, its abundance should increase in response to heat shock and decrease during recovery from heat shock. To test this possibility, we grew cells in rich medium to midlog phase, heat shocked them for 30 min at 37°C, and then allowed them to readapt to 25°C for either 6 or 24 h. We chose two time points during the readaptation period to ensure that cells had fully recovered from the heat shock. Figure 1E tracks HSF activity, as measured by expression of an HSE–lacZ reporter gene, before, during, and after exposure to thermal stress. As expected, a brief heat shock results in the induction of β-galactosidase activity. During the recovery period, the level of β-galactosidase activity gradually returns to preheat-shocked levels, though the kinetics depends both on the dilution of the β-galactosidase enzyme that was produced during the shock and on the decrease in transcriptional activation by HSF. We observed a similar trend with respect to complex III formation (Figure 1D); complex III was induced by heat shock and became less abundant during recovery. Again, the propensity to form complex III was affected by the physiological state of the cells from which the HSF was extracted, with the cells showing higher activity also forming more complex III. Yet, the correlation between in vivo activity and complex III is imperfect; we therefore sought additional evidence that might support or refute the idea that complex III might be an active form of HSF.
In addition to heat shock, other agents also induce the stress response. Here, we examined ethanol and hydrogen peroxide to determine whether they might also induce complex III formation. To test their effects, we grew cells to midlog phase and exposed them either to 6% ethanol or to 125 μM H2O2 for 30 min. Figure 1F demonstrates that these chemical agents also induce complex III formation, although to different degrees.
Together, the data presented above are consistent with the idea that complex III may represent an active conformation of HSF. Data presented below, identifying the triggers that can induce HSF to form complex III in vitro and examining the effects of additional mutations, further support this idea.
Two Adjacent DNA-bound Trimers Can Convert to Complex III
Kinetic analysis of the formation of complexes I, II, and III (Carlson, 1998) indicated that complexes I and II form before complex III and then decrease in abundance as complex III appears. This suggests that complexes I and II may serve as precursors to complex III. Our kinetic analyses also showed that whole-cell extracts from nonshocked cells begin to form more complex III after long incubations, suggesting that some form of biochemical “stress” might activate HSF in vitro. Therefore, we sought to define experimental conditions that would test these ideas rigorously.
To determine whether nonshocked extracts could respond to heat shock in vitro and thus generate complex III in response to stress, we performed a series of DNA-binding reactions at different temperatures. As shown in Figure 2A, relatively little complex III forms when the DNA-binding reaction is incubated at 25°C for 30 min, but increasing quantities of complex III form with increasing reaction temperatures. Thus, an in vitro heat shock generates conditions that favor the formation of complex III. This finding offers the possibility of determining the biochemical conditions that lead to complex III formation but also complicates the analysis, because the quantity of complex III is dependent not only on the treatments to which the cells were subjected before extract preparation but also on the conditions of the DNA-binding assay.
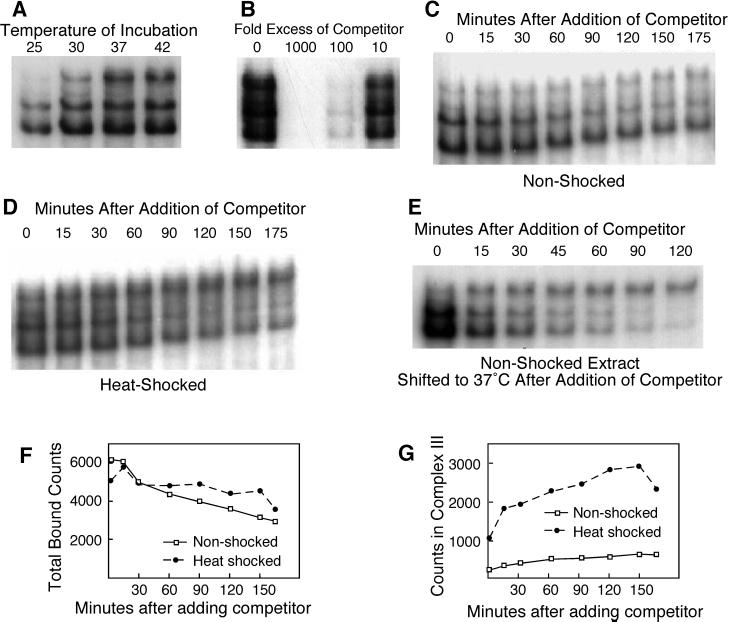
Figure 2.
Complex III can be generated in vitro after the binding of HSF to DNA. (A) DNA-binding assays of extract from cells grown at 25°C. The DNA-binding reactions were incubated at the temperatures indicated. (B) DNA-binding assays of extract from heat-shocked cells, incubated with excess, unlabeled BS6T as a competitor. (C) DNA-binding assays of nonshocked extract, incubated at 25°C for 30 min and then analyzed after the addition of 1000-fold excess of cold competitor. (D) DNA-binding assays of heat-shocked extract, similar to C. (E) Conversion of preexisting complexes I and II into complex III. A DNA-binding reaction using extract from nonshocked cells was incubated with labeled probe for 30 min, a 1000-fold excess of competitor was added, and the incubation was shifted to 37°C. (F and G) Phosphorimager analysis of the gels shown in C and D.
Although it seemed logical that complex III should form by the independent binding of “activated” HSF trimers to DNA, our kinetic data (our unpublished observations) suggested that complex III actually formed by modification of preexisting complexes I or II. To determine whether complex III can form from preexisting complexes I and II, we performed a two-step incubation experiment. First, we allowed HSF to bind to labeled HSE6T probe for a time sufficient to form primarily complexes I and II, and then we added a 1000-fold excess of unlabeled HSE6T to block further binding and sampled the DNA-binding reaction over time. (This concentration of unlabeled HSE6T is sufficient to eliminate binding to the labeled probe when both are added simultaneously [Figure 2B].)
When we used nonshocked extracts for this assay (Figure 2C), we observed primarily a decrease in bound counts, consistent with the dissociation of HSF from the labeled probe (Figure 2F, quantitated by phosporimager). Nonetheless, complexes I and II disappeared more rapidly than did complex III. Indeed, there was some production of complex III even as the other complexes disappeared (Figure 2G), consistent with the idea that extracts incubated under these conditions can undergo a biochemical stress that affects HSF. The nature of this stress is described below.
When we used heat-shocked extracts for this assay (Figure 2D), we observed a similar decrease in bound counts (Figure 2F), but with a marked difference in the behavior of complexes I and II versus complex III. Complexes I and II disappeared somewhat more rapidly than they did when we used nonshocked extracts, whereas the amount of complex III increased significantly (Figure 2G). This result can be accounted for most easily by the conversion of complex I or II (or both) into complex III. That is, the difference in the rate of disappearance of complexes I and II for the two experiments most likely reflects simple dissociation for nonshocked extracts and dissociation plus conversion to complex III for heat-shocked extracts.
As a last experiment to confirm the conversion of complexes I and/or II into complex III, we asked whether preformed complexes I and II, from nonshocked extracts, could be induced to convert to complex III by an in vitro heat shock. As before, we incubated nonshocked extracts with labeled HSE6T and added competitor but then shifted the reaction to 37°C. As shown in Figure 2E, this dramatically increased the appearance of complex III, which formed at the expense of complexes I and II. This result indicates that complex III forms from preexisting complexes I and/or II.
These data do not distinguish which complex serves as the precursor to complex III. To address this issue, we considered the possibility that the cooperative interaction between the two trimers that form complex II might be necessary to form complex III. To disturb the cooperative interaction, we introduced 5 bp between nGAAn motifs 3 and 4 in our probe. This new probe (HSE6T + 5) not only affects the spacing between trimer binding sites but also alters the helical orientation between them. Cooperativity should be abolished. Figure 3A shows that this 5-bp insertion still allows the formation of complex I but reduces the abundance of complex II to ∼10% of normal and eliminates complex III. Thus, complex III, like complex II, requires cooperative interactions for its formation. It is therefore likely that complex II serves as the precursor to complex III.
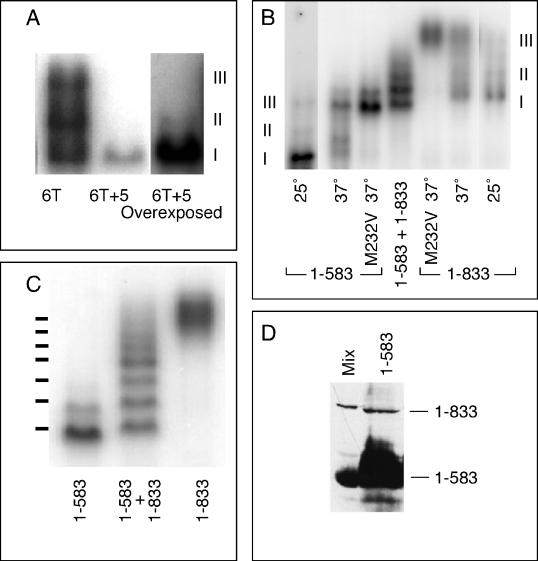
Figure 3.
Complex III is composed of two HSF trimers bound cooperatively to DNA. (A) DNA-binding reactions using as the probe BS6T (6T; left lane) or BS6T + 5, which differs from BS6T only in having 5 bp inserted between the first three and the last three AGAAG repeats (6T + 5). Because the 6T + 5 probe was of lower specific radioactivity, the right lane shows an overexposure to show the band of complex II. (B) DNA-binding reactions run with Tris-glycine buffer to identify the positions of complexes I, II, and III for HSF1–583 and HSF1–833. Nonshocked extracts show primarily complex I (lanes 1 [left lane] and 7 [right lane]), heat shock induces the formation of complex III (lanes 2 and 6), heat shock of the M232V mutant forms of these HSFs generates solely complex III (lanes 3 and 5), and coexpression of both long and short forms of HSF allows formation of heteromultimers (lane 4). (C) DNA-binding reactions with extracts from cells carrying the M232V versions of full-length HSF (1–833; right lane), HSF truncated at residue 583 by insertion of a TAA stop codon (1–583; left lane), or both (center lane) (a repeat of lanes 3–5 of B, but with the gel run for a longer time). The positions of the seven bands predicted if complex III is an HSF hexamer are shown to the left of the gel. (D) Western blot of a mixture of extracts from cells carrying full-length HSF or HSF1–583 (Mix) and of an extract from cells carrying only the plasmid-expressing HSF1–583 (1–583). The sample of HSF1–583 was overloaded to allow detection of the readthrough product.
Complex III Is Comprised of Two HSF Trimers in an Alternate Conformation
The slower mobility of complex III relative to complex II could be caused by one of several possibilities. Complex III could represent 1) the binding of a third trimer of HSF, 2) the binding of one or more non-HSF proteins, or 3) a change in the conformation of HSF. To address the first possibility, we used the technique that has been traditionally used to determine the subunit structure of enzymes: heteromultimer formation with “fast” and “slow” allelic variants. For a fast electrophoretic variant, we used a truncated HSF allele, HSF1–583, analogous to the method used by Sorger and Nelson (1989) to demonstrate the trimeric nature of HSF. This analysis was facilitated by the fact that the truncated HSF forms complex III under the same conditions as does full-length HSF (see Figures 3B and 6A) and by the finding that Tris-glycine electrophoresis buffer destabilizes complexes I and II (or converts them into complex III) so that complex III alone can be studied using HSFM232V from heat-shocked cells (Figure 3B).
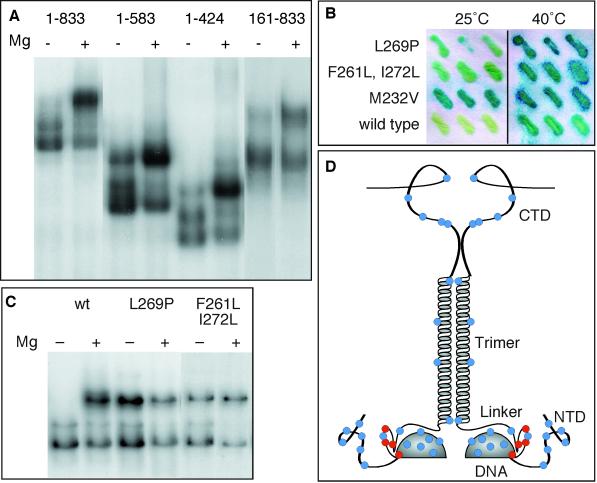
Figure 6.
Mapping the regions on which the O2− response depends. (A) DNA-binding reactions using whole-cell extracts from cells expressing the indicated HSF derivatives were treated with 10 mM MgCl2 to stimulate O2− production and to induce formation of complex III. (B) β-Galactosidase assays of yeast carrying HSFL269P, HSFF261L,I272L, HSFM232V, or wild-type HSF are shown. Cells were grown in triplicate at 25°C with or without a 2-h heat shock at 40°C and then analyzed by a β-galactosidase filter-lift assay. (C) DNA-binding reactions using whole-cell extracts from cells expressing wild-type HSF (wt), HSFL269P, or HSFF261L,I272L are shown. (D) A schematic summary of mutations that do (red dots) or do not (blue dots) affect complex III formation is shown. Mutations in the N-terminal domain (NTD) were T43C, S69C, S89C, S111C, A128C, and S151C. Mutations in the DNA-binding domain (DNA) were S181C, T189C, E196C, E208C, S221C, M232V, and Q247C. Mutations in the flexible loop between the DNA-binding domain and the flexible linker were L269P and K271C and the double mutant F261L, I272L. A mutation in the linker (Linker) was S295C. Mutations in the trimerization domain (Trimer) were I347C, M381C, Y356C, and L400C. Mutations in the C-terminal domain (CTD) were A543C, S548C, S568C, S626C, S655C, and S788C.
We coexpressed full-length HSFM232V with the M232V version of HSF1–583, heat shocked the cells, and assayed the extracts by the gel mobility shift DNA-binding assay, using Tris-glycine as the gel buffer. As shown in Figure 3C, the coexpressed HSFs display six distinct bands, the lowest of which represents the HSF M232V:1–583 homomultimer, and a seventh, more diffuse complex that corresponds to the homomultimer of full-length HSFM232V. We obtained the same result if we simply mixed extracts containing the two HSFs, because of reassociation of trimers (our unpublished observations). The HSF M232V:1–583 sample (Figure 3C, left lane) reveals several bands whose mobilities are similar to those of heteromultimers with full-length HSFM232V, despite the fact that HSFM232V:1–583 is expressed from a gene into which a TAA stop codon was inserted. We have shown previously (Kopczynski et al., 1992) that some stop codons in our strain of yeast can be subject to low-frequency translational readthrough (Kopczynski et al., 1992); immunoblot analysis confirmed that this is the case for this stop codon as well (Figure 3D). Therefore, the “pure HSF1–583 control” not only identifies the mobility of HSF1–583 homomultimers but also serves to illustrate that altering the ratio of full-length to truncated HSF derivatives shifts the distribution of heteromultimers in a predictable manner.
Because the bands are not fully resolved at the top of the distribution, it is difficult to determine, unambiguously, how many bands there are. From the relative masses of the two HSF polypeptides, one can predict the positions of the various heteromultimers, not only for complexes containing six HSF subunits, but also for those containing more. Only the prediction based on six subunits matches the observed mobilities of the bands (Figure 3C). These data are most easily accommodated by a model in which complex III contains only six HSF subunits, i.e., two trimers.
To investigate the possibility that other proteins might associate with HSF to generate complex III, we used amine-specific cross-linking, coimmunoprecipitation, and epitope-tagging of HSF but were unable to demonstrate the presence of another protein besides HSF in the complexes. We note that the hsp70 proteins that bind HSF are unlikely candidates, because they are dissociated from HSF by the ATP present in the DNA-binding reactions (Bonner et al., 2000). However, these are negative results and thus carry relatively little weight. The strongest evidence that complex III does not contain additional proteins is the finding (see below) that HSF that has been purified from nonstressed cells (and should therefore lack this putative protein) can nonetheless form complex III upon in vitro activation. Although these results do not exclude the possibility that another protein exists that we cannot detect, we think it more likely that complex III represents an alternate conformation by which two trimers can bind DNA cooperatively.
Characterization and Purification of an HSF-activating Factor
The availability of a gel mobility assay for a conformational change in HSF offers the possibility of defining the biochemical conditions that trigger this change. Could there be a factor in heat-shocked cells that modifies HSF, thereby increasing its propensity to form complex III? In an initial experiment (Carlson et al., 1999), we found that material from heat-shocked cells, when added to extract from nonshocked cells, stimulates the formation of complex III in vitro. It therefore seemed possible that we could use the formation of complex III to identify an HSF-activating factor. Because it is known that HSF is subject to phosphorylation (Sorger and Pelham, 1988) and that phosphorylation at some sites seems to be responsible for downregulation of HSF activity (Høj and Jakobsen, 1994; Xia et al., 1998), whereas phosphorylation at other sites may be responsible for activation (Liu and Thiele, 1996), we thought it likely that this HSF-activating factor might be a kinase or a phosphatase. We began the fractionation of heat-shocked extracts using gentle methods and assaying the fractions by adding them to a DNA-binding reaction using nonshocked HSF. It soon became clear that gentle methods were not necessary. Fractionation of heat-shocked extracts on Affi-Gel blue, phenyl-Sepharose, and heparin agarose revealed that the factor failed to bind to these resins (Carlson, 1998). By contrast, it proved to be susceptible to dialysis (molecular weight cutoff, 6000–8000) and resistant to boiling, to treatment with N-ethylmaleimide, and to extraction with phenol/chloroform. It fractionated in the included volume on Sephadex G25. Together, these observations argue that the HSF-activating factor is small and not a peptide. It is, however, ethanol precipitable, thus offering a convenient means of concentrating activity.
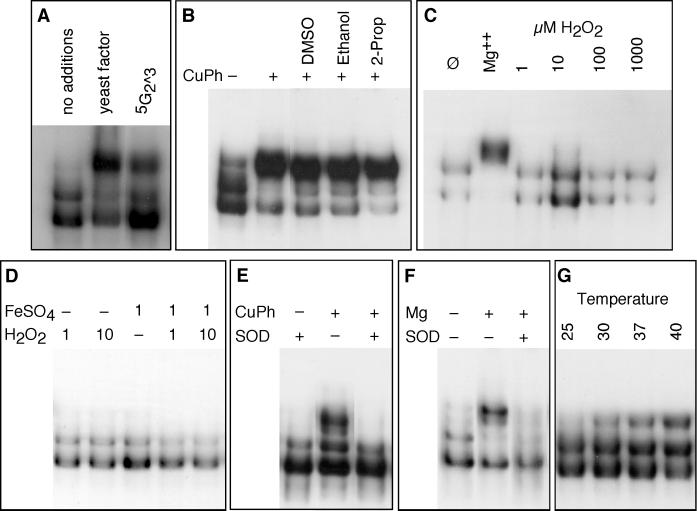
A partially purified fraction, when analyzed by mass spectroscopy and UV absorption, suggested that a nucleotide of mass 448 might be the active material. The nucleotide guanosine-5′-monophosphate-2′3′-cyclic monophosphate (5G2^3) has such a mass. Commercially prepared 5G2^3 exhibited activity (Figure 4A). Because the activity of 5G2^3 required surprisingly high concentrations, we suspected that the activity might be either a breakdown product of the nucleotide or a contaminant in the commercial preparation.
Figure 4.
Complex III is induced by superoxide or heat. (A) DNA-binding reactions with whole-cell extract to which was added yeast factor or 10 mM 5G2^3. (B) DNA-binding reactions using purified HSF treated with CuPh, with the addition of 20 mM DMSO, ethanol, or 2-propanol (2-Prop) as ·OH scavengers. (C) DNA-binding reactions using purified HSF treated with 10 mM MgCl2 or H2O2 at the indicated concentrations. (D) DNA-binding reactions using purified HSF treated with Fenton reagent (H2O2 + FeSO4) or with H2O2 or FeSO4 alone. (E) DNA-binding reactions using purified HSF treated with CuPh with or without the addition of human erythrocyte superoxide dismutase (SOD). (F) DNA-binding reactions using purified HSF treated with 5 mM MgCl2 with or without the addition of human erythrocyte SOD. (G) DNA-binding reactions using purified HSF incubated at the indicated temperatures.
Our most highly purified preparation exhibited a main component of mass 175 and lacked the component of mass 448. Collision-induced breakdown analysis of this component (our unpublished observations) was most consistent with a magnesiated ene-diol form of a pentose (the intermediate in the interconversion of ribose and ribulose) in which the sugar is coordinated with a Mg+2 ion. Ribulose alone was unable to affect the conformation of HSF, but upon extended incubation with MgCl2, it formed an ethanol-precipitable complex that induced the formation of complex III (our unpublished observations). The incubation period probably reflects isomerization to the Mg-trapping, ene-diol form. It is likely that both ribose/ribulose and Mg+2 are present in the commercial preparation of 5G2^3, the former as a breakdown product.
It is unclear from these findings whether Mg+2, the ene-diol pentose, or the coordinated complex is responsible for stimulating the formation of complex III—or whether they are physiologically significant. Ene-diol sugars have been shown to autoxidize and generate superoxide anion (O2−) (Thornalley and Stern, 1984; Thornalley et al., 1984; Thornalley, 1985; Benov and Fridovich, 1998). Is it possible that superoxide stimulates HSF directly?
To test this possibility, we performed our DNA-binding reactions with purified HSF in the presence of copper-phenanthroline (CuPh), which undergoes cyclic oxidation–reduction to generate O2−. As shown in Figure 4B, CuPh induced formation of complex III very efficiently. This result indicates that HSF is sensitive to reactive oxygen species. However, after O2− has been generated, it undergoes a dismutation reaction, to generate H2O2, that can be acted on by CuPh to generate the hydroxyl radical (·OH). It is therefore necessary to determine which of these reactive oxygen species acts on HSF. We tested the effects of radical scavengers that are sensitive to ·OH (DMSO, ethanol, and 2-propanol); they were unable to block the effect of CuPh (Figure 4B). H2O2 had no effect on HSF, even at very high concentrations (Figure 4C). The same was true for H2O2 plus FeSO4 (the Fenton reagent), which stimulates production of ·OH (Figure 4D), despite the fact that the same concentrations of FeSO4 and H2O2 readily oxidized phenol (our unpublished observations). By contrast, superoxide dismutase (both human and E. coli) blocked the effect of CuPh (Figure 4E, human SOD). These results indicate that HSF responds directly to O2− but not to hydrogen peroxide or the hydroxyl radical. To determine whether O2− or its protonated form ·HO2 is the active species, we examined the pH dependence of CuPh; between pH 6.0 and 7.9, there was no difference in how well CuPh induced complex III (our unpublished observations). We conclude that it is superoxide anion itself that acts on HSF.
Because the material we purified from cells also contained Mg+2, we tested the ability of Mg+2 to induce the formation of complex III. It could, both with crude extracts and with purified protein, although at higher concentrations than those at which free Mg+2 is typically found in vivo (Figure 4, C and F). How might Mg+2 work to induce formation of complex III? Because we had recovered Mg+2 in our preparation of putative HSF-activating factor, we considered the possibility that it may function in our DNA-binding reactions to stimulate the formation of superoxide. To test this idea, we treated the DNA-binding reactions with Mg+2 or with Mg+2 and SOD. SOD blocked the ability of Mg+2 to induce formation of complex III (Figure 4F). It therefore seems likely that Mg+2 acts indirectly on HSF, by stimulating the production of O2−, which in turn acts on HSF.
These results indicate that HSF can form complex III upon exposure to O2−. To determine whether purified HSF also forms complex III in response to heat, we incubated purified HSF with DNA at elevated temperature (Figure 4G). Indeed, in vitro heat shock stimulated the formation of complex III. Apparently, HSF is directly responsive to elevated temperature.
The ability of HSF to respond directly to elevated temperature seems to obviate the need for a temperature-dependent signaling system to activate HSF during stress. Yet, our fractionation studies recovered Mg+2 as a possible HSF activator. Is it likely that Mg+2 might act in vivo as a signal of stress? To be useful in such a role, it seems that Mg+2 would need to be released from sites of sequestration so that it could activate HSF. To test the idea of Mg+2 release, we performed in vitro heat shocks with both purified HSF and crude extracts in the presence of 10 mM EDTA to chelate any Mg+2 that might be released. EDTA had no effect on the stimulation of complex III by elevated temperature (our unpublished observations), suggesting that free Mg+2 is not released during this in vitro analogue of heat shock. We therefore think it more likely that the stimulation of O2− production by Mg+2 is a property of our in vitro DNA-binding conditions; as such, it may be useful for analyzing the transition to complex III but reveals no more about the in vivo situation than do the data shown above for CuPh.
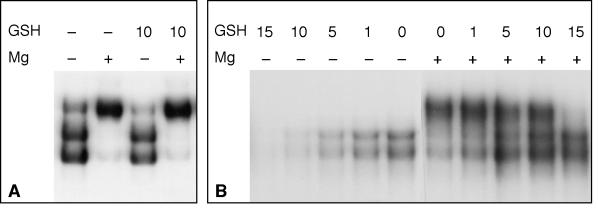
Glutathione Counteracts the Effects of Superoxide
In vivo, treatment with H2O2 induces HSF activity and causes the formation of complex III (Figure 1F), but HSF does not respond to H2O2 in vitro (Figure 4C). This suggests that the interplay between HSF activation and oxidative stress is more complex in vivo than it is in our simplified system. Cells contain a number of enzymes that serve to maintain the redox balance, among them SOD, catalase, and enzymes that rely on the tripeptide glutathione for their reducing power. It is therefore conceivable that the effect of H2O2 in vivo reflects its ability to alter the redox balance of cells, perhaps via an effect on glutathione. Because yeast HSF contains no cysteine residues, it is unlikely that reduced glutathione (GSH) can act on it directly. This prediction is met in the finding that GSH had no effect on purified HSF and did not block the induction of complex III by Mg+2-induced production of O2− (Figure 5A). However, in whole-cell extracts, GSH did influence the behavior of HSF. Figure 5B shows that GSH can counteract the effect of Mg+2 and, in the absence of high concentrations of Mg+2, can prevent the binding of HSF to DNA. These results suggest that there are likely to be GSH-dependent enzymes in the extract, undoubtedly part of the normal redox balance system, that can influence the behavior of HSF. This may explain the differing effects of H2O2 in vivo and in vitro: after H2O2 oxidizes the redox system in vivo, it may be difficult to prevent the buildup of superoxide that may be produced by normal physiological sources.
Figure 5.
Glutathione affects the behavior of HSF in whole-cell extract. DNA-binding reactions using purified HSF (A) or whole-cell extract (B) were treated with 10 mM MgCl2 and/or reduced GSH at the indicated millimolar concentrations.
The Response to O2− Maps to the Evolutionarily Conserved Portion of the Linker Domain
Yeast HSF contains an N-terminal transcriptional activation domain (TAD) and two C-terminal transcriptional activation domains (Sorger, 1990; Bonner et al., 1992; Chen et al., 1993). Neither of these shows significant sequence similarity to HSFs from other species. Sequence conservation is restricted to the DNA-binding domain and trimerization domain. Because a major component of the regulatory response to heat shock also maps to this conserved region (Bonner et al., 1992), we sought to determine whether the sensitivity to O2−, and formation of complex III, might also map to this region.
We performed gel mobility shift DNA-binding assays with HSF derivatives that were truncated at residues 583 and 424 (HSF1–583 and HSF1–424) and with an HSF derivative from which residues 1–160 had been removed (HSF161–833). The C-terminal deletions HSF1–424 and HSF1–583 behaved exactly like full-length HSF (Figure 6A), demonstrating that material C-terminal to the trimerization domain is unnecessary for complex III formation. The N-terminal deletion HSF161–833 showed a more complex behavior. Like the C-terminal deletions, it showed a significant increase in the abundance of complex III after addition of Mg+2, suggesting that the critical O2− response element lies between residues 161 and 424. However, HSF161–833 showed a detectable amount of complex III even without Mg+2 and appeared to be unable to form complex II. This is consistent with the idea that complexes II and III represent the inactive and active forms of HSF, respectively, because it has been shown by Sorger (1990) that deletion of residues 1–161 renders HSF constitutively active. From the data, we suggest that the N-terminal domain functions to stabilize complex II but does not actually contain the O2− response element.
To obtain additional mapping data, we examined mutations that we recovered during the course of a variety of mutagenesis experiments (Torres and Bonner, 1995; our unpublished observations). Although most mutations that we have recovered do not affect HSF phenotypically, two additional DNA-binding domain mutations besides M232V display elevated activity in the absence of heat shock. Both the single mutation L269P and the double-mutation F261L, I272L elevate expression of an HSE–lacZ reporter gene under normal growth conditions, although less than does M232V (Figure 6B). When analyzed by gel mobility shift DNA-binding assay, both mutations also induce the constitutive formation of complex III (Figure 6C). By contrast, a variety of mutations isolated as part of a structural investigation of HSF (Bonner, unpublished observations) had no effect on complex III formation. The mutations that we have analyzed are listed in the legend to Figure 6D, which summarizes the results of these analyses in the context of the known structure of HSF.
These genetic data map a critical element for the formation of complex III to the region immediately downstream of the minimal DNA-binding domain—a region that is essential for viability (Flick et al., 1994). This region, which contains a number of highly conserved residues in HSF, forms a flexible loop in the NMR structure of Vuister et al. (1994), where it forms a close contact with Met232. This region has been considered to be part of the flexible linker that joins the DNA-binding and trimerization domains (Flick et al., 1994), but our results suggest that this flexible loop might be a functional part of the DNA-binding domain, because it appears to play a role in modulating the structure of DNA-bound trimers.
DISCUSSION
It has long been thought that the activation of HSF upon heat shock must involve a conformational change that “unmasks” the transcriptional activation domains of HSF. There have been a number of analyses of HSF conformational changes (Rabindran et al., 1993; Westwood and Wu, 1993; Zuo et al., 1994), but these have generally examined metazoan HSF free in solution and have provided information primarily on the monomer–trimer transition. The observations of Jurivich et al. (1992) that salicylate can induce human HSF to trimerize and bind DNA without concomitant activation of hsp70 expression suggest that there is likely to be a second aspect of HSF activation beyond trimerization. Our observations provide information about this second step, at least for Saccharomyces HSF.
The argument that complex III represents an active form of HSF is based on three kinds of evidence. The first is correlative, as it must be for an assay based on electrophoretic mobility. The propensity of wild-type HSF to form complex III in vitro depends, at least in part, on the physiological state of the cells from which the HSF was extracted. Heat shock favors the formation of complex III; recovery from heat shock disfavors it. Chemical stresses that affect the heat shock response also favor the formation of complex III. These correlations are imperfect, however. In part, this may reflect the fact that HSF can be induced to form complex III by exposure to superoxide, which is generated spontaneously in the DNA-binding reactions that are exposed to atmospheric oxygen. The imperfect correlation may also reflect the fact that reduced glutathione, present in whole-cell extracts, counteracts the induction of complex III in vitro.
The second line of evidence is that HSF can be induced to form complex III in vitro in response to agents that are known to induce HSF activity in vivo: heat and oxidation. If the mechanism of HSF activation were a direct effect of these stress agents, rather than a complex signal transduction cascade, this is the very result that would be predicted. The third line of evidence that complex III represents an active conformation of HSF is genetic. We have observed that several mutations that elevate HSF activity in vivo also increase the formation of complex III in vitro. Together, these three lines of evidence suggest that complex III may, indeed, represent a conformation that HSF can adopt upon activation.
Our data do not reveal the nature of the conformational change that gives rise to complex III. However, they do provide two important pieces of information. First, the change in conformation is detectable only when two HSF trimers are bound cooperatively to DNA; singly bound trimers show no detectable differences in our gel mobility shift assay. This suggests that the conformational change represents a change in the nature of the cooperative interaction between trimers. This is intriguing, because it helps explain how differences in the architecture of HSEs can exhibit differences in biological activity that are not easily explained simply by differences in binding affinity. For example, the HSEs of the yeast CUP1 gene (Sewell et al., 1995; Liu and Thiele, 1996; Santoro et al., 1998) and of the human IL1β gene (Cahill et al., 1996) do not provide for cooperative interactions between trimers and display activities that are distinct from traditional HSEs. In contrast, traditional HSEs of natural heat shock genes typically have binding sites for two or more HSF trimers (Nover, 1987). In both yeast and Drosophila, these cooperative interactions have been shown to be either important (yeast) or critical (Drosophila) for normal heat shock–inducible transcription (Cohen and Meselson, 1988; Amin et al., 1994; Bonner et al., 1994).
Our data also map the regions that are important for complex III formation. The N-terminal “masking” domain (Sorger, 1990), the flexible loop joining the DNA-binding domain to the linker domain, and residue Met232 all seem to play roles in the transition between complexes II and III. Interestingly, these all share a certain physical proximity. The structural data for the DNA-binding domain (Harrison et al., 1994; Vuister et al., 1994) indicate that the N-terminal domain joins the DNA-binding domain near the flexible loop, which in turn is capable of closely approaching Met232. Unfortunately, the elegant cocrystal of Littlefield and Nelson (1999) uses the minimal DNA-binding domain and lacks the flexible loop. It is therefore unable to offer insights into the nature of this portion of the HSF protein. In the absence of more precise structural data, we imagine that the flexible loop adopts one conformation in complexes I and II and a different conformation in complex III.
Our data indicate that the transition to complex III can be induced in vitro by stimulation of purified HSF by heat or by the superoxide anion O2−. This indicates that HSF is directly responsive to the two stresses that are best known as inducers of the heat shock response: heat and oxidative stress. These findings are primarily consistent with those of Zhong et al. (1998), who have shown that Drosophila HSF also responds directly to heat and to oxidation. The difference that yeast HSF responds to O2− whereas Drosophila HSF responds to H2O2 (Zhong et al., 1998) may be caused by methodological differences or by differences in the physiology or in the sequences of HSF, but the similarity is striking. The production of O2− is coupled in vivo to the production of H2O2 via the dismutation of superoxide into H2O2. The major source of cellular O2− is mitochondrial metabolism, via which, it has been estimated, 1–2% of electrons are “spilled” into the generation of superoxide (Grisham, 1992). The mitochondrial Mn-dependent superoxide dismutase rapidly converts much of this to H2O2. The generation of superoxide will increase during stresses that affect mitochondrial activity. Thus, the same basic physiological stress creates both species of reactive oxygen.
The finding that HSF is directly responsive to superoxide (or, in the case of Drosophila HSF, its byproduct H2O2) provides a mechanistic explanation for the finding that the heat shock response is induced by such stresses as recovery from anoxia, hypoxia, and ischemic stroke. For aerobic organisms, all of these stresses have the same result: a burst of superoxide production. Via the dismutation of O2− to H2O2 and the subsequent conversion to the hydroxyl radical (·OH), these treatments lead to significant cellular damage—the classic “reperfusion injury.” It has long been thought that the induction of a heat shock response was the result of the ·OH-induced protein damage, stimulating an unfolded protein response. These findings, both ours and those of Zhong et al. (1998), indicate that HSF is activated directly by the oxidative stress and may therefore occur before there is extensive protein damage. Thus, the induction of the heat shock response by anoxic stresses is immediate and potentially protective, rather than a mere reaction to damage previously done.
In analyses of protein damage by reactive oxygen species, it is usually the ·OH that causes damage after production of O2− (Goscin and Fridovich, 1972; Que et al., 1980; Davies et al., 1987; Gieseg et al., 1993). Hydroxylation of aromatic amino acid residues can usually be prevented by ·OH scavengers, but not by superoxide dismutase. It is therefore surprising that it is O2− itself that activates HSF. Superoxide seems to work directly, and probably reversibly, on yeast HSF. It is unlikely that O2− simply binds to HSF, because at least some HSF remains in its “complex III-competent” form after extraction from stressed cells. More likely, O2− modifies one or more amino acid residues, in which case conversion of activated HSF back to the inactive form must entail the reversal of this modification.
It is possible that the inactivation of HSF during recovery from stress requires two activities: hsp70 proteins to help refold HSF and, as suggested by our data, one or more glutathione-dependent enzymatic activities. It is striking that high concentrations of GSH could completely prevent nonshocked HSF from binding to DNA, as judged by our gel mobility shift assay. This suggests that there may be an “inactive” from of HSF that has a very low affinity for DNA, as is the case in metazoans. This finding is consistent with the observations of Giardina and Lis (1995), who have shown that, in vivo, yeast HSF is rapidly and efficiently removed from its DNA-binding site upon the return of heat-shocked cells to normal growth temperature. On the basis of our findings, we suspect that this may occur via the GSH-dependent deactivation of HSF.
ACKNOWLEDGMENTS
This work was supported by grant GM-51853 from the National Institutes of Health.
REFERENCES
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- Amin J, Fernandez M, Ananthan J, Lis JT, Voellmy R. Cooperative binding of heat shock transcription factor to the Hsp70 promoter in vivo and in vitro. J Biol Chem. 1994;269:4804–4811. [PubMed] [Google Scholar]
- Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baler R, Welch WJ, Voellmy R. Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J Cell Biol. 1992;117:1151–1159. doi: 10.1083/jcb.117.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baler R, Zou J, Voellmy R. Evidence for a role of Hsp70 in the regulation of the heat shock response in mammalian cells. Cell Stress Chaperones. 1996;1:33–39. doi: 10.1379/1466-1268(1996)001<0033:efaroh>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benov L, Fridovich I. Superoxide dependence of the toxicity of short chain sugars. J Biol Chem. 1998;273:25741–25744. doi: 10.1074/jbc.273.40.25741. [DOI] [PubMed] [Google Scholar]
- Bonner JJ. Vectors for the expression and analysis of DNA-binding proteins in yeast. Gene. 1991;104:113–118. doi: 10.1016/0378-1119(91)90475-q. [DOI] [PubMed] [Google Scholar]
- Bonner JJ, Ballou C, Fackenthal DL. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol. 1994;14:501–508. doi: 10.1128/mcb.14.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner, J.J., Carlson, T., Fackenthal, D.L., Paddock, D., Storey, K., and Lea, K. (2000). Complex regulation of the yeast heat shock transcription factor. Mol. Biol. Cell 11. [DOI] [PMC free article] [PubMed]
- Bonner JJ, Heyward S, Fackenthal DL. Temperature-dependent regulation of a heterologous transcriptional activation domain fused to yeast heat shock transcription factor. Mol Cell Biol. 1992;12:1021–1030. doi: 10.1128/mcb.12.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazzell C, Ingolia TD. Stimuli that induce a yeast heat shock gene fused to beta-galactosidase. Mol Cell Biol. 1984;4:2573–2579. doi: 10.1128/mcb.4.12.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Cahill CM, Waterman WR, Xie Y, Auron PE, Calderwood SK. Transcriptional repression of the prointerleukin 1β gene by heat shock factor 1. J Biol Chem. 1996;271:24874–24879. [PubMed] [Google Scholar]
- Carlson T. Mechanisms of HSF Activity Regulation in Saccharomyces cerevisiae. Ph.D. Thesis. Bloomington, IN: Indiana University; 1998. [Google Scholar]
- Carlson T, Christian N, Bonner JJ. A role for RNA metabolism in inducing the heat shock response. Gene Expr. 1999;7:283–292. [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Barlev NA, Westergaard O, Jakobsen BK. Identification of the C-terminal activator domain in yeast heat shock factor: independent control of transient and sustained transcriptional activity. EMBO J. 1993;12:5007–5018. doi: 10.1002/j.1460-2075.1993.tb06194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RS, Meselson M. Periodic interactions of heat shock transcriptional elements. Nature. 1988;332:856–858. doi: 10.1038/332856a0. [DOI] [PubMed] [Google Scholar]
- Davies K, Delsignore ME, Lin SW. Protein damage and degradation by oxygen radicals. II. Modification of amino acids. J Biol Chem. 1987;262:9902–9907. [PubMed] [Google Scholar]
- Flick KE, Gonzalez L, Harrison CJ, Nelson HCM. Yeast heat shock transcription factor contains a flexible linker between the DNA-binding and trimerization domains—implications for DNA binding by trimeric proteins. J Biol Chem. 1994;269:12475–12481. [PubMed] [Google Scholar]
- Giardina C, Lis JT. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15:2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseg SP, Simpson JA, Charlton TS, Duncan MW, Dean RT. Protein-bound 3,4-dihydroxyphenylalanine is a major reductant formed during hydroxyl radical damage to proteins. Biochemistry. 1993;32:4780–4786. doi: 10.1021/bi00069a012. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goscin SA, Fridovich I. The role of superoxide radical in a nonenzymatic hydroxylation. Arch Biochem Biophys. 1972;153:778–783. doi: 10.1016/0003-9861(72)90398-0. [DOI] [PubMed] [Google Scholar]
- Grisham MB. Reactive Metabolites of Oxygen and Nitrogen in Biology and Medicine. Austin, TX: R. G. Landes; 1992. [Google Scholar]
- Gross DS, English KE, Collins KW, Lee SW. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol. 1990;216:611–631. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HCM. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- Høj A, Jakobsen BK. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 1994;13:2617–2624. doi: 10.1002/j.1460-2075.1994.tb06552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen BK, Pelham HR. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol. 1988;8:5040–5042. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in yeast. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Jurivich DA, Sistonen L, Kroes RA, Morimoto RI. Effect of sodium salicylate on the human heat shock response. Science. 1992;255:1243–1245. doi: 10.1126/science.1546322. [DOI] [PubMed] [Google Scholar]
- Kopczynski JB, Raff AC, Bonner JJ. Translational readthrough at nonsense mutations in the HSF1 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1992;234:369–378. doi: 10.1007/BF00538696. [DOI] [PubMed] [Google Scholar]
- Littlefield O, Nelson HCM. A new use for the “wing” of the “winged” helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- Liu X-D, Thiele DJ. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996;10:592–603. doi: 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- Mosser DD, Kotzbauer PT, Sarge KD, Morimoto RI. Proc. Natl. Acad. Sci. USA 87: 3748–3752. 1990. In vitro activation of heat shock transcription factor DNA-binding by calcium and biochemical conditions that affect protein conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Wiederrecht G, Okuda A, Parker CS. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell. 1990;62:807–817. doi: 10.1016/0092-8674(90)90124-w. [DOI] [PubMed] [Google Scholar]
- Nover L. Expression of heat shock genes in homologous and heterologous systems. Enzyme Microb Technol. 1987;9:129–192. [Google Scholar]
- Que GG, Downey KM, So AG. Degradation of deoxyribonucleic acid by a 1, 10-phenanthroline-copper complex: the role of hydroxyl radicals. Biochemistry. 1980;19:5987–5991. doi: 10.1021/bi00567a007. [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- Ritossa FM. A new puffing pattern induced by heat shock and DNP in Drosophila. Experientia. 1962;18:571–573. [Google Scholar]
- Ritossa FM. Experimental activation of specific loci in polytene chromosomes of Drosophila. Exp Cell Res. 1964;35:601–607. doi: 10.1016/0014-4827(64)90147-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santoro N, Johansson N, Thiele DJ. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol. 1998;18:6340–6352. doi: 10.1128/mcb.18.11.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell AK, Yokoya F, Yu W, Miyawaga T, Murayama T, Winge DR. Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J Biol Chem. 1995;270:25079–25086. doi: 10.1074/jbc.270.42.25079. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990;62:793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Lewis MJ, Pelham HRB. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987;329:81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Nelson HC. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. Monosaccharide autoxidation in health and disease. Environ Health Perspect. 1985;64:297–307. doi: 10.1289/ehp.8564297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ, Stern A. The production of free radicals during the autoxidation of monosaccharides by buffer ions. Carbohydr Res. 1984;134:191–204. doi: 10.1016/0008-6215(84)85037-5. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ, Wolff S, Crabbe J, Stern A. The autoxidation of glyceraldehyde and other simple monosaccharides under physiological conditions catalyzed by buffer ions. Biochim Biophys Acta. 1984;797:276–287. doi: 10.1016/0304-4165(84)90131-4. [DOI] [PubMed] [Google Scholar]
- Torres FAG, Bonner JJ. Genetic identification of the site of DNA contact in the yeast heat shock transcription factor. Mol Cell Biol. 1995;15:5063–5070. doi: 10.1128/mcb.15.9.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuister GW, Kim S-J, Orosz A, Marquardt J, Wu C, Bax A. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Biol. 1994;1:605–614. [PubMed] [Google Scholar]
- Westwood JT, Wu C. Activation of Drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol. 1993;13:3481–3486. doi: 10.1128/mcb.13.6.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Guo Y, Vilaboa N, Zuo J, Voellmy R. Transcriptional activation of heat shock factor HSF1 probed by phosphopeptide analysis of factor 32P-labeled in vivo. J Biol Chem. 1998;273:8749–8755. doi: 10.1074/jbc.273.15.8749. [DOI] [PubMed] [Google Scholar]
- Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]
- Zuo J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- Zuo J, Baler R, Dahl G, Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol Cell Biol. 1994;14:7557–7568. doi: 10.1128/mcb.14.11.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]