Abstract
Glucan-binding protein B (GbpB) from Streptococcus mutans has been shown to induce protective immunity to dental caries in experimental models. Having recently sequenced the gbpB gene, our objective in this study was to identify immunogenic regions within the GbpB sequence for use in subunit vaccines. Potential regions of immunogenicity were sought by use of a matrix-based algorithm (EpiMatrix) to estimate the binding characteristics of peptides derived from the GbpB sequence by using a database of known major histocompatibility complex class II binding alleles. Screening the entire sequence revealed several peptides with estimated high binding probabilities. Two N-terminal 20-mer peptides (SYI and QGQ) subtending two of these regions were synthesized. A preliminary experiment, in which these peptides were synthesized in the multiple antigenic peptide format and were used to subcutaneously immunize Sprague-Dawley rats twice at a 21-day interval, revealed that the SYI peptide induced a higher percentage of responses to the inciting peptide as well as to intact GbpB, as measured by enzyme-linked immunosorbent assay. The effect of immunization with the SYI peptide construct on the cariogenicity of S. mutans was then investigated by immunizing weanling Sprague-Dawley rats twice at a 9-day interval with SYI or with phosphate-buffered saline. All rats were then orally infected with S. mutans strain SJ. After a 78-day infection period, the SYI-immunized groups had significant reductions in dental caries on both smooth and occlusal surfaces compared with the sham-immunized group. Thus, these experiments indicated that at least one linear sequence, derived from the N-terminal third of GbpB, was sufficiently immunogenic to induce a protective immune response in this experimental rat model for dental caries.
The ability of cariogenic mutans streptococci to accumulate in the dental biofilm is thought to be a consequence of the glucosyltransferase-mediated synthesis of glucans (5). These polymers mediate accumulation by providing binding sites for glucan-binding proteins associated with mutans streptococci. Epitopes associated with glucan binding would theoretically be primary targets for immunological attack, provided that the relevant sequences are located in molecular areas that can be accessible to antibody. Several mutans streptococcal proteins with glucan-binding activities have been described (16, 17, 19). One of these components, glucan-binding protein B (GbpB), has been shown to induce protective immune responses against experimental dental caries following systemic (24) or mucosal (20) immunization. Furthermore, there is evidence that the expression of GbpB is directly related to mutans streptococcal biofilm formation (11). These observations suggest that GbpB epitopes may have value in dental caries vaccine constructs.
GbpB has recently been cloned and sequenced (11). The deduced expressed sequence encompasses a single polypeptide chain which is 431 residues in length. Analysis of the primary sequence revealed a leucine zipper domain, but GbpB bore no sequence homology to glucan-binding domains of glucosyltransferases (1, 10) or Streptococcus mutans glucan-binding protein A (2, 3, 6). This prevented the specific recognition and immunological targeting of GbpB domains of putative glucan-binding function by use of subunit vaccine approaches that had been employed successfully with synthetic (23, 27) or recombinant (7) peptide constructs derived from glucosyltransferase glucan-binding domains. Interestingly, the GbpB sequence bears significant homology to peptidoglycan hydrolases from other gram-positive microorganisms (14, 18), and comparative genomic analysis of the gbpB region suggests a functional relationship between genes involved in cell shape and cell wall maintenance (11). Attempts to knock out the gbpB gene have also supported the notion that expression of GbpB is essential for the organism (12). Although these associations have suggested a role for GbpB which is separate from glucan binding, they have not as yet provided specific information about domains of putative function.
GbpB is exceptionally immunogenic in animals (24) and humans (21). Understanding the molecular basis of this immunogenicity potentially affords another direction through which the function of this protein, and ultimately S. mutans accumulation, may be intercepted. The immunogenicity of GbpB is based, in part, on the presentation of processed GbpB peptides on the surface of antigen-presenting cells in the context of major histocompatibility complex (MHC) class II molecules to T lymphocytes involved in the process leading to antibody formation (4, 9). Matrix-based algorithms have been used in T-cell epitope prediction to prospectively identify conserved class II-restricted MHC ligands in the protein sequence (15). Application of this approach to GbpB sequences in order to identify such peptides could suggest peptide constructs which could be used to focus the dental caries-protective responses seen with the intact protein. GbpB peptides associated with these regions could then be synthesized and evaluated for immunogenicity, reactivity with the parent protein, and, ultimately, induction of caries-protective immunity. Subunit vaccines which block functional domains have an additional advantage in that, if properly selected, they would not induce immunity to irrelevant or unwanted epitopes.
In this study, the GbpB sequence was analyzed for regions associated with peptides known to bind to the MHC II complex by using a matrix-based algorithm that estimates binding probabilities. Potentially immunogenic peptides suggested by this analysis were then synthesized, and their immunogenicity was confirmed in Sprague-Dawley rats. The extent of reactivity of the antipeptide responses with the parent GbpB protein were also evaluated. Finally, the ability of these peptides to induce protective immunity in the experimental rat model for dental caries was measured.
MATERIALS AND METHODS
Identification of MHC binding regions.
Peptides presented in conjunction with class II MHC molecules are derived from GbpB that has been processed in the phagosome of the antigen-processing cell. The peptides bind to MHC molecules on the surface of these cells in a linear fashion. The binding is determined by the interaction of the peptide's amino acid side chains with the binding pockets in the MHC molecule. The characteristics of peptides that are likely to bind to a given MHC can be directly deduced from pooled sequence data of MHC alleles, resulting in an estimated binding probability. Thus, in order to identify potential B-cell epitopes within the GbpB sequence which could be used for design of subunit vaccines, a matrix-based algorithm for epitope prediction (EpiMatrix) was used by EpiVax, Inc., (Providence, R.I.) to search the primary amino acid sequence for known MHC class II binding motifs. These motif-matching algorithms analyze the GbpB sequence against each MHC class II allele to indicate regions of sequence that contain clusters of binding motifs. Those sequences with sufficiently high estimated binding probabilities predict MHC ligand.
Peptide constructs.
SYI (KSNAATSYINAIINSKSVSD; GbpB residues 113 to 132) and QGQ (KHKLITIQGQVSALQTQQAG; GbpB residues 57 to 71 [residues KHKLI are irrelevant to GbpB sequence]) were selected for synthesis based on the estimated high probability of MHC class II binding identified in the matrix-based approach described above. Peptides were synthesized (Applied Diagnostics, Foster City, Calif.) as multiple antigenic peptide (MAP) constructs, using the stepwise solid-phase method of Merrifield (13), on a core matrix of lysines to yield macromolecules with four peptides per molecule, following the method of Tam (26). Purity (>90%) was assessed by using high-performance liquid chromatography, amino acid analysis, and molecular-weight determination by mass spectrometry.
GbpB.
GbpB was purified from S. mutans strain SJ by ion-exchange chromatography on MonoQ HR 5/5 (Pharmacia) in the presence of urea as described previously (22). Bacteria were cultivated overnight at 37°C under anaerobic conditions in sucrose-free defined medium as previously described (25). GbpB prepared in this manner migrates to a position of approximately 60 kDa in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (22).
ELISA.
Serum immunoglobulin G (IgG) and salivary IgA antibodies were tested by enzyme-linked immunosorbent assay (ELISA). Polystyrene microtiter plates (Flow Laboratories) were coated with 2.5 μg of SYI or QGQ/ml or 0.5 μg of S. mutans GbpB/ml. Antibody activity was then measured by incubation with 1:400 and 1:4,000 dilutions of sera or 1:4 dilutions of saliva. Plates were then developed for IgG antibody with rabbit anti-rat IgG, followed in sequence by alkaline phosphatase goat anti-rabbit IgG (Biosource Inc.) and p-nitrophenylphosphate (Sigma Chemical Co., St. Louis, Mo.). A mouse monoclonal antibody to rat α chain (Zymed, South San Francisco, Calif.) was used with biotinylated goat anti-mouse IgG (Zymed) and then avidin-alkaline phosphatase (ICN Biomedicals, Inc., Aurora, Ohio) followed by p-nitrophenylphosphate to reveal levels of salivary IgA antibody to peptides. Reactivity was recorded as absorbance (A405) in a microplate reader (Biotek Instruments, Winooski, Vt.). Serum data are reported as absorbance. Salivary IgA antibody data are reported as ELISA units (EU), which were calculated relative to levels of antibody in reference salivas from rats twice immunized with SYI. Saliva dilutions producing an A405 of 0.8 were considered 100 EU.
Immunogenicity of peptides.
Forty-five-day-old female Sprague-Dawley CD strain rats (Charles River Laboratories, Wilmington, Mass.) were used for injection. Four groups of six rats were injected subcutaneously (s.c.) in the vicinity of the salivary glands with 50 μg each of SYI or QGQ peptide construct or with 10 μg of GbpB, or they were sham immunized with buffer alone. The initial injection included complete Freund adjuvant (Difco Laboratories, Detroit, Mich.); one subsequent injection 21 days later included incomplete Freund adjuvant. Animals were bled prior to injection and 14 days after the second injection. In this experiment, rats were first momentarily anesthetized with a gas mixture of 50% carbon dioxide and 50% oxygen and then anesthetized by intraperitoneal injection of a mixture (0.65 ml/kg of body weight) of 3 parts ketamine (Ketaset [100 mg/ml]; Fort Dodge Laboratories, Fort Dodge, Iowa) and 7 parts xylazine (Rompun [20 mg/ml]; Bayer Corp., Shawnee Mission, Kans.). Saliva secretion was stimulated by s.c. injection of 0.6 ml of carbachol (0.1 mg/ml in saline; Sigma Chemical Co.) per kilogram of rat weight. After fluid collection, rats were injected s.c. first with 0.1 ml of atropine sulfate/kg (0.4 mg/ml; American Pharmaceutical Partners, Inc., Los Angeles, Calif.) and then with yohimbine (Yobine [2.0 mg/ml]; Lloyd Laboratories, Shenandoah, Iowa) at a volume equal to 1.4 times that used for anesthesia. Sera from coagulated and centrifuged blood were stored frozen at −20°C until measurement of antibody activity. Clarified saliva was stored at −70°C.
Protection experiment.
Two groups (n = 13 per group) of 25-day-old female Sprague-Dawley rats were singly caged. Rats were injected s.c. in the vicinity of the salivary gland with 50 μg of SYI MAP peptide construct or phosphate-buffered saline (PBS) (control animals). Antigen was incorporated with complete Freund adjuvant. Nine days later, rats were reinjected with PBS or with SYI at the same dose in incomplete Freund adjuvant. Six days after the second injection, sera and saliva were collected under anesthesia as described above. Fifteen days after the second injection, rats were placed in tubs (6 rats/tub), given cariogenic diet 2000 (56% confectioners' sugar [8]), and orally infected with approximately 108 S. mutans SJ32 for three consecutive days. Rats were again singly caged after the infection protocol was completed and were continued on diet 2000 for the duration of the experiment. Blood and saliva were collected 78 days after initial infection, followed by sacrifice. In preparation for the scoring of dental caries, rat skulls were defleshed by dermastidide beetles, followed by a rinse with 70% ethanol.
Bacterial recoveries.
The mutans streptococcal flora was assessed at 70 days after infection as previously described (22). After systematic swabbing of teeth, sonication, and plating of appropriate dilutions on mitis salivarius agar (MS; total streptococci) and MS agar with 0.2 mg of streptomycin sulfate/ml (MSS; S. mutans strain SJr), plates were incubated for 48 h at 37°C in 80% N2, 10% CO2, and 10% H2. S. mutans CFU were then enumerated microscopically on MSS agar.
Caries assessment.
The extent and depth of carious lesions in all rat molar teeth (caries score) were microscopically evaluated by a modified Keyes method as previously described (29). Caries scores were determined separately on smooth and occlusal dental surfaces.
Statistical analysis.
The differences in the median values among the treatment groups were analyzed by one-way analysis of variance, followed by the Tukey pairwise multiple comparison test when data were normally distributed. Alternatively, data were analyzed by Kruskal-Wallis one-way analysis of variance on ranks, followed by Dunn's multiple comparison procedure when nonparametric distributions were encountered (e.g., bacterial CFU and caries scores).
RESULTS
MHC class II binding studies.
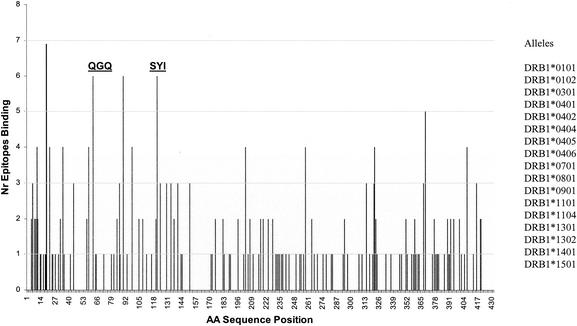
Motif-matching algorithms were run with the GbpB sequence against several sets of known MHC class II alleles. Figure 1 illustrates a representative screening against DRB1 alleles in which the number of motif matches within this set of alleles is plotted against the respective position in the protein sequence. By using the EpiMatrix algorithm, four regions within the protein sequence were identified which had at least six matches within the DRB1 allele data set. One of these regions, beginning at residue 16 (Fig. 1), fell within the 27-residue signal peptide domain. Since an epitope at this position would not be expected to be useful for vaccine purposes, this region was not considered further. Three other regions within the N-terminal third of the molecule had high estimated binding probabilities. Since regions beginning at residue 62 and residue 121 were sequentially the most separated of these three sites, they were selected for subsequent synthetic-peptide design. Analysis with other sets of known alleles also identified these two locations as being regions with higher estimated binding probabilities (data not shown). Subsequent to these analyses, two 20-mer peptides (QGQ and SYI) whose sequences included the predicted binding epitopes following GbpB residues 62 and 121, respectively, were synthesized. Each peptide was synthesized as a MAP construct containing four copies of the respective sequence.
FIG. 1.
MHC class II motif prediction. A matrix-based MHC class II motif-matching algorithm was used to search the primary sequence of GbpB for known MHC class II binding motifs associated with MHC class II alleles. Shown is the result of the application of this algorithm for screening GbpB sequence against a set of 17 known MHC binding motifs (listed at right) associated with alleles at the MHC class II DRB1 locus. The ordinate represents the number (Nr) of motif matches associated with a given peak, while the abscissa represents the GbpB sequence position. The locations of the sequences used to make synthetic peptides for use in this study are indicated by “QGQ” and “SYI.” Regions of binding interest extend from 6 to 11 places to the right of peak residue 62 (associated with QGQ) and peak residue 121 (associated with SYI).
Immunogenicity of peptides.
Rats were injected twice s.c. in the salivary gland vicinity with SYI or QGQ MAP peptide or native GbpB protein in adjuvant. Sera taken 35 days after the first injection were analyzed by ELISA for levels of serum IgG antibody to each peptide construct and to GbpB (Fig. 2).
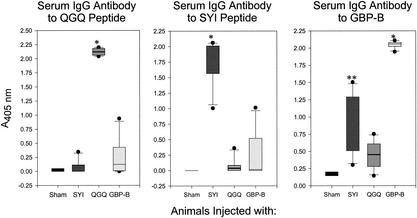
FIG. 2.
Box plots of serum IgG antibody to GbpB peptide constructs QGQ and SYI. Serum IgG antibody activities against QGQ (left panel) and SYI (middle panel) peptide constructs and GbpB protein (right panel) were measured by ELISA. Sham-immunized and SYI-, QGQ-, and GbpB-immunized groups are indicated on the abscissae of each plot and represent the immune experience of 4 to 6 rats per group at 35 days after the first of two s.c. immunizations. Antibody reactivity is indicated on the ordinate as A405. Antibody levels are not directly comparable between panels. Group data indicated by box plots with a single asterisk are significantly different from data for the respective sham group (P < 0.001); double asterisks indicate a P value of 0.071.
All rats injected with the QGQ peptide responded with significantly elevated levels (P < 0.001) of serum antibody to the QGQ peptide, whereas no significant responses to QGQ epitopes were seen in sham-immunized rats or rats injected with SYI. Interestingly, sera from two of the four rats injected with GbpB protein also reacted with QGQ.
All rats injected with the SYI peptide demonstrated significantly elevated levels (P < 0.01) of serum IgG antibody to the inciting SYI peptide construct, in contrast to what was seen with sham- or QGQ-injected rats. Again, serum IgG from one of the four rats injected with the parent GbpB protein also reacted with the SYI peptide.
All sera were also evaluated by ELISA using plates coated with native GbpB. Rats from SYI (6 of 6 animals) or QGQ (4 of 6 animals) peptide-injected groups reacted with the parent GbpB protein, although the levels of serum IgG antibody reactive with GbpB from peptide-injected rats were lower than the anti-GbpB levels from intact GbpB-injected rats. Taken together, these results supported the immunogenicity of these peptides predicted by using the bioinformatics approach. Furthermore, they also suggested that the linear epitope(s) found on both the SYI and QGQ peptide constructs was similar to the respective epitope(s) on the intact GbpB protein.
Caries-protective effect of immunization with SYI.
We hypothesized that the induction of significant GbpB-reactive immune responses by constituent peptides could be protective since GbpB could consistently induce protection in the experimental rat model for dental caries. The SYI peptide was selected to test this assumption since this peptide induced more-consistent and somewhat higher immune responses reactive with GbpB than did the QGQ peptide.
Two groups of rats were either sham immunized or immunized with the SYI peptide construct by s.c. injection in the vicinity of the salivary gland. Twenty-four days after the first immunization, both groups were infected with streptomycin-resistant S. mutans. Sera collected at the end of the 78-day infection period were analyzed for IgG and IgA antibody to the peptide construct and to GbpB (Fig. 3). As expected, immunization with the peptide induced significant levels (P < 0.001) of serum antibody in both isotypes to the inciting SYI peptide in all rats. Also, consistent with the results of the previous experiment, SYI immunization induced IgG antibody to intact GbpB in all rats (P < 0.001). Most SYI-injected rats also demonstrated levels of serum IgA antibody to GbpB (P < 0.001). Salivas were collected prior to infection and at the end of the experiment and analyzed by ELISA for IgA antibody to SYI and GbpB (Table 1). Several (5 of 13 animals) SYI-immunized rats demonstrated induction of salivary IgA antibody to both the peptide and the intact protein at either time point, although the levels were not significantly different from the levels for the sham-immunized group under the conditions of measurement.
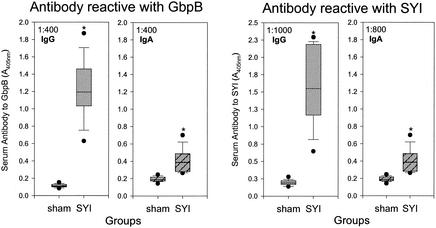
FIG. 3.
Box plots of serum IgG and IgA antibody to GbpB and SYI from rats in the protection experiment. The left two panels indicate IgG or IgA antibody activity reactive with S. mutans GbpB, and the right two panels indicate IgG or IgA antibody activity reactive with SYI peptide construct. Serum antibody activity was measured by ELISA at the dilution indicated in the upper left corner of each panel. Antibody reactivity is indicated on the ordinate as A405. Each plot represents the immune experience of 13 rats per group 3 months after the first of two s.c. immunizations. Asterisks indicate that the antibody reactivity in the SYI-immunized group was significantly different from that of the sham group for the respective antigen and antibody isotype (P < 0.001).
TABLE 1.
Salivary IgA antibody to SYI peptide construct and GbpB protein
| Test antigen | Group | EU (mean ± SE) |
|---|---|---|
| GbpB protein | Sham immunized | 7.1 ± 5.1 |
| SYI immunized | 31.1 ± 16.5 | |
| SYI peptide | Sham immunized | 3.6 ± 2.4 |
| SYI immunized | 12.5 ± 5.9 |
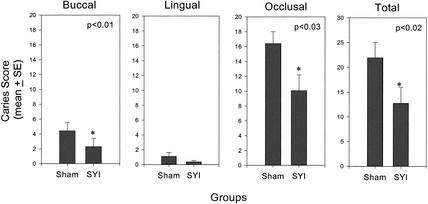
Challenge of sham- and SYI-immunized rats with S. mutans SJr resulted in infection of all rats with these cariogenic streptococci. The mean levels of streptomycin-resistant S. mutans SJr recovered from the SYI-immunized group were lower than the mean levels from the sham-immunized group at both 8 and 65 days after infection was initiated, although the differences did not achieve statistical significance because of the variation in bacterial recoveries. The extent of dental caries was measured on molar surfaces 78 days after initial infection with S. mutans. Caries scores on smooth (buccal; P < 0.01) and occlusal (P < 0.03) surfaces as well as total (P < 0.02) caries scores of SYI peptide-immunized rats were significantly lower than those of sham-immunized and infected rats (Fig. 4). These data indicate that at least one epitope on SYI was capable of inducing a caries-protective response in this model.
FIG. 4.
Dental caries after 78 days of infection with S. mutans SJr. The buccal, lingual, occlusal, and total molar caries scores from sham- and SYI-immunized groups are shown. Columns indicate the mean caries scores, and error bars show two standard errors. P values are indicated in each panel where the caries experience of the SYI-immunized group is significantly lower than that of the sham-immunized group.
DISCUSSION
Mucosal immune responses to S. mutans GbpB appear in many children (21), suggesting that this protein has significant immunogenicity, even when presented early in life. Experimental immunization with GbpB induced protective immunity to S. mutans infection after local s.c. (24) or intranasal immunization (20). Thus, it could be hypothesized that S. mutans GbpB would be an effective component of a childhood vaccine for dental caries. The experiments described in this study sought to identify the structural basis for these protective effects in order to potentially improve protective immune responses by the designing of subunit vaccines which target functional domains. Given that S. mutans GbpB was initially revealed through its affinity for α-1-6-linked glucan (20), a glucan-binding domain could be expected as one functional target. However, affinity measurements with α-1-6-linked dextran or de novo-synthesized glucan indicated that this affinity is very low (data not shown). In addition, amino acid sequence homologies between S. mutans GbpB and glucan- or carbohydrate-binding domains of other proteins, such as S. mutans GbpA, S. mutans glucosyltransferases, or Clostridial toxins could not be found, making it problematic, at this point, to design subunit vaccines which interfere with the glucan binding of S. mutans GbpB. On the other hand, the potential exists for the targeting of other functional domains of S. mutans GbpB, since this protein does appear to share sequence homology with putative peptidoglycan hydrolases (11). Efforts are under way to verify the cell wall-synthetic activity of S. mutans GbpB and establish respective structure-function relationships.
Since targeting immune responses to S. mutans GbpB to functional domains is not yet possible, we sought sequences which could be associated with the inherent immunogenicity of GbpB, based on the potential of peptides to serve as MHC class II ligand. Recognition of such MHC ligands by T lymphocytes is dependent on the presentation of the T-cell epitope in the context of MHC molecules. S. mutans GbpB peptides that are presented in this context have been processed in the cytoplasm and bind to MHC molecules in a linear fashion (4), a binding which is constrained by the amino acid side chains of the peptide. Using motif- and matrix-based algorithms (15) that predict the relative degree of binding to a given MHC class II allele with a sequence length of 10 or 20 GbpB amino acids, we found several predicted binding regions within the GbpB sequence (Fig. 1). The N-terminal region contained the greatest number of putative binding sequences within the protein. This result was not surprising since this region contains the greatest sequence heterogeneity. This region is also highly conserved. Only one of the nine gene mutations seen among 44 amplitypes of S. mutans strains that lead to changes at the protein level occurred in the N-terminal region of GbpB (11). Thus, subunit vaccines drawn from this region could be expected to react with GbpB from most if not all gbpB gene products. Interestingly, two of the most prominent predicted binding peptides (residues 55 to 71 and 88 to 107) occurred within an observed leucine zipper motif (residues 65 to 93).
Local s.c. injection of either peptide QGQ or SYI, which were selected on the basis of probability of strong binding to MHC class II alleles, induced levels of IgG antibody to the respective peptide which could still be detected in most rats at serum dilutions above 1:105 and in all rats at dilutions above 1:104. These elevated responses support the predictive ability of the bioinformatic approach. Furthermore, serum IgG antibody from most QGQ-injected rats in the immunization experiment (4 of 6 animals) and all SYI-injected rats in either the immunization (6 of 6 animals) or the immunization-protection experiment (12 of 12 animals; P < 0.001) reacted with epitopes on the native GbpB protein (Fig. 2 and 3). Such data would suggest that the epitopes represented by these two GbpB subsequences are displayed at or near the surface of the protein. Interestingly, only a minority of the rats immunized with native GbpB demonstrated significant serum IgG antibody reactivity to either peptide construct. This could suggest that the cytoplasmic processing of GbpB does not favor peptides of these two sequences or that other processed GbpB peptides are preferentially used as MHC class II ligands in the rat. Alternatively, the differences between the extent of anti-peptide and anti-native protein antibody binding to peptide could be related to antibody affinity. In contrast, the immunological complexity of the immune response to GbpB results in far higher aggregate IgG antibody specificities to native GbpB in protein-injected animals than in peptide-injected animals (Fig. 2).
The immune response induced by immunization with the SYI peptide construct was sufficient to modify the extent of dental disease caused by cariogenic S. mutans. For example, significant differences in buccal and occlusal caries scores were observed between SYI- and sham-immunized groups (Fig. 4). Interestingly, the percentage reductions in caries on smooth (approximately 50%) and occlusal (approximately 40%) surfaces were nearly as great as those seen after immunization with the native protein (smooth, approximately 65%; occlusal, approximately 50%) (24). Since this experimental model is designed to favor disease (high doses of S. mutans infection in the presence of 56% dietary sucrose), some dental caries is expected even in the presence of antibody. Thus, the similarity between caries reductions after immunization with the multiepitopic native GbpB (24) and the monoepitopic SYI peptide is remarkable, highlighting the importance of the immune response to epitope(s) contained within this sequence. These data suggest that sufficient salivary anti-SYI antibody was present at the time of infection, despite the inability to detect such antibody in the IgA class in some rats. The presence and protective potential of salivary IgG antibody to GbpB could also contribute to protection, although such levels were not measured in this experiment.
GbpB epitopes separate from those found on SYI also have the potential to induce caries-protective responses. Clearly, epitopes expressed on QGQ induce GbpB-reactive responses. In addition, the bioinformatic approach suggested several other GbpB peptide sequences with potential to lead to induction of antibody activity. These remain to be evaluated in the experimental protection model, as do sequences based on heretofore unidentified functional domains. The potential exists to design di- or multiepitopic subunit GbpB vaccines containing sequences which would target different features of the molecule, as has been done for other vaccines (28). Furthermore, the protective features of SYI immunization make it now possible to design multiepitopic subunit vaccines targeting both the catalytic function of glucosyltransferase and immunogenic (possibly associated with function) epitopes of GbpB. These approaches should enhance the effectiveness of subunit-based dental caries vaccines.
Acknowledgments
This work was supported by Public Health Service grant DE-06153 from the National Institute of Dental and Craniofacial Research and by a student research grant from Harvard Medical School (Z.P.).
Editor: V. J. DiRita
REFERENCES
- 1.Abo, H., T. Masumura, T. Kodama, H. Ohta, K. Fukui, K. Kato, and H. Kagawa. 1991. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase). J. Bacteriol. 173:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banas, J. A., H. C. Potvin, and R. N. Singh. 1997. The regulation of Streptococcus mutans glucan-binding protein A expression. FEMS Microbiol. Lett. 154:289-292. [DOI] [PubMed] [Google Scholar]
- 3.Banas, J. A., R. R. Russell, and J. J. Ferretti. 1990. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect. Immun. 58:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Germain, R. N., and D. H. Margulies. 1993. The biochemistry and cell biology of antigen processing and presentation. Annu. Rev. Immunol. 11:403-450. [DOI] [PubMed] [Google Scholar]
- 5.Hamada, S., and H. D. Slade. 1980. Biology, immunology and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazlett, K. R., J. E. Mazurkiewicz, and J. A. Banas. 1999. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect. Immun. 67:3909-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jespersgaard, C., G. Hajishengallis, Y. Huang, M. W. Russell, D. J. Smith, and S. M. Michalek. 1999. Protective immunity against Streptococcus mutans infection in mice after intranasal immunization with the glucan-binding region of S. mutans glucosyltransferase. Infect. Immun. 67:6543-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keyes, P. H., and H. V. Jordan. 1964. Periodontal lesions in the Syrian hamster. III. Findings related to an infectious and transmissible component. Arch. Oral Biol. 9:377. [DOI] [PubMed] [Google Scholar]
- 9.Kubo, R. T., A. Sette, H. M. Grey, E. Appella, K. Sakaguchi, N. Z. Zhu, D. Arnott, N. Sherman, J. Shabanowitz, and H. Michel. 1994. Definition of specific peptide motifs for four major HLA-A alleles. J. Immunol. 152:3913-3924. [PubMed] [Google Scholar]
- 10.Lis, M., T. Shiroza, and H. K. Kuramitsu. 1995. Role of C-terminal direct repeating units of the Streptococcus mutans glucosyltransferase-S in glucan binding. Appl. Environ. Microbiol. 61:2040-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattos-Graner, R. O., S. Jin, W. F. King, T. Chen, D. J. Smith, and M. J. Duncan. 2001. Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect. Immun. 69:6931-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattos-Graner, R. O., P. Zucchi, D. J. Smith, and M. J. Duncan. 2002. Mutant analysis of the gene encoding glucan binding protein B indicates an essential role in Streptococcus mutans. J. Dent. Res. 81:A40.
- 13.Merrifield, R. B. 1963. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 85:2149-2154. [Google Scholar]
- 14.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts. C. G. P., G. E. Meister, S. M. Jesdale, J. Lieberman, J. A. Berzofosky, and A. S. DeGroot. 1996. Identification of HIV peptide epitopes by novel algorithm. AIDS Res. Hum. Retrovir. 12:593-610. [DOI] [PubMed] [Google Scholar]
- 16.Russell, R. R. B. 1979. Glucan-binding proteins of Streptococcus mutans serotype c. J. Gen. Microbiol. 112:197-201. [DOI] [PubMed] [Google Scholar]
- 17.Sato, Y., Y. Yamamoto, and H. Kizaki. 1997. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 65:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert, K., A. M. Bichlmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 19.Smith, D. J., H. Akita, W. F. King, and M. A. Taubman. 1994. Purification and antigenicity of a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 62:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, D. J., R. L. Heschel, J. Melvin, W. F. King, M. B. B. Periera, and M. A. Taubman. 1997. Streptococcus mutans glucan binding proteins as dental caries vaccines, p. 367-377. In A. J. Husband, K. W. Beagley, R. L. Clancy, A. M. Collins, A. W. Cripps, and D. L. Emery (ed.), Mucosal solutions: advances in mucosal immunology, vol 2. University of Sydney Press, Sydney, Australia.
- 21.Smith, D. J., W. F. King, H. Akita, and M. A. Taubman. 1998. Association of salivary immunoglobulin A antibody and initial mutans streptococcal infection. Oral Microbiol. Immunol. 13:278-285. [DOI] [PubMed] [Google Scholar]
- 22.Smith, D. J., W. F. King, and R. Godiska. 2001. Passive transfer of IgY antibody to Streptococcus mutans glucan binding protein B can be protective for experimental dental caries. Infect. Immun. 69:3135-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, D. J., B. Shoushtari, R. L. Heschel, W. F. King, and M. A. Taubman. 1997. Immunogenicity and protective potential of peptides derived from a hypothetical second GTF catalytic domain. Infect. Immun. 65:4424-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, D. J., and M. A. Taubman. 1996. Experimental immunization of rats with a Streptococcus mutans 59-kilodalton glucan-binding protein protects against dental caries. Infect. Immun. 64:3069-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Socransky, S. S., J. L. Dzink, and C. M. Smith. 1985. Chemically defined medium for oral microorganisms. J. Clin. Microbiol. 22:303-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam, J. P. 1988. Synthetic peptide vaccine design: synthesis and properties of high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 85:5409-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taubman, M. A., C. J. Holmberg, and D. J. Smith. 1995. Immunization of rats with synthetic peptide constructs from the glucan binding or catalytic regions of mutans streptococcal glucosyltransferase protects against dental caries. Infect. Immun. 63:3088-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taubman, M. A., C. J. Holmberg, and D. J. Smith. 2001. Diepitopic construct of functionally relevant complementary peptides enhances immunogenicity, reactivity with glucosyltransferase, and protection against dental caries. Infect. Immun. 69:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taubman, M. A., and D. J. Smith. 1974. Effects of local immunization with Streptococcus mutans on induction of salivary immunoglobulin A antibody and experimental dental caries. Infect. Immun. 9:1079-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]